Abstract
Seasonal and pandemic influenza is a cause of morbidity and mortality worldwide. Most people infected with influenza virus display mild-to-moderate disease phenotypes and recover within a few weeks. Influenza is known to cause persistent alveolitis in animal models; however, little is known about the molecular pathways involved in this phenotype. We challenged C57BL/6 mice with influenza A/PR/8/34 and examined lung pathologic processes and inflammation, as well as transcriptomic and epigenetic changes at 21 to 60 days after infection. Influenza induced persistent parenchymal lung inflammation, alveolar epithelial metaplasia, and epithelial endoplasmic reticulum stress that were evident after the clearance of virus and resolution of morbidity. Influenza infection induced robust changes in the lung transcriptome, including a significant impact on inflammatory and extracellular matrix protein expression. Despite the robust changes in lung gene expression, preceding influenza (21 days) did not exacerbate secondary Staphylococcus aureus infection. Finally, we examined the impact of influenza on miRNA expression in the lung and found an increase in miR-155. miR-155 knockout mice recovered from influenza infection faster than controls and had decreased lung inflammation and endoplasmic reticulum stress. These data illuminate the dynamic molecular changes in the lung in the weeks after influenza infection and characterize the repair process, identifying a novel role for miR-155.
Influenza virus, a member of the family Orthomyxoviridae, is a highly contagious respiratory virus, infecting between 5% and 20% of the US population and accounting for an average of 30,000 deaths annually. Influenza virus is identified by its serotype, based on antibody responses to the hemagglutinin and neuraminidase surface glycoproteins. H1N1, H2N2, and H3N2 strains have caused significant pandemics in humans in the past 100 years, and the 2009 novel H1N1 was reported to have had infectivity rates as high as 45% in young adults in the United States, imparting a significant disease burden.1 In 2009, approximately 55 million persons were infected in the United States with 274,000 hospitalizations and 12,470 deaths.2 Global infection rates were estimated at 24%, more than a billion persons, with as many as 200,000 deaths.3 These data indicate the breadth of the toll that influenza infection takes on humankind.
On infection, influenza replicates quickly in the upper and lower respiratory tracts. This initial infection can go somewhat unnoticed by the immune system, and viral replication peaks around 48 hours after infection. In patients not immunocompromised, the immune system sufficiently controls infection, and clinical symptoms are limited to 2 to 8 days.4 Histologically, acute influenza infection involves widespread epithelial damage. Multifocal destruction and desquamation of tracheal, bronchial, and alveolar epithelium were reported. At certain stages of influenza-induced pneumonia, normal alveolar cells may be partially or completely undetectable. In later stages of disease, hyaline membranes form course and fine strands lining the alveolar spaces. After the initial influenza virus injury, regeneration of the bronchiolar and alveolar epithelia is evident with the presence of squamous cell metaplasia and alveolar type II pneumocyte hyperplasia.5
Most influenza infections are not fatal; rather most patients appear to fully recover within 2 weeks of infection. Accordingly, there tends to be an assumption that the changes and damage that occur in response to infection are transient. However, epithelial hyperplasia and parenchymal histologic changes were documented as long as 90 days after influenza infection in mice.6 These data suggest a potentially long-lasting impact of influenza infection on the lung. Little molecular data are known about the long-term recovery from influenza infection in mice or humans. To address this gap in knowledge, we set out to define the molecular hallmarks of persistent lung injury after influenza infection.
Materials and Methods
Mice
Wild-type (WT), C57BL/6 mice were purchased from Taconic Farms (Germantown, NY). miR-155−/− mice and C57BL/6 control mice were purchased from The Jackson Laboratory (Bar Harbor, ME). All studies were performed using 8- to 10-week-old male mice housed under pathogen-free conditions. All animal studies were approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
Influenza Infection
Influenza A/PR/8/34 H1N1 was propagated in chicken eggs as previously described.7 Mice were infected with 150 plaque-forming units of influenza virus in 50 μL of sterile phosphate-buffered saline (PBS) by oropharyngeal aspiration (control mice received PBS vehicle). Lungs were harvested at the indicated time points after infection. Viral titer was determined by Madin-Darby kidney plaque assay, and viral genome load was determined by RT-PCR for viral M protein expression as previously described.8, 9
Analysis of Lung Injury and Inflammation
Mouse lungs were lavaged with 1 mL of sterile PBS. Bronchoalveolar lavage (BAL) was then used for evaluation of inflammatory cells by differential cell counting. The right cranial lung lobe was homogenized in sterile PBS for analysis of cytokine production by Bio-plex (Bio-Rad, Hercules, CA) multiplex cytokine assay. The middle and caudal lung lobes were snap-frozen in liquid nitrogen and homogenized by mortar and pestle to generate lung powder for RNA isolation. RNA was isolated using an Absolutely RNA kit (Agilent, Santa Clara, CA) and was reverse transcribed to cDNA using an iScript cDNA kit (Bio-Rad). Gene expression analysis was performed using TaqMan primers and probes (Applied Biosystems, Foster City, CA). The left lung was fixed with 10% neutral-buffered formalin before dehydration and paraffin embedding. Paraffin blocks were sectioned at 5 μm and stained with hematoxylin and eosin. To assess collagen deposition, sections were deparaffinized and stained using the Masson's trichrome stain (Sigma-Aldrich, St. Louis, MO). Briefly, sections were heated in Bouin's solution (56°C, 15 minutes), washed in tap water, and stained in Weigert's Hematoxylin Solution (5 minutes). Sections were washed in running tap water, rinsed in deionized water, and then placed in Biebrich scarlet-acid fuschin (5 minutes). Slides were rinsed, placed in phosphotungstinic/molybdic acid for 5 minutes, and then placed in Aniline Blue for 5 minutes. Slides were washed in 1% acetic acid (2 minutes), dehydrated through alcohol, cleared in xylene, and mounted.
Flow Cytometry
Alternatively, the right lung was digested with collagenase as described.8, 10 Dispersed single-cell preparations were re-stimulated with 50 ng/mL PMA and 750 ng/mL ionomycin for 5 hours at 37°C. Cells were then stained with fluorescent-conjugated antibodies to CD4 and CD8, followed by permeabilization and staining for interferon (IFN)-γ, and IL-17A (BD Biosciences, Franklin Lakes, NJ). Stained cells were analyzed using a FACSAria apparatus (BD Biosciences).
Determination of Endoplasmic Reticulum Stress
After deparaffinization, rehydration, and permeabilization of embedded tissue sections, samples were blocked with 5% bovine serum albumin/PBS for 1 hour at room temperature and incubated with primary antibodies (Cell Signaling, Danvers, MA; and Abcam, Cambridge, United Kingdom) in 5% bovine serum albumin/PBS (dilution 1:250) overnight at 4°C. The sections were rinsed multiple times with PBS and incubated with secondary antibodies conjugated with fluorophores (Alexa 488 or 647 at dilution 1:500). After the incubation with secondary antibody, the slides were again rinsed with PBS and counterstained with DAPI (dilution 1:4000) and mounted with Aquapolymount. The images were acquired using a Zeiss LSM 510 META Confocal Laser Scanning Imaging System (Zeiss, Dublin, CA). Images were captured at ×40 magnification in oil immersion. The image files were converted to Tiff format using Photoshop CS5 (Adobe, San Jose, CA). Brightness and contrast were adjusted equally in all images. Fluorescent images were quantified using ImageJ software version 1.50 (NIH, Bethesda, MD). For assessment of active caspase-3 or activating transcription factor (ATF)6α three bronchioles were randomly selected per mouse, and intensity values were pooled from samples. These values were normalized to the intensity of DAPI (blue nuclei).
RNA and miRNA Microarray
Whole-lung RNA isolated as described above was checked for integrity with an Agilent bioanalyzer. RNA microarray was performed by the University of Pittsburgh Genomics and Proteomics Core Laboratory with the Illumina (San Diego, CA) mouse WG6 array chip according to the manufacturer's instructions. Microarray data were analyzed with the J5 score threshold as previously described.11 The threshold value is set as the absolute value of the J5 score that a gene must be above to be considered to be differentially expressed between groups. J5 score is calculated by dividing the mean difference between group 1 and 2 by the average absolute mean difference for all genes in the data set. Positive J5 values indicate overexpression, whereas negative J5 values indicate underexpression. The choice of J5 score was based on its better performance than traditional methods.12 miRNA microarray was performed by the Louisiana State University Microarray and Genome Bioinformatics center, using the Agilent miRNA microarray (version 1) according to the manufacturer's protocols. Data were analyzed using Agilent Genespring GX software version 11. Microarray data are deposited at the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession number GSE92938).
miRNA/mRNA Interaction Analysis
mRNA and miRNA data sets were probed for interactions with the mirConnX, a web tool that integrates condition-specific expression data with prior information to construct a miRNA-mRNA regulatory network.13 A prior, static network was constructed that consisted of computationally predicted transcriptional regulations (motif scanning of the sequence) and after-transcriptional regulations (miRNA target predictions), as well as known interactions supported by literature evidence. Next, a dynamic network based on mRNA and miRNA expression data were generated by using one of the following metrics: Pearson, Spearman, or Kendall correlation. Then the nonspecific, static network, which provided the directionality of the regulatory edges, was combined with the dynamic network derived from given data sets, which suggests the sign (activation or suppression) of the interactions. Finally, we searched for network motifs (feedforward loops and feedback loops) in the posterior regulatory network. Additional miR-155 targets were identified by using the web tool miRTarBase, an online collection of literature-supported miRNA targets.
Bacterial Superinfection
Mice were infected with influenza A or control PBS vehicle treated as outlined above. Twenty-one days after infection, mice were challenged with 5 × 107 colony-forming units of Staphylococcus aureus (USA300) in 50 μL of sterile PBS by oropharyngeal aspiration. S. aureus was cultured in CCY-modified medium overnight to stationary growth phase. After lung harvest, bacterial burden was determined by plating lung homogenate in serial dilution on CCY-modified medium agar plates.
Statistical Analysis
Data are presented as the group means ± SEM. Significance was tested using unpaired t-test (for 2 means) or one-way analysis of variance, followed by Tukey post hoc test. Data were analyzed with the Excel 2007 software package (Microsoft, Redman, WA). All experiments were repeated a minimum of twice at the indicated number.
Results
Influenza Infection Results in Persistent Lung Inflammation after Viral Clearance
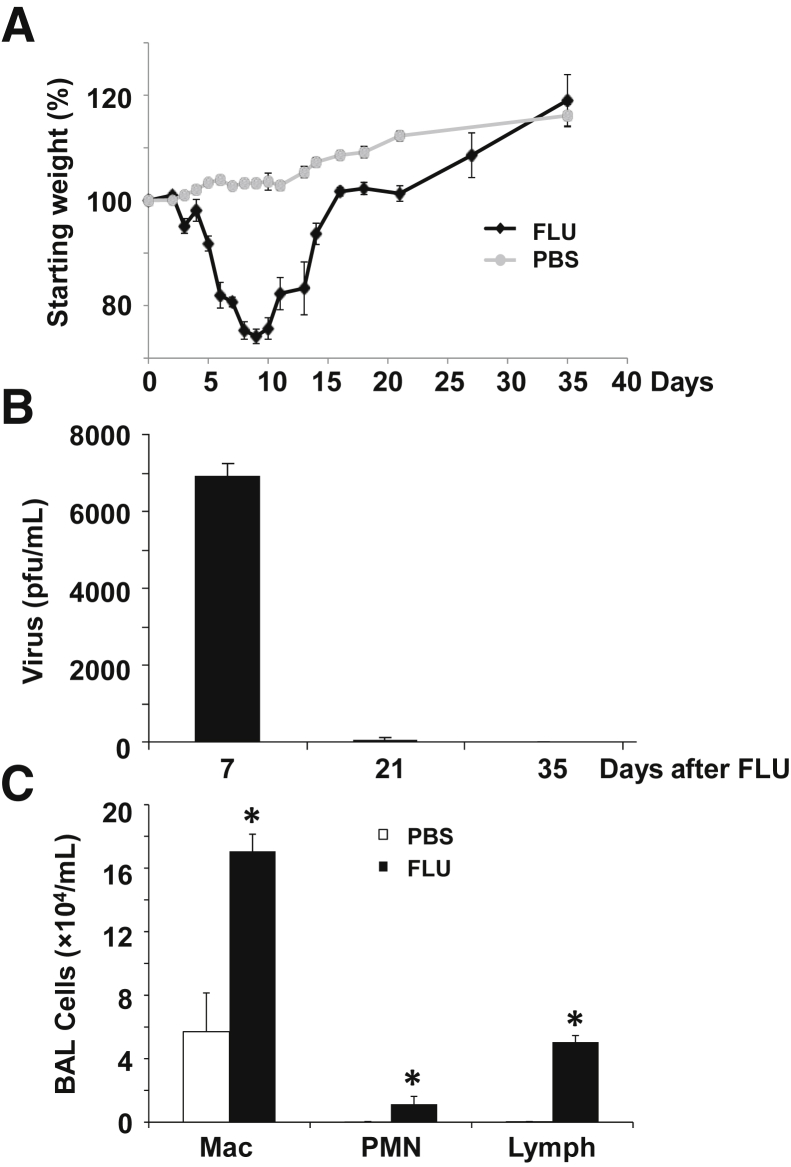
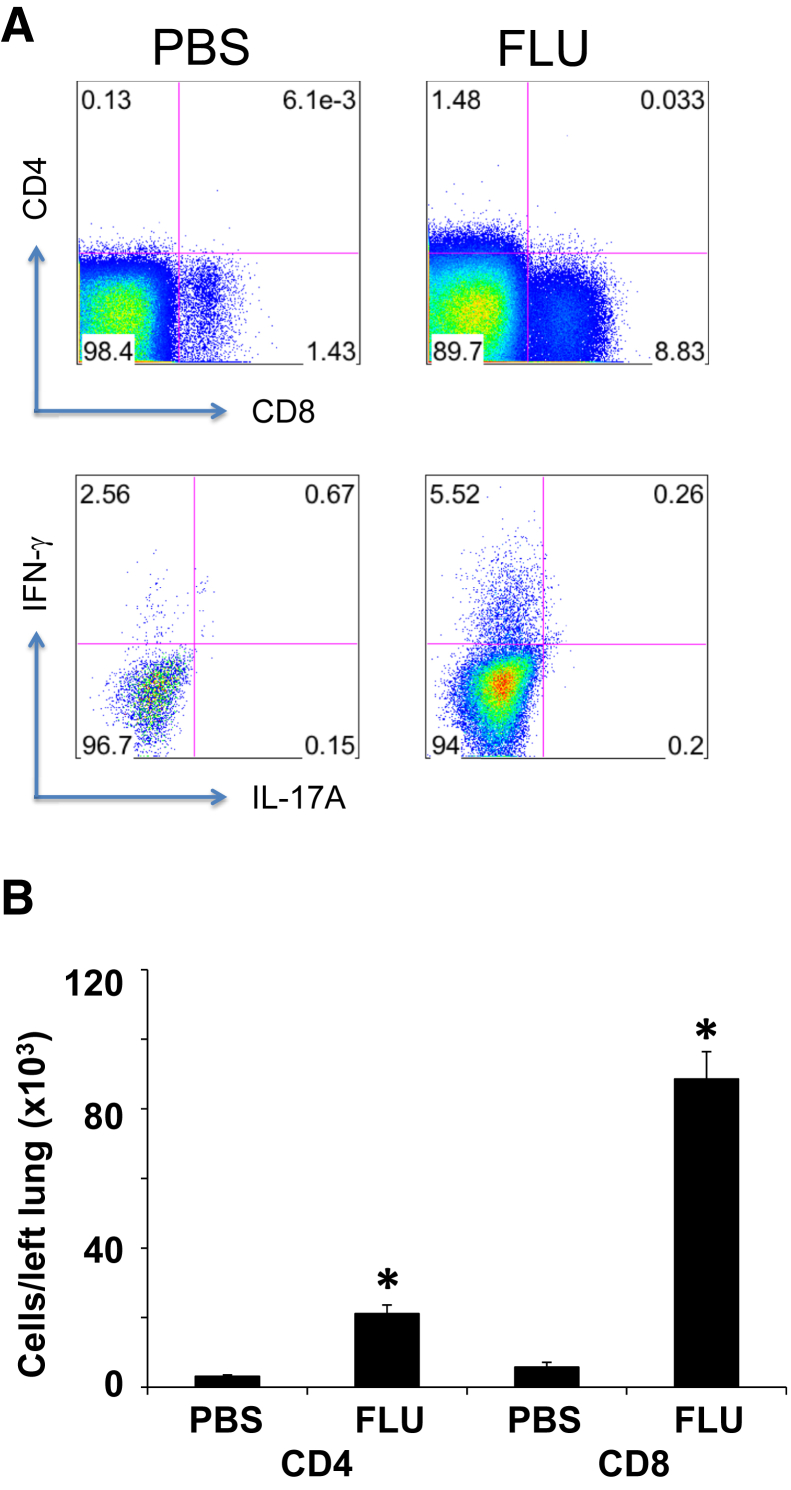
As noted above, the original observation that influenza infection results in alveolitis that lasts for months was made many years ago. However, little is known about the molecular mechanisms governing this lung reparative process. To investigate this further, WT C57BL/6 mice were infected with influenza A/PR/8/34 (H1N1) or PBS vehicle, and lung inflammation and histopathologic process were analyzed. Influenza infection resulted in significant weight loss, and mice recovered to their initial starting weight within 2 weeks of challenge (Figure 1A). We then focused our analyses on day 21 after infection when viral clearance had been achieved (Figure 1B) and mice were phenotypically behaving similar to control animals. Viral clearance was confirmed by RT-PCR for influenza matrix protein (data not shown; at day 21 all CT equaled 40, not detected). Three weeks after influenza infection, mice maintained elevated inflammatory cell counts in the BAL (Figure 1C). Numbers of macrophages, neutrophils, and lymphocytes were significantly elevated in influenza-infected mice compared with control mice. To better evaluate the lymphocyte populations present after influenza, we analyzed mouse lung homogenate by flow cytometry. Both CD4+ and CD8+ T cells were elevated 3 weeks after influenza infection compared with control mice (Figure 2). Further, IFN-γ (Type 1) lymphocytes were specifically increased. These data demonstrated that, despite clearance of influenza virus, pulmonary inflammation persisted at day 21 after lung infection.
Figure 1.
Influenza infection induces persistent lung inflammation after viral clearance. C57BL/6 mice were infected with influenza A/PR/8/34 or control PBS vehicle for the indicated time points. A: Morbidity as measured by weight loss. B: Lung viral titer measured by plaque assay. C: BAL inflammatory cells determined by differential counts. n = 4–15 FLU (A); n = 3–18 PBS (A); n = 4 (B); n = 4 PBS (C); n = 8 FLU (C). ∗P < 0.05, FLU versus PBS. BAL, bronchoalveolar lavage; FLU, influenza; Mac, macrophage; PBS, phosphate-buffered saline; pfu, plague-forming unit; PMN, polymorphonuclear leukocyte.
Figure 2.
Characterization of T cells in the lung at 21 days after influenza infection. C57BL/6 mice were infected with influenza A/PR/8/34 or control PBS vehicle for 21 days. A: Representative flow cytometry plots indicating the frequency of CD4+, CD8+, IFN-γ+, and IL-17A+ cells. B: Quantification of CD4+ and CD8+ lymphocytes persisting in the lung. n = 6 (B). ∗P < 0.05, FLU versus PBS. FLU, influenza; IFN, interferon; PBS, phosphate-buffered saline.
Influenza Infection Results in Alveolitis and Epithelial Metaplasia for up to 2 Months
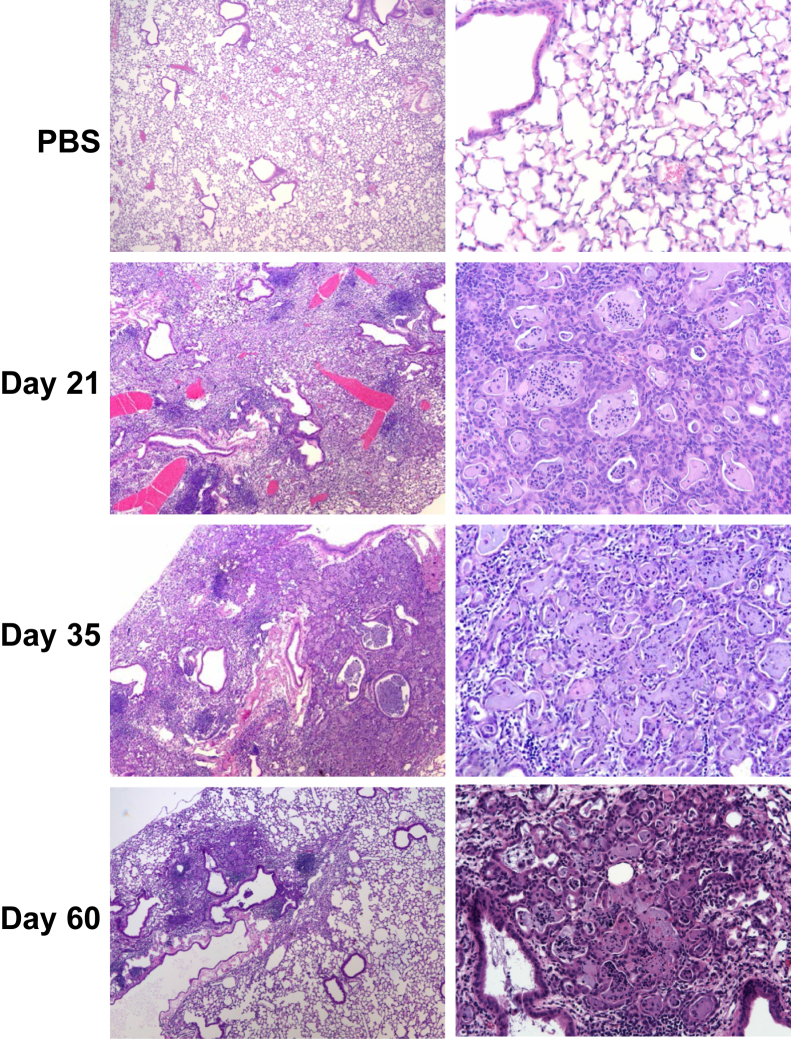
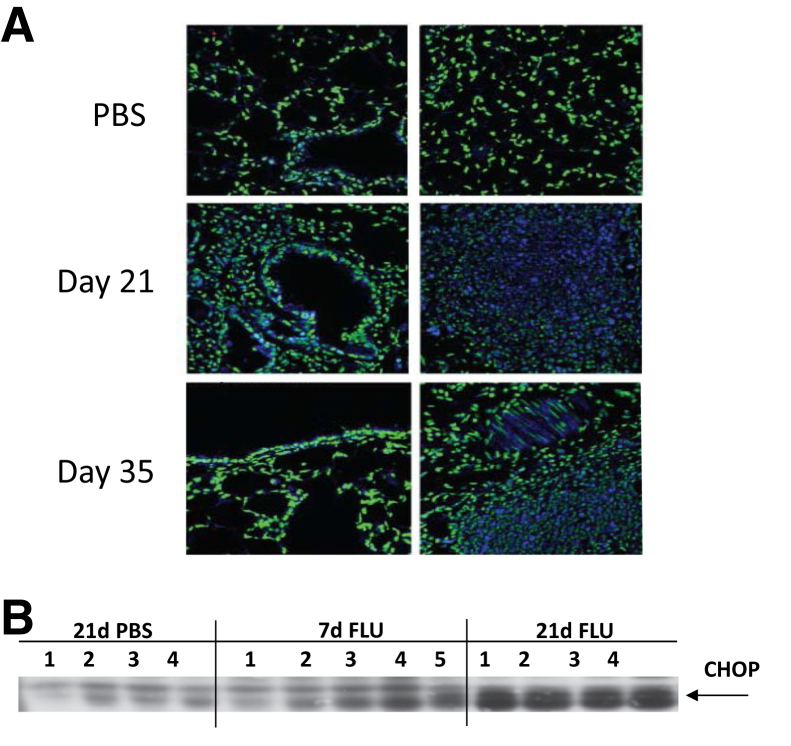
Next, we investigated whether the lung inflammation observed at day 21 after infection persists throughout recovery from influenza challenge. Mice were infected with influenza A/PR/8/34 (H1N1) or PBS vehicle, and lung samples were collected up to 60 days after infection. Interestingly, at 21 days after infection bronchus-associated lymphoid tissue was observed in the lung (dense lymphocytic staining). In addition, regions of the lung parenchyma displayed alveolar epithelial metaplasia and airspace occlusion (Figure 3). This phenotype persisted at both day 35 and 60 after infection, albeit incidence was reduced as resolution from lung injury progressed. Considering the remarkable degree of alveolar remodeling/injury, we examined whether influenza infection induced persistent endoplasmic reticulum (ER) stress in the lung. ER stress was expected to occur during periods of active viral replication; however, lasting ER stress induced by acute influenza infection has not been described. Immunofluorescence staining for ATF6 showed ER stress in foci of alveolar injury in influenza-infected lungs at day 21 and 35 after infection (Figure 4A). Further, Western blot analysis of lung homogenate showed increased expression of C/EBP homologous protein in influenza-infected mice compared with controls at 21 days after infection (Figure 4B). Increased protein expression of these ER stress markers indicated persistent epithelial injury after viral clearance.
Figure 3.
Persistent epithelial metaplasia and alveolitis after influenza infection. C57BL/6 mice were infected with influenza A/PR/8/34 or control PBS vehicle for the indicated time points (representative images). Original magnification: ×40 (left column); ×200 (right column). PBS, phosphate-buffered saline.
Figure 4.
Influenza infection induces lasting endoplasmic reticulum stress in the lung parenchyma. C57BL/6 mice were infected with influenza A/PR/8/34 or control PBS vehicle for the indicated time points. A: Immunofluorescence for ATF6 (green = nuclei, blue = ATF6, representative images). B: Western blot analysis of CHOP expression. n = 4–5 (B). Original magnification, ×200. ATF6, activating transcription factor 6; CHOP, C/EBP homologous protein; FLU, influenza; PBS, phosphate-buffered saline.
Influenza Infection Results in Lasting Changes in the Lung Transcriptome
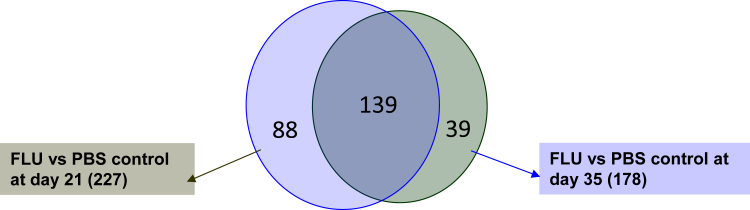
Because of the robust pathologic changes observed in the lung after influenza infection, we examined the impact of influenza on the lung transcriptome. Whole-lung RNA was collected at 21 and 35 days after infection with influenza or PBS vehicle and was subjected to microarray analysis. With the use of a cutoff J5 score of 10.0, 227 genes were found to be differentially regulated in the lung 21 days after influenza infection. At 35 days after infection 178 genes were found to be differentially regulated (J5 of 7.5), most of which were similar to those detected on day 21 (Figure 5). Influenza infection resulted in altered gene expression of many inflammatory mediators, including cytokines, receptors, and antimicrobial peptides in the lung (Table 1). Lactotransferrin, Cxcl9 (monokine induced by γ-IFN), and peptidoglycan recognition protein 1 were the most overexpressed genes in influenza-challenged mice at day 21, and these were persistently elevated at day 35. Evaluation of the lung histopathologic process suggested significant alveolar remodeling, in addition to inflammation. Next, we examined lung injury markers and extracellular matrix protein expression. Influenza infection resulted in up-regulation of many genes associated with tissue remodeling (Table 2). These included chloride channel accessory 3 (Clca3), elastin, and keratin 14 as most increased by influenza at day 21 after infection. A variety of cytokeratins and collagens were induced by influenza infection compared with control, further illustrating the complex molecular processes occurring in the recovering lung. These data indicated that the lung transcriptome was significantly altered by acute viral challenge in a manner that persists for several weeks after infection.
Figure 5.
Influenza infection results in a persistently altered lung transcriptome. C57BL/6 mice were infected with influenza A/PR/8/34 or control PBS vehicle for the indicated time points. Whole-lung RNA was analyzed by microarray. Differentially expressed gene numbers are depicted (J5 score >10 for day 21, J5 score >7.5 for day 35). n = 4. FLU, influenza; PBS, phosphate-buffered saline.
Table 1.
Differential Inflammatory Gene Expression Induced by Influenza Infection
| Gene symbol | Flu d21 | Flu d35 |
|---|---|---|
| Ltf | 42.503 | 10.068 |
| Cxcl9 | 34.21 | 15.521 |
| Pglyrp1 | 20.315 | 11.955 |
| Prg2 | 17.106 | 20.312 |
| Cxcl13 | 14.99 | 11.354 |
| Cxcr3 | 12.656 | 5.464 |
| Ccl9 | 12.23 | 11.827 |
| Ccl21a | 11.702 | 5.739 |
| Plunc | 11.443 | 14.455 |
| Ccl21c | 10.919 | 3.588 |
| Ccr5 | 10.804 | 3.522 |
| Lcn2 | 8.785 | 7.547 |
| Cxcl10 | 8.301 | 2.101 |
| Saa3 | 8.168 | 9.345 |
| Socs3 | 7.223 | ND |
| Ccl6 | 6.828 | 3.254 |
| Casp1 | 6.452 | ND |
| Ccl3 | 6.443 | ND |
| Ccl4 | 6.416 | ND |
| Ccl19 | 6.335 | ND |
| Ccl17 | 5.77 | 6.251 |
| Il10ra | 5.722 | ND |
| Clec11a | 5.649 | ND |
| Ccr6 | 5.437 | 2.053 |
| Fstl1 | 4.537 | ND |
| Cx3cr1 | 4.439 | ND |
| Il27ra | 4.306 | ND |
| Ccl5 | 4.024 | ND |
| Cxcr4 | −4.118 | ND |
| Il6ra | −4.224 | −3.316 |
| Nod1 | −4.284 | ND |
| Il1b | −4.457 | ND |
| Il12a | −5.113 | −4.868 |
| Clec1a | −6.905 | −6.038 |
| Camp | −12.714 | −3.002 |
| S100A8 | −15.762 | −8.756 |
| S100A9 | −16.034 | −7.669 |
J5 scores at 21 and 35 days after influenza infection.
Flu, influenza; ND, not detected.
Table 2.
Differential Matrix and Profibrotic Gene Expression Induced by Influenza Infection
| Gene symbol | Flu d21 | Flu d35 |
|---|---|---|
| Clca3 | 37.315 | 4.093 |
| Eln | 25.301 | ND |
| Krt14 | 24.406 | 10.008 |
| Krt17 | 17.648 | 8.104 |
| Slpi | 15.72 | 11.784 |
| Spp1 | 15.435 | 13.395 |
| Mmp2 | 12.67 | 2.299 |
| Fn1 | 11.071 | ND |
| Tnc | 10.374 | 2.977 |
| Mmp14 | 9.703 | 2.421 |
| Itgae | 9.51 | 4.718 |
| Areg | 9.49 | 7.661 |
| Igf1 | 9.443 | 6.66 |
| Krt15 | 8.974 | 3.44 |
| Col1a1 | 7.972 | 2.526 |
| Aqp3 | 7.456 | 4.365 |
| Krt5 | 7.01 | 3.759 |
| Tgfb3 | 6.65 | 2.482 |
| Col28a1 | 6.081 | 2.553 |
| Muc5ac | 5.711 | 2.211 |
| Timp1 | 4.557 | 5.304 |
| Col6a2 | 4.82 | ND |
| Col6a3 | 4.67 | 4.681 |
| Itgb7 | 4.619 | 4.756 |
| Muc1 | 4.561 | 3.223 |
| Krt18 | 4.553 | 3.323 |
| Adamts12 | 4.435 | ND |
| Col6a1 | 4.345 | ND |
| Col5a1 | 4.321 | ND |
| Col15a1 | 4.287 | 3.014 |
| Tgfb1 | 4.175 | ND |
| Aqp4 | 4.08 | 2.303 |
J5 scores at 21 and 35 days after influenza infection.
Flu, influenza; ND, not detected.
Influenza Infection Does Not Increase Susceptibility to Secondary Bacterial Pneumonia during the Reparative Phase
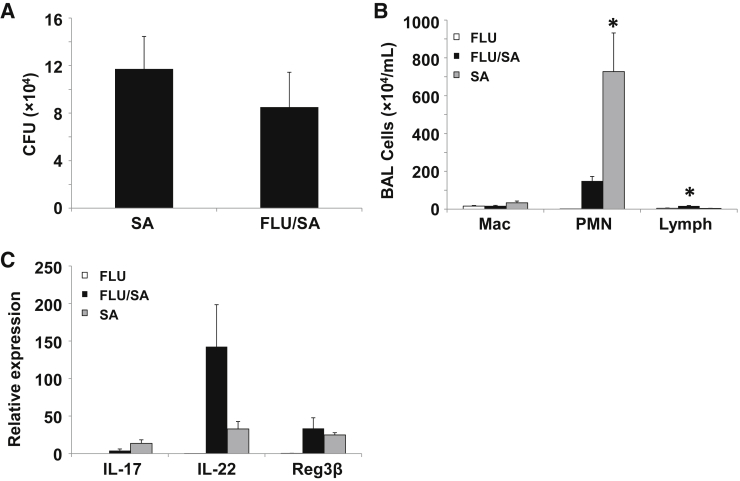
Work by our laboratory and others has demonstrated that preceding influenza infection results in increased susceptibility to subsequent bacterial challenge in mice.10, 14, 15, 16, 17 A primary mechanism by which influenza impairs host defense against extracellular bacteria was through type I IFN-mediated suppression of Type 17 immunity.10, 14 Type I IFN levels peak during the first week of influenza infection and diminish as viral clearance occurs.14 Data presented herein show many changes in lung structure and inflammation 21 days after influenza infection. To determine whether the lung alveolitis and epithelial metaplasia affect host defense against secondary bacterial pneumonia, mice were infected with S. aureus (USA300, MRSA) for 24 hours at 21 days after infection with influenza or PBS vehicle. Unlike findings at earlier time points after influenza challenge, superinfected mice were not more susceptible to S. aureus infection (Figure 6A). Interestingly, mice infected with S. aureus after previous administration of only PBS vehicle displayed marked neutrophil recruitment to the lung, which was significantly suppressed in superinfected animals (Figure 6B). Finally, S. aureus challenge resulted in increased IL-17 and IL-22 expression typical of Type 17 immune activation. Superinfection with influenza resulted in decreased IL-17 expression (similar to other models) but enhanced IL-22 production (Figure 6C). No differences in expression of the antimicrobial peptide, regenerating islet-derived protein-3 β were observed, suggesting intact antimicrobial responses in the lung.
Figure 6.
Influenza infection fails to exacerbate secondary bacterial pneumonia at 21 days after infection. C57BL/6 mice were infected with influenza A/PR/8/34 or control PBS vehicle for 21 days followed by Staphylococcus aureus for 24 hours. A: Bacterial burden in lung homogenate. B: BAL inflammatory cells determined by differential counts. C: Inflammatory gene expression by quantitative RT-PCR. n = 7 SA (A); n = 8 FLU/SA (A); n = 8 (B and C). ∗P < 0.05 versus both of the other infection groups. BAL, bronchoalveolar lavage; CFU, colony-forming unit; FLU, influenza; Mac, macrophage; PBS, phosphate-buffered saline; PMN, polymorphonuclear leukocyte; SA, Staphylococcus aureus.
Influenza Infection Induces Persistent Epigenetic Alterations in the Lung
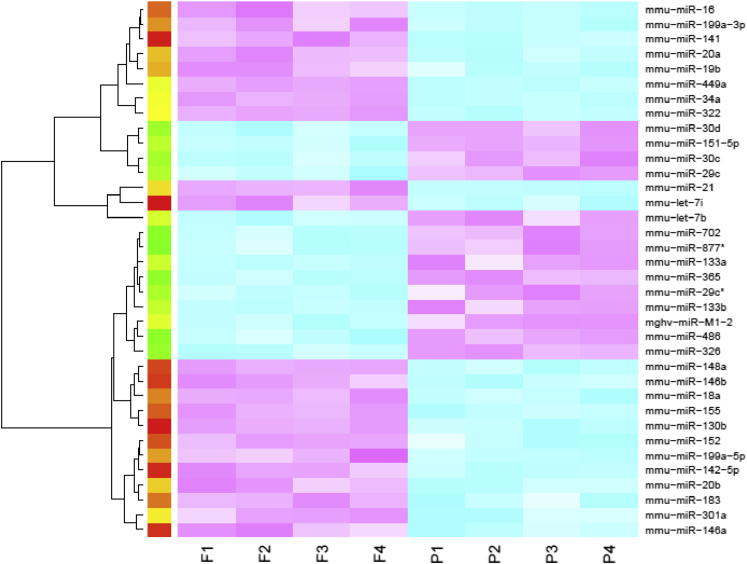
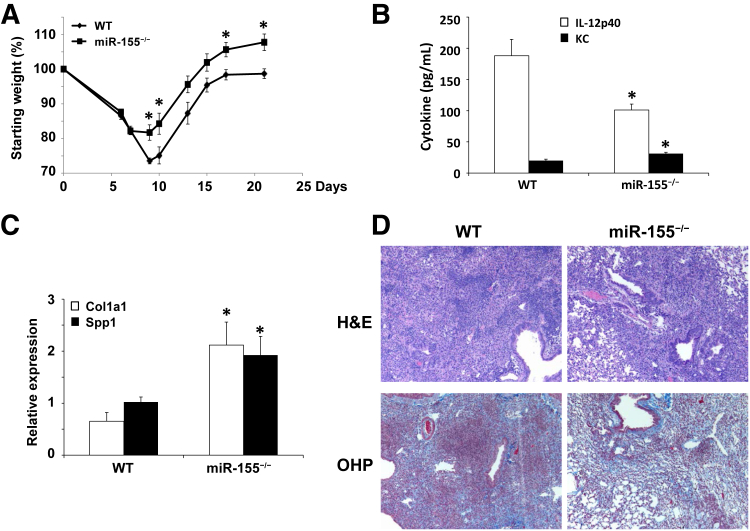
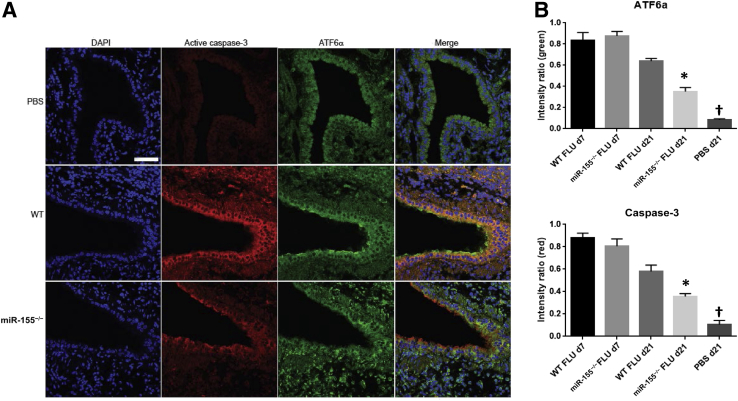
Altered epigenetic regulation was described in interstitial lung disease.18, 19 Considering the lung pathologic process associated with the resolution from influenza infection and the dramatic changes in the lung transcriptome, we examined the impact of influenza infection on miRNA expression in the lung 21 days after infection. Mice were infected with influenza A/PR/8/34 (H1N1) or PBS vehicle for 21 days, and miRNA microarray was performed. Thirty-six miRNAs were differentially expressed (1.5-fold cutoff) between influenza-infected and control lungs (Figure 7). Several of the identified miRNAs were previously identified as altered by influenza infection.20, 21 Specifically, miR-155 was studied for a role in regulation of tissue remodeling. In our samples, miR-155 was increased by influenza infection compared with control mice. Next, we examined whether there was evidence of miR-155–regulated gene expression during recovery from influenza infection. We performed miRNA-mRNA network analysis and identified 26 known target genes for miR-155 that were differentially expressed in the lung at 21 days after influenza infection (Table 3). These data suggested epigenetic control of the host repair process after infection. To determine whether miR-155 played a direct role in lung injury and repair induced by influenza infection, WT and miR-155−/− mice at 7 and 21 days after infection with influenza A/PR/8/34 (H1N1) or PBS vehicle were studied. Interestingly, miR-155−/− mice displayed similar morbidity as WT mice up to 7 days after infection. However, miR-155−/− mice did not continue to lose weight and rebounded significantly faster than WT mice (Figure 8A). No differences were found in BAL cells or protein at day 7 or 21 after influenza infection (data not shown). Inflammatory cytokines were not significantly different at day 7 after infection; however, only two, IL-12p40 and KC (Cxcl1), were altered at day 21 (Figure 8B). Viral clearance was not significantly different between the groups (data not shown). Lung gene expression of profibrotic genes collagen type-1a1 and secreted phosphoprotein-1 were significantly increased in miR-155−/− mice compared with controls (Figure 8C). Next, we examined lung histopathologic process and collagen deposition in WT and miR-155−/− mice at 21 days after infection. miR-155−/− mice had decreased lung parenchymal injury compared with WT animals (Figure 8D). Despite decreased parenchymal cellularity, collagen deposition was similar in both groups. Finally, because of the improved recovery from influenza infection in miR-155−/− mice, we examined if ER stress was limited compared with WT controls. Indeed, influenza infection induced robust epithelial ER stress in WT mice 21 days after infection (Figure 9); miR-155−/− mice had significantly reduced ER stress staining (ATF6α and active caspase-3). These data demonstrated a role for miR-155 epigenetic regulation of lung injury and repair after influenza infection.
Figure 7.
Differential miRNA expression at 21 days after influenza infection. C57BL/6 mice were infected with influenza A/PR/8/34 or control PBS vehicle for 21 days. Whole-lung miRNA were analyzed by microarray (1.5-fold differential cutoff). Blue indicates underexpressed; red, overexpressed. n = 4. PBS, phosphate buffered saline.
Table 3.
miR-155/mRNA Interactions at 21 Days after Influenza Infection
| Gene symbol | miR-155 action | J5 score |
|---|---|---|
| Sla | Activates | 6.537 |
| Ddit4 | Activates | 4.978 |
| Ikbke | Activates | 3.657 |
| Aicda | Activates | 2.887 |
| Sdcbp | Activates | 2.676 |
| Cldn1 | Activates | 2.655 |
| Inpp5d | Activates | 2.606 |
| Trp53 | Activates | 2.010 |
| Pea15a | Suppresses | −2.087 |
| Trim32 | Suppresses | −2.382 |
| Clock | Suppresses | −2.547 |
| Osr1 | Suppresses | −2.715 |
| Agtrap | Suppresses | −2.855 |
| Phf17 | Suppresses | −3.012 |
| Rheb | Suppresses | −3.064 |
| Atxn1l | Suppresses | −3.28 |
| Wnk1 | Suppresses | −3.522 |
| Tcf4 | Suppresses | −3.605 |
| Tle4 | Suppresses | −3.628 |
| Camta1 | Suppresses | −3.676 |
| Rora | Suppresses | −3.809 |
| Kras | Suppresses | −4.078 |
| Meis1 | Suppresses | −5.999 |
| Ghr | Suppresses | −6.274 |
| Sox11 | Suppresses | −6.478 |
| Fgf7 | Suppresses | −8.167 |
Figure 8.
miR-155 regulates lung injury and morbidity during influenza infection recovery. C57BL/6 or miR-155−/− mice were infected with influenza A/PR/8/34 or control PBS vehicle for 21 days. A: Morbidity as measured by weight loss. B: Inflammatory cytokine expression in lung homogenate as assayed by multiplex cytokine assay as described in Materials and Methods. C: Profibrotic gene expression by quantitative RT-PCR. D: Lung injury and collagen content by OHP staining, representative images. n = 6 (A–C). ∗P < 0.05 versus WT. Original magnification, ×100. H&E, hematoxylin and eosin; KC, Cxcl1; OHP, hydroxyproline; WT, wild-type.
Figure 9.
Influenza infection-induced ER stress is attenuated in miR-155−/− mice. C57BL/6 or miR-155−/− mice were infected with influenza A/PR/8/34 or control PBS vehicle for 21 days. A: ER stress staining for active caspase 3 (red), ATF6 (green), and nuclei (DAPI, blue). B: Quantification of ER stress staining by blinded scoring (V.A.). n = 4. ∗P < 0.05 versus WT FLU 21 days; †P < 0.05 versus all groups. Scale bar = 50 μm. Original magnification, ×200. ATF6, activating transcription factor 6; ER, endoplasmic reticulum; FLU, influenza; PBS, phosphate-buffered saline; WT, wild-type.
Discussion
Persistent alveolitis and collagen deposition in the lung after influenza infection was first described >30 years ago.22 However, to date this robust remodeling/repair process has not been thoroughly investigated at the molecular level. Our data demonstrate persistent lung inflammation, epithelial injury and metaplasia, and genetic reprogramming in the weeks after primary influenza infection. Specifically, ER stress and transcriptomic and epigenetic changes remain well after viral clearance and recovery from disease-induced morbidity. Inflammation is commonly associated with epithelial injury and disrepair in the lung. Consistent with this, we found that inflammation persisted to 21 days after infection. These findings are in line with advance temporal imaging studies that demonstrate increased lung capillary leak at 21 days after infection.23, 24 Further, others have shown disruption in lung antigen-presenting cell populations after primary influenza infection.25 Although we cannot say which event is provoking the other (inflammation leading to extended disrepair or vice versa), the lung remains in an extended state of frustrated inflammation and repair, resulting in increased fibrotic gene expression.
We have previously shown that acute influenza infection induces ER stress in lung epithelial cells.26 Our finding that ER stress is present in epithelial cells during recovery from influenza infection is consistent with the finding that DNA damage and apoptotic cells are also persistently increased after infection.27 It is likely that persistent ER stress contributes to the epithelial metaplasia and honeycombing that is seen through 60 days after infection. It is known that parenchymal repair occurs through the migration of distal airway stem/progenitor cells to sites of severe influenza injury. Although these cells are considered to be lineage negative, they are identified by the expression of Krt5, ΔNP63, and α6β4 integrin during their migratory and proliferative phase.28, 29, 30 Once at sites of injury, they form structures known as pods that are associated with alveolar regeneration.28 Successful lung repair requires these progenitor cells to differentiate to type II phenotype and subsequent conversion to type I cells. The inability of these cells to differentiate leads to the honeycomb-like structures.29 Recently, it was shown that these Krt5 pods are not observed during lung injury induced by less virulent X31 influenza in mice, suggesting that the severity of lung injury may dictate the cellular repair process.31 It is possible that the persistent ER stress observed herein may modulate the ability of the progenitor cells to differentiate and to form alveolar structure.
Our data demonstrate that influenza infection induces a persistent profibrotic gene expression pattern in the lung. Further, lung pathologic process is reminiscent of that seen with interstitial lung disease. Note that the experiments reported here use the mouse-adapted experimental A/PR/8/34 H1N1 strain of influenza. This strain has differences from some circulating human influenza strains and may only be representative of severe or pandemic influenza infections. Consistent with this, it was shown that both adults32, 33 and children34 recovering from severe influenza outbreaks experienced necrotizing bronchiolitis, alveolitis, intimal thickening, and fibrosis. Highly pathogenic influenza (H5N1) was shown to cause lung fibrosis in patients 30 days after infection.35 Recently, histopathologic fibroproliferative changes were observed in a patient with severe H7N9 influenza infection.36 Further, influenza was shown to induce transforming growth factor (TGF)-β activation, a known profibrotic mediator, via Tlr3-dependent αvβ6 integrin.37 Similarly, our data show increased TGF-β1 and TGF-β3 expression after influenza challenge. These data suggest that primary influenza infection may prime the lung for greater responses to fibrotic stimuli.
miRNA involvement in fibrosis is emerging and likely complex. Evidence is mounting that certain families of miRNAs function as global regulators of cellular differentiation. The let-7 family has a crucial role in development and may act as a tumor suppressor, because it is down-regulated in a number of cancers.38 In the epithelium, let-7 is considered a good candidate for epithelial gene signature,39 and, similar to the miR-200 family, let-7 down-regulation leads to loss of epithelial characteristics and fibrotic gene expression.40, 41 Although loss of miRNA may lead to loss of epithelial function, increases in certain miRNAs were associated with fibrogenesis. miR-21 was demonstrated to promote fibroblast survival, and its inhibition decreased scarring in a mouse model of myocardial infarction.42, 43 Inhibition of miR-21 also attenuated bleomycin-induced fibrosis.44 In the case of pulmonary fibrosis, miR-155 was shown to be increased after TGF-β treatment of epithelial cells, causing disruption of tight junctions.45 Further, miR-155 and miR-29 were shown to be significantly increased after bleomycin injury and important in fibroblast survival.45, 46 miR-155 was shown to regulate keratinocyte growth factor (KGF; FGF7), resulting in reduced expression.45 KGF is known to play a protective role in pulmonary fibrosis.47 miR-155 is also known to regulate TGF-β signaling by affecting SMAD2, and miR-155 knockout mice have altered collagen deposition late in life.48, 49 These findings suggest a potential role for miRNA modulation in controlling gene expression in epithelial cells and, thus, regulating fibrosis. Our data demonstrate a role for miR-155 in negatively regulating KGF during lung repair after influenza infection, suggesting a pathogenic role. Further, miR-155 regulates a number of transcriptional regulators in our data set, likely indicating a broad role for epigenetic regulation of the host repair process.
Influenza-induced alteration in miRNA expression is an active area of research. Acute miRNA expression changes after influenza infection were characterized by microarray and deep sequencing.20, 21 These studies identified miR-29, miR-30, and let-7 family member alterations consistent with our data. Further work examined miR-30, let-7, and miR-21 expression changes 15 days after influenza infection.50 We chose to focus on miR-155 in our study because of the significant amount of interest in this miRNA in fibrosis and also in host defense. Pandemic 2009 H1N1 influenza was shown to induce miR-155 in the lung.51 Recently, miR-155−/− mice were shown to be protected against influenza, S. aureus superinfection via rescue of Type 17 immunity.52 These mice had improved bacterial clearance without an impact on viral clearance, similar to our data. In addition, miR-155 antagomir treatment improved bacterial clearance during superinfection. Our data suggest that the transcription factor Rora may be regulated by miR-155, and this interaction may explain the enhanced Type 17 immunity seen in miR-155−/− mice during superinfection. The impact of miR-155 deletion on the recovery from influenza infection was not examined in that study. Another group has shown that mir-155 regulates influenza virus–induced CD8+ T-cell function and subsequent viral clearance.53 However, we did not see a viral clearance defect 7 days after infection in our study. Our data demonstrate that miR-155−/− mice recover faster than WT controls after influenza infection, with decreased lung inflammation, ER stress, and pathologic process, despite increased collagen expression. These data indicate that miR-155 likely regulates the lung repair process after influenza injury through interactions with many gene pathways.
Work by our group and others have demonstrated that preceding influenza infection increases susceptibility to secondary S. aureus pneumonia.10, 14, 15, 16, 17 Epidemiologic examination from both seasonal and pandemic influenza outbreaks confirms this severe and often lethal synergy between these two pathogens.54 The window of susceptibility to secondary bacterial pneumonia after influenza infection ranges from 5 to 10 days after challenge. During this time, secondary infection results in increased lung inflammation, impaired Type 17 antibacterial immunity, decreased antimicrobial peptide production, and increased mortality.55 Interestingly, influenza infection did not exacerbate S. aureus burden at 21 days after infection. Despite similar bacterial clearance, airspace inflammation induced by S. aureus was attenuated in influenza superinfected mice. Similar to earlier time points, IL-17 production in response to S. aureus remained attenuated in superinfected animals; however, IL-22 production was enhanced by preceding influenza infection. IL-22 is known to induce antimicrobial peptides in the lung in response to bacteria.56 Our transcriptomic data indicated that influenza increased expression of several antimicrobial peptide genes (Pglyrp1, Plunc, Lcn2) at 21 and 35 days after infection, and Reg3β expression induced by S. aureus was no longer attenuated by preceding influenza. We have previously shown that Lcn2 promotes clearance of S. aureus during influenza superinfection.17 It is possible that the lung repairing after influenza is able to competently control S. aureus via antimicrobial peptide functions, despite decreased phagocyte recruitment.
Conclusions
Our data provide a glimpse into the complicated molecular events occurring during recovery from influenza infection. There is significant interest in epithelial reprogramming in the interstitial lung disease field. Our data demonstrate a clear persistent impact of influenza on the lung epigenetic and transcriptomic profile after completion of viral clearance and resolution of morbidity. These programmatic changes may alter the lung epithelial response to secondary challenges, be it from pathogen or environmental stimuli. Additional study is required to determine which of these pathways are normative repair processes and which may be pathogenic in nature.
Acknowledgment
Microarray data analysis was performed by Dr. James Lyons-Weiler (University of Pittsburgh Genomics Research Core).
Footnotes
See related Commentary on page 697
Supported by University of Pittsburgh NIH Clinical and Translational Science Award grants UL1RR024153 and UL1TR000005 (J.F.A.); NIH grants R01HL107380 (J.F.A.), R01HL122760 (D.A.P.), R21AI117569 (D.A.P.), R01HL122383 (V.A.), R01HL079331 (Y.M.W.J.-H.), R01LM012087 (P.V.B.), and R01LM009657 (P.V.B.); and Parker B. Francis Foundation Fellowship (J.F.A.).
Disclosures: None declared.
References
- 1.Zimmer S.M., Crevar C.J., Carter D.M., Stark J.H., Giles B.M., Zimmerman R.K., Ostroff S.M., Lee B.Y., Burke D.S., Ross T.M. Seroprevalence following the second wave of Pandemic 2009 H1N1 influenza in Pittsburgh, PA, USA. PLoS One. 2010;5:e11601. doi: 10.1371/journal.pone.0011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shrestha S.S., Swerdlow D.L., Borse R.H., Prabhu V.S., Finelli L., Atkins C.Y., Owusu-Edusei K., Bell B., Mead P.S., Biggerstaff M., Brammer L., Davidson H., Jernigan D., Jhung M.A., Kamimoto L.A., Merlin T.L., Nowell M., Redd S.C., Reed C., Schuchat A., Meltzer M.I. Estimating the burden of 2009 pandemic influenza A (H1N1) in the United States (April 2009-April 2010) Clin Infect Dis. 2011;52 Suppl 1:S75–S82. doi: 10.1093/cid/ciq012. [DOI] [PubMed] [Google Scholar]
- 3.Van Kerkhove M.D., Hirve S., Koukounari A., Mounts A.W., HIN1pdm serology working group Estimating age-specific cumulative incidence for the 2009 influenza pandemic: a meta-analysis of A(H1N1)pdm09 serological studies from 19 countries. Influenza Other Respir Viruses. 2013;7:872–886. doi: 10.1111/irv.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peteranderl C., Herold S., Schmoldt C. Human influenza virus infections. Semin Respir Crit Care Med. 2016;37:487–500. doi: 10.1055/s-0036-1584801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taubenberger J.K., Morens D.M. The pathology of influenza virus infections. Annu Rev Pathol. 2008;3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jakab G.J. Sequential virus infections, bacterial superinfections, and fibrogenesis. Am Rev Respir Dis. 1990;142:374–379. doi: 10.1164/ajrccm/142.2.374. [DOI] [PubMed] [Google Scholar]
- 7.Braciale T.J. Immunologic recognition of influenza virus-infected cells. I. Generation of a virus-strain specific and a cross-reactive subpopulation of cytotoxic T cells in the response to type A influenza viruses of different subtypes. Cell Immunol. 1977;33:423–436. doi: 10.1016/0008-8749(77)90170-8. [DOI] [PubMed] [Google Scholar]
- 8.Crowe C.R., Chen K., Pociask D.A., Alcorn J.F., Krivich C., Enelow R.I., Ross T.M., Witztum J.L., Kolls J.K. Critical role of IL-17RA in immunopathology of influenza infection. J Immunol. 2009;183:5301–5310. doi: 10.4049/jimmunol.0900995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van der Velden J., Janssen-Heininger Y.M., Mandalapu S., Scheller E.V., Kolls J.K., Alcorn J.F. Differential requirement for c-Jun N-terminal kinase 1 in lung inflammation and host defense. PLoS One. 2012;7:e34638. doi: 10.1371/journal.pone.0034638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kudva A., Scheller E.V., Robinson K.M., Crowe C.R., Choi S.M., Slight S.R., Khader S.A., Dubin P.J., Enelow R.I., Kolls J.K., Alcorn J.F. Influenza A inhibits Th17-mediated host defense against bacterial pneumonia in mice. J Immunol. 2011;186:1666–1674. doi: 10.4049/jimmunol.1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pociask D.A., Scheller E.V., Mandalapu S., McHugh K.J., Enelow R.I., Fattman C.L., Kolls J.K., Alcorn J.F. IL-22 is essential for lung epithelial repair following influenza infection. Am J Pathol. 2013;182:1286–1296. doi: 10.1016/j.ajpath.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jordan R., Patel S., Hu H., Lyons-Weiler J. Efficiency analysis of competing tests for finding differentially expressed genes in lung adenocarcinoma. Cancer Inform. 2008;6:389–421. doi: 10.4137/cin.s791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang G.T., Athanassiou C., Benos P.V. mirConnX: condition-specific mRNA-microRNA network integrator. Nucleic Acids Res. 2011;39:W416–W423. doi: 10.1093/nar/gkr276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee B., Robinson K.M., McHugh K.J., Scheller E.V., Mandalapu S., Chen C., Di Y.P., Clay M.E., Enelow R.I., Dubin P.J., Alcorn J.F. Influenza-induced type I interferon enhances susceptibility to gram-negative and gram-positive bacterial pneumonia in mice. Am J Physiol Lung Cell Mol Physiol. 2015;309:L158–L167. doi: 10.1152/ajplung.00338.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson K.M., Choi S.M., McHugh K.J., Mandalapu S., Enelow R.I., Kolls J.K., Alcorn J.F. Influenza A exacerbates Staphylococcus aureus pneumonia by attenuating IL-1beta production in mice. J Immunol. 2013;191:5153–5159. doi: 10.4049/jimmunol.1301237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson K.M., Lee B., Scheller E.V., Mandalapu S., Enelow R.I., Kolls J.K., Alcorn J.F. The role of IL-27 in susceptibility to post-influenza Staphylococcus aureus pneumonia. Respir Res. 2015;16:10. doi: 10.1186/s12931-015-0168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson K.M., McHugh K.J., Mandalapu S., Clay M.E., Lee B., Scheller E.V., Enelow R.I., Chan Y.R., Kolls J.K., Alcorn J.F. Influenza A virus exacerbates Staphylococcus aureus pneumonia in mice by attenuating antimicrobial peptide production. J Infect Dis. 2014;209:865–875. doi: 10.1093/infdis/jit527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandit K.V., Milosevic J. MicroRNA regulatory networks in idiopathic pulmonary fibrosis. Biochem Cell Biol. 2015;93:129–137. doi: 10.1139/bcb-2014-0101. [DOI] [PubMed] [Google Scholar]
- 19.Pandit K.V., Milosevic J., Kaminski N. MicroRNAs in idiopathic pulmonary fibrosis. Transl Res. 2011;157:191–199. doi: 10.1016/j.trsl.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 20.Li Y., Chan E.Y., Li J., Ni C., Peng X., Rosenzweig E., Tumpey T.M., Katze M.G. MicroRNA expression and virulence in pandemic influenza virus-infected mice. J Virol. 2010;84:3023–3032. doi: 10.1128/JVI.02203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng X., Gralinski L., Ferris M.T., Frieman M.B., Thomas M.J., Proll S., Korth M.J., Tisoncik J.R., Heise M., Luo S., Schroth G.P., Tumpey T.M., Li C., Kawaoka Y., Baric R.S., Katze M.G. Integrative deep sequencing of the mouse lung transcriptome reveals differential expression of diverse classes of small RNAs in response to respiratory virus infection. MBio. 2011;2:e00198-11. doi: 10.1128/mBio.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jakab G.J., Astry C.L., Warr G.A. Alveolitis induced by influenza virus. Am Rev Respir Dis. 1983;128:730–739. doi: 10.1164/arrd.1983.128.4.730. [DOI] [PubMed] [Google Scholar]
- 23.Gotts J.E., Abbott J., Matthay M.A. Influenza causes prolonged disruption of the alveolar-capillary barrier in mice unresponsive to mesenchymal stem cell therapy. Am J Physiol Lung Cell Mol Physiol. 2014;307:L395–L406. doi: 10.1152/ajplung.00110.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin L., Xu S., Cheng J., Zheng D., Limmon G.V., Leung N.H., Rajapakse J.C., Chow V.T., Chen J., Yu H. Spatiotemporal quantification of cell dynamics in the lung following influenza virus infection. J Biomed Opt. 2013;18:046001. doi: 10.1117/1.JBO.18.4.046001. [DOI] [PubMed] [Google Scholar]
- 25.Strickland D.H., Fear V., Shenton S., Wikstrom M.E., Zosky G., Larcombe A.N., Holt P.G., Berry C., von Garnier C., Stumbles P.A. Persistent and compartmentalised disruption of dendritic cell subpopulations in the lung following influenza A virus infection. PLoS One. 2014;9:e111520. doi: 10.1371/journal.pone.0111520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberson E.C., Tully J.E., Guala A.S., Reiss J.N., Godburn K.E., Pociask D.A., Alcorn J.F., Riches D.W., Dienz O., Janssen-Heininger Y.M., Anathy V. Influenza induces endoplasmic reticulum stress, caspase-12-dependent apoptosis, and c-Jun N-terminal kinase-mediated transforming growth factor-beta release in lung epithelial cells. Am J Respir Cell Mol Biol. 2012;46:573–581. doi: 10.1165/rcmb.2010-0460OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li N., Parrish M., Chan T.K., Yin L., Rai P., Yoshiyuki Y., Abolhassani N., Tan K.B., Kiraly O., Chow V.T., Engelward B.P. Influenza infection induces host DNA damage and dynamic DNA damage responses during tissue regeneration. Cell Mol Life Sci. 2015;72:2973–2988. doi: 10.1007/s00018-015-1879-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar P.A., Hu Y., Yamamoto Y., Hoe N.B., Wei T.S., Mu D., Sun Y., Joo L.S., Dagher R., Zielonka E.M., Wang de Y., Lim B., Chow V.T., Crum C.P., Xian W., McKeon F. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell. 2011;147:525–538. doi: 10.1016/j.cell.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaughan A.E., Brumwell A.N., Xi Y., Gotts J.E., Brownfield D.G., Treutlein B., Tan K., Tan V., Liu F.C., Looney M.R., Matthay M.A., Rock J.R., Chapman H.A. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 2015;517:621–625. doi: 10.1038/nature14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuo W., Zhang T., Wu D.Z., Guan S.P., Liew A.A., Yamamoto Y., Wang X., Lim S.J., Vincent M., Lessard M., Crum C.P., Xian W., McKeon F. p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature. 2015;517:616–620. doi: 10.1038/nature13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanegai C.M., Xi Y., Donne M.L., Gotts J.E., Driver I.H., Amidzic G., Lechner A.J., Jones K.D., Vaughan A.E., Chapman H.A., Rock J.R. Persistent pathology in influenza-infected mouse lungs. Am J Respir Cell Mol Biol. 2016;55:613–615. doi: 10.1165/rcmb.2015-0387LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hers J.F., Masurel N., Mulder J. Bacteriology and histopathology of the respiratory tract and lungs in fatal Asian influenza. Lancet. 1958;2:1141–1143. doi: 10.1016/s0140-6736(58)92404-8. [DOI] [PubMed] [Google Scholar]
- 33.Hers J.F., Mulder J. Broad aspects of the pathology and pathogenesis of human influenza. Am Rev Respir Dis. 1961;83(Pt 2):84–97. doi: 10.1164/arrd.1961.83.2P2.84. [DOI] [PubMed] [Google Scholar]
- 34.Kitamura S., Higuchi M. Pathological studies on viral pneumonia and interstitial pneumonitis caused by unknown etiology. Acta Pathol Jpn. 1972;22:859–876. doi: 10.1111/j.1440-1827.1972.tb00768.x. [DOI] [PubMed] [Google Scholar]
- 35.Qiao J., Zhang M., Bi J., Wang X., Deng G., He G., Luan Z., Lv N., Xu T., Zhao L. Pulmonary fibrosis induced by H5N1 viral infection in mice. Respir Res. 2009;10:107. doi: 10.1186/1465-9921-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang J.B., Li H.Y., Liu J.F., Lan C.Q., Lin Q.H., Chen S.X., Zhang H.Y., Wang X.H., Lin X., Pan J.G., Weng H. Histopathological findings in a critically ill patient with avian influenza A (H7N9) J Thorac Dis. 2015;7:E672–E677. doi: 10.3978/j.issn.2072-1439.2015.11.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jolly L., Stavrou A., Vanderstoken G., Meliopoulos V.A., Habgood A., Tatler A.L., Porte J., Knox A., Weinreb P., Violette S., Hussell T., Kolb M., Stampfli M.R., Schultz-Cherry S., Jenkins G. Influenza promotes collagen deposition via alphavbeta6 integrin-mediated transforming growth factor beta activation. J Biol Chem. 2014;289:35246–35263. doi: 10.1074/jbc.M114.582262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson C.D., Esquela-Kerscher A., Stefani G., Byrom M., Kelnar K., Ovcharenko D., Wilson M., Wang X., Shelton J., Shingara J., Chin L., Brown D., Slack F.J. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 39.Shell S., Park S.M., Radjabi A.R., Schickel R., Kistner E.O., Jewell D.A., Feig C., Lengyel E., Peter M.E. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci U S A. 2007;104:11400–11405. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pandit K.V., Corcoran D., Yousef H., Yarlagadda M., Tzouvelekis A., Gibson K.F., Konishi K., Yousem S.A., Singh M., Handley D., Richards T., Selman M., Watkins S.C., Pardo A., Ben-Yehudah A., Bouros D., Eickelberg O., Ray P., Benos P.V., Kaminski N. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2010;182:220–229. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peter M.E. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–852. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thum T., Gross C., Fiedler J., Fischer T., Kissler S., Bussen M., Galuppo P., Just S., Rottbauer W., Frantz S., Castoldi M., Soutschek J., Koteliansky V., Rosenwald A., Basson M.A., Licht J.D., Pena J.T., Rouhanifard S.H., Muckenthaler M.U., Tuschl T., Martin G.R., Bauersachs J., Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 43.Dong S., Cheng Y., Yang J., Li J., Liu X., Wang X., Wang D., Krall T.J., Delphin E.S., Zhang C. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J Biol Chem. 2009;284:29514–29525. doi: 10.1074/jbc.M109.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu G., Friggeri A., Yang Y., Milosevic J., Ding Q., Thannickal V.J., Kaminski N., Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pottier N., Maurin T., Chevalier B., Puissegur M.P., Lebrigand K., Robbe-Sermesant K., Bertero T., Lino Cardenas C.L., Courcot E., Rios G., Fourre S., Lo-Guidice J.M., Marcet B., Cardinaud B., Barbry P., Mari B. Identification of keratinocyte growth factor as a target of microRNA-155 in lung fibroblasts: implication in epithelial-mesenchymal interactions. PLoS One. 2009;4:e6718. doi: 10.1371/journal.pone.0006718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cushing L., Kuang P.P., Qian J., Shao F., Wu J., Little F., Thannickal V.J., Cardoso W.V., Lu J. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol. 2011;45:287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakamoto S., Yazawa T., Baba Y., Sato H., Kanegae Y., Hirai T., Saito I., Goto T., Kurahashi K. Keratinocyte growth factor gene transduction ameliorates pulmonary fibrosis induced by bleomycin in mice. Am J Respir Cell Mol Biol. 2011;45:489–497. doi: 10.1165/rcmb.2010-0092OC. [DOI] [PubMed] [Google Scholar]
- 48.Louafi F., Martinez-Nunez R.T., Sanchez-Elsner T. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-{beta} J Biol Chem. 2010;285:41328–41336. doi: 10.1074/jbc.M110.146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez A., Vigorito E., Clare S., Warren M.V., Couttet P., Soond D.R., van Dongen S., Grocock R.J., Das P.P., Miska E.A., Vetrie D., Okkenhaug K., Enright A.J., Dougan G., Turner M., Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan K.S., Choi H., Jiang X., Yin L., Seet J.E., Patzel V., Engelward B.P., Chow V.T. Micro-RNAs in regenerating lungs: an integrative systems biology analysis of murine influenza pneumonia. BMC Genomics. 2014;15:587. doi: 10.1186/1471-2164-15-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Z., Hao R., Li P., Zhang X., Liu N., Qiu S., Wang L., Wang Y., Xue W., Liu K., Yang G., Cui J., Zhang C., Song H. MicroRNA expression profile of mouse lung infected with 2009 pandemic H1N1 influenza virus. PLoS One. 2013;8:e74190. doi: 10.1371/journal.pone.0074190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Podsiad A., Standiford T.J., Ballinger M.N., Eakin R., Park P., Kunkel S.L., Moore B.B., Bhan U. MicroRNA-155 regulates host immune response to postviral bacterial pneumonia via IL-23/IL-17 pathway. Am J Physiol Lung Cell Mol Physiol. 2016;310:L465–L475. doi: 10.1152/ajplung.00224.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gracias D.T., Stelekati E., Hope J.L., Boesteanu A.C., Doering T.A., Norton J., Mueller Y.M., Fraietta J.A., Wherry E.J., Turner M., Katsikis P.D. The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat Immunol. 2013;14:593–602. doi: 10.1038/ni.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rynda-Apple A., Robinson K.M., Alcorn J.F. Influenza and bacterial superinfection: illuminating the immunologic mechanisms of disease. Infect Immun. 2015;83:3764–3770. doi: 10.1128/IAI.00298-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson K.M., Kolls J.K., Alcorn J.F. The immunology of influenza virus-associated bacterial pneumonia. Curr Opin Immunol. 2015;34:59–67. doi: 10.1016/j.coi.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aujla S.J., Chan Y.R., Zheng M., Fei M., Askew D.J., Pociask D.A., Reinhart T.A., McAllister F., Edeal J., Gaus K., Husain S., Kreindler J.L., Dubin P.J., Pilewski J.M., Myerburg M.M., Mason C.A., Iwakura Y., Kolls J.K. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med. 2008;14:275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]