Abstract
The catabolic enzyme myo-inositol oxygenase (MIOX) is expressed in proximal tubules and up-regulated in the diabetic state. Previously, we reported its transcriptional and translation regulation by high glucose (HG), osmolytes, and fatty acids. However, its epigenetic regulation is unknown. Bisulfite sequencing revealed that both human and mouse MIOX promoters, enriched with CpG sites, are hypomethylated and unmethylated under HG ambience and hyperglycemic states associated with increased MIOX expression. Eletrophoretic mobility shift assays revealed increased binding of unmethylated oligos with nucleoproteins of cells maintained under HG. In addition, a strong binding of specificity protein (Sp)-1 transcription factor with MIOX promoter was observed under HG, especially with unmethylated Sp-1 oligo. Specificity of binding was established by supershift assays and treatment with the Sp-1 inhibitor mithramycin. Promoter analysis revealed an increase in luciferase activity under HG, which was reduced after mutation of the Sp-1–binding site. Sp1 siRNA treatment reduced mRNA and protein expression of Sp-1 and MIOX and generation of reactive oxygen species derived from NADPH oxidase (NOX)-4 and mitochondrial sources. In addition, there was reduced expression of hypoxia-inducible factor-1α relevant in the pathogenesis of diabetic nephropathy. Sp1 siRNA treatment reduced fibronectin expression, an extracellular matrix protein that is increased in diabetic nephropathy and tubulopathy. HG-induced MIOX expression was also reduced with the treatment of apelin-13, which deacetylates histones. Overall, these findings highlight the epigenetic regulation of MIOX in the pathogenesis of diabetic tubulopathy.
Diabetic nephropathy is a well-recognized complication in patients with diabetes mellitus, a metabolic disorder in which sustained hyperglycemia leads to perturbations in the homeostasis of all the compartments of the kidney (ie, glomerulus, tubulointerstitium, and the vasculature).1, 2 In this scenario virtually every cell type of the kidney, albeit with different developmental lineages, including glomerular podocytes, mesangial, and endothelial cells, tubular epithelia, interstitial fibroblasts, and vascular endothelia, are adversely affected by the hyperglycemic injury.2, 3, 4, 5, 6, 7, 8 The extent of injury and the pathologic lesions may be variable among various cell types, but disruption in their cross talk with perturbations in their collective interdependent homeostasis would ultimately lead to progressive renal failure.8 Renal lesions in types 1 and 2 are more or less similar, except for minor variations, and the typical glomerular lesions in diabetic nephropathy are characterized by thickening of basement membrane and formation of Kimmelstiel-Wilson mesangial nodules.2, 3 Nevertheless, the tubulointerstitial compartment is also invariably affected and is regarded as part and parcel of pathologic lesions of diabetic nephropathy.2, 3, 6, 7 Interestingly, the extent of tubulointerstitial injury has been reported to correlate with the degree of deterioration in the renal functions.7, 9
The pathogenetic mechanisms that affect glomerular compartment in diabetic nephropathy have been extensively investigated.2, 4, 5, 6, 7, 8, 10, 11, 12, 13 Briefly, they include increased flux of glucose intermediaries, activation of protein kinase C and transforming growth factor (TGF)-β–Smad–mitogen-activated protein kinase pathways and of the renin-angiotensin system, generation of reactive oxygen species (ROS), and ultimately an aberrant expression of extracellular matrix (ECM) proteins. Incidentally, the ROS are regarded as the common denominator in the pathogenesis of diabetic nephropathy, especially that of the glomerulus.2, 10, 11, 12, 13 Some of these mechanisms may also be applicable to the tubulointerstitial compartment, where increased cellular flux of glucose and initiation of a multitude of signaling events lead to increased fibrosis and thickening of the tubular basement membrane, which conceivably contribute to a compromise in renal functions.6, 7, 9 This underscores the importance of tubular pathobiology in the context of diabetic nephropathy; thus, it has attracted the attention of some investigators in recent years, and their emphasis has been to delineate new pathogenetic mechanisms and to identify novel molecules that would be relevant to the biology of hyperglycemia-induced injury in diabetes.6, 7
A few years back we identified myo-inositol oxygenase (MIOX), which is expressed in renal proximal tubular epithelium and up-regulated in states of hyperglycemia.14 Since then a number of studies have investigated its biological characteristics in the context of diabetic nephropathy.14, 15, 16, 17, 18 The promoter analyses of MIOX indicates the presence of osmotic response elements (OsREs), carbohydrate response elements (ChRE), and oxidant response elements (OxREs).15, 16 In addition, sterol response elements have been localized within its promoter.17 The OsRE, ChRE, and OxRE modulate MIOX expression in diabetic states and under high glucose ambience.14, 15, 16, 17, 18 The sterol response elements modulate expression of MIOX in states of obesity, metabolic syndrome, and insulin resistance.17, 18 In addition, posttranslational modifications, such as, serine and threonine phosphorylation, also modulate its enzymatic activity in states of diabetes and obesity.16, 18 Interestingly, MIOX also modulates mitochondrial dynamics in diabetic states.19 All these transcriptional or posttranslational events fundamentally up-regulate MIOX expression, and as a result there is an increase in the oxidant stress in tubular epithelia and synthesis of ECM proteins, and these changes are considered reminiscent of what has been described in the glomerulus.2, 20
Recently, besides transcriptional and posttranslational regulation, the epigenetic modification of various genes has been incriminated to modulate their expression and thereby the pathophysiology of various organ systems.21, 22, 23 Although epigenetic regulation, including DNA methylation, histone lysine acetylation, and methylation, has been investigated in diabetic states using epigenomic profiling approaches, focus on a particular gene related specially to renal proximal tubules in the diabetic state has been lacking.24, 25, 26, 27, 28, 29, 30, 31 After a detailed analyses of MIOX promoter, we noted that it is populated with CpG islands or enriched with CpG dinucleotides. The presence of such characteristics within its promoter would suggest that MIOX is amenable to epigenetic regulation by various transcription factors because promoters of various genes include regions enriched with CG sites.32, 33, 34 There are a multitude of transcription factors that can bind to the CpG sites and modulate the epigenetic regulation of various genes, and the notable transcription factors include specificity protein (Sp)-1 and Kruppel-like factor, both related to zinc-finger proteins that regulate various biological processes.35, 36 We investigated the epigenetic regulation of MIOX while focusing on Sp-1 transcription factor under high glucose ambience to study its implications in diabetic tubulopathy.
Materials and Methods
Reagents
HK-2 cells (renal proximal tubular cell line) were purchased from ATCC (Manassas, VA); TRIzol Reagent (catalog number 15596026) from Invitrogen Corporation (Carlsbad, CA); EZ DNA Methylation-Gold Kit (catalog number D5005) from Zymo Research (Irvine, CA); Dulbeco's modified Eagle's medium (DMEM), poly-(deoxyinosinic-deoxycytidylic) acid sodium salt (catalog number P4929), mouse anti–β-actin (catalog number A5441), anti-mouse IgG (catalog number A9917), anti-rabbit IgG (catalog number A0545), and anti-fibronectin (catalog number F3648) antibody from Sigma-Aldrich (St Louis, MO); fetal bovine serum, antibiotic solution, and Power SYBR Green PCR Master Mix (catalog number 4367659), and DAPI (catalog number D1306) from Life Technologies (Carlsbad, CA); plasmid promoter vector pGEMT, pGL4.16, pRL-CMVRenilla, FuGENE6, and Dual-Luciferase kit from Promega (Madison, WI); Lipofectamine 2000 reagent, dihydroethidium (DHE; catalog number D7008), TO-PRO-3 iodide (catalog number T-3605), T4 Polynulceotide Kinase (10 U/μL), Supersignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific, Waltham, MA); anti–hypoxia-inducible factor (HIF)-1α (catalog number sc-10790) and anti–Sp-1 (catalog number sc-59), anti–NOX-4 antibody (catalog number sc-21860), Sp-1 Oligo, and Sp1 siRNA from Santa Cruz (Santa Cruz, CA); ATP[γ-32P] (PerkinElmer Inc, Waltham, MA); and apelin-13 and mithramycin A (Cayman Chemical, Ann Arbor, MI). Anti-MIOX polyclonal antibody was prepared in our laboratory using recombinant mouse MIOX as the immunogen.14
Animals
Six-week-old C57BL/6J mice were purchased from Jackson Laboratories (Bar Harbor, ME), and they were maintained at the Northwestern animal facilities. The animals were acclimatized for 1 week in rooms maintaining temperature of 22°C and 50% humidity with a 12-hour light/dark cycle. Animals had access to water and food ad libitum. The mice received i.p. injections of 50 mg/kg of streptozotocin dissolved in 0.1 mol/L sodium citrate buffer (pH 4.5) for 5 consecutive days. The control mice were injected with citrate buffer only. Blood glucose was measured 3 days after the last injection, and the mice with blood glucose levels >250 mg/dL were considered diabetic. The animals were sacrificed 2 weeks after the onset of diabetes. All animal studies were approved by the Northwestern University Animal Care and Use Committee.
Immunohistochemical Studies of Renal Tissues
Kidneys from control and diabetic mice were harvested, and 3- to 4-mm thick sections were prepared. They were fixed in 10% buffered formalin, dehydrated in graded concentrations (50% to 100%) of ethanols, and then transferred into xylene and embedded in paraffin. For immunohistochemistry, 4-μmol/L-thick sections were prepared and mounted on glass slides. The tissue sections were air-dried overnight. They were then deparaffinized, hydrated, and processed for antigen retrieval.18, 19, 20 After antigen retrieval, the sections were washed with Tris-buffered saline with 0.025% Triton X-100 (TBS-T) for 5 minutes. The endogenous peroxidase activity in the tissues was quenched by immersing glass slides in 3% H2O2 for 3 minutes. Tissue sections were then immersed in 3% bovine serum albumin for 1 hour and incubated overnight with primary antibody at 4°C. After washing with TBST, the sections were incubated with horseradish peroxidase–conjugated anti-rabbit IgG (immPRESS, Vector Laboratories, Burlingame, CA) for 1 hour. The sections were subjected to another TBS-T wash and incubated with horseradish peroxidase substrate diaminobenzadine for color development. Sections were rewashed and counterstained with hematoxylin to delineate the cellular details. Sections were dehydrated in graded series of ethanols, treated with xylene, and coverslip mounted after immersion in a drop of mounting media. The sections were then evaluated and photographed.
Cell Culture Studies
HK-2 cells were grown in low-glucose DMEM supplemented with 5% heat-inactivated fetal calf serum that contained 100 U/mL of penicillin and 100 μg/mL of streptomycin. Cells were maintained in a humidified atmosphere of 5% CO2 and 95% air at 37°C. The cells were then seeded on the coverslips placed in 6-well culture plates at a density of 1 × 106 cells per well. They were maintained to achieve 80% confluency. The cells were then treated with 5 mmol/L, 30 mmol/L d-glucose or l-glucose, 200 nmol/L mithramycin, or 300 pmol/L apelin-13 individually or in combination for 24 to 48 hours. The cells were harvested for various morphologic and biochemical studies.
In another set of experiments, cells were transfected with 50 μmol/L of Sp1 siRNA. Scrambled siRNA served as a negative control. The transfection was also performed in 6-well culture dishes using 4 μL of Lipofectamine 2000 reagent. A mixture of siRNA and lipofectamine was prepared in DMEM devoid of serum and antibiotics, and it was mixed gently by vortexing and then incubated at 22°C for 30 minutes to allow lipid-siRNA complex formation. The growth cell culture media was replaced with media lacking serum and antibiotics, and approximately 200 μL of medium that contained lipid-siRNA complex was added to the wells of culture dishes. The cultures were incubated at 37°C in a 5% CO2 and 95% air for 6 hours after which culture media was replaced with DMEM that contained serum and antibiotics. The cells were treated with 5 mmol/L or 30 mmol/L d-glucose or l-glucose for 24 hours and then processed for morphologic and immunofluorescence studies and also subjected to Western blotting and RT-PCR analyses.
For morphologic studies, HK-2 cells (1 × 106) seeded on the coverslips were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) (pH 7.4) for 20 minutes at room temperature, followed by washing three times with 1× PBS. They were permeabilized for 10 minutes by treating with 0.25% Triton X100 in PBS. After blocking with 1% bovine serum albumin in PBS containing Triton X100 for 30 minutes, the cells were then incubated with anti-fibronectin, anti–Sp-1, or anti-MIOX antibodies in blocking buffer for 12 to 15 hours at 4°C. After washing cells with PBS, they were incubated with fluorescein isothiocyanate–conjugated secondary antibodies in 1% bovine serum albumin for 1 hour at 22°C in the dark. After rewashing with PBS three times, cells were either stained with DAPI (blue) or TO-PRO-3 iodide (red) to demarcate the nuclei. The coverslips were then inverted and mounted on the glass slides after placing a drop of mounting media. The coverslips were sealed with polymeric resin, and then the cells were examined.
Isolation of DNA and Bisulfite Modification
DNA was isolated from glucose-treated and control HK-2 cells and diabetic mouse kidney cortices enriched with tubules. Approximately 20 mg of cortex or pelleted HK-2 cells were homogenized in 550 μL of lysis buffer (50 mmol/L Tris-HCL, 10 mmol/L EDTA, 100 mmol/L NaCl, 1% SDS) containing 1 μg/μL of RNAase1. The homogenates were incubated at 55°C for 2 hours. They were thoroughly mixed with 500 μL of Tris-saturated phenol by rigorous vortexing. The mixture was centrifuged at 12,283 × g for 10 minutes at room temperature. Supernatant was transferred into a fresh Eppendorf microfuge and extracted twice with 500 μL of chloroform to remove phenol. DNA was precipitated with the addition of 500 μL of isopropanol, and the mixture was kept at for 1 hour at 4°C. The DNA was pelleted by microfuging the precipitate. The pelleted DNA was washed with 70% ethanol, air dried, and dissolved in nuclease free water.
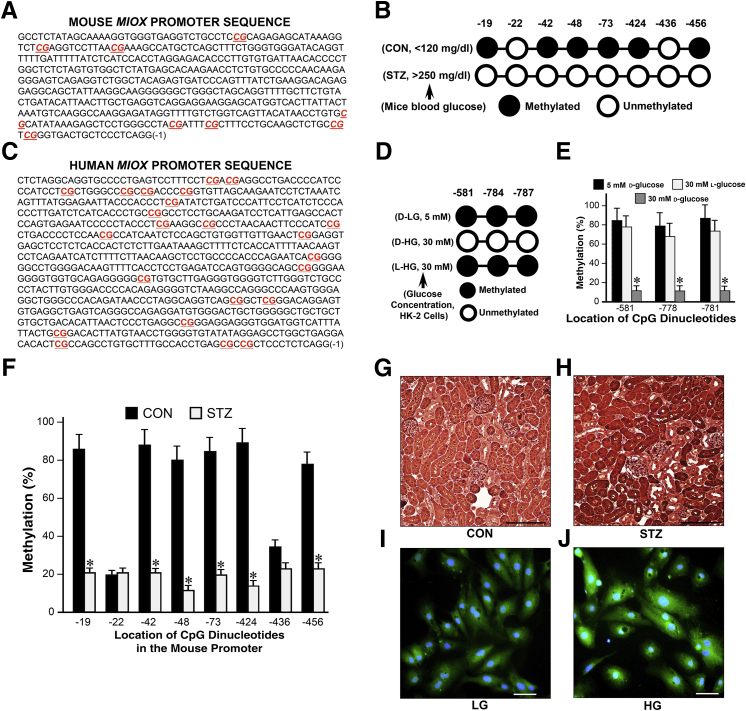
The isolated DNA (2 μg) was subjected to bisulfite modification using the EZ DNA Methylation-Gold Kit and by following the procedure originally described by Frommer et al.37 The principle being that sodium bisulfite in an acidic environment will convert all cytosine residues in single-stranded DNA to uracil, excluding the methylated cytosine.38 The bisulfite-treated DNA was loaded onto spin columns provided in the kit and eluted with 20 μL of elution buffer. An aliquot of this purified and bisulfite-treated DNA was used to determine the frequency of methylation of CpG residues within MIOX promoter, using bisulfite-specific PCR primers. The frequency of methylation of CpG residues within the MIOX promoter under low- and high-glucose conditions was assessed after bisulfite-specific PCR. For amplification of human and mouse MIOX promoter, 2 μL of bisulfite-treated DNA was amplified in a reaction mixture that contained 4 μmol/L primers, 1× PCR buffer, 0.4 mmol/L dNTP mix, and 1 U Taq polymerase (1 U/μL) in a total volume of 25 μL made up with nuclease free water. The nucleotide sequences of various primers were as follows: human MIOX promoter [forward: 5′-TGTGGTTGAATTTTAGGT-3′ (-828), reverse: 5′-CACCCCAAATTACATAAATA-3′ (-72)] and mouse MIOX promoter [forward: 5′-GTTTTTATAGTAAAAGGTGG-3′ (-491), reverse: 5′-CCTAAAAAAACAAACACC-3′ (-2)]. The primer locations are underscored in Figure 1.
Figure 1.
Distribution of CpG dinucleotide in the human and mouse MIOX promoters and their methylation profile under high-glucose (HG) ambience and a diabetic state. A–C: Human and mouse MIOX promoters are enriched with CG dinucleotides (A and C, red). In silico analyses predict that the mouse Miox gene only has CpG dinucleotides present at -19, -42, -48, -73, -424, -436, and -456 bp sites, and CpG islands are absent (A and B), whereas the human MIOX gene has multiple CpG islands (C). B, D, E, and F: Bisulfite treatment reveals that in the proximal mouse Miox promoter, six of eight CpG dinucleotides are methylated, whereas in humans (HK-2 cells) only three CpG dinucleotides are methylated under basal conditions. The efficiency of bisulfite treatment with conversion of cytosine to thymidine is approximately 95% (E and F). Under high-d-glucose (D-HG) ambience (30 mmol/L), human MIOX promoter analyses reveal that the CpG dinucleotide at -581, -784, and -787 bp are unmethylated compared with the control cells maintained under low-d-glucose (D-LG) or 30 mmol/L l-glucose ambience (D and E). In the mouse Miox promoter, all eight CpG dinucleotides are unmethylated in streptozotocin (STZ)–induced diabetic state (B and F). G–J: HG ambience and diabetic state may cause unmethylation or demethylation of MIOX promoter, which leads to increased transcription and expression of MIOX. Data are expressed as means ± SEM. n = 3 independent experiments (E and F). ∗P < 0.05 versus the respective control. Scale bars: 200 μm (G and H); 20 μm (I and J). CON, control; LG, low glucose.
For amplification, the following conditions were used: 94°C for 5 minutes, 39 cycles at 94°C for 30 seconds, 55°C for 45 seconds, 70°C for 2 minutes, and final extension at 70°C for 7 minutes. The PCR products were fractionated on a 1.5% agarose gels, and the gel bands were extracted using QIA quick gel extraction kit (Qiagen, Hilden, Germany). The extracted DNA product was cloned in pGEM-T Vector System I (Promega). After transformation in DH5α strain of Escherichia coli, the transfectants were plated on ampicillin agar plates. At least 10 positive colonies of each sample were amplified in Luria-Bertani media for preparation and purification of plasmid DNA that contains MIOX promoter. For sequencing of plasmid DNA, T7 promoter and SP6 promoter primers were used.
Generation of Reporter Construct and Dual Luciferase Reporter Assay
Human genomic DNA was used as a template for generation for MIOX promoter construct spanning Sp-1 site. A common reverse primer (5′-GACTCTCGAGTTGTTAGGGCGGCCTTCG-3′) and two forward primers (5′-GGGGGTACCAGTCCTTTCCTCGACGAGGCCT-3′ and 5′-GGGGGTACCAGTCCTTTCCTTGACGAGGCCT-3′) covering the -799 and -572 regions upstream of open reading frame were used. Restriction sites for KpnI and XhoI (underlined and bolded) were inserted in forward and reverse primers. For preparation of constructs having mutation in the Sp-1 site (underlined, italicized, and bolded) of the MIOX promoter, we used latter forward primers for replacing nucleotide C with T (CGA → TGA). The amplified PCR products were purified and cloned into pGL4.16 vector. Plasmids DNA were purified and orientation of inserts was confirmed by sequencing. Reporter vector 3–specific primer and insert primer spanning the MIOX promoter region were used for sequencing. The MIOX promoter reporter construct was transfected into exponentially growing HK-2 cells (1 × 105 cells) in 24-well culture plates in Opt-MEM I medium and allowed to grow for 12 to 15 hours. Transfection was performed using 1.5 μL of FUGENE 6, 1 μg of DNA (reporter plasmid), and 25 ng (pRL-TK, Renilla-thymidine kinase) as an internal control per well. Procedures used for transfection has been detailed previously.18 The cells were then treated with 5 mmol/L and 30 mmol/L d-glucose for 24 hours followed by harvesting for promoter activity assay. The cells were lyzed with 25 μL per well of lysis buffer, and dual luciferase-reporter activity was determined immediately using a TD 20/20 Luminometer, as per manufacturer instructions. Promoter activity was expressed as relative luciferase unit (ie, firefly luciferase activity was normalized with Renilla luciferase activity).
Western Blotting
Cultured cells were washed with ice cold PBS (pH 7.5) and harvested for preparation of cell lysates. Cells were lyzed with radio immunoprecipitation buffer that contained cocktail of protease inhibitors (Sigma-Aldrich). Protein content in the lysates was measured using the Bradford method. Equal amount of protein (30 μg) was loaded in each well, and proteins were fractionated by 12% SDS-PAGE followed by electroblotting onto polyvinylidene difluoride membranes (Millipore, Billerica, MA). Membranes were immersed in 5% nonfat dry milk dissolved in TBS-T buffer (50 mmol/L Tris, pH 7.6, 150 mmol/L NaCl, and 1% Tween 20) for 1 hour at 22°C. Membranes were immersed in TBS-T buffer that contained anti–Sp-1 (1:200), anti-MIOX (1:500), anti-fibronectin (1:1000), anti–NOX-4 (1:500), or anti–HIF-1α (1:200) antibodies and 2% fat-free dry milk for 12 to 15 hours at 4°C. The membrane blots were then washed with TBS-T thrice and incubated with horseradish peroxidaseanti–conjugated secondary antibody for anti-mouse or anti-rabbit IgG (1:10, 000) for 2 hours at 22°C. After washing with TBST, the autoradiograms were developed for the detection of bands using an enhanced chemiluminence kit (Thermo Fisher Scientific). Equal loading of proteins was confirmed by stripping the membranes and probing with anti–β-actin antibody (1:2000).
RNA Isolation and Real-Time PCR
Total RNA was isolated from cultured HK-2 cells using TRIzol (Invitrogen) method. The cDNA was synthesized using one microgram of RNA and Go Script reverse transcription system (Promega), and it was used to quantify the mRNA levels of various genes using Step One Plus System Real Time PCR (Applied Biosystems, Foster City, CA). The PCR reaction mixture included 1 μg of cDNA, 50 nmol/L sense and antisense primers, and 1× FAST SYBR Green (a total of 10 μL). For amplifying target and internal control regions, the reaction conditions used were as follows: 94°C for 2 minutes, followed by 39 cycles of 94°C for 20 seconds, 60°C for 15 seconds, 72°C for 15 seconds, and the final extension of 4 minutes at 72°C. The β-actin was used as an internal control for normalization of gene expression, and the relative abundance of mRNA of each gene was calculated by a comparative Ct method. The peaks were analyzed for generation of a single PCR product during amplification. All the data were derived from quadruplicate PCR reactions. The primers used were as follows: MIOX: forward: 5′-TGTCTTCACCACCTACAAGCTC-3′, reverse: 5′-GGCCTCCATGACTGTCATTTTC-3′; Sp1: forward: 5′-TCATCACAAGCCAGTTCCAG-3′, reverse: 5′-GAGCATCAGACCCCTGTAGC-3′; HIF1a: forward: 5′-ATGTAATGCTCCCCTCACCC-3′, reverse: 5′-CCTGAATCTGGGGCATGGTA-3′; NOX4: forward: 5′-CAGAAGGTTCCAAGCAGGAG-3′, reverse: 5′-GGCCAGGAACAGTTGTGAAG-3′; fibronectin: forward: 5′-CAACACCGTGACAGGAGAGA-3′, reverse: 5′-TCCCTCCTCACTCAGCTCAT-3′; and β-actin: forward, 5′-GGTCATCACCATTGGCAATGAG-3′, reverse: 5′-TACAGGTCTTTGCGGATGTCC-3′.
Nuclear Protein Isolation and Electrophoretic Mobility Shift Assay
Nuclear proteins were isolated from HK-2 cells subjected to low (5 mmol/L) or high (30 mmol/L) glucose ambience. Briefly, cells were scarped off from the 100-mm culture dishes and washed with PBS. They were first homogenized in a cytoplasmic extraction buffer (10 mmol/L HEPES, 10 mmol/L KCl, 0.1 mmol/L EDTA, and 0.1 mmol/L EGTA) and incubated for 15 minutes on ice. Then 12.5 μL of 10% NP-40 per 400 μL of lysis buffer was added and vortexed vigorously for 10 seconds to lyse the cells. Lysates were then centrifuged at 4643 × g for 1 minute to sediment the nuclei, and the supernatants were discarded. The pellets were incubated in nuclear extraction buffer (20 mmol/L HEPES, 400 mmol/L NaCl, 1 mmol/L EDTA, and 1 mmol/L EGTA) for 30 minutes on ice with intermittent vigorous vortexing to lyze nuclei. Nuclear extracts were collected as supernatant after 5 minutes of centrifugation at 12,463 × g at 4°C, and the protein concentration in various samples was measured by Biorad Bradford Method.
The nuclear extracts prepared above from HK-2 cells were used for electrophoretic mobility shift assays (EMSA) to assess the putative binding of the Sp-1 transcription factor. Double-stranded probe with Sp1 consensus sequence was purchased from Santa Cruz (catalog number sc-2502). The oligos of differentially methylated segment (DMS) regions were custom synthesized at Integrated DNA Technologies (Coralville, IA). The sequence of Sp1 and DMS region oligos were as follows: Sp1: 5′-ATTCGATCGGGGCGGGGCGAGC-3′; DMS1: 5′-AGTCCTTTCCTCGACGAGGCCT-3′ (CG residues at -787 and -784); and DMS2: 5′-GAAGGCCGCCCTAACAA-3′ (CG residues at -581). CpG dinucleotides within oligo used for EMSA are in bold. The underlined CG sites were found to be unmethylated under high glucose treatment and are also the putative binding sites for transcription factor Sp-1. Double-stranded probes (10 pmol/L) were used for 5′-terminal end-labeling reaction using ATP[γ-32P] using T4 polynucleotide kinase (Promega). For binding, reaction was performed in a volume of 20 μL that contained 1× binding buffer (200 mmol/L HEPES, 4 mmol/L EDTA, 4 mmol/L dithiothreitol, 50% glycerol), 1 μg/μL poly dI:dC, 10% NP-40, 1 pmol/μL of labeled probe, and 12 μg of nuclear extract. The reaction was performed at 37°C for 15 minutes. The reaction mixture that contained DNA-protein complexes was immediately placed on ice-cold water. After adding the DNA loading dye buffer, the reaction mixture was loaded onto 7.5% nondenaturating polyacrylamide gels. The gels were dried and exposed to X-ray film to prepare the autoradiograms. The specificity of binding of DMS oligo probes with Sp-1 protein was assessed by performing supershift assay. Nuclear extracts prepared from glucose-treated cells were incubated with antibody against Sp-1 (Santa Cruz) for 15 minutes at 22°C before incubation with radiolabeled probes. The shift in the DNA-protein-antibody complex migration in the gel was visualized after the preparation of the autoradiogram. In addition, the specificity of Sp-1 binding was assessed by treating the nuclear extracts with gene-selective Sp-1 inhibitor, mithramycin (200 nmol/L), before incubation with oligos.
For preparation of methylated oligos, CpG methytransferase (M. SssI; catalog number MO226S; New England Biolabs, Ipswich, MA) was used. The reaction mixture included 1× NEB buffer (50 mmol/L NaCl, 10 mmol/L Tris-HCL, pH 7.9, 10 mmol/L MgCl2, 1 mmol/L dithiothreitol), 160 μmol/L S-adenosyl methionine, and 10 pmol/L of double-stranded oligo in a total volume of 20 μL, and incubation lasted for 1 hour at 37°C. The reaction was terminated by incubating the mixture at 65°C for 20 minutes. The oligos were then purified using MicrospinG-50 columns (GE Healthcare, Chicago, IL). After radiolabeling of methylated oligos, they were used for EMSA as described above.
Assessment of ROS in Cells
Cellular mitochondrial oxidant stress was determined using DHE. HK-2 cells were seeded on coverslips placed in 6-well plates (1 × 106 cells per well) and exposed to low (5 mmol/L) or high (30 mmol/L) glucose ambience or Sp1 siRNA (50 μmol/L) treatment after which the cells were washed with PBS and incubated with 20 μmol/L of DHE diluted in PBS for 15 minutes at 37°C in dark. Cells were rewashed three times with PBS. The coverslips were then gently taken out of culture dish wells, inverted, and mounted on glass slides after placing a drop of cold glycerol mounting media (Dako, Glostrup, Denmark). The edges of coverslips were sealed as described above. They were examined with a Zeiss microscope equipped with UV epiillumination.
For flow cytometric analyses, the cells were washed in PBS and detached gently from the culture plates using BD Accutase cell detachment solution followed by washing twice with PBS for 5 minutes each. The cells were then collected in FACS tubes. They were stained with 20 μmol/L of DHE for 15 minutes at 37°C in dark. After rewashing twice with PBS, the cells were suspended in 300 μL of PBS. After which, mean fluorescent intensity of the DHE was measured using standard operational procedures of flow cytometry and using FACSDiva software version 6.1.3 (Becton Dickinson, Franklin Lakes, NJ).
Statistical Analysis
Results were expressed as means ± SD of four independent experiments after statistical analyses. The t-test was used to compare the data between groups. P < 0.05 was considered statistically significant.
Results
Distribution of CpG Dinucleotide within the Promoter Regions of Human and Mouse MIOX Genes
For differential methylation analyses of MIOX promoters, CpG dinucleotides within the MIOX gene were mapped. Both the human and mouse MIOX promoters were enriched with CG dinucleotides (Figure 1, A and C). To delineate the location of CpG islands versus CpG dinucleotides in silico analyses39 (Bioinformatics, www.bioinformatics.org/sms2/cpg_islands.html, last accessed December 2, 2016) were performed. The analyses predicated that compared with the mouse proximal promoter region the human MIOX gene had multiple CpG islands, defined as observed CpG/expected CpG >0.6 and GC content >55% within the nucleotide region spanning >200 bp (Figure 1C). However, in the mouse Miox gene, only CpG dinucleotides were present at -19, -42, -48, -73, -424, -436, and -456 bp, and CpG islands were absent (Figure 1, A and B).
Methylation Profile of MIOX Gene in Human and Mouse
The control isolated DNA after bisulfite treatment revealed that in the proximal mouse Miox promoter six of eight CpG dinucleotides were methylated, whereas in human HK-2 cells only three CpG dinucleotides were methylated (Figure 1, A–D). Ten clones of each sample were sent for sequencing, and their analyses indicated 95% efficiency of bisulfite treatment with conversion of cytosine to thymidine (Figure 1, E and F). Under high d-glucose ambience (30 mmol/L) human MIOX DNA analyses revealed the CpG dinucleotide at -581, -784, and -787 bp were unmethylated compared with the control cells maintained under low d-glucose or 30 mmol/L l-glucose ambience (Figure 1, D and E), whereas all eight CpG dinucleotides were noted to be unmethylated in the mouse Miox promoter in the diabetic state (Figure 1, B and F). This finding suggested that the hyperglycemia leads to hypomethylation of the Miox promoter and may be responsible for overexpression of MIOX in the diabetic state or under high-glucose ambience. The fact that we observed an increased expression of MIOX under high-glucose ambience and in streptozotocin diabetic state would be supportive of this contention (Figure 1, G–J).
Identification of Transcription Factor Sp-1 Binding Site in the Human MIOX Promoter and Effect of in Vitro Methylation on the Binding with Nucleoproteins
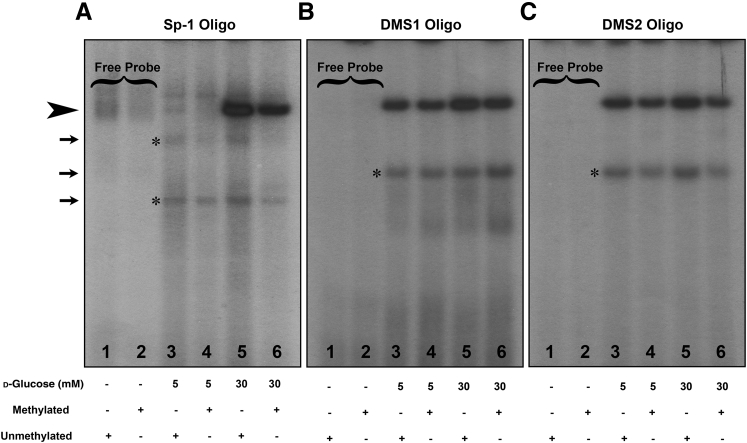
The overexpression of MIOX may also be modulated by various transcription factors that bind to the MIOX gene promoter, as described in our previous publications.14, 16, 18 In this regard, further in silico detailed analyses (TRANSFAC version 8.3) searching for additional transcription factors' binding sites predicated that Sp-1, a zinc finger transcription factor, could also potentially bind to the GC regions of the MIOX gene that are masked by differential methylation and are susceptible to its regulation when the gene is demethylated, such as under high-glucose ambience. To attest to this contention, we assessed the binding of Sp-1 and 2 nucleotide oligos spanning putative DMSs of the human MIOX promoter by EMSAs after in vitro methylation. The use of methylated Sp-1 and DMS oligos also gave us the opportunity to directly assess the effect of in vitro methylation on the binding of oligos with nucleoproteins as their targets because methylation plays an important role in their binding to the promoters.33, 40 The oligos were custom synthesized, and DMS1 oligo included CpG sites -784 and -787, whereas DMS2 oligo only included -581, and they were used for EMSAs. An increased binding of Sp-1 was observed under high-glucose ambience (30 mmol/L) when the oligo is differentially demethylated or unmethylated (Figure 2A). No binding of methylated or demethylated oligos was observed under low-glucose ambience (Figure 2A). Likewise, an increased binding of unmethylated DMS1 and DMS2 oligos was observed under high-glucose ambience (Figure 2, B and C). Although binding was seen under low-glucose ambience, increased binding with unmethylated oligos (Sp-1, DMS1, and DMS2) under high-glucose ambience strongly suggests that methylation and demethylation modulate MIOX expression under high-glucose ambience and conceivably in the hyperglycemic state.
Figure 2.
Identification of transcription factor specificity protein (Sp)-1–binding site in the human MIOX promoter and effect of in vitro methylation on the binding with nucleoproteins. Eletrophoretic mobility shift assays using Sp-1 and differentially methylated segment (DMS) oligos (DMS1 includes CpG sites -784 and -787 and DMS2 includes -581). A: An increased binding of Sp-1 is observed under high-glucose ambience (30 mmol/L) when the Sp-1 oligo is differentially demethylated or unmethylated (arrowhead). No binding of methylated or demethylated oligos is observed under low-glucose (5 mmol/L) ambience. B and C: Likewise an increased binding of unmethylated DMS1 and DMS2 oligos is observed under high-glucose ambience (arrowhead). The binding is also seen under low-glucose ambience; nevertheless, binding with unmethylated oligos (Sp-1, DMS1, and DMS2) under high-glucose ambience is much stronger. The arrows and asterisks indicate the nonspecific bands in the autoradiograms.
Specificity of Sp-1 Binding with Human MIOX Gene and Modulation of Its Promoter Activity
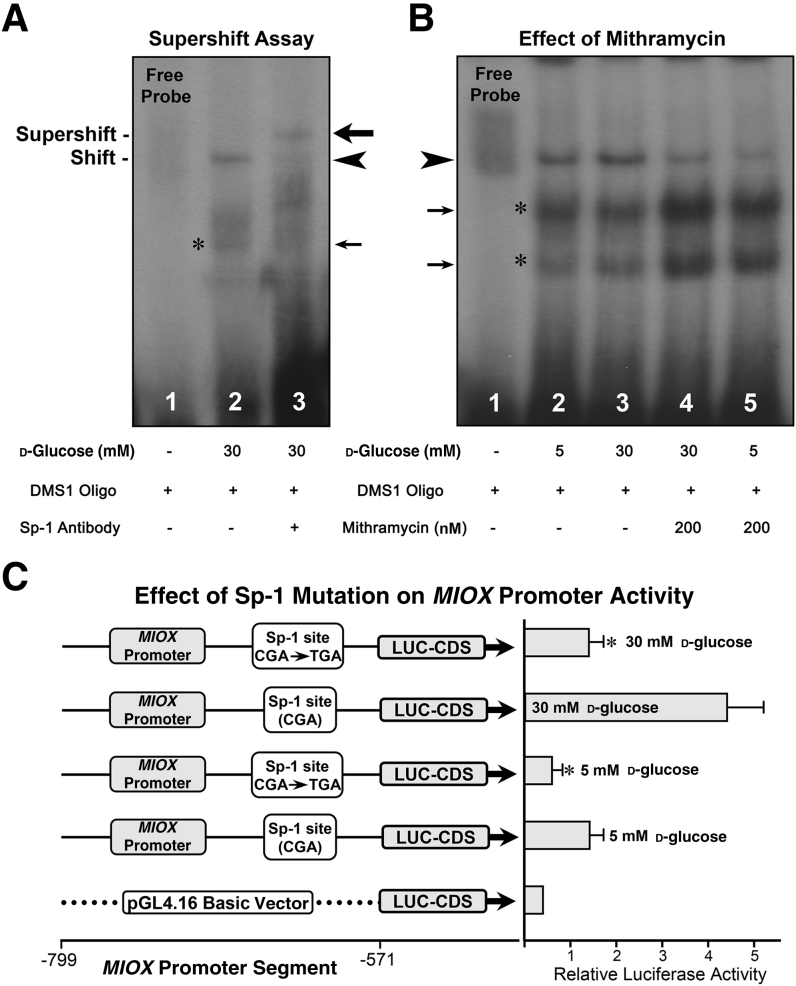
The specificity of binding of DMS1 and DMS2 oligos was assessed by using anti–Sp-1 antibody (Figure 3). Both unmethylated oligos yielded similar results, and the results of DMS1 are included. Nuclear proteins were isolated from HK-2 cells treated with 30 mmol/L d-glucose and used for EMSAs. A distinct band was seen when nuclear extracts were incubated with DMS1 (Figure 3A). After incubation with the Sp-1 antibody, the band shifted to higher position (supershift), suggesting the formation of a DMS1-DNA:nuclear protein:Sp-1 antibody complex and thus authenticating the Sp-1–binding specificity (Figure 3A). Further validation of Sp-1 binding was performed with the Sp1 gene selective inhibitor mithramycin. HK-2 cells were treated with 200 mmol/L mithramycin in the presence of low- and high-glucose ambience. The nuclear proteins were isolated and subjected to EMSAs. An increase in the intensity of the band was observed with the incubation of DMS1 oligo under high-glucose ambience (Figure 3B). Co-treatment of HK-2 cells with mithramycin considerably reduced the intensity of the band under high-glucose ambience, whereas it was drastically reduced under low-glucose (5 mmol/L) ambience (Figure 3B). These results validate the specificity of Sp-1 binding with the MIOX gene promoter, and the binding affinity increases under high-glucose ambience accompanied with hypomethylation of its CpG sites. The next question we addressed was whether Sp-1 modulates the MIOX promoter activity. Mutational analyses were performed using reporter constructs and luciferase assays. Two reporter constructs were generated, where one had a normal binding site for Sp-1 and in the other CGA site was replaced with TGA (Figure 3C). The constructs were transfected into HK-2 cells maintained under low- (5 mmol/L) and high- (30 mmol/L) d-glucose ambience, and relative luciferase activities were determined. The basic empty pGL4 vector had very little basal activity. Under low-glucose ambience a mild relative luciferase activity was observed, and it was notably reduced with a single base pair mutation (C → T) in the MIOX gene. Under high-glucose ambience, a remarkable increase in the promoter activity was observed (Figure 3C). The activity was markedly reduced with mutation in the Sp-1 biding site, suggesting that Sp-1 transcription factor modulates MIOX promoter activity.
Figure 3.
Specificity of specificity protein (Sp)-1–binding with human MIOX gene and modulation of its promoter activity. A: The specificity of binding of differentially methylated segment (DMS)-1 oligo was assessed by using anti–Sp-1 antibody in eletrophoretic mobility shift assays. A distinct band is seen when nuclear extracts are incubated with DMS1 (arrowhead). On subsequent incubation with the antibody, the band shifts to higher position (supershift), suggesting the formation of a DMS1-DNA:nuclear protein:Sp-1 antibody complex, and binding seems to be specific (big arrow). B: Validation of Sp-1 binding was assessed with the use of the Sp1 gene–specific inhibitor mithramycin. An increase in the intensity of the band is observed with the incubation of DMS1 oligo under high-glucose (30 mmol/L) ambience (arrowhead). Co-treatment of HK-2 cells with 200 nm of mithramycin notably reduces the intensity of the band under high-glucose ambience, whereas its intensity is drastically reduced under low glucose (5 mmol/L) ambience. C: Whether Sp-1 modulates the MIOX promoter activity was assessed by mutational analyses, where mutant reporter constructs were generated. Under low-glucose ambience, a mild relative luciferase (LUC) activity is observed, and it is notably reduced with a single base pair mutation (C → T) within the MIOX promoter. Under high-glucose ambience, a remarkable increase in the promoter activity is observed, and it is markedly reduced with the mutation in the Sp-1–binding site. The asterisks and small arrows in A and B indicate nonspecific bands. Data are expressed as means ± SEM. n = 4 independent experiments (C). ∗P < 0.05 versus the respective control. CDS, coding segment.
Status of MIOX, Sp-1, NOX-4, HIF-1α, and Fibronectin Expression under High-Glucose Ambience and after Sp1 Gene Disruption
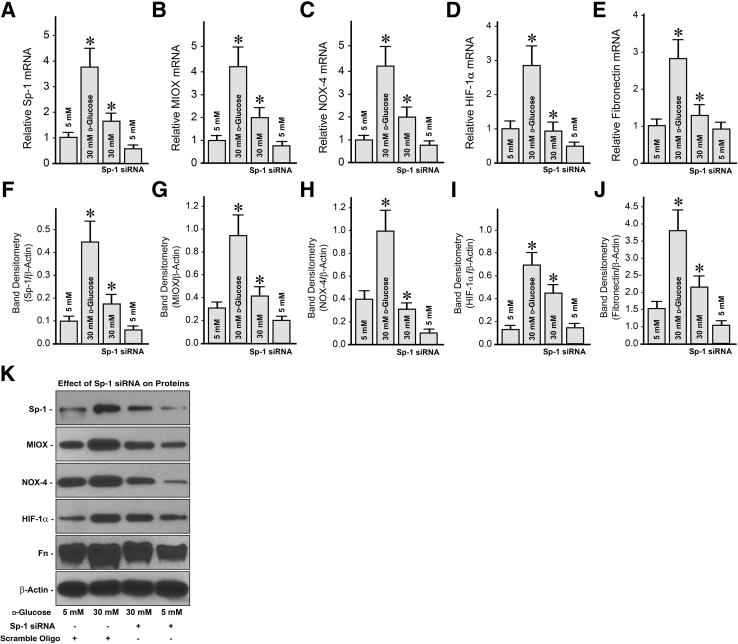
Since Sp-1 modulates the promoter activity of MIOX studies were initiated to assess the effect of Sp1 gene-disruption on MIOX and other relevant readouts that are affected under high-glucose ambience. Both mRNA and protein expression studies were performed (Figure 4). Under high-glucose (30 mmol/L) ambience, both Sp-1 and MIOX expression was increased compared with the control low-glucose (5 mmol/L) ambience (Figure 4, A, B, F, G, and K). It was notably reduced after Sp1 siRNA treatment. The other significant readouts relevant to the pathophysiology of diabetes include cellular redox, reflective of the expression of tubular NOX-4, and hypoxia, reflected of the expression of transcription factor HIF-1α (Figure 4, C, D, H, I, and K). The expression of both NOX-4 and HIF-1α was increased under high-glucose ambience. After Sp1 siRNA treatment, expression of both mRNA and protein was considerably decreased. The final readout evaluated was fibronectin, a prototype of ECM proteins that are known to be increased in the tubulointerstitial compartment in diabetic nephropathy. Interestingly, a remarkable increase in the fibronectin cellular mRNA and protein expression was observed under high-glucose ambience, and it was reduced with the inclusion of Sp1 siRNA in the culture media of HK-2 cells (Figure 4, E, J, and K). The expression of Sp-1, MIOX, and the other readouts was unchanged following treatment of cells with scramble oligo.
Figure 4.
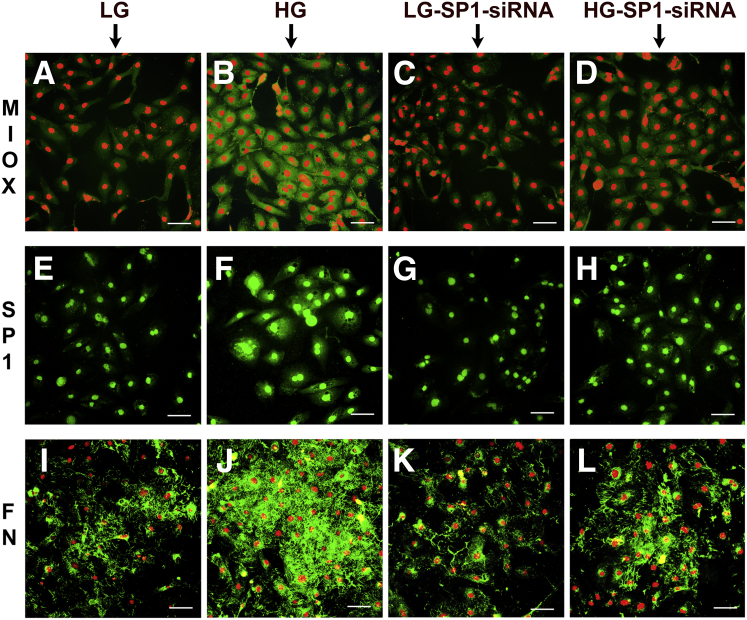
Effect of Sp1 gene disruption on the expression of specificity protein (Sp)-1, myo-inositol oxygenase (MIOX), NADPH oxidase (NOX)-4, hypoxia-inducible factor (HIF)-1α, and fibronectin (Fn) under high-glucose ambience. Under high-glucose (30 mmol/L) ambience, Sp-1 and MIOX expression increases compared with the control low-glucose (5 mmol/L) ambience and is reduced by the Sp1 siRNA treatment (A, B, F, G, and K). The other relevant parameters, such as tubular NOX-4 and HIF-1α, increase under high-glucose ambience and decrease after Sp1 siRNA treatment (C, D, H, I, and K). Fibronectin was used as a distal readout. Fibronectin cellular mRNA and protein expression increases under high-glucose ambience and reduces with the treatment of Sp1 siRNA (E, J, and K). The scramble oligo has no significant effect in any of the readouts. Data are expressed as means ± SEM. n = 4 independent experiments (A–J). ∗P < 0.05 versus the respective control.
The effect of Sp1 gene disruption of Sp-1, MIOX, and the final readout (ie, fibronectin) was also delineated in the cellular versus nuclear versus extracellular compartments by immunofluorescence microscopy (Figure 5). The expression of MIOX was markedly increased in the cytoplasm of HK-2 cells under high-glucose ambience, and it was reduced after Sp1 siRNA treatment (Figure 5, B and D). A mild decrease was also observed after Sp1 siRNA treatment in cells subjected to low-glucose ambience (Figure 5, A and C). The localization of Sp-1 was confined to the nuclear compartment (Figure 5E), and it increased considerably under high-glucose ambience (Figure 5F). Interestingly, some of the cells were hypertrophied with enlarged nuclei under high-glucose ambience. The expression of Sp-1 decreased after treatment with Sp1 siRNA in cotreatment with low glucose (Figure 5G) and along in treatment with high glucose (Figure 5H). The fibronectin expression was confined to the extracellular compartment (Figure 5, I–L). A marked increased in the de novo synthesis of fibronectin was observed under high-glucose environment (Figure 5, J and I), and it was reduced after Sp1 siRNA treatment (Figure 5L). No significant differences in the fibronectin expression were observed in cells treated with Sp1 siRNA while maintained under a basal low-glucose environment (Figure 5, I and K).
Figure 5.
Effect of Sp1 gene disruption on the cellular morphologic findings related to the expression of myo-inositol oxygenase (MIOX), specificity protein (Sp)-1, and fibronectin (Fn) under high-glucose ambience. B and D: The expression of MIOX markedly increases in the cytoplasm of HK-2 cells under high-glucose (HG) ambience and reduces after Sp1 siRNA treatment. A and C: A mild decrease is also observed after Sp1 siRNA treatment in cells subjected to low-glucose (LG) ambience. E and F: The expression of Sp-1 confines to the nuclear compartment and increases considerably under HG ambience in the hypertrophic cells with enlarged nuclei. G and H: Decrease in Sp-1 expression after siRNA treatment in cotreatment with LG and HG. I–L: The Fn expression markedly increases under HG environment (I and J) and reduces after Sp1 siRNA treatment (L). I and K: No difference in the Fn expression is observed in cells maintained under basal LG environment and treated with Sp1 siRNA. Scale bars = 40 μm (A–L).
Effect of Sp1 siRNA on High Glucose–Induced Perturbations in Mitochondrial Redox in Renal Tubular Cells
The generation of ROS could be related to the perturbations in the NOX-4 system or in the mitochondrial homeostasis. The changes in NOX-4 are described above. In addition, we investigated whether there are perturbations in the mitochondrial homeostasis with the generation of ROS. We used DHE staining to assess the status of mitochondrial-derived ROS. Compared with the control, increased DHE staining was observed under high-glucose ambience, and this increase was also reflected in flow cytometric analyses (Figure 6, A, B, E, F, and G). With the treatment of Sp1 siRNA, the staining was considerably lessened, and there was a reduced generation of ROS, as assessed by flow cytometric analyses (Figure 6, B, C, E, G and H). No significant differences in the DHE staining or generation of ROS were observed in cells maintained under low-glucose ambience and treated with Sp1 siRNA (Figure 6, A, D, E, F, and I).
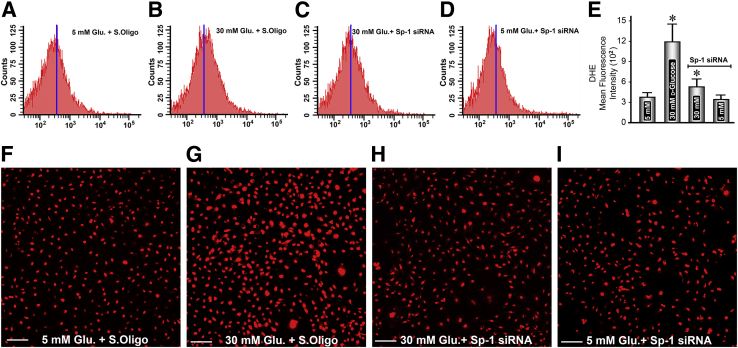
Figure 6.
Effect of Sp1 siRNA on high-glucose–induced perturbations in mitochondrial redox in renal tubular cells. Compared with the control, an increased staining of dihydroethidium (DHE) is observed under high-glucose ambience, and this increase is also reflected in flow cytometric analyses (A, B, F, and G). Sp1 siRNA treatment considerably reduces the staining and the mitochondrial oxidant stress (B, C, G, and H). No significant differences in the DHE staining or generation of reactive oxygen species are observed in cells maintained under low-glucose ambience and treated with Sp1 siRNA (A, D, E, F, and I). Data are expressed as means ± SEM (E). n = 4 independent experiments (E). ∗P < 0.05 versus the respective control. Scale bars = 80 μm (F–I). Glu, glucose; S.Oligo, scramble oligo.
Effect of Apelin-13 on MIOX Expression in HK-2 Cells Subjected to High-Glucose Ambience
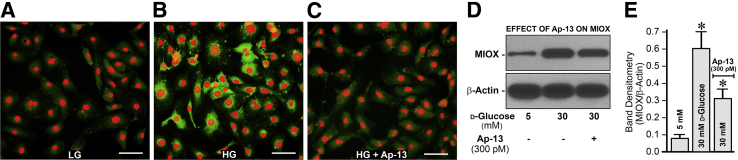
Besides methylation that influences the pathobiology of MIOX in diabetic tubulopathy, some preliminary studies were also performed to assess whether the up-regulated expression of MIOX is related to other forms of epigenetic regulation (ie, hyperacetylation). The processes of histone acetylation and chromatin opening play an important role in epigenetic regulation of various genes and thereby change their expression in various disease processes.24, 25, 41, 42 In line with this contention, HK-2 cells were treated with apelin-13, an inhibitor of histone hyperacetylation, under high-glucose ambience. Apelin-13 treatment notably reduced the MIOX expression as indicated by immunofluorescence microscopy (Figure 7, A–C) and immunoblot analyses (Figure 7, D and E), suggesting that the acetylation of MIOX gene modulates its expression besides methylation in states of high-glucose ambience and conceivably in hyperglycemia.
Figure 7.
Effect of apelin (Ap)-13 on myo-inositol oxygenase (MIOX) expression in HK-2 cells subjected to high-glucose (HG) ambience. Treatment of HK-2 cells with and without apellin-13 (A), an inhibitor of histone hyperacetylation, considerably reduces the MIOX expression under high-glucose ambience, as assessed by immunofluoresence microscopy (B and C) and immunoblot analyses (D and E). Data are expressed as means ± SEM. n = 4 independent experiments (E). ∗P < 0.05 versus the respective control. Scale bars = 40 μm (A–C). LG, low-glucose.
Discussion
The results of this investigation highlight the significance of epigenetic regulation of MIOX, an enzyme expressed in the proximal tubules, and it has been described to be relevant in the pathobiology of diabetic nephropathy or tubulopathy. Conceivably, a compromise in the integrity of tubules or the tubulointerstitial compartment is readily reflected in the decreasing renal functions—an idea originally proposed by Adalbert Bohle and later marshaled by Nath and others.7, 9, 43, 44 Unfortunately, tubular pathobiology in the context of diabetic nephropathy has not received as much attention as that for glomerulus. Certainly, the molecules that are expressed in the proximal tubules, such as glucose transporters and sodium glucose transporters, have been given adequate considerations in the pathophysiology of diabetes, especially that relates to glucose handling.31, 45, 46, 47, 48 In view of this, it would be reasonable to explore the biological characteristics of MIOX because it is a tubular-specific gene that seems to be relevant to the pathobiology of diabetic tubulopathy, and its up- or down-regulation is likely to influence the homeostasis of the tubulointerstitial compartment of the kidney.
Transcriptional or translational regulation of MIOX by high-glucose ambience, organic osmolytes, and fatty acids has been extensively studied by us and other investigators.14, 15, 16, 17, 18, 19, 20 However, its epigenetic regulation remained unexplored. The epigenetic regulation is known to modulate the gene expression and thus has been implicated in the pathogenesis of various disease processes.22, 23 Various studies that highlight the epigenetic regulation of some of the genes expressed in the proximal tubules, especially the glucose transporters, have been adequately reported in the literature, but their relevance was investigated in nonrenal cell lines.46, 47 However, a recent study by Marumo et al31 addressed the issue of epigenetic regulation of glucose transporters and certain other genes expressed in proximal tubular cells. The conclusions drawn from this study were not uniform because certain genes were hypomethylated in control mice, whereas other genes were hypomethylated in the diabetic mice, thus making a correlation with gene expression somewhat enigmatic. Nevertheless, in view of the vast amount of literature data concerning epigenetic regulation of gene expression and because the MIOX promoter is enriched with CpG dinucleotides (Figure 1), we anticipated that most likely a methylation and demethylation process plays an important role in the modulation of its expression. The fact that we consistently observed the demethylation of CpG dinuncleotides both in vitro and in vivo in the MIOX promoters under high-glucose ambience with up-regulated MIOX expression gives reasonable credence to our contention (Figure 1). In this regard, the general consensus that methylation of the promoter and gene expression are inversely related would certainly support our contention.22
The next question that needs to be addressed is if and how the methylation or demethylation process affects binding of transcription factors to the MIOX promoter on masking and unmasking of the CpG sites and the modulation of MIOX's expression. It is well recognized that modification in the DNA sequence by methylation or demethylation leads to changes in the access of cellular machinery for transcription factors, thereby affecting the gene expression.24, 25 To analyze the effect of methylation and demethylation of oligos on the binding with MIOX promoter, we prepared oligos corresponding to DMSs that encompass CpG residues. They were in vitro methylated with CpG methytransferase as indicated in Materials and Methods. In addition, a corresponding methylated Sp-1 oligo was prepared because among the multitude of transcription factors Sp-1 exhibits a high-affinity binding to the DNA sequences populated with CpG sites. Interestingly, we observed a relatively strong binding of unmethylated oligo (Sp-1, DMS1, or DMS2) under high-glucose ambience (Figure 2), suggesting that methylation or demethylation status of oligo or of promoter sequence modulates the DNA-protein interaction and thereby the gene expression. Another key observation worth pointing out is that Sp-1 oligo whether methylated or unmethylated only binds to nucleoproteins under high-glucose (30 mmol/L) ambience, thereby underscoring this transcription factor's significance in the pathobiology of diabetes. The fact that there was a supershift in the electrophoretic mobility assays and inhibition of DMS1 oligo binding with nucleoproteins by mithramycin, a selective inhibitor of Sp-1, reinforces the idea that the Sp-1 transcription factor is intricately involved in the regulation of MIOX in states of hyperglycemia or high-glucose ambience (Figure 3). To further validate that Sp-1 is modulating the transcription of the MIOX promoter, mutational analyses were performed. With the mutation in Sp-1 sites, a marked decrease in the MIOX promoter activity was observed, thus authenticating the relevance of this transcription factor in the pathobiology of MIOX (Figure 3).
Conceivably, high-glucose ambience or hyperglycemia by hypomethylating the CpG sites provide an environment that is most conducive for the binding of Sp-1 transcription factor with the MIOX promoter and thereby modulating downstream events. Although Sp-1 modulates the MIOX activity, the upstream events (eg, increased transcription of Sp-1) could further accentuate the net effect on the downstream events. A precedent for this notion is derived from our earlier work in which another transcription factor (ie, sterol element–binding protein 1) has increased transcription and binds to the MIOX promoter in states of hyperlipidemia and hyperglycemia.18 With these observations in perspective, the status of Sp-1 under high-glucose ambience was assessed, and an increased mRNA and protein expression of Sp-1 concomitant with increased expression of MIOX was observed (Figure 4). Morphologic examination clearly established increased cytoplasmic and nuclear localization of MIOX and Sp-1, respectively (Figure 5). Moreover, the fact that both the cytoplasmic and nuclear localization of the MIOX gene and Sp1 transcription could be reduced by siRNA treatment indicated that the effects are specifically mediated by the Sp-1 transcription factor. The fact that the high-glucose–induced hypomethylation yields favorable conditions for the binding of Sp-1, whereas there is an increased expression of Sp-1 at the same time, would translate into a scenario where there would be a highly boosted expression of MIOX and accentuated activity of the downstream events. Among the downstream signaling events, the oxidant stress and the generation of ROS are regarded as the common denominators in the pathogenesis of diabetic complications, including diabetic nephropathy.2, 7, 12, 20, 49, 50, 51 The two major sources for the generation of intracellular ROS include the NADPH oxidase and mitochondrial compartment, and the consensus is that they are derived from both sources in the setting of diabetic nephropathy.2, 12, 20, 50, 51 The observations that there was an increased expression of NOX-4, a tubular NADPH oxidase, and increased DHE staining under high-glucose ambience supports the literature data (Figures 4 and 6). More importantly, the observation of novel significance is that there is diminution in the generation of ROS by siRNA treatment, which would suggest that these effects are proximally mediated by the Sp-1 transcription factor.
Besides the generation of the ROS, there is concomitant hypoxia and pseudohyoxia in kidneys in states of hyperglycemia, as originally proposed by Williamson et al,52 and it has attracted the attention of various investigators working in the field of renovascular complications of diabetes.52, 53, 54 According to Williamson et al,52,pp.801 that “increased cytosolic ratio of free NADH/NAD+ caused by hyperglycemia would lead to relative pseudohypoxia, characteristic of poorly controlled diabetes.” As a consequence of hypoxia or pseudohypoxia there would be an activation of transcription factor referred to as HIF-1α. Conceivably, induction of HIF-1α is regarded as an adaptive response, and in fact some of the investigators believe that its activation may have an ameliorative effect on diabetic nephropathy.53 Interestingly, previous studies have documented that NADPH oxidases (Nox4/Nox1) regulate HIF expression.55 Our results indicating a dose-dependent increase in HIF-1α expression (Figure 4) support the notion that it may be a protective and an adaptive response to a high-glucose environment. Collectively, this would mean that the biology of hypoxia or pseudohypoxia and ROS generation may be intertwined in states of high-glucose ambience. Of note, the relevant observation here is that the Sp-1 transcription factor modulates these above described key downstream events. Another important downstream event that typifies the characteristic of diabetic nephropathy includes increased synthesis of ECM proteins, which as a consequence leads to nodular glomerulosclerosis and tubulointerstitial fibrosis and ultimately progresses to end-stage renal disease.2, 3, 6, 56 In this regard, we investigated the status of fibronectin, a prototype of ECM protein that is up-regulated under the influence of the profibrogenic cytokine TGF-β1 in diabetes.57 Likewise, we observed an increase in the expression of fibronectin in HK-2 cells in a high-glucose environment (Figures 4 and 5). This increase was notably reduced by the siRNA treatment, thus establishing the notion that Sp-1 plays a pivotal role in modulating all the critical downstream events that lead to ECM accumulation in states of high-glucose ambience or hyperglycemia, where there is an up-regulation of the tubular-specific enzyme MIOX. An interesting question can be raised of whether TGF-β, a multifunctional cytokine, caused hypomethylation of the MIOX because it is known that TGF-β signaling modulates methylation and demethylation of several target genes.58 This possibility is unlikely in our scenario because TGF-β signaling is downstream of the MIOX-mediated events and such TGF-β–related processes mainly pertain to neoplastic cells.58
Having established the role of methylation in the modulation of MIOX, we explored whether any other epigenetic event(s) can regulate MIOX transcription and thereby its expression in a hyperglycemic environment. Among the known events, the acetylation and deacetylation of histone and nonhistone proteins are well-known to modulate gene expression and regulate a wide variety of biological processes.59, 60 One of the well-described molecules that modulate hyperacetylation directly or indirectly in states of hyperglycemia is apelin-13, an adipokine that exerts its effects after binding with its G protein–coupled receptor, named APJ.41, 61 Apparently, apelin-13 inhibits diabetes-induced histone hyperacetylation by up-regulating histone deacetylase-1 levels in the renal cortex, and its ameliorative effect on diabetic nephropathy has been alluded to in several studies in mice models of diabetes.41, 62, 63 In addition, in the renal ischemia and reperfusion model, a notable decrease in acetylation of the H3K23 and H3K56 sites have been reported after apelin treatment, which is most likely mediated via the up-regulation of histone deacetylases.64 In addition, other studies have reported that administration of apelin-13 alleviates endoplasmic stress and improves fatty acid oxidation and mitochondrial homeostasis.65, 66 Overall, it appears from these studies that apelin certainly has beneficial effects in states of hyperglycemia; however, these studies describe its global effects on deacetylation without focusing on a given particular gene that is relevant to diabetic nephropathy or tubulopathy. In this regard, we performed some experiments to have a preview of the events if the apelin-13 could exert a specific effect on MIOX in states of high-glucose ambience. Treatment of apelin-13 reduced the cellular expression of high-glucose–induced MIOX in HK-2 cells, as assessed by Western blot analysis and immunofluorescence microscopy (Figure 7). These observations, although preliminary, support the conclusions drawn in earlier reported studies describing the global decetylation effect of apelin-13,41, 62, 63, 64, 65, 66 but here we document its effect specifically targeted on the expression of MIOX under high-glucose ambience. The detailed mechanisms that involve acetylation and deacetylation of MIOX will be the subject of future investigations.
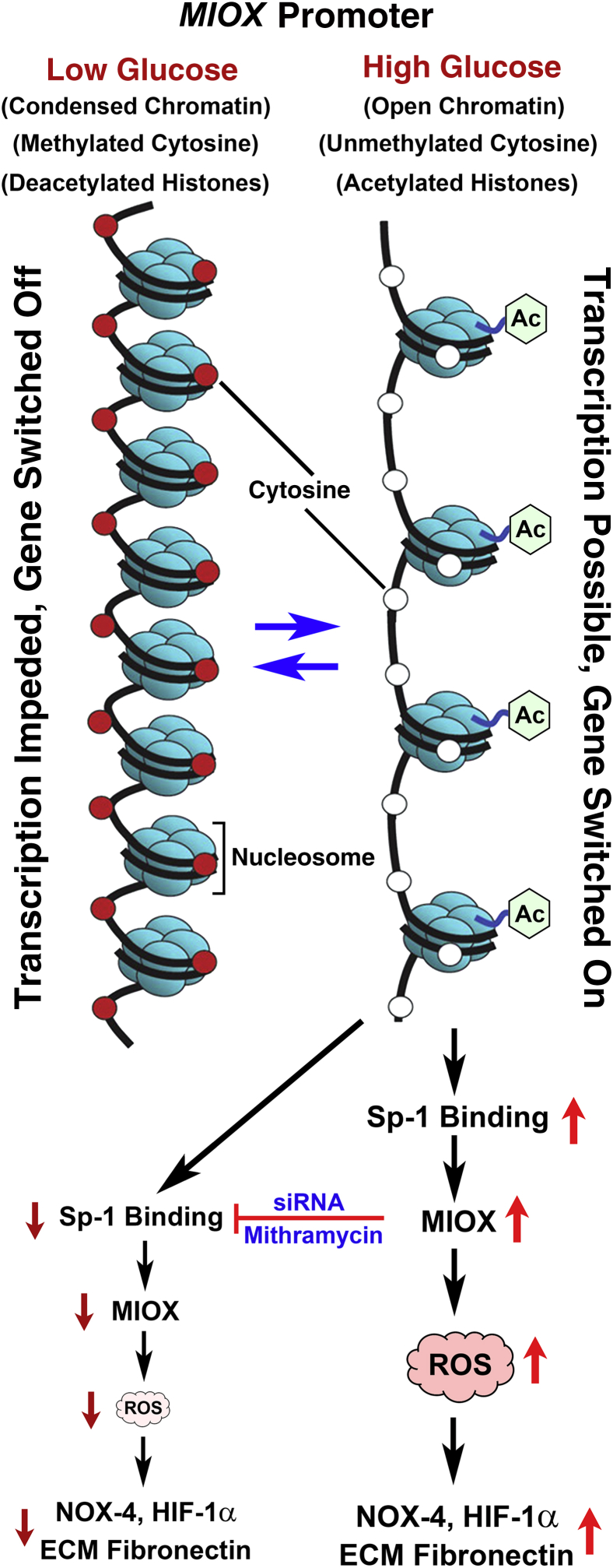
In summary, this investigation describes the epigenetic regulation of MIOX, an enzyme relevant to the pathogenesis of diabetic tubulopathy. The two critical processes that modulate the epigenetic regulation of MIOX include demethylation and deacetylation. Interestingly, these processes have opposing directional influence on the MIOX, that is, demethylation or unmethylation increases MIOX gene expression, whereas acetylation, not deacetylation, proportionately increases its expression. The events highlighting these two epigenetic processes are depicted in Figure 8 along with the downstream effects that are relevant to the pathogenesis of diabetic tubulopathy. Finally, it is hoped that the information provided in this communication would yield impetus to future studies to further investigate the pathobiology of MIOX and to develop therapeutic targets for the amelioration of diabetic nephropathy and tubulopathy.
Figure 8.
Schematic depicting the sequence of events related to methylation and acetylation that modulate the expression of MIOX and downstream events under high-glucose ambience. Besides methylation, histone acetylation and chromatin opening also modulate the expression of MIOX. High-glucose ambience leads to demethylation of cytosine residues, leading to hypomethylation of certain regions of the MIOX promoter. In addition, the high-glucose ambience leads to acetylation of histones. Both of these nuclear events lead to increased binding of specificity protein (Sp)-1, expression of MIOX, generation of reactive oxygen species (ROS), and de novo synthesis of extracellular matrix (ECM) proteins, such as fibronectin. HIF-1α, hypoxia-inducible factor 1α; NOX-4, NADPH oxidase-4.
Footnotes
Supported by NIH grant DK60635 (Y.S.K.).
Disclosures: None.
References
- 1.Gregg E.W., Li Y., Wang J., Burrows N.R., Ali M.K., Rolka D., Williams D.E., Geiss L. Changes in diabetes-related complications in the United States, 1990-2010. N Engl J Med. 2014;370:1514–1523. doi: 10.1056/NEJMoa1310799. [DOI] [PubMed] [Google Scholar]
- 2.Kanwar Y.S., Sun L., Xie P., Liu F., Chen S. A glimpse of various pathogenetic mechanisms of diabetic nephropathy. Annu Rev Pathol. 2011;6:395–423. doi: 10.1146/annurev.pathol.4.110807.092150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Najafian B., Alpers C.E., Fogo A.B. Pathology of human diabetic nephropathy. Contrib Nephrol. 2011;170:36–47. doi: 10.1159/000324942. [DOI] [PubMed] [Google Scholar]
- 4.Lin J.S., Susztak K. Podocytes: the weakest link in diabetic kidney disease? Curr Diab Rep. 2016;16:45. doi: 10.1007/s11892-016-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goligorsky M.S., Chen J., Brodsky S. Workshop: endothelial cell dysfunction leading to diabetic nephropathy: focus on nitric oxide. Hypertension. 2001;37:744–748. doi: 10.1161/01.hyp.37.2.744. [DOI] [PubMed] [Google Scholar]
- 6.Phillips A.O., Steadman R. Diabetic nephropathy: the central role of renal proximal tubular cells in tubulointerstitial injury. Histol Histopathol. 2002;17:247–252. doi: 10.14670/HH-17.247. [DOI] [PubMed] [Google Scholar]
- 7.Vallon V., Thomson S.C. Renal function in diabetic disease models: the tubular system in the pathophysiology of the diabetic kidney. Annu Rev Physiol. 2012;74:351–375. doi: 10.1146/annurev-physiol-020911-153333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu J., Lee K., Chuang P.Y., Liu Z., He J.C. Glomerular endothelial cell injury and cross talk in diabetic kidney disease. Am J Physiol Renal Physiol. 2015;308:F287–F297. doi: 10.1152/ajprenal.00533.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nath K.A. Tubulointerstitial changes as a major determinant in the progression of renal damage. Am J Kidney Dis. 1992;20:1–17. doi: 10.1016/s0272-6386(12)80312-x. [DOI] [PubMed] [Google Scholar]
- 10.Wu J., Zhang R., Torreggiani M., Ting A., Xiong H., Striker G.E., Vlassara H., Zheng F. Induction of diabetes in aged C57B6 mice results in severe nephropathy: an association with oxidative stress, endoplasmic reticulum stress, and inflammation. Am J Pathol. 2010;176:2163–2176. doi: 10.2353/ajpath.2010.090386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhattacharjee N., Barma S., Konwar N., Dewanjee S., Manna P. Mechanistic insight of diabetic nephropathy and its pharmaco-therapeutic targets: an update. Eur J Pharmacol. 2016;791:8–24. doi: 10.1016/j.ejphar.2016.08.022. [DOI] [PubMed] [Google Scholar]
- 12.Forbes J.M., Coughlan M.T., Cooper M.E. Oxidative stress as a major culprit in kidney disease in diabetes. Diabetes. 2008;57:1446–1454. doi: 10.2337/db08-0057. [DOI] [PubMed] [Google Scholar]
- 13.Gorin Y., Block K. Nox4 and diabetic nephropathy: with a friend like this who needs enemies. Free Radic Biol Med. 2013;61:130–142. doi: 10.1016/j.freeradbiomed.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nayak B., Xie P., Akagi S., Yang Q., Sun L., Wada J., Thakur A., Danesh F.R., Chugh S.S., Kanwar Y.S. Modulation of renal-specific oxidoreductase/myo-inositol oxygenase by high-glucose ambience. Proc Natl Acad Sci U S A. 2005;102:17952–17957. doi: 10.1073/pnas.0509089102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prabhu K.S., Arner R.J., Vunta H., Reddy C.C. Up-regulation of human myo-inositol oxygenase by hyperosmotic stress in renal proximal tubular epithelial cells. J Biol Chem. 2005;280:19895–19901. doi: 10.1074/jbc.M502621200. [DOI] [PubMed] [Google Scholar]
- 16.Nayak B., Kondeti V.K., Xie P., Sun L., Viswakarma N., Raparia K., Kanwar Y.S. Transcriptional and post-translational modulation of myo-inositol oxygenase by high glucose and related pathobiological stresses. J Biol Chem. 2011;286:27594–27611. doi: 10.1074/jbc.M110.217141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang H.H., Chao H.N., Walker C.S., Choong S.Y., Phillips A., Loomes K.M. Renal depletion of myo-inositol is associated with its degradation in animal models of metabolic disease. Am J Physiol Renal Physiol. 2015;309:F755–F763. doi: 10.1152/ajprenal.00164.2015. [DOI] [PubMed] [Google Scholar]
- 18.Tominaga T., Dutta R.K., Joladarashi D., Doi T., Reddy J.K., Kanwar Y.S. Transcriptional and translational modulation of myo-inositol oxygenase (MIOX) by fatty acids: implications in renal tubular injury induced in obesity and diabetes. J Biol Chem. 2016;291:1348–1367. doi: 10.1074/jbc.M115.698191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhan M., Usman I.M., Sun L., Kanwar Y.S. Disruption of renal tubular mitochondrial quality control by myo-inositol oxygenase in diabetic kidney disease. J Am Soc Nephrol. 2015;26:1304–1321. doi: 10.1681/ASN.2014050457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun L., Dutta R.K., Xie P., Kanwar Y.S. myo-Inositol oxygenase overexpression accentuates generation of reactive oxygen species and exacerbates cellular injury following high glucose ambience: a new mechanism relevant to the pathogenesis of diabetic nephropathy. J Biol Chem. 2016;291:5688–5707. doi: 10.1074/jbc.M115.669952. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Fukuda H., Sano N., Muto S., Horikoshi M. Simple histone acetylation plays a complex role in the regulation of gene expression. Brief Funct Genomic Proteomic. 2006;5:190–208. doi: 10.1093/bfgp/ell032. [DOI] [PubMed] [Google Scholar]
- 22.Jin B., Li Y., Roberston K.D. DNA methylation: superior or subordinate in the epigenetic hierarchy. Genes Cancer. 2011;2:607–617. doi: 10.1177/1947601910393957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Handy D.E., Castro R., Loscalzo J. Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation. 2011;123:2145–2156. doi: 10.1161/CIRCULATIONAHA.110.956839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villeneuve L.M., Reddy M.A., Natarajan R. Epigenetics: deciphering its role in diabetes and its chronic complications. Clin Exp Pharmacol Physiol. 2011;38:401–409. doi: 10.1111/j.1440-1681.2011.05497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy M.A., Park J.T., Natarajan R. Epigenetic modifications in the pathogenesis of diabetic nephropathy. Semin Nephrol. 2013;33:341–353. doi: 10.1016/j.semnephrol.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai M., Bompada P., Atac D., Laakso M., Groop L., De Marinis Y. Epigenetic regulation of glucose-stimulated osteopontin (OPN) expression in diabetic kidney. Biochem Biophys Res Commun. 2016;469:108–113. doi: 10.1016/j.bbrc.2015.11.079. [DOI] [PubMed] [Google Scholar]
- 27.Reddy M.A., Natarajan R. Epigenetics in diabetic kidney disease. J Am Soc Nephrol. 2011;22:2182–2185. doi: 10.1681/ASN.2011060629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan H., Reddy M.A., Deshpande S., Jia Y., Park J.T., Lanting L.L., Jin W., Kato M., Xu Z.G., Das S., Natarajan R. Antioxid Redox Signal. 2016;24:361–375. doi: 10.1089/ars.2015.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z., Miao F., Paterson A.D., Lachin J.M., Zhang L., Schones D.E., Wu X., Wang J., Tompkins J.D., Genuth S., Braffett B.H., Riggs A.D., DCCT/EDIC Research Group. Natarajan R. Epigenomic profiling reveals an association between persistence of DNA methylation and metabolic memory in the DCCT/EDIC type 1 diabetes cohort. Proc Natl Acad Sci U S A. 2016;113:E3002–E3011. doi: 10.1073/pnas.1603712113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ko Y.A., Mohtat D., Suzuki M., Park A.S.D., Izquierdo M.C., Han S.Y., Kang H.M., Si H., Hostetter T., Pullman J.M., Fazzari M., Verma A., Zgeng D., Greally J.M., Susztak K. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol. 2013;14:R108. doi: 10.1186/gb-2013-14-10-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marumo T., Yagi S., Kawarazaki W., Mishimoto M., Ayuzawa N., Watanabe A., Ueda K., Hirahashi J., Hishikawa K., Sakurai H., Shiota K., Fujita T. Diabetes induces aberrant DNA methylation in the proximal tubules of the kidney. J Am Soc Nephrol. 2015;26:2388–2397. doi: 10.1681/ASN.2014070665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wariishi S., Miyahara K., Toda K., Ogoshi S., Doi Y., Ohnishi S., Mitsui Y., Tui Y., Kawai C., Shizutta Y. A SP1 binding site in the GC-rich region is essential for a core promoter activity of the human endothelial nitric oxide synthase gene. Biochem Biophys Res Commun. 1995;216:729–735. doi: 10.1006/bbrc.1995.2682. [DOI] [PubMed] [Google Scholar]
- 33.Campanero M.R., Armstrong M.I., Flemington E.K. CpG methylation as a mechanism for the regulation of E2F activity. Proc Natl Acad Sci U S A. 2000;97:6481–6486. doi: 10.1073/pnas.100340697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deaton A.M., Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaczynski J., Cook T., Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206.1–206.8. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolell K.J., Crawford D.L. Evolution of Sp transcription factors. Mol Biol Evol. 2002;19:216–222. doi: 10.1093/oxfordjournals.molbev.a004074. [DOI] [PubMed] [Google Scholar]
- 37.Frommer M., McDonald L.E., Millar D.S., Collins C.M., Watt F., Gregg G.W., Molloy P.L., Paul C.L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yebra M.J., Bhagwat A.S. A cytosine methyltransferase converts 5-methylcytosine in DNA to thymine. Biochemistry. 1995;34:14752–14757. doi: 10.1021/bi00045a016. [DOI] [PubMed] [Google Scholar]
- 39.Oshchepkov D.Y., Levitsky V.G. In silico predication of transcription factor-binding sites. Methods Mol Biol. 2011;760:251–267. doi: 10.1007/978-1-61779-176-5_16. [DOI] [PubMed] [Google Scholar]
- 40.Guo J.U., Su Y., Shin J.H., Shin J., Li H., Xie B., Zhong C., Hu S., Le T., Fan G., Zhu H., Chang Q., Gao Y., Ming G.L., Song H. Distribution, recognition and regulation of non-CpG methylation in adult mammalian brain. Nat Neurosci. 2014;17:215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen H., Li J., Jiao L., Petersen R.B., Li J., Peng A., Zheng L., Huang K. Apelin inhibits the development of diabetic nephropathy by regulating histone acetylation in Akita mouse. J Physiol. 2014;592:505–521. doi: 10.1113/jphysiol.2013.266411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verdone L., Caserta M., Di Mauro E. Role of histone acetylation in the control of gene expression. Biochem Cell Biol. 2005;83:344–353. doi: 10.1139/o05-041. [DOI] [PubMed] [Google Scholar]
- 43.Bohle A. Importance of the renal tubulo-interstitium for kidney function. Klin Wochenschr. 1982;60:1186–1190. doi: 10.1007/BF01716721. [DOI] [PubMed] [Google Scholar]
- 44.Nath K.A. The tubulo-interstitium in progressive renal disease. Kidney Int. 1998;54:992–994. doi: 10.1046/j.1523-1755.1998.00079.x. [DOI] [PubMed] [Google Scholar]
- 45.Rahmoune H., Thompson P.W., Ward J.M., Smith C.D., Hong G., Brown J. Glucose transporters in human renal proximal tubular cells isolated from urine of patients with non-insulin-dependent diabetes. Diabetes. 2005;54:3427–3434. doi: 10.2337/diabetes.54.12.3427. [DOI] [PubMed] [Google Scholar]
- 46.Martin M.G., Wang J., Solorzano-Vargas R.G., Lam J.T., Turk E., Wright E.M. Regulation of the human Na+-glucose cotransporter gene, SGLT1, by HNF-1 and Sp-1. Am J Physiol Gastrointest Liver Physiol. 2000;278:G591–G603. doi: 10.1152/ajpgi.2000.278.4.G591. [DOI] [PubMed] [Google Scholar]
- 47.Ruegg J., Cai W., Karimi M., Kiss N.B., Swedenberg E., Larsson C., Ekstrom T.J., Pongratz I. Epigenetic regulation of glucose transporter 4 by estrogen receptor-β. Mol Encrinol. 2011;25:2017–2028. doi: 10.1210/me.2011-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vallon V. The mechanisms and therapeutic potential of SGLT2 inhibitors in diabetes mellitus. Annu Rev Med. 2015;66:255–270. doi: 10.1146/annurev-med-051013-110046. [DOI] [PubMed] [Google Scholar]
- 49.Pitocco D., Tesauro M., Alessandro R., Ghirlanda G., Cardillo C. Oxidative stress in diabetes: implications for vascular and other complications. Int J Mol Sci. 2013;14:21525–21550. doi: 10.3390/ijms141121525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ha H., Lee H.B. Reactive oxygen species amplify glucose signaling in renal cells cultured under high glucose and in diabetic kidney. Nephrology (Carlton) 2005;10(Suppl):S7–S10. doi: 10.1111/j.1440-1797.2005.00448.x. [DOI] [PubMed] [Google Scholar]
- 51.Ha H., Lee H.B. Reactive oxygen species and matrix remodeling in diabetic kidney. J Am Soc Nephrol. 2003;14:S246–S249. doi: 10.1097/01.asn.0000077411.98742.54. [DOI] [PubMed] [Google Scholar]
- 52.Williamson J.R., Chang K., Frangos M., Hasan K.S., Ido Y., Kawamura T., Nyengaard J.R., Enden M.V., Tilton R.G. Hyperglycemic pseudohypoxia & diabetic complications. Diabetes. 1993;42:801–813. doi: 10.2337/diab.42.6.801. [DOI] [PubMed] [Google Scholar]
- 53.Nordquist L., Friederich-Persson M., Fasching A., Liss P., Shoji K., Nangaku M., Hansell P., Palm F. Activation of hypoxia-inducible factors prevents diabetic nephropathy. J Am Soc Nephrol. 2015;26:328–338. doi: 10.1681/ASN.2013090990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takiyama Y., Haneda M. Hypoxia in diabetic kidneys. Biomed Res Int. 2014;2014:837421. doi: 10.1155/2014/837421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Block K., Gorin Y., Hoover P., Williams P., Chelmicki T., Clark R.A., Abboud H.E. NADPH oxidases regulate HIF-2α protein expression. J Biol Chem. 2007;282:8019–8026. doi: 10.1074/jbc.M611569200. [DOI] [PubMed] [Google Scholar]
- 56.Kolset S.O., Reinholt F.P., Jenssen T. Diabetic nephropathy and extracellular matrix (ECM) J Histochem Cytochem. 2012;60:976–986. doi: 10.1369/0022155412465073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schena F.P., Gesualdo L. Pathogenetic mechanisms of diabetic nephropathy. J Am Soc Nephrol. 2005;16:S30–S33. doi: 10.1681/asn.2004110970. [DOI] [PubMed] [Google Scholar]
- 58.Khin S.S., Kitazawa R., Kondo T., Idei Y., Fujimoto M., Haraguchi R., Mori K., Kitazawa S. Epigenetic alteration by DNA promoter hypermethylation of growth factor-β (TGF-β) signaling in cancer. Cancers (Basel) 2011;3:982–993. doi: 10.3390/cancers3010982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Verdone L., Agricola E., Caserta M., Di Mauro E. Histone acetylation in gene regulation. Brief Funct Genomic Proteomic. 2006;5:209–221. doi: 10.1093/bfgp/ell028. [DOI] [PubMed] [Google Scholar]
- 60.Spange S., Wagner T., Heinzel T., Kramer O.H. Acetylation of non-histone proteins modulates cellular signaling at multiple levels. Int J Biochem Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 61.O'Dowd B.F., Heiber M., Chan A., Heng H.H.Q., Tsui L.C., Kennedy J.L., Shi X., Petronis A., George S.R., Nguyen T. A human gene that shows identity with the gene encoding the angiotensin receptor is located on chromosome 11. Gene. 1993;136:355–360. doi: 10.1016/0378-1119(93)90495-o. [DOI] [PubMed] [Google Scholar]
- 62.Castan-Laurell I., Dray C., Attane C., Duparc T., Knauf C., Valet P. Apelin, diabetes and obesity. Endocrine. 2011;40:1–9. doi: 10.1007/s12020-011-9507-9. [DOI] [PubMed] [Google Scholar]
- 63.Day R.T., Cavaglieri R.C., Feliers D. Apelin retards the progression of diabetic nephropathy. Am J Physiol Renal Physiol. 2013;304:F788–F800. doi: 10.1152/ajprenal.00306.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen H., Wan D., Wang L., Peng A., Xiao H., Petersen R.B., Liu C., Zheng L., Huang K. Apelin protects against acute renal injury by inhibiting TGF-β1. Biochim Biophys Acta. 2015;1852:1278–1287. doi: 10.1016/j.bbadis.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 65.Chen H., Zheng C., Zhang X., Li J., Li J., Zhenf L., Huang K. Apelin alleviates diabetes-associated endoplasmic reticulum stress in the pancreas of Akita mice. Peptides. 2011;32:1634–1639. doi: 10.1016/j.peptides.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 66.Attane C., Foussal C., Le Gonidec S., Benani A., Daviaud D., Wanecq E., Guzman-Ruiz R., Dray C., Bezaire V., Rancoule C., Kuba K., Ruiz-Gayo M., Levade T., Penninger J., Burcelin R., Penicaud L., Valet P., Castan-Laurell I. Apelin treatment increases complete fatty acid oxidation, mitochondrial oxidative capacity, and biogenesis in muscle of insulin-resistant mice. Diabetes. 2012;61:310–320. doi: 10.2337/db11-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]