Abstract
Ezrin–radixin–moesin (ERM) proteins regulate the organization and function of specific cortical structures in polarized epithelial cells by connecting filamentous (F)-actin to plasma membrane proteins. The contribution of ERM proteins to these structures depends on a conformational change to an active state in which the C-terminal region interacts with F-actin and the N-terminal domain interacts with membrane ligands. The specific ligands necessary for stabilizing ERM proteins at the membrane are not known. By generating mice deficient for ERM-binding phosphoprotein 50/Na+/H+ exchanger regulatory factor 1 (EBP50/NHERF1), which binds the N-terminal domain of ERM proteins, we found that EBP50 is required for the maintenance of active ERM proteins at the cortical brush border membranes (BBM) of polarized epithelia. In EBP50(–/–) mice, ERM proteins were significantly decreased specifically in BBM from kidney and small intestine epithelial cells, whereas they remained unchanged in the cytoplasm. In wild-type animals, EBP50 was localized to the BBM compartment where it was processed by cleavage of the ERM-binding motif. In BBM, active ERM proteins formed distinct complexes with full-length EBP50 and with F-actin, suggesting a switch mechanism in which proteolytically processed EBP50 would release ERM proteins to complex with F-actin. The structural defects found in the EBP50(–/–) intestinal microvilli were reminiscent of those described in ezrin(–/–) mice, suggesting a role for EBP50 in organizing apical epithelial membranes.
Keywords: mutant (knockout) mice, brush border membranes, intestine, kidney
The compartmented, polarized architecture of epithelia is a basic property of higher life forms that makes possible unidirectional fluid and solute transport, the bases for epithelial tissue reabsorptive and secretory functions throughout the body. In the epithelial cell sheet, there is a barrier separating the apical cell surface from the basolateral membranes that has two components: the polarized cells themselves and the junction strands that circumferentially band each cell at the most apical point of the lateral intercellular space. The disruption of epithelial polarity appears as an early step in the development of the epithelial neoplasm, which is the preponderant form of human cancer (1). Ezrin, radixin, moesin, and the neurofibromatosis 2 tumor-suppressor protein form the ERM subfamily of proteins that belongs to the larger protein band 4.1 family (2). Band 4.1 members link membrane proteins to the cytoskeleton and perform structural and regulatory roles at the polarized cell cortex. The ERM proteins are structured into three functional domains: an N-terminal FERM (four point one, ERM) domain, an extended coiled-coil region and a short C-terminal domain. Through the FERM domain, ERM proteins associate with intracellular molecules such as ERM-binding phosphoprotein 50 (EBP50) and Rho GDP-dissociation inhibitor or with transmembrane proteins such as CD44 (reviewed in ref. 2), and the abundant microvillar neutral endopeptidase 24.11 (NEP) (3). Through conserved C termini, ERM proteins bind directly to F-actin (4). In addition, ERM proteins form intra- and intermolecular homo- or heterotypic associations by interactions between their N- and C-terminal domains (5). The disruption of the intramolecular interaction by phosphorylation of the C-terminal conserved T567/T564/T568 residue in ezrin/radixin/moesin, respectively (6), is the key step in regulating the ability of ERM proteins to interact with other molecules.
Na+/H+ exchanger regulatory factor 1 (NHERF1)/EBP50 was independently identified as a regulator factor for Na+/H+ exchanger 3 (NHE3) (7) and as an ERM-binding phosphoprotein of 50 kDa (8). EBP50 is a 358-residue adapter molecule that has two PDZ (PSD-95/Disk-large/ZO-1 homology) domains and a C-terminal ERM-binding (EB) region. EBP50 can bind to many proteins through its PDZ domains. Most of the ligands, such as cystic fibrosis transmembrane conductance regulator (9, 10) and the type IIa Na/Pi cotransporter Npt2 (11), bind to the PDZ1 domain and only a few proteins, such as β-catenin (12), appear to interact specifically with the PDZ2 domain. Through its EB motif, EBP50 binds to the FERM domain of ERM proteins (8). Through its interactions with a large number of proteins, EBP50 has been involved in a broad array of biological systems although the in vivo significance of these interactions is unknown.
We took the genetic approach of disrupting the EBP50 gene in mice to define which of these interactions are important in vivo. Our study indicates that EBP50 is required for the maintenance of active phosphorylated ERM proteins at the apical membrane in the specialized epithelia containing microvilli [brush border membranes (BBM)]. In EBP50-deficient animals, the destabilizing effect of EBP50 on ezrin is most likely the cause of the observed structural defects in the intestinal epithelium.
Methods
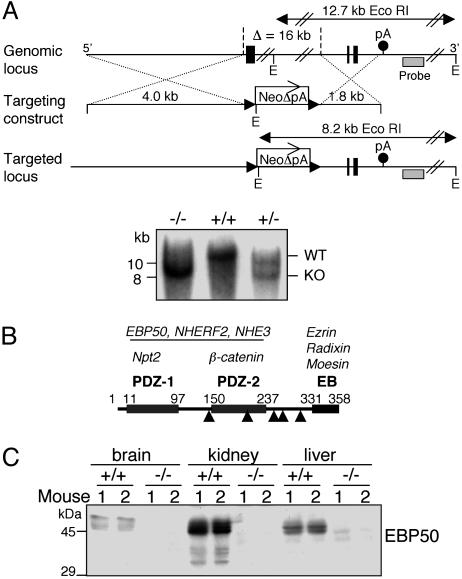
Generation of EBP50/NHERF1-Deficient Mice. The targeting construct for homologous recombination in the NHERF1/EBP50 genomic locus on mouse chromosome 11 was assembled in pBSK vector (Stratagene). The genomic DNA of the NHERF1/EBP50 gene was obtained by screening a 129/SvJ mouse genomic DNA bacterial artificial chromosome library (Research Genetics) with a probe derived by PCR from exon 1 of mouse full-length EBP50 cDNA (IMAGE EST clone 2192738 [American Type Culture Collection (ATCC)]. A bacterial artificial chromosome clone containing the full EBP50 genomic locus was identified and purchased from Research Genetics. The 4.0-kb 5′ fragment and 1.8-kb 3′ fragment flanking the genomic region targeted for recombination were PCR amplified by using proofreading LA Taq DNA polymerase (Takara, Otsu, Shiga, Japan) and primers with restriction sites. The neomycin-resistance (neoR) gene lacking the polyadenylation signal and flanked by LoxP sites to allow its Cre-mediated excision after homologous recombination was inserted between the EBP50 5′ and 3′ genomic fragments. The diphtheria toxin α gene was placed downstream of the EBP50 3′ genomic fragment to provide negative selection against random insertion of the targeting construct in the mouse genome. The targeting construct was electroporated in embryonic stem cells from the same mouse strain as the genomic DNA used for homologous recombination, and two positive embryonic stem cell clones identified by Southern hybridization were injected in C57BL/6J blastocysts. The probe used for Southern blotting was amplified by PCR from the bacterial artificial chromosome clone containing the EBP50 genomic locus (Fig. 1A). Three chimeric mice were obtained from each embryonic stem cell clone and further crossed to C57BL/6J mice (The Jackson Laboratory). Heterozygous pups EBP50 (+/–) were obtained from chimeric mice resulting from both embryonic stem cell clones.
Fig. 1.
Generation of EBP50-deficient mice. (A) Targeted disruption of the EBP50 locus. A fragment of the wild-type locus (Top) containing the first and last two exons (solid boxes) and the polyadenylation signal (pA) of EBP50 is shown. The targeting construct (Middle) consisting of a 4.0-kb 5′ genomic fragment, a 1.8-kb 3′ genomic fragment, and the neoR gene without the polyadenylation signal (NeoΔpA) replaces a 16-kb genomic fragment containing exons 1–4 of the EBP50 gene in the targeted mutated locus (Bottom). The solid arrowheads flanking the neoR gene represent LoxP sites. The positions of the Southern blot probe (hatched boxes), EcoRI sites (E), and the EcoRI wild-type and mutated genomic fragments (double-pointed arrows) are shown. The Southern analysis is shown beneath. (B) Schematic representation of the EBP50 domain structure. The two PDZ domains (1 and 2) and the C-terminal ERM-binding (EB) region are shown with the respective boundaries in amino acids. Solid arrowheads indicate EBP50 mRNA splice sites that delimit EBP50's six exons. Some known interacting proteins for the various domains are listed in italics above each domain. The dimerization with itself or NHERF2 and the interaction with NHE3 most likely extend through both PDZ domains. (C) Western blot analysis of EBP50 from whole organs homogenized in TNN buffer. The organs were collected from two mice per each genotype.
Cells, Splice Isoforms, and Plasmids. Opossum kidney cells (a gift of Patricia Preisig, University of Texas Southwestern Medical Center, Dallas) were grown in a 1:1 mixture of low-glucose DMEM/Ham's F-12 medium supplemented with 10% FCS. U-87 MG and U937 cells (ATCC) were grown in DMEM and RPMI medium 1640, respectively, supplemented with 10% FCS. EBP50 splice isoforms were detected by using reverse transcription and PCR protocols (13) from human brain, kidney, and placenta mRNA (Stratagene) and from RNA extracted from the U-87 MG and U937 cell lines. The PCR fragment including all of the splice sites of EBP50 RNA was obtained with the primers F-EBP-140 (5′-AGGCCGACAAGAGCCACCCG-3′) and R-EBP-3′-UTR (5′-GAGGACGGGAACACATTCACC-3′). Ezrin-NT, comprising the N-terminal 309 residues of ezrin that form a functional FERM domain, was inserted in pGEX-6P-1 vector (Amersham Pharmacia) and purified from bacteria as glutathione S-transferase (GST) fusion protein (13). The C terminus of human Npt2 (residues 562–639), containing the PDZ motif, was obtained by reverse transcription followed by PCR from human kidney mRNA and further inserted in the pGEX-6P-1 vector.
Protein Analysis. The protocols for transfection, GST fusion protein pull-down assays, Western blotting, and immunoprecipitation were described elsewhere (13). The gel-filtration analysis of protein extracts from BBM was performed as described (14). Organ homogenates in TNN buffer [50 mM Tris·HCl (pH 7.4)/150 mM NaCl/5 mM EDTA/0.5% Nonidet P-40] were clarified by centrifugation (total lysate). Antibodies were obtained as follows: EBP50 (Calbiochem); EBP50 C terminus (a gift of Vijaya Ramesh, Massachusetts General Hospital, Charlestown); ezrin–radixin (C-19), moesin (C-15), and NEP (H-321) (Santa Cruz Biotechnology); phospho-ERM (Cell Signaling Technology, Beverly, MA); NHE3 (a gift of Mark Knepper, National Heart, Lung, and Blood Institute, Bethesda); vinculin (Sigma); and actin and villin (Chemicon). The NHERF2 antibody was custom-made (Covance Research Products, Denver, PA) and purified by using mouse NHERF2 (EST GI001B12 from Open Biosystems, Huntsville, AL) fused to GST as antigen. Human kidney and brain tissue lysates were purchased from ProSci, Poway, CA.
BBM Purification. BBM from kidney cortex and intestine were prepared as described (15) with minor modifications. Briefly, kidney cortex and small intestine isolated from EBP50 (+/+) and (–/–) mice were homogenized in 5 ml of buffer [300 mM d mannitol/5 mM EGTA/12 mM Tris·HCl (pH 7.4)] and 7 ml of MgCl2 to give a final concentration of 12 mM was added. After 15 min of incubation on ice, the samples were centrifuged at 4,000 × g for 15 min at 4°C (P1). The supernatant was centrifuged at 10,000 × g for 30 min at 4°C (S2). The remaining pellet was resuspended in 10 ml of 150 mM d mannitol/2.5 mM EGTA/6 mM Tris·HCl (pH 7.4)/12 mM MgCl2, incubated on ice for 15 min, and centrifuged at 4,000 × g for 15 min at 4°C (P3). The supernatant was recovered and centrifuged at 10,000 × g for 30 min at 4°C (S4). The BBM pellet (P4) was resuspended in TNN buffer.
Histology and Electron Microscopy. Formalin-fixed, paraffin-embedded sections were stained with periodic acid/Schiff reagent by using standard protocols. For the transmission electron microscopy analysis, the samples were fixed with 3% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.3), postfixed with 1% osmium tetroxide, and stained en bloc with 1% uranyl acetate. Samples were dehydrated in increasing concentrations of ethanol, infiltrated, embedded in Epon medium, and polymerized at 70°C for 2 days. Ultrathin sections were stained with uranyl acetate and lead citrate and examined in a JEM 1010 transmission electron microscope (JEOL). Digital images were obtained by using AMT Image System (Advanced Microscopy Techniques, Danvers, MA).
Results
Generation of EBP50-Null Mice. To study the role of EBP50 in vivo, we generated mice with targeted disruption of the EBP50 gene (Fig. 1A). Because EBP50 is structured in domains that are encoded by distinct exons (Fig. 1B), a partial disruption of an isolated exon could potentially allow the generation of residual dominant-negative forms. To eliminate this possibility, we devised a strategy to disrupt most of the EBP50 coding sequence. Because the EBP50 locus is relatively large, we used a polyadenylation site-trapping method to enhance the efficiency of recombination by placing the 3′ genomic arm of the targeting construct upstream of the polyadenylation site of the EBP50 gene and downstream of the neoR gene lacking its own polyadenylation site (Fig. 1A). The size of the genomic sequence targeted for deletion was 16 kb, and it comprised 775 nucleotides from the promoter region, exons 1–4 (encompassing residues 1–263 of mouse EBP50) and introns 1–3. In comparison, in the NHERF1/EBP50-deficient mouse generated by Shenolikar et al. (16), only exon 1 (residues 1–142) of the coding region was eliminated. The presence of the mutant and wild-type allele in mice was confirmed by both Southern blotting (Fig. 1A) and PCR (not shown). RT-PCR analysis and sequencing of mutant mRNA revealed that the remaining introns 4 and 5 of EBP50 were spliced, whereas exons 5 and 6 were transcribed after the stop codon of the neoR mRNA, as part of the 3′ UTR. No internal translation-initiation codon was present in exons 5 and 6 or in the 15-codon linker that separated these exons from the stop codon of the neoR gene. The absence of EBP50 products translated from exons 5 and 6 in EBP50(–/–) mice was also confirmed by Western blotting of kidney extracts with an antibody recognizing the C terminus of EBP50 (not shown). The transmission of the mutant allele was Mendelian in both sexes. In contrast to the high mortality rate in females observed in the NHERF1-deficient mice established by Shenolikar et al. (16), we did not observe gross phenotypic abnormalities in our mice. On rare occasions, EBP50(–/–) females developed progressive general weakness and died. However, we confirmed the decrease of phosphatemia reported by Shenolikar et al. (16) in EBP50(–/–) mice, detecting phosphate values of 7.4 ± 0.8 mg/dl in EBP50(+/+) versus 6.3 ± 0.3 mg/dl in EBP50(–/–) mice (for n = 6 mice per group, P = 0.05).
Protein analysis in various organs confirmed the absence of EBP50 from EBP50(–/–) animals (Fig. 1C). EBP50 migrated as a triplet of ≈50,000 molecular weight (MW), most likely representing phosphorylation species of full-length EBP50. In the EBP50(+/+) kidney, the anti-EBP50 antibody specifically detected three bands of lower MW that, as shown later, represent cleavage products of EBP50.
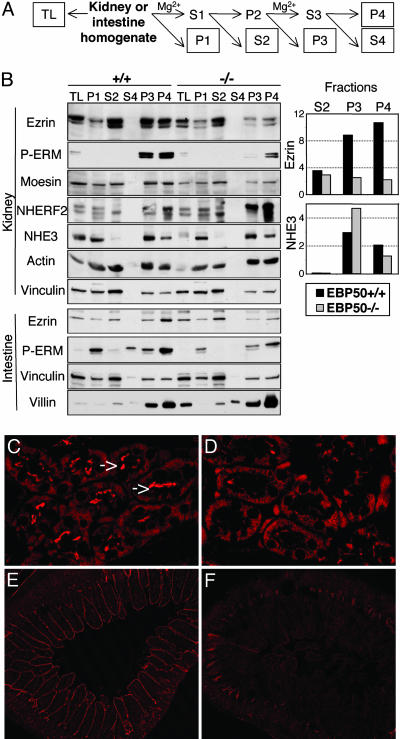
ERM Proteins Are Decreased in EBP50(–/–) BBM. We examined the influence of EBP50 on the distribution of ERM proteins, which are postulated to exist in active and inactive conformations, presumably at the cell cortex and in the cytoplasm of epithelial polarized cells, respectively (reviewed in ref. 2). To separate the cytoplasmic and membrane compartments, we used the differential centrifugation method yielding BBM (15), derived from the apical membranes rich in microvilli (like a brush) of epithelial cells (Fig. 2A). Employing markers for the various cellular compartments, we found that S2 corresponds to the cytoplasmic fraction (phosphatase and tensin homologue marker, not shown), and P3 and P4 contain membrane-rich structures associated with both membrane-resident (NHE3) and cytoskeletal (actin, vinculin, and villin) proteins (Fig. 2B). Using this fractionation method, we found that ERM proteins are significantly reduced in the BBM P3 and P4 fractions from kidney and intestine homogenates of EBP50(–/–) mice (Fig. 2B). This finding implies that EBP50 is required for proper recruitment and compartmentalization of ERM proteins at the cell cortex. In contrast, the cytoplasmic ezrin, which represents the dormant inactive pool of ezrin, appeared unchanged in EBP(+/+) and (–/–) S2 fractions. On the other hand, NHERF2, the homologue of EBP50 (17), was increased in EBP50(–/–) kidney membrane fractions, most likely compensating for EBP50 loss. Other proteins known to bind to EBP50, such as β-catenin (not shown) or NHE3 (also previously reported in ref. 16), presented unchanged levels in EBP50(–/–) kidney membrane fractions, further underscoring the importance of EBP50 for ERM compartmentalization. The recruitment of actin in EBP50(–/–) BBM fractions was not impaired (Fig. 2B), suggesting that recruitment may take place in the presence of small amounts of ezrin in these fractions or perhaps through an ezrin-independent mechanism. In this sense, villin, which binds to actin and organizes the microvillus core bundle, was up-regulated in EBP50(–/–) small intestine BBM (Fig. 2B), probably compensating for the decrease of ezrin at this level.
Fig. 2.
Decreased expression levels of ERM proteins in EBP50(–/–) BBM. (A) Schematic protocol of BBM preparation. The protein concentration was measured in each boxed fraction, and samples containing 30 μg of proteins [except from S4 (supernatant 4), which contained only 1.5 μg of proteins] were loaded on polyacrylamide gels. TL, total tissue lysates. P4 (pellet 4) corresponds to the BBM fraction. (B) Western blot analysis of kidney and small intestine BBM fractionation samples from EBP50 (+/+) and (–/–) mice was performed with the indicated antibodies. The quantification analysis (Right) was performed with the imagej program (National Institutes of Health) and shows the expression levels of ezrin and NHE3 normalized to vinculin in kidney fractions. For ezrin, only the upper bands, which represent ezrin, were quantified. Similar results were obtained at least seven times for BBM prepared from either male or female mice. (C–F) The expression of phosphorylated ezrin was analyzed with phospho-ERM antibody by immunofluorescence of kidney (C and D) and jejunum (E and F) sections from EBP50(+/+) (C and E) and (–/–) (D and F) 5-week-old littermates. The samples were imaged with a Zeiss LSM 510 confocal microscope by using the HeNe 543-nm laser and identical imaging parameters. Note the higher apical expression of phosphorylated ezrin (arrows) in wild-type kidney tubules and intestinal villi.
The activation by phosphorylation of ERM proteins was revealed with an antibody recognizing the C-terminal phospho-T567/T564/T568 residue. ERM proteins appeared to be phosphorylated in the membrane fractions (Fig. 2B), confirming in vivo the proposed existence of dormant-cytoplasmic and membrane-active states of ERM proteins (2). In BBM (Fig. 2B) and histological sections (Fig. 2 C–F), phosphorylated ERM were significantly reduced in EBP50(–/–) kidney and intestinal epithelia compared with wild-type, implying that EBP50 is necessary to stabilize active phosphorylated ERM proteins at the apical membrane.
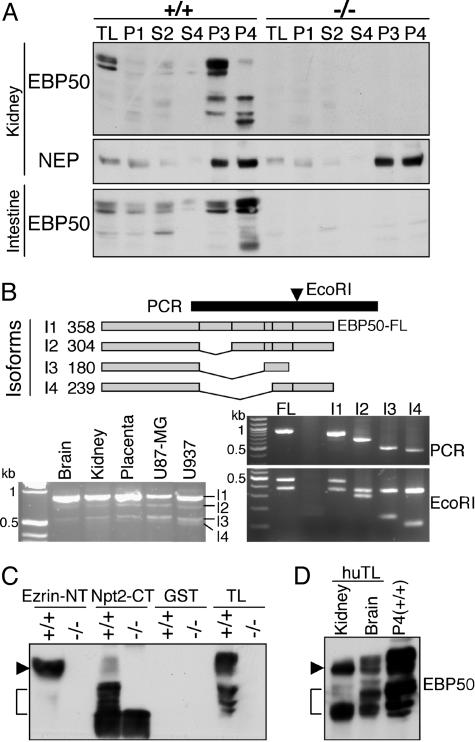
Proteolysis of the EBP50 EB Motif in BBM. EBP50 appeared in EBP50(+/+) P3 and P4 fractions from kidneys and was also concentrated in these fractions from intestine (Fig. 3A). This pattern pointed to restricted membrane localization for EBP50 as opposed to ezrin in these specialized epithelia. In kidney, the P3 fraction contained full-length EBP50 whereas the P4 fraction contained mainly the low-MW fragments that we initially observed in kidney total lysates (see Fig. 1C). In intestine, the P4 fraction contained the highest amount of both full-length EBP50 and low-MW fragments (Fig. 3A). As expected, there were no EBP50 products in fractions from EBP50(–/–) mice, whereas NEP, used as a positive control for the BBM fractions, was present in similar amounts in (+/+) and (–/–) preparations.
Fig. 3.
The EB region of EBP50 is cleaved in BBM. (A) The Western blot analysis of EBP50 in BBM fractions from EBP50(+/+) and (–/–) mice was carried out on the same samples as in Fig. 2B. Note the presence of EBP50 full-length and low-MW products in membrane fractions from EBP50(+/+) mice. (B)(Upper) EBP50 splice isoforms (I1 to I4) are schematically drawn, and the corresponding size in amino acids is indicated on the left. The black bar indicates the PCR used to identify the isoforms, and the arrowhead marks the position of the EcoRI site used for restriction-fragment-length polymorphism. The PCR on reverse transcribed mRNA extracted from various human tissues and cell lines is shown (Lower Left). The PCR fragments corresponding to the isoforms indicated on the left were subcloned from the U937 PCR product (Middle Right) and analyzed for RFLP with EcoRI (Bottom Right). FL, control full-length EBP50. (C) Pull-down assay with GST ezrin-NT and Npt2-CT fusion proteins (5 μg) of proteins (300 μg) from pooled P3 and P4 EBP50(+/+) or (–/–) kidney BBM fractions. Thirty micrograms of proteins from P3 and P4 fractions are included (TL). Note the specific precipitation of EBP50 FL (arrowhead) by ezrin-NT and of low-MW products (bracket) by Npt2-CT. (D) Western analysis showing EBP50 FL (arrow) and low-MW products (bracket) in human tissue lysates (huTL) in comparison with mouse P4 kidney fraction.
The low-MW forms of EBP50 could be splice isoforms or cleavage products. To discriminate between these possibilities, we first examined whether there are splice EBP50 mRNA variants (Fig. 3B). Extracted RNAs from various human tissues or cell lines were reverse transcribed, and the resultant cDNAs were used as templates for PCR amplification with primers external to all splice sites (Fig. 3B). In addition to the most abundant species represented by a fragment at the expected size of full-length EBP50, there were several fragments of lower MW (Fig. 3B Lower Left). Three fragments were purified, subcloned, and sequenced (Fig. 3B Right). These fragments corresponded to splice variants: I2, I3, and I4, in which exon 1 is connected with exon 3, 4, or 5, respectively (Fig. 3B Upper). All isoforms contained the PDZ1 domain and disrupted the PDZ2 domain. Isoforms I2 and I4 preserved the frame and thus retained the EB region.
To identify the EBP50 domains present in the low-MW fragments of EBP50, these fragments were precipitated with the Npt2-C terminus (Npt2-CT), which binds to the EBP50–PDZ1 domain, and with the ezrin-NT, which binds to the EBP50–EB region (Fig. 3C). Only the Npt2-CT precipitated the low-MW fragments, implying that these fragments contain the PDZ1 domain but lack the EB region of EBP50. Because most EBP50 splice isoforms retain the EB region (Fig. 3B), it is unlikely that the three or four low-MW fragments we observed represent splice isoforms. More likely, these fragments are cleavage products rendered unable to interact with ezrin by proteolytic removal of the C-terminal EB region. These fragments appeared also to be present in human normal tissue lysates (Fig. 3D), suggesting that the processing of EBP50 may have physiological importance in both mice and humans.
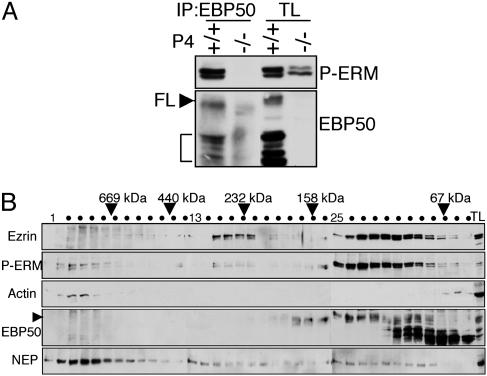
Phosphorylated ERM Proteins Form Distinct Complexes with EBP50 and with Actin. The analysis of BBM fractions indicated that the only possibility for EBP50 to associate with ERM proteins is in the membrane fractions that contain both ERM proteins and EBP50. Furthermore, the interaction should take place between active phosphorylated ERM proteins and EBP50. We confirmed this assumption by showing immunoprecipitation of phosphorylated ERM proteins by EBP50 in EBP50(+/+) kidney P4 fractions (Fig. 4A). Similarly, EBP50 associated with phosphorylated ERM proteins or with phosphorylated ezrin in kidney P3 fractions or in intestine P4 fractions, respectively (not shown).
Fig. 4.
EBP50 interacts with phosphorylated ERM proteins. (A) Coimmunoprecipitation of phosphorylated ERM proteins (P-ERM) with EBP50 from EBP50(+/+) and (–/–) kidney BBM P4 fractions. Full-length EBP50, FL and arrowhead; cleavage products, bracket. (B) Gel-filtration analysis of protein complexes from EBP50(+/+) kidney BBM P4 fractions with antibodies indicated on the left. Arrowhead indicates FL EBP50 and TL, total lysate. Note coelution of phosphorylated ERM proteins with FL EBP50 in a low-MW peak and with actin in a high-MW peak.
Because ERM proteins bind through their C terminus to actin (4), we investigated whether EBP50 that associates with ERM proteins through their N terminus (8) is present in complexes with F-actin. The distribution of these proteins in complexes was analyzed by gel filtration in EBP50(+/+) kidney P4 fractions (Fig. 4B). Ezrin eluted mainly in two peaks, a higher-MW peak (≥232,000), most likely containing oligomers (18), and an extended, lower-MW peak >67,000. Moesin had a similar elution pattern (not shown). The same membranes probed with phospho-ERM antibody showed a different profile consisting of a high-MW peak (>669,000), in which phosphorylated ERM proteins cofractionated with polymerized actin, and a low-MW peak (between 67,000 and 158,000), in which phosphorylated ERM proteins cofractionated with full-length EBP50. The oligomeric forms of ezrin (232,000-MW peak) were not reactive with the phospho-ERM antibody, indicating that ezrin is inactive in this conformation.
Full-length EBP50 presented a plateau profile between 67,000 and 158,000 (Fig. 4B) that, moving from right to left, consisted of heterodimers with its cleavage products and homodimers (F.C.M. and M.-M.G., unpublished results) followed by complexes with phosphorylated ERM proteins. Because of the proximity of molecular weights between monomeric ezrin (78,000), EBP50 hetero- and homodimers, and the dual ezrin–EBP50 complex, their respective peaks overlapped. Taken together, these results indicate that, at the membrane, phosphorylated ERM proteins associated with both full-length EBP50 and actin, but in different complexes. This finding suggests that EBP50 may not connect ezrin to the membrane directly.
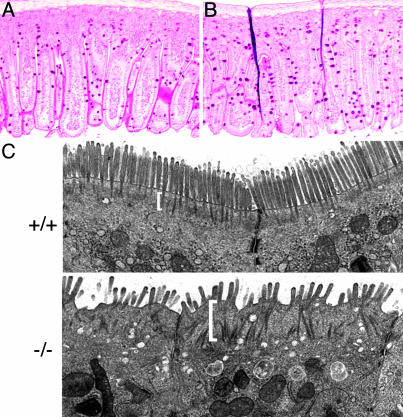
Defects of Intestinal Villi in EBP50(–/–) Mice. Macroscopic and histological examination did not reveal gross abnormalities in EBP50(–/–) kidneys (not shown). However, the small intestine, where ezrin is the only ERM protein expressed in the epithelium, presented a significantly higher mechanical fragility in EBP50(–/–) animals compared with wild type. The EBP50(–/–) intestine tore easily upon flushing or sectioning. Histological analysis showed an increase in the number of goblet cells in EBP50(–/–) intestinal sections with no apparent disorganization of the columnar epithelial cells (Fig. 5 A and B). In villi and in crypts, the number of periodic acid/Schiff reagent-positive goblet cells was more than two times higher in EBP50(–/–) versus wild type (5.96 ± 0.16/(–/–) villus versus 2.53 ± 0.14/(+/+) villus, P < 0.0001 and 3.61 ± 0.15/(–/–) crypt versus 1.72 ± 0.2/(+/+) crypt, P < 0.001). The ultrastructural examination of the apical brush border of intestinal epithelial cells revealed well-ordered brush-like microvilli for wild-type cells and disorganized microvilli for EBP50(–/–) cells (Fig. 5C). The actin-rich terminal web region that provides a platform into which the microvilli are anchored (Fig. 5C, brackets) was well defined in wild-type cells. In contrast, EBP50(–/–) terminal webs were dispersed, wide, and difficult to demarcate, suggesting an alteration of the organization of the apical cytoskeleton.
Fig. 5.
Morphology of intestinal villi in EBP(–/–) mice. (A and B) periodic acid/Schiff reagent (PAS) staining of intestinal villi from EBP50(+/+)(A) and (–/–)(B) littermates showing a higher number of PAS+ goblet cells in (–/–) villi. (×100.) (C) Transmission electron microscopy of ileum epithelial cells from 5-week-old littermates shows EBP50(+/+) ordered, rod-like microvilli emerging from a structured terminal web region (bracket) contrasting with the EBP50(–/–) disorganized microvilli and thick terminal web. (×12,000.)
Discussion
Among the multitude of roles attributed to EBP50 from in vitro studies, we aimed to identify its essential functions in the context of the whole organism. Disruption of the EBP50 gene in mice pointed to a structuring role that EBP50 fulfills by stabilizing ERM proteins at the apical membrane of polarized epithelia. The lack of EBP50 specifically decreased the levels of ERM proteins in the BBM of the kidney proximal tubules and of the small intestine. The morphological defects found in EBP50(–/–) intestine are reminiscent of defects described in ezrin(–/–) mice, although the defects are not as dramatic (19). Unlike the total absence of ezrin that determines abnormal villus morphogenesis in ezrin(–/–) animals (19), the persistence of low levels of ezrin in the intestinal BBM from EBP50(–/–) mice might explain the normal development of the villi in these animals. However, these decreased ezrin levels were not sufficient for the proper organization of the apical membrane of the columnar epithelial cells, as reflected by the disorganization of the microvilli and the effacement of the actin-rich terminal web region in EBP50(–/–) animals. In EBP50(–/–) kidney BBM, low levels of ERM proteins in phosphorylated state were still present, including the active form engaged in high-MW complexes (not shown). This observation argues against the assumption that EBP50 is the only protein stabilizing ERM proteins at the membrane. It is possible that NHERF2, the homologue of EBP50 (17), which we found up-regulated in EBP50(–/–) kidney BBM, compensates for the absence of EBP50. The lack of NHERF2 in the intestinal epithelium (20) may explain the more profound defects found in this organ when EBP50 is lost. The apical down-regulation of ezrin may not be the cause of the increased number of goblet cells present in the EBP50(–/–) intestinal epithelium. It is conceivable that the disrupted interaction between EBP50 and the apical ion transporters, cystic fibrosis transmembrane conductance regulator and NHE3 (7, 9, 10), in EBP50(–/–) mice is responsible for this defect.
Previous studies have postulated that ERM proteins reside in an inactive state in the cytoplasm and in an active ligand-bound state at the membrane (2). Phosphorylation would be necessary to activate ezrin (6), and Hayashi et al. have elegantly shown by immunofluorescence using an antibody that recognizes phosphorylated ERM proteins that the phosphorylated forms could be visualized only at the apical membrane of epithelial cells (21). By fractionating BBM, we have confirmed the membrane localization of phosphorylated ERM proteins, and we have shown the interaction of ERM proteins with EBP50. The complexes between phosphorylated ERM proteins and EBP50 appeared to be binary at a 1:1 stoichiometric ratio by gel filtration analysis, whereas phosphorylated ERM proteins coeluted with polymerized actin in high-MW complexes. The distribution of ERM proteins in the distinct complexes could be controlled by cleavage of the EB region from EBP50 that would release active ERM proteins from the complexes with EBP50. Released ERM proteins might then enter in complex with actin and transmembrane proteins such as NEP, which also coelutes in the high-MW complex (see Fig. 4B), or undergo dephosphorylation and convert to the monomeric or oligomeric dephosphorylated forms that we identified by gel filtration. A study on the distribution of ezrin by fluorescence recovery after photobleaching supports our model of sequential association–release events for membrane-localized ezrin (22). In this study (22), ezrin adopted three energy states at the membrane corresponding to a mobile fraction perhaps represented by the inactive forms freely diffusing from and to the cytoplasm, to a fraction with intermediate mobility that we relate in our study to the EBP50-bound population, and to a slowly moving population probably corresponding to the actin-bound ezrin.
The question of how EBP50 is localized at the apical membrane remains. Two scenarios are possible. Recently, Saotome et al. (19) found, by immunohistochemistry, diffuse localization of EBP50 in ezrin(–/–) intestinal cells in contrast to the apical distribution of EBP50 in ezrin(+/+) cells. Although in this latter study, BBM were not prepared for a quantitative analysis of this effect, the possibility cannot be excluded that ezrin and EBP50 function together in organizing the apical membrane, and that their expression at the apical membrane is interdependent. In support of this hypothesis are our gel filtration data showing that phosphorylated ezrin and EBP50 associate in binary complexes at a 1:1 stoichiometric ratio. It is thus possible that both partners stabilize each other in the formation of this complex at the membrane. A second scenario would involve PDZ domain-mediated interactions between EBP50 and transmembrane molecules (8). This hypothesis is supported by our finding that the cleavage products of EBP50, which contain PDZ domains but no C-terminal EB region, were detected only in membrane fractions and not in cytoplasmic fractions. However, we have not identified EBP50 in high-MW complexes by gel filtration. Rather, EBP50 is found in a dimerized form, whether with itself or with its cleavage products (Fig. 4B and data not shown). It is possible that the high-MW complexes are labile, and, because EBP50 has a strong propensity to dimerize, it may convert to the dimer form upon membrane disruption by detergent. It is also possible that the cleavage of the EB region quickly removes full-length EBP50 from these complexes. Although we do not know the cleavage sites or the proteases responsible for the cleavage of EBP50, the possibility of regulation of EBP50 engagement in complexes by proteolysis is worth considering. The cleavage products of EBP50 appeared to be tissue-specific, occurring in BBM of polarized epithelia but not in other types of tissue (Fig. 1C and data not shown). They may function as dominant-negative forms to disrupt PDZ-mediated complexes with full-length EBP50. For example, they bound better than the full-length molecule to the PDZ motif containing Npt2 transporter (Fig. 3C). Furthermore, the proposed release mechanism of active ERM proteins by EBP50 cleavage may explain the dynamic states of ERM proteins. Last, by dissociating the ability of EBP50 to connect transmembrane proteins to ERM proteins, the processing of EBP50 may modulate the regulation of the activity of NHE3, which requires the EB C terminus of EBP50 for proper function (23). The EB region was also required for the recycling of the κ opioid receptor (24), and it would be interesting to investigate whether cleavage of the EB region in BBM regulates the trafficking of the transmembrane ion transporters that associate with EBP50.
Acknowledgments
We thank Chingwen Yang for the work with mouse embryonic stem cells, Jia-Hui Dong for blastocyst injections, Hermann Steller for logistical support, Margaret Kripke for support of the gel filtration studies, Henry Adams and Lucian Chirieac for valuable help with histological examination, and Kenneth Dunner for his expertise in electron microscopy. The animal breeding was partially supported by National Cancer Institute Grant CA16672.
Author contributions: M.-M.G. designed research; F.C.M., Y.T., E.L.K., and M.-M.G. performed research; F.C.M. and M.-M.G. analyzed data; and M.-M.G. wrote the paper.
Abbreviations: ERM, ezrin–radixin–moesin; EBP50/NHERF1, ERM-binding phosphoprotein 50/Na+/H+ exchanger regulatory factor 1; NEP, neutral endopeptidase 24.11; PDZ, PSD-95/Disc-large/ZO-1; EB, ERM-binding; BBM, brush border membranes; MW, molecular weight.
References
- 1.Mullin, J. M. (2004) Sci. STKE 2004, pe2. [DOI] [PubMed] [Google Scholar]
- 2.Bretscher, A., Edwards, K. & Fehon, R. G. (2002) Nat. Rev. Mol. Cell Biol. 3, 586–599. [DOI] [PubMed] [Google Scholar]
- 3.Iwase, A., Shen, R., Navarro, D. & Nanus, D. M. (2004) J. Biol. Chem. 279, 11898–11905. [DOI] [PubMed] [Google Scholar]
- 4.Turunen, O., Wahlstrom, T. & Vaheri, A. (1994) J. Cell. Biol. 126, 1445–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gary, R. & Bretscher, A. (1995) Mol. Biol. Cell 6, 1061–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsui, T., Maeda, M., Doi, Y., Yonemura, S., Amano, M., Kaibuchi, K. & Tsukita, S. (1998) J. Cell Biol. 140, 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinman, E. J., Steplock, D., Wang, Y. & Shenolikar, S. (1995) J. Clin. Invest. 95, 2143–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reczek, D., Berryman, M. & Bretscher, A. (1997) J. Cell Biol. 139, 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall, R. A., Ostedgaard, L. S., Premont, R. T., Blitzer, J. T., Rahman, N., Welsh, M. J. & Lefkowitz, R. J. (1998) Proc. Natl. Acad. Sci. USA 95, 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Short, D. B., Trotter, K. W., Reczek, D., Kreda, S. M., Bretscher, A., Boucher, R. C., Stutts, M. J. & Milgram, S. L. (1998) J. Biol. Chem. 273, 19797–19801. [DOI] [PubMed] [Google Scholar]
- 11.Gisler, S. M., Stagljar, I., Traebert, M., Bacic, D., Biber, J. & Murer, H. (2001) J. Biol. Chem. 276, 9206–9213. [DOI] [PubMed] [Google Scholar]
- 12.Shibata, T., Chuma, M., Kokubu, A., Sakamoto, M. & Hirohashi, S. (2003) Hepatology 38, 178–186. [DOI] [PubMed] [Google Scholar]
- 13.Georgescu, M.-M., Kirsch, K. H., Akagi, T., Shishido, T. & Hanafusa, H. (1999) Proc. Natl. Acad. Sci. USA 96, 10182–10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sumitomo, M., Iwase, A., Zheng, R., Navarro, D., Kaminetzky, D., Shen, R., Georgescu, M. M. & Nanus, D. M. (2004) Cancer Cell 5, 67–78. [DOI] [PubMed] [Google Scholar]
- 15.Biber, J., Stieger, B., Haase, W. & Murer, H. (1981) Biochim. Biophys. Acta 647, 169–176. [DOI] [PubMed] [Google Scholar]
- 16.Shenolikar, S., Voltz, J. W., Minkoff, C. M., Wade, J. B. & Weinman, E. J. (2002) Proc. Natl. Acad. Sci. USA 99, 11470–11475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yun, C. H., Oh, S., Zizak, M., Steplock, D., Tsao, S., Tse, C. M., Weinman, E. J. & Donowitz, M. (1997) Proc. Natl. Acad. Sci. USA 94, 3010–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berryman, M., Gary, R. & Bretscher, A. (1995) J. Cell Biol. 131, 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saotome, I., Curto, M. & McClatchey, A. I. (2004) Dev. Cell 6, 855–864. [DOI] [PubMed] [Google Scholar]
- 20.Ingraffea, J., Reczek, D. & Bretscher, A. (2002) Eur. J. Cell Biol. 81, 61–68. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi, K., Yonemura, S., Matsui, T. & Tsukita, S. (1999) J. Cell Sci. 112, 1149–1158. [DOI] [PubMed] [Google Scholar]
- 22.Coscoy, S., Waharte, F., Gautreau, A., Martin, M., Louvard, D., Mangeat, P., Arpin, M. & Amblard, F. (2002) Proc. Natl. Acad. Sci. USA 99, 12813–12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinman, E. J., Steplock, D., Donowitz, M. & Shenolikar, S. (2000) Biochemistry 39, 6123–6129. [DOI] [PubMed] [Google Scholar]
- 24.Li, J. G., Chen, C. & Liu-Chen, L. Y. (2002) J. Biol. Chem. 277, 27545–27552. [DOI] [PubMed] [Google Scholar]