Abstract
An Escherichia coli cell-free expression system is encapsulated in a phospholipid vesicle to build a cell-like bioreactor. Large unilamellar vesicles containing extracts are produced in an oil–extract emulsion. To form a bilayer the vesicles are transferred into a feeding solution that contains ribonucleotides and amino acids. Transcription–translation of plasmid genes is isolated in the vesicles. Whereas in bulk solution expression of enhanced GFP stops after 2 h, inside the vesicle permeability of the membrane to the feeding solution prolongs the expression for up to 5 h. To solve the energy and material limitations and increase the capacity of the reactor, the α-hemolysin pore protein from Staphylococcus aureus is expressed inside the vesicle to create a selective permeability for nutrients. The reactor can then sustain expression for up to 4 days with a protein production of 30 μM after 4 days. Oxygen diffusion and osmotic pressure are critical parameters to maintain expression and avoid vesicle burst.
Keywords: α-hemolysin, cell-free protein expression, membrane-anchoring polypeptide
In his logical theory of automata, J. von Neumann (1) compared computing machines and living organisms. The self reproduction of automata was discussed and linked to a Turing-like principle (2). In parallel, the biological sciences raised the question of how to engineer a minimal self-reproducing cell (3). Building a protocell gives clues to how self-replicating systems emerge but also may help researchers to engineer artificial self-replicating machines. Although theoretical models have been proposed (4–6), approaches to reducing the problem complexity are not straightforward. Based on the concept of minimal cell and one possible definition of life (7, 8), a critical step in building an artificial cell is the construction of an enclosed space displaying exchange and use of external energy/nutrients through a semipermeable membrane. In fact, the simple encapsulation of active ingredients into a phospholipid bilayer can be considered as an important transition (9) and a major step in making an artificial cell.
Two complementary approaches are in general considered to build an artificial cell. The bottom-up approach starts with the construction of a minimal cell from the molecular level, the RNA world being one of the main models (10). With the top-down approach, scientists try to reach a minimal cell by reducing the genome of bacteria to a minimum set of genes or proteins (7, 11, 12). In this paper, an approach is presented where the first step consists in assembling a mesoscopic bioreactor by encapsulation of a cell-free expression extract in phospholipid vesicles.
To express proteins in vitro, transcription-translation cell-free systems of wheat germ and Escherichia coli are usually used. Expression stops after a few hours because of energy and nutrients consumption (13, 14). Accumulation of the hydrolyzed forms of ATP and GTP is the main factor of this limitation (15). To solve this problem, large-scale continuous systems have been constructed where a buffered solution containing the nutrients for energy and materials feeds the reaction compartment through a dialysis polymeric membrane (16). The extract itself cannot be used as a feeding solution because of the presence of proteins that hydrolyze ATP and GTP.
In this work, an E. coli extract has been used to carry out in vitro transcription and translation of plasmid genes. Outside of the nonaffordable reconstitution of Shimizu and coworkers (17), an extract is the best available system to express protein in vitro. We chose DNA as the substrate of genetic information instead of RNA because it gives more possibilities to program and develop functions inside the cell-like bioreactor. Furthermore, it is important to show that the two universal steps of genetic expression, transcription and translation, can be performed in an artificial compartment. To go from a homogeneous to a heterogeneous system displaying two distinct aqueous phases, our first challenge was to encapsulate the extract into a vesicle composed of a phospholipid bilayer in nondenaturing conditions for the extract. Phospholipids are the main constituents of biological membranes (18). No other biological or synthetic barrier provides as many possibilities or as much flexibility to functionalize and establish controllable exchanges between the two phases. We present a method to encapsulate the extract efficiently and carry out in vitro transcription and translation inside large unilamellar vesicles made of l-α-lecithin. It is striking to see that formation of vesicles is still possible with a solution as complex and as dense as a cell-free extract, full of proteins that interfere with phospholipids for the formation of interfaces. Such vesicles are transferred into a feeding solution composed of a buffer with the nutrients (mainly ribonucleotides and amino acids). Composition of both phases is the same except for the high protein concentration of the extract. This asymmetry causes a high osmotic pressure that reduces considerably the yield of vesicle formation and their stability in time. To overcome these difficulties, a fine-tuning was found between the extract and the feeding mixture. In such a configuration, expression stops after 5 h.
Our next goal was to establish a more reliable exchange between the two phases. We used the internal expression of a protein to improve the capacity and lifetime of the bioreactors. In large-scale continuous systems (16), extracts and feeding solution are both stirred. For our vesicular reactor, exchange of nutrients is realized by diffusion; the main problem was then to find a protein that makes a nonspecific pore into the phospholipids bilayer without perturbing or lysing the membrane and that has a compatible channel size. We show that the expressed α-hemolysin toxin is functional and create such a selective leak. Expression of the toxin boosts the expression up to 4 days at the level of 1 mg/ml synthesized proteins. For expression, effective diffusion of oxygen is essential. The vesicles have thus to be within 1 mm from the feeding solution–air interface.
Lastly, we indicate how one can further functionalize the membrane by incorporating a small polypeptide that can act as an anchor for other proteins. Such an anchor can be used to bind biopolymers to the membrane that can induce mechanical stress, a step toward possible fission of the vesicles. Such a long-lived bioreactor that sustains expression for 4 days is a first step toward the assembly of a synthetic minimal cell but also a testing chamber to develop and test synthetic genomes.
Materials and Methods
Constructions and Cell-Free Expression. Cloning was performed by routine procedures (19). The sequences of enhanced GFP (eGFP), enhanced yellow fluorescent protein (eYFP), and firefly luciferase were amplified by PCR and inserted into the vector pIVEX2.3d (Roche) between NcoI and SacI (plasmid pIVEX2.3d-eGFP, pIVEX2.3d-eYFP, and pIVEX2.3d-Luc). Sequence of the α-hemolysin gene without the signal peptide was amplified by PCR from the genomic DNA of Staphylococcus aureus (American Type Culture Collection 10832) and inserted into the vector pIVEX2.3d between NcoI and SacI (plasmid pIVEX2.3d-α-hemolysin). The α-hemolysin-eGFP fusion gene was obtained in three steps: a linker containing the ApaI and XhoI sites was introduced into the vector pIVEX2.3d between the sites NcoI and SacI. The α-hemolysin and eGFP genes were amplified by PCR and introduced between the sites NcoI–ApaI and XhoI–SacI, respectively (plasmid pIVEX2.3d-α-hemolysin-eGFP). eGFP was replaced by enhanced cyan fluorescent protein (eCFP) to obtain the plasmid pIVEX2.3d-α-hemolysin-eCFP. An oligonucleotides coding for the 18L peptide (20) was inserted between the sites NcoI and XhoI of plasmid pIVEX2.3d-α-hemolysin-eGFP to obtain the plasmid pIVEX2.3d-18L-eGFP (GGTATAAAGAAGTTTCTGGGAAGTATATGGAAGTTTATAAAGGCATTTGTAGGG, coding for the amino acid sequence GIKKFLGSIWKFIKAFVG). All of the constructions were verified by sequencing.
The E. coli extract (RTS500, Roche) and its feeding solution were used as provided by the supplier. Briefly, and as indicated by the manufacturer (16, 21), the extract contains the following: a buffer that maintains pH between 7.4 and 8, the crude extract [ribosomes (70S), tRNA, translation initiation, elongation, and termination factors], the T7 RNA polymerase, the 20 amino acids between 10 and 100 μM, the 4 ribonucleotides ATP, GTP, UTP, and CTP between 0.2 and 2 mM, 8–15 mM magnesium salt, 100–250 mM potassium salt, an ATP regenerating system, and sulfhydryl compounds (2-mercaptoethanol or DTT). The feeding solution contains the same components except the crude extract, tRNA, the kinase for the ATP regenerating system, and the RNA polymerase. Experiments were carried out at room temperature (25°C). Reactions were done in two different conditions: either 100% of extract was encapsulated in the vesicles and transferred into the feeding solution, or, to reduce the osmotic pressure effect, the extract was diluted one time in feeding solution (50% extract–50% feeding) and the feeding solution was supplemented with 4% extract.
Data Acquisition. For kinetics measurements of eGFP in bulk solution, samples were deposited between two glass coverslips with a 300-μm spacer. Fluorescence was collected through a ×40 objective and amplified with a photo multiplier tube (Hamamatsu GaAsp H7421-40) mounted on a microscope equipped with a 75-W Xe lamp and the proper filter sets (Olympus IX-70). Data acquisition was done by using a PC counter board with a labview interface (National Instruments). Luminescence was measured with a photomultiplier tube (Type P10PC, Electron Tube) and the same data acquisition system. For the vesicles kinetics measurements, the same setup was used with an intensified linear digital camera (Intensified Retiga, QImaging), pictures were analyzed with imagej (National Institutes of Health). For bulk and vesicle measurements, the light source was blocked between acquisitions to avoid photobleaching.
Vesicle Preparation. Egg Lecithin (Sigma) was dissolved in mineral oil at 5 mg/ml, heated at 50°C, and sonicated in a bath. After overnight incubation at room temperature, a precipitate forms, and only the clear supernatant was used to prepare the vesicles as described below. BSA–rhodamine isothiocyanate (RITC) (Sigma) and fluorescein-12-UTP (Roche) were used to test vesicle leakage.
Selective permeability of the vesicles is obtained with the α-hemolysin toxin from S. aureus. Permeability of the vesicles was not obtained with short antimicrobial peptides like Magainin I and II. Fluorescein-UTP leakage was observed with Cecropin A at a minimum concentration of 20 μM, but results were inhomogeneous: a few percent of vesicles were leaking; the majority were not.
Results and Discussion
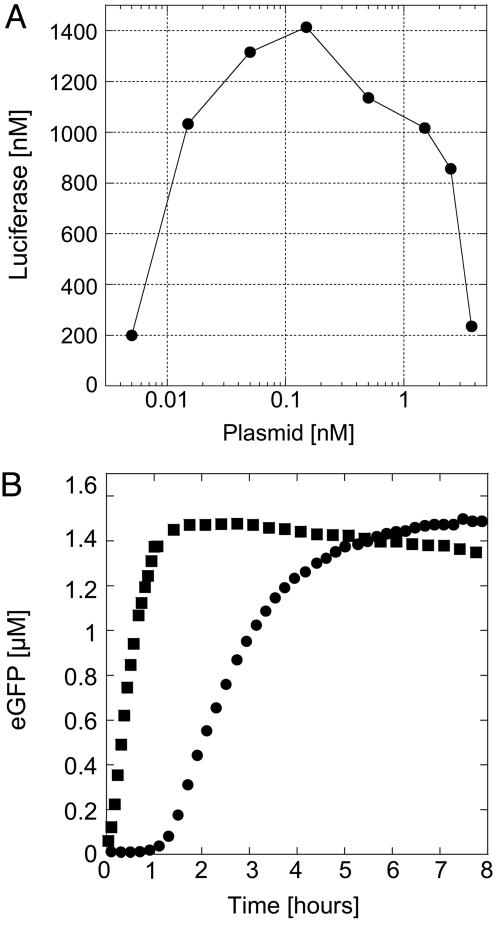
Measurement of Expression in Nonencapsulated Volume. Expression with the E. coli extract is first measured in a volume of 10 μl at room temperature (25°C) with the firefly luciferase and eGFP as reporter proteins. After 2 h of incubation, a maximum yield of 1.5 μM protein is achieved at plasmid concentration between 0.1 and 1 nM, depending on the batch of extract (Fig. 1A). Below 0.1 nM plasmid expression is not linear with template concentrations. Above 1 nM plasmid, protein production decreases probably because of saturation of the translation machinery (14). In all of the experiments plasmid concentration is 0.5 nM. To measure the time course of expression, 12 μl of reaction are deposited between two glass coverslips with a spacer of half a millimeter, making a droplet 6 mm in diameter. When the fluorescence intensity from eGFP is measured near the border of the droplet (few tens of micrometers), protein production is sharp and stops after 1.5 h (Fig. 1B). When it is measured in the center of the droplet (3 mm from the border), expression starts after 1 h and is much slower, lasting 5 h. Because oxygen is required for expression and for eGFP functionality, its diffusion time from the surrounding atmosphere has to be short enough, imposing distances of hundred micrometers. In the large-scale continuous systems, both compartments are constantly stirred to increase the diffusion of oxygen. In our microscopic system, all of the exchanges are done by diffusion. Fluorescence intensity measured in the center of the sample is in fact a superposition of weak local expression and diffusion of highly produced eGFP from the border of the droplet.
Fig. 1.
Characterization of the extract. (A) Firefly luciferase production in the E. coli extract measured after2hasa function of the pIVEX2.3d-Luc plasmid concentration. (B) Time course of expression of eGFP in the extract, 0.5 nM pIVEX2.3d-eGFP plasmid. Twelve microliters of reaction was deposited between two glass coverslips, forming a droplet 6 mm in diameter. The fluorescence signal was measured at the air–sample interface (squares) and in the center of the sample (circles).
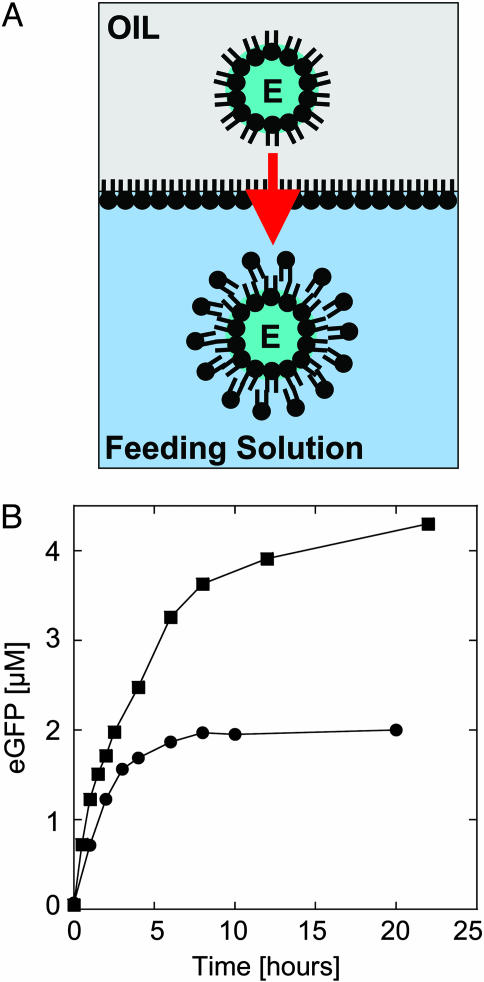
Encapsulation of the Extract. Our next step was to find a way to encapsulate the cell-free expression system into a vesicle. A cytoplasmic extract is a complex solution of ∼100 mg/ml protein, 50–100 mM salts, and 10–20 mM ions. Although numerous methods of encapsulation have been described for simple aqueous solutions to carry out biochemical reactions (11, 22–24), no methods have been found to realize both transcription and translation into vesicles. With a two-step protocol, the cytoplasmic extract can be efficiently and quickly encapsulated (Fig. 2A). First, the reaction is assembled in a volume of 10–20 μl in a tube. One microliter of this solution is added to another tube containing 200 μl of mineral oil with dissolved phospholipids. With a gentle vortex for a few seconds, the small aqueous droplet is broken and forms an extract–oil emulsion. After a few minutes, the microdroplets are stabilized by a monolayer of phospholipids at the oil–extract interface (Fig. 2 A). In the second step, 50 μl of the emulsion is placed on top of the feeding solution where the vesicles will be transferred. A monolayer of phospholipids forms at the interface of the biphasic solution and offers a perfect configuration to make a bilayer with the droplets passing through by centrifugation (Fig. 2 A). Vesicles 1 to a few tens of micrometers in diameter are formed after centrifugation and recovered in the feeding solution. Osmotic pressure is the main factor affecting the yield of vesicle formation. To increase the number of vesicles formed and their stability in time by reducing the osmotic pressure, one must, on one hand, dilute the extract with the feeding solution, and on the other hand, add some extract in the feeding solution where the vesicles are transferred. With the material used in this study, a good working point is when the reaction to encapsulate is made of 50% extract–50% feeding solution and the feeding solution is supplemented with 4% of extract. From 50 μl of the emulsion, one can recover a few hundreds vesicles and aggregates of vesicles in 25-μl feeding solution. We chose to study large vesicles (>10 μm in diameter), which are numerous. Vesicles are formed easily with a mix of phosphatidylcholine, mainly of 16 and 18 carbons. A similar technique was used recently to engineer asymmetric vesicles (25). This encapsulation mechanism could be a model for cellular compartment formation (26). Indeed, natural processes like a shear flow can break aqueous droplet and lead to this configuration. With this process, the desired material is confined into the vesicles, and no desalting or further separation is required.
Fig. 2.
Encapsulation of a cell-free expression extract in a vesicle and expression of eGFP. (A) The extract–oil emulsion is added on top of the feeding solution; the microdroplets are stabilized with a monolayer of phospholipids while another monolayer forms at the interface of the biphasic solution. E, extract. Vesicles are formed after centrifugation through the interface (the arrow indicates the direction of centrifugation). (B) Expression of eGFP, 0.5 nM pIVEX2.3d-eGFP plasmid, inside a vesicle under different osmotic pressure: 100% extract encapsulated into the vesicles and feeding (squares), and 50% extract–50% feeding encapsulated into the vesicles and feeding supplemented with 4% extract (circles).
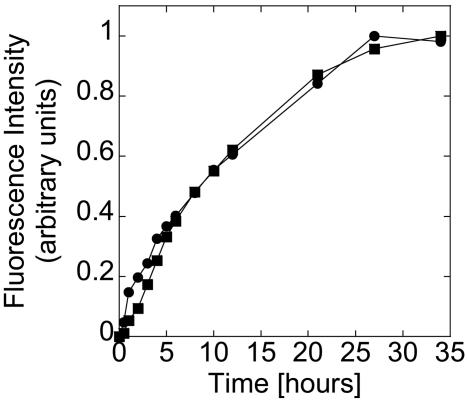
Expression of eGFP. We studied first the expression of eGFP inside the vesicles to see whether the encapsulation process deteriorates the extract. To the contrary, protein production is observed for 5 h instead of 2 h in large volume (Fig. 2B), and the concentration of eGFP is twice as high. Because we used phospholipids of 16 and 18 carbons, permeability of nucleotides and amino acids is very low and thus supplementation of the bioreactor is limited, especially at 25°C where the bilayer is in a crystalline phase (27). However, exchange of nutrients is favored by the osmotic pressure that promotes the spontaneous formation of transient pores, as demonstrated by Taupin and coworkers (28). To test this hypothesis, the same experiment was performed under much higher osmotic pressure. For that, we used undiluted extract and nonsupplemented feeding solution. The result shows an increase of the production time, lasting >10 h. Concentration of eGFP in the vesicles (4.5 μM) is three times higher than in bulk (Fig. 2B). Osmotic pressure is thus an important effect and can, if not compensated, induce vesicle burst. As reported recently (29), osmotic pressure could be also an important aspect for the emergence of protocell.
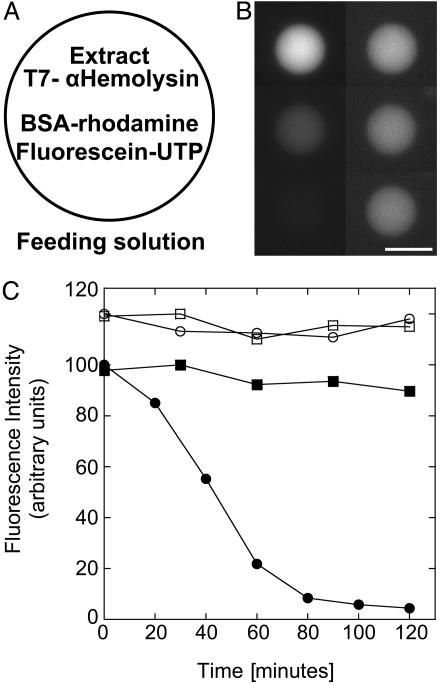
Expression of α-Hemolysin. Permeability of vesicle under osmotic stress allows a prolongation of expression for up to 5 h when vesicles are surrounded by a feeding solution. The next step was to make the membrane more permeable by using a pore in a specific range of molecular mass cut-off so as to keep the encapsulated biosynthetic machinery inside and feed from the surrounding nucleotides and amino acids. The α-hemolysin protein from S. aureus is the ideal candidate to permeabilize selectively the membrane. This molecule, expressed as a soluble monomer, assembles into a heptamer at the membrane to form a pore 1.4 nm in diameter corresponding to a molecular mass cut-off of 3 kDa (30, 31). Pore formation can occur at an extremely low concentration; they are stable and do not perturb the membrane. To test the permeability of the vesicles, two fluorescent molecules are used: the BSA protein labeled with rhodamine (60 kDa) and the ribonucleotide UTP labeled with fluorescein (1 kDa). The extract with the plasmid encoding the toxin and the two fluorescent probes (BSA-rhodamine and fluorescein-UTP) are encapsulated in the vesicles (Fig. 3A). Whereas the BSA-rhodamine signal is constant (within a weak bleaching), leakage of fluorescein-UTP is observed after 20 min (Fig. 3 A and B). After 2 h of incubation, 95% of the dye has diffused out of the vesicle. This finding indicates that the synthesized toxin is functional and that ribonucleotides and amino acids can diffuse in and out while the extract is kept inside the vesicles. More than 90% of the vesicles show complete leakage of fluorescein-UTP. As a control, the firefly luciferase is expressed instead of the toxin, and no leakage is observed (Fig. 3C). Leakage is also observed when a minimum of 500 nM pure α-hemolysin protein is added, instead of expressing it, into the feeding solution (data not shown).
Fig. 3.
Selective permeability of the membrane with expression of α-hemolysin toxin inside the vesicle. (A) Schematic of a vesicle transferred in the feeding solution containing the extract, the plasmid pIVEX2.3d-α-hemolysin (0.5 nM), BSA-RITC (6 μM), and fluorescein-UTP (35 μM). (B) Time sequence of the vesicle fluorescence of BSA-RITC (Right) and fluorescein-UTP (Left) after 10, 70, and 120 min (Top to Bottom). (Scale bar, 20 μm.) (C) Kinetics of fluorescence of the vesicle: filled circles, fluorescein-UTP; filled squares, BSA-RITC. Negative control (data shifted up): expression of firefly luciferase instead of α-hemolysin in the same conditions (0.5 nM pIVEX2.3d-Luc plasmid), fluorescein-UTP (open circles) and BSA-RITC (open squares).
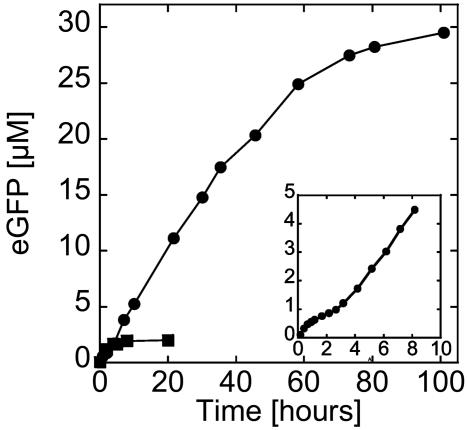
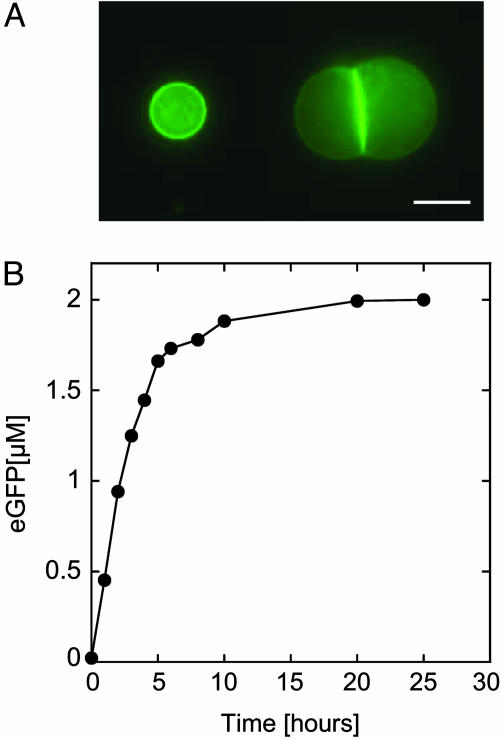
A Long-Lived Bioreactor. To visualize the toxin and quantify the pore concentration as a function of time, a fusion α-hemolysin-eGFP was engineered with the reporter in the C terminus of the pore. The first important result is that expression is observed for >4 days with a maximum protein production between 30 and 40 μM (Fig. 4). To show the improvement introduced by the toxin, bulk and eGFP expression in vesicles without α-hemolysin are added to the figure. A one order of magnitude increase in protein production and duration of expression is thus obtained. Accumulation of the toxin at the membrane and in bulk is observed (Fig. 5A). The protein permeabilizes the membrane and induces its own expression prolongation. The encapsulation in a vesicle produces a positive feedback where more pores means more protein production. To prolong expression, pore production must be large enough during the first hours, the time during which the extract can run independently of supplementation. A few hundreds of nanomolar protein production is necessary to obtain autoinduced expression prolongation. This finding may explain why the rate of protein production slows down after 2 h and increases once the pores ensure an efficient feeding of the vesicle (Fig. 4 Inset). Expression stabilizes after 100 h, in part because of the degradation of the feeding solution and probably also because of the accumulation of byproducts. This duration is the longest observed, and depending on the vesicle, the dispersion goes from 1 to 4 days.
Fig. 4.
Kinetics of expression of α-hemolysin-eGFP inside a vesicle. Filled circles: 0.5 nM pIVEX2.3d-α-hemolysin-eGFP. (Inset) Blow up of the first 10 h of expression of α-hemolysin-eGFP. For comparison, the time course of expression of eGFP inside a vesicle without α-hemolysin under low osmotic pressure is shown (filled squares, curve from Fig. 2B; 0.5 nM pIVEX2.3d-eGFP).
Fig. 5.
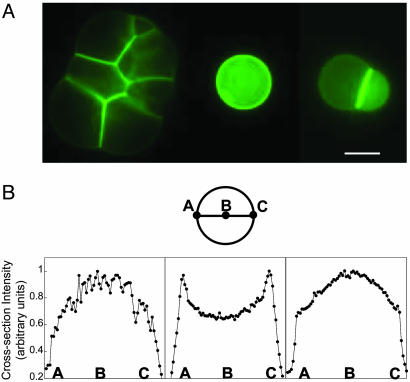
α-Hemolysin-eGFP inside vesicles and fluorescence intensity along a cross section. (A) Expression of α-hemolysin fused to eGFP after a few tens of hours inside an aggregate of vesicles (Left), a single vesicle (Center), and a doublet (Right). The E. coli extract is encapsulated in the vesicles with the plasmid pIVEX2.3d-α-hemolysin-eGFP (0.5 nM) surrounded by a feeding solution. (Scale bar, 20 μm.) (B) Diameter section of a single-vesicle image schematic and sections after 70 min (Left), 550 min (Center), and 4,400 min (Right).
We analyzed the profile of a single vesicle 20 μm in diameter to follow the repartition of the tagged toxin between membrane and solution inside the vesicle and to quantify the concentration of pores. A diameter section of the vesicle image gives the fluorescence-intensity profile (Fig. 5B). This profile is compared to vesicles made with rhodamine phospholipids as a model fluorescent sphere, without dyes in solutions. During the first hour, the pore protein stays mainly in solution (Fig. 5B Left). Then, the toxin accumulates at the membrane and its proportion becomes predominant after 10 h, reaching a maximum (Fig. 5B Center). At this time, we estimate that >75% of the 5 μM expressed proteins are on the membrane. Twenty percent of the membrane is occupied by pores, corresponding to 2,000 pores per μm2 and an average distance of 10 nm between the heptamers. After 10 h, the proteins accumulate predominantly in the solution (Fig. 5B Right). After 4 days, 75% of the membrane is occupied by pores. One can also add the pure toxin into the feeding solution to prolong the expression (data not shown). However, we noticed that prolongation of the expression is better with a continuous expression of the toxin because of a possible molecular jamming that reduces the diffusion of substrates through the channels. We have observed that, when some BSA-rhodamine (6 μM) is added to the extract, it accumulates near the membrane.
Finally, we show that coexpression of different proteins is possible inside the vesicles. For example, we coexpressed eYFP and α-hemolysin fused to eCFP (Fig. 6). As described previously, plasmid concentrations have to be adjusted to avoid saturation of the translation machinery (14).
Fig. 6.
Coexpression of α-hemolysin-eCFP (squares; 0.25 nM pIVEX2.3d-α-hemolysin-eCFP) and eYFP (circles; 0.25 nM pIVEX2.3d-eYFP) inside a vesicle.
First Step in Membrane Functionalization. An important aspect of this system is the possibility to anchor proteins at the membrane. In the cell, incorporation is a complex process involving many different proteins. In our system, a way to anchor proteins at the membrane is to use the artificial peptide 18L that spontaneously inserts itself into the phospholipids bilayer (20). This polypeptide of 18 aa forms an α-helix with a balance of hydrophilic–hydrophobic residues. To test incorporation, eGFP was tagged at its N terminus with the 18L peptide, expressed in the vesicle, and successfully incorporated into the membrane (Fig. 7A). As reported in ref. 20, under certain conditions this peptide improves diffusion of small molecules through the membrane, but clearly in our system the effect is small (Fig. 7B). Successful anchoring of the tagged eGFP shows that biopolymers and motor proteins can be bound to the membrane to perturb the bilayer mechanically.
Fig. 7.
18L-eGFP inside vesicles and kinetics of expression. (A) Fluorescence images of a single vesicle and a doublet with 18L-eGFP after 5 h. The E. coli extract (50% extract–50% feeding) is encapsulated in the vesicles with the plasmid pIVEX2.3d-18L-eGFP (0.5 nM) surrounded by a feeding solution supplemented with 4% extract. (Scale bar, 15 μm.) (B) Kinetics of the expression.
Summary and Conclusions
Our results show that two main problems have been solved in the process of building an artificial cell. Transcription and translation have been brought at the scale of the cell, and energy/nutrients limitations have been solved through the internal expression of a membrane pore and by exchange with the environment. Molecules from bacteriophage, bacteria, and eukaryotic cells were used in this study based on the universal biosynthetic machinery, showing how diverse the assembly of a synthetic cell can be. This bioreactor allows one to build various functional entities, using all of the possible genetic modules. Our first step was to anchor proteins to the membrane. One can imagine encapsulating “minigenomes” encoding for complete pathways. The next two questions are these: How can we improve the capacity of this system and how can we engineer an elementary genome to get closer to a minimal cell? Design of a small program has to include autoregulation and repression to ensure a strict bookkeeping of the resources. Because cell-free extracts have been optimized to have a low level of ribonucleases and proteases, a faster degradation of messengers and proteins has to be introduced to express a few genes in parallel and avoid any saturation of the machinery (14). In addition, new genes have to be included inside the vesicle to transform the bilayer into a functional interface by translocating simple membrane protein. In particular, a step toward a minimal cell would be to induce vesicle fission. In its actual form, the main disadvantage of the cellular extract is its lack of modularity to design genetic networks. The T7 RNA polymerase is the only functional transcription factor for the E. coli used, restricting the capacity of the system. One must develop cell-free extracts capable of transcription with multiple polymerase; wheat germ extracts offer good possibilities (14).
Such small, enclosed laboratories can be useful to study chemical reactions in confined geometries (32). Enclosing molecules in a small volume has important applications for chemical reactions, biological screening, and evolutionary and singlemolecule studies (33–35). Furthermore, building an artificial cell that can perform specific tasks is of great interest for pharmacology and medical diagnostics (5).
Acknowledgments
We thank Alexander Feigel, Zoher Gueroui, Hanna Salman, Gregoire Bonnet, and Jeremy Godefroy for their active interest and Pablo Meyer for technical help. V.N. and A.L. acknowledge the support of the Revson Foundation and the NEC Research Institute, respectively.
Author contributions: V.N. and A.L. designed research, performed research, analyzed data, and wrote the paper.
Abbreviations: eCFP, enhanced cyan fluorescent protein; eGFP, enhanced GFP; eYFP, enhanced yellow fluorescent protein; RITC, rhodamine isothiocyanate.
References
- 1.von Neumann, J. (1966) Theory of Self-Reproducing Automata (Univ. of Illinois Press, Champaign).
- 2.Turing, A. M. (1936) Proc. London Math. Soc. Ser. 2 42, 230–265. [Google Scholar]
- 3.Ganti, T. (2003) The Principles of Life (Oxford Univ. Press, New York).
- 4.Jay, D. G. & Gilbert, W. (1987) Proc. Natl. Acad. Sci. USA 84, 1978–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pohorille, A. & Deamer, D. (2002) Trends Biotechnol. 20, 123–128. [DOI] [PubMed] [Google Scholar]
- 6.Szostak, J., Bartel, D. P. & Luisi, P. L. (2001) Nature 409, 387–390. [DOI] [PubMed] [Google Scholar]
- 7.Luisi, P. L. (2002) Anat. Rec. 268, 208–214. [DOI] [PubMed] [Google Scholar]
- 8.Luisi, P. L. (1998) Origins Life Evol. Biosphere 28, 613–622. [DOI] [PubMed] [Google Scholar]
- 9.Morowitz, H. J. (1992) Beginning of Cellular Life (Yale Univ. Press, New Haven, CT).
- 10.Gesteland, R. F., Cech, T. & Atkins, J. F. (1999) The RNA World (Cold Spring Harbor Lab. Press, Plainview, NY).
- 11.Oberholzer, T., Nierhaus, K. H. & Luisi, P. L. (1999) Biochem. Biophys. Res. Commun. 261, 238–241. [DOI] [PubMed] [Google Scholar]
- 12.Smith, H. O., Hutchison, C. A., III, Pfannkoch, C. & Venter, J. C. (2003) Proc. Natl. Acad. Sci. USA 100, 15440–15445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawarasaki, Y., Nakano, H. & Yamane, T. (1994) Biosci. Biotechnol. Biochem. 58, 1911–1913. [DOI] [PubMed] [Google Scholar]
- 14.Noireaux, V., Bar-Ziv, R. & Libchaber, A. (2003) Proc. Natl. Acad. Sci. USA 100, 12672–12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atkinson, D. E. (1968) Biochemistry 7, 4030–4034. [DOI] [PubMed] [Google Scholar]
- 16.Spirin, A. S., Baranov, V. I., Ryabova, L. A., Ovodov, S. Y. & Alakhov, Y. B. (1988) Science 242, 1162–1164. [DOI] [PubMed] [Google Scholar]
- 17.Shimizu, Y., Inoue, A., Tomari, Y., Suzuki, T., Yokogawa, T., Nishikawa, K. & Ueda, T. (2001) Nat. Biotechnol. 19, 751–755. [DOI] [PubMed] [Google Scholar]
- 18.Finean, J. B., Coleman, R. & Michell, R. H. (1984) Membranes and Their Cellular Functions (Blackwell Scientific, Oxford).
- 19.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 20.Polozov, I. V., Anantharamaiah, G. M., Segrest, J. P. & Epand, R. M. (2001) Biophys. J. 81, 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zubay, G. (1973) Annu. Rev. Genet. 7, 267–287. [DOI] [PubMed] [Google Scholar]
- 22.Gebicki, J. M. & Hicks, M. (1976) Chem. Phys. Lipids 16, 142–160. [DOI] [PubMed] [Google Scholar]
- 23.Angelova, M. & Dimitrov, D. (1986) Faraday Discuss. Chem. Soc. 81, 303–311. [Google Scholar]
- 24.New, R. R. C. (1990) Liposomes: A Practical Approach (IRL, Oxford).
- 25.Pautot, S., Frisken, B. J. & Weitz, D. A. (2003) Proc. Natl. Acad. Sci. USA 100, 10718–10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanczyc, M. M., Fujikawa, S. M. & Szostak, J. (2003) Science 302, 618–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakrabarti, A. C., Breaker, R. R., Joyce, G. F. & Deamer, D. W. (1994) J. Mol. Evol. 39, 555–559. [DOI] [PubMed] [Google Scholar]
- 28.Taupin, C., Dvolaitzky, M. & Sauterey, C. (1975) Biochemistry 14, 4771–4775. [DOI] [PubMed] [Google Scholar]
- 29.Chen, I. A., Roberts, R. W. & Szostak, J. W. (2004) Science 305, 1474–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song, L., Hobaugh, M. R., Shustak, C., Cheley, S., Bayley, H. & Gouaux, J. E. (1996) Science 274, 1859–1866. [DOI] [PubMed] [Google Scholar]
- 31.Bayley, H. (1997) Sci. Am. 277, 62–67. [DOI] [PubMed] [Google Scholar]
- 32.Chiu, D. T., Wilson, C. F., Ryttsen, F., Stromberg, A., Farre, C., Karlsson, A., Nordholm, S., Gaggar, A., Modi, B. P., Moscho, A., et al. (1999) Science 283, 1892–1895. [DOI] [PubMed] [Google Scholar]
- 33.Thorsen, T., Roberts, R. W., Arnold, F. H. & Quake, S. R. (2001) Phys. Rev. Lett. 86, 4163–4166. [DOI] [PubMed] [Google Scholar]
- 34.Tawfik, D. S. & Griffiths, A. D. (1998) Nat. Biotechnol. 16, 652–656. [DOI] [PubMed] [Google Scholar]
- 35.Boukobza, E., Sonnenfeld, A. & Haran, G. (2001) J. Phys. Chem. B 105, 12165–12170. [Google Scholar]