Abstract
Cytosolic thymidine kinase 1, TK1, is a well known cell-cycle-regulated enzyme of importance in nucleotide metabolism as well as an activator of antiviral and anticancer drugs such as 3′-azido-3′-deoxythymidine (AZT). We have now determined the structures of the TK1 family, the human and Ureaplasma urealyticum enzymes, in complex with the feedback inhibitor dTTP. The TK1s have a tetrameric structure in which each subunit contains an α/β-domain that is similar to ATPase domains of members of the RecA structural family and a domain containing a structural zinc. The zinc ion connects β-structures at the root of a β-ribbon that forms a stem that widens to a lasso-type loop. The thymidine of dTTP is hydrogen-bonded to main-chain atoms predominantly coming from the lasso loop. This binding is in contrast to other deoxyribonucleoside kinases where specific interactions occur with side chains. The TK1 structure differs fundamentally from the structures of the other deoxyribonucleoside kinases, indicating a different evolutionary origin.
Keywords: crystal structures, deoxynucleotide metabolism, prodrug activation
A balanced supply of deoxyribonucleotides is essential for all living organisms for repair and replication of nuclear, as well as mitochondrial, DNA. The deoxyribonucleotides are synthesized by two main routes, the de novo and salvage pathways. In de novo synthesis, deoxyribonucleotides are formed from ribonucleotides in a regulated manner by the enzyme ribonucleotide reductase (1). Deoxyribonucleoside kinases (dNKs) catalyze the first phosphorylation step of deoxyribonucleosides in the salvage pathway (2). Humans have four such kinases: two cytosolic enzymes, thymidine kinase 1 (TK1) and deoxycytidine kinase, and two mitochondrial enzymes, thymidine kinase 2 (TK2) and deoxyguanosine kinase, with different substrate specificities. TK1 differs from the others in several respects. It has a narrow specificity, phosphorylates only deoxythymidine (dT) and deoxyuridine, and is strictly cell-cycle-regulated. TK1 activity is low or absent in resting cells, starts to occur in late G1 cells, increases in S phase (coinciding with the increase in DNA synthesis), and disappears during mitosis (3–6).
The synthesis of isotopically labeled dT and the demonstration of its incorporation into DNA after cellular uptake and phosphorylation were landmark discoveries in cell biology, providing a way to define the various phases of the cell cycle (7, 8). Recent studies based on these methods have provided insight into the role of TK1 and TK2 for the cytosolic and mitochondrial dTTP pools (9). The researchers demonstrated a rapid exchange of dTTP between the two compartments. In cells with active TK1, [3H]dT salvage competed effectively with de novo synthesis, and most of the mitochondrial pool of labeled dTTP was imported from the cytosol, indicating low contribution from TK2. In TK1-deficient cells, however, cytosolic labeled dTTP was derived from the mitochondrial pools because of phosphorylation of labeled dT by the mitochondrial TK2. That TK1 plays an important physiological role is further emphasized in studies of Tk–/– knockout mice. Although viable, the TK1-deficient mice had an abnormal immune system, developed fatal kidney disease, and had increased mutation rates (10).
In chemotherapeutic treatment of cancer and viral diseases, deoxynucleoside kinases are key enzymes in the activation of otherwise nontoxic prodrugs, the nucleoside analogs. Well known examples are the phosphorylation by herpes virus 1–thymidine kinase (TK) of the guanosine analog acyclovir and the TK1-catalyzed phosphorylation of the anti-HIV drug 3′-azido-3′-deoxythymidine (AZT or zidovudine) (11, 12). TK1 is also used in cancer diagnostics (13).
Based on the amino acid sequence, deoxynucleoside kinases may be arranged in two families: the TK1 family and a family containing all of the others, the dNKs (12). The dNKs form a homologous family with sequence similarities throughout the whole peptide chain, a family to which also insect and herpes viral dNKs belong (12). Herpes virus 1–TK was the first member of the family for which the structure was determined (14, 15). Other members of the dNK family are generally smaller but have similar core structures, and their different substrate specificity can be related to a few differences at the active site (12, 16–19). TK1 has sequence similarities to the dNKs in a P loop but lacks other significant conserved sequence motifs. Furthermore, the other dNKs are dimers, whereas the active form of TK1 is a tetramer (20).
TK1-like sequences are found in a broad variety of organisms from plants to humans, as well as in poxvira and in several bacteria, e.g., the Gram-positive Bacillus subtilis and Staphylococcus aureus and the Gram-negative Escherichia coli. Recently, Ureaplasma urealyticum TK (Uu-TK) was cloned and expressed and characterized biochemically (21). U. urealyticum is a mycoplasma species with a known genome of ≈600 genes (22). These bacteria colonize the urogenital tract and are associated with infertility, altered sperm motility, and pneumonia in neonates (23). Initial characterization of Uu-TK demonstrated that it has similar enzyme kinetic properties to human TK1 (hTK1) regarding the narrow specificity for deoxyuridine and dT and with dTTP serving as a feedback inhibitor (21). No genes for the de novo synthesis of deoxyribonucleotides have been found in the U. urealyticum genome. Therefore, this bacterium has to rely solely on salvage for synthesis of DNA precursors. Thus, Uu-TK is a good target for antibacterial drugs blocking bacterial TK1 but not hTK1.
Despite its metabolic importance and pharmaceutical role in prodrug activation, no structure of the TK1-like enzymes has so far been reported. Here we describe the structures of hTK1 as well as Uu-TK, which show a high degree of similarity and contain a unique Zn domain with a lasso-like loop covering the nucleoside-binding site. Availability of the 3D structure of TK1 is valuable for the design of new TK1-selective and efficient antiviral, antibacterial, and anticancer prodrugs and specific antibodies for clinical diagnostics.
Methods
Protein Expression and Purification. C-terminally truncated hTK1 lacking 41 C-terminal amino acids was created by inserting a stop codon after amino acid 193 by the recombinant PCR technique with the plasmid pGEX-2T-LyTK1val106 as described in ref. 24. LyTK1Val106 is derived from the tk1 gene from human lymphocytes (25), where codon 106 encodes a valine instead of a methionine as reported for the original tk1 clone (26). Amino acid 106 has been shown to have high impact on the enzymatic and oligomerization properties of TK1, with the valine 106 TK1 form behaving essentially as the endogenous TK1 (25, 27). Comprehensive sequencing studies verify that valine is, in fact, the naturally occurring amino acid at site 106 (25, 28, 29). Expression and purification of TK1 was performed as described in ref. 25, except that Triton X-100 replaced 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate in all buffers.
The U. urealyticum tdk gene was cloned, and native protein was expressed and purified as described in ref. 21. For production of selenomethionine (SeMet)-substituted Uu-TK, the Uu-TK construct was transformed into the chemical competent, methionine auxotrophic E. coli strain B834(DE3) (Novagen). The bacterial culture was grown in SelenoMet media (Molecular Dimensions, Apopka, FL) containing 21.6 gl–1 base mix, 5.1 gl–1 nutrient mix, 50 μg ml–1 l-methionine, and 50 μgml–1 carbenicillin at 37°C to an OD of 0.6 at 600 nm. The SelenoMet media was changed to contain SeMet before induction with 1 mM isopropyl β-d-galactopyranoside for 6 h at 25°C, and SeMet-substituted Uu-TK was purified essentially as described in ref. 21.
Crystallization. Crystals of the recombinant truncated hTK1 cocrystallized with dTTP were grown with the hanging drop vapor diffusion method by using Hampton Screen I (Hampton Research, Aliso Viejo, CA). The final crystallization conditions at 15°C were: 0.1 M Na citrate (pH 5.6), 20% 2-propanol, and 14–16% polyethylene glycol 4000. The crystallization solution was mixed in equal volumes with 7–13 mg/ml protein that contained 5 mM dTTP. Both native and SeMet-substituted Uu-TK were crystallized by using the hanging drop vapor diffusion method at 15°C. Native protein solution consisted of 8 mg/ml Uu-TK and 5 mM dTTP, whereas reservoir solution contained 19% polyethylene glycol 2000, 0.2 M LiCl, 5 mM DTT, and 0.1 M Mes (pH 6.0). SeMet-substituted protein crystallized in similar conditions. To increase the size of SeMet crystals, streak-seeding (30–32) was carried out. The crystallization solution with 20–30% glycerol added was used as cryoprotectant.
Structure Determination. Crystals of hTK1 were soaked in 10 mM KAu(CN)2 for 40 min and back-soaked for 30 s in a cryosolution containing reservoir solution plus 20% glycerol before flash-freezing in liquid nitrogen. Synchrotron data were indexed by using mosflm and were scaled with scala (33). Crystals belonged to the space group C2 and contained two tetramers in the asymmetric unit (subunits A–H). Twelve gold sites were located by using shelx (34) and were refined in autosharp (35, 36), where two new sites were added. After heavy-atom site refinement, density modification and solvent flattening were performed. The initial electron density map was obtained with dm (33, 37). Maps were improved by using eightfold noncrystallographic symmetry (NCS) averaging in DM, and the major part of the structure was built in this map by using o (38). After rigid body refinement, refinement was continued by using a native data set to 2.4-Å resolution. Eightfold NCS was used during restraint refinement in refmac5 (33, 39). A total of 165 residues could be built into the electron density in the best-defined subunit, subunit G. Statistics for the data collection and refinement are shown in Table 1.
Table 1. Data collection and refinement statistics for hTK1 and Uu-TK.
| Native hTK1 | hTK1 at Au peak | Native Uu-TK | SeMet-substituted Uu-TK | |
|---|---|---|---|---|
| Space group unit cell | C2 | C2 | P21 | P21 |
| a, Å | 157.7 | 156.6 | 63.8 | 64.0 |
| b, Å | 123.2 | 123.1 | 110.8 | 111.0 |
| c, Å | 116.0 | 115.4 | 73.0 | 73.1 |
| β, ° | 130.2 | 130.0 | 109.9 | 109.7 |
| Content of the asymmetric unit | 2 tetramers | 2 tetramers | 1 tetramer | 1 tetramer |
| Resolution, Å | 2.4 (2.53-2.4) | 3.5 (3.69-3.5) | 2.5 (2.54-2.5) | 3.0 (3.16-3.0) |
| Completeness, % | 99.0 (98.9) | 100 (100) | 99.7 (99.3) | 98.7 (98.7) |
| Rmeas, % | 10.8 (37.5) | 12.9 (19.9) | 13.5 (26.6) | 6.8 (18.4) |
| I/σI | 5.4 (2.1) | 4.4 (3.5) | 10.2 (2.8) | 10.0 (4.6) |
| Redundancy | 3.5 (3.5) | 6.8 (6.9) | 4.4 (4.0) | 9.8 (9.8) |
| No. of observed reflections | 224,588 | 145,327 | 146,317 | 186,680 |
| No. of unique reflections | 63,895 | 21,250 | 33,042 | 19,066 |
| European Synchrotron Radiation Facility beam line | ID14eh2 | ID14eh4 | ID14eh1 | BM14 |
| Wavelength, Å | 0.933 | 1.040 | 0.934 | 0.976 |
| Temperature, K | 100 | 100 | 100 | 100 |
| R, % | 20.1 | 22.2 | ||
| Rfree, % | 23.2 | 28.2 | ||
| rms deviation | ||||
| Bond length, Å | 0.009 | 0.011 | ||
| Bond angle, ° | 1.259 | 1.364 | ||
| Mean B value, Å2 | 34.7 | 46.2 |
Numbers in parentheses refer to the outer resolution bin.
Native data from Uu-TK were collected at the European Synchrotron Radiation Facility and processed and scaled by using denzo and scalepac (40). A complete single anomalous diffraction dataset was collected at the peak wavelength. The data were processed with mosflm (41) and scaled with separated anomalous pairs with scala (33). Crystals belonged to the monoclinic space group P21 and contained one tetramer in the asymmetric unit (subunits A–D). By using data to 4 Å, 12 heavy-atom positions were located with shelx (34) and refined in autosharp (35, 36). Electron density was improved by cyclic averaging before model-building. From this map, most of the polypeptide chain could be tracked and assigned with correct amino acids. After simulated annealing refinement, performed in cns (42), most of the polypeptide could be built. Further refinement with initially strict fourfold noncrystallographic symmetry restraints was carried out in refmac5 (39). Restraints were loosened in the latest rounds of the refinement cycle, and native data to 2.5-Å resolution were used. All model-building was made by using o (38). Each monomer consisted of 223 amino acids, of which residues 11–217 were visible in the electron density (subunits B and D). Monomers A and C lacked residues 50–67 in a flexible loop. Data-collection and refinement statistics are shown in Table 1.
The sequence alignment was performed in clustalw (43), and the picture was made by using alscript (44). The figures were made with pymol (45).
Determination of Zn2+ Bound to TK. The Zn2+ content of truncated hTK1 and Uu-TK was determined after desalting the enzyme preparations used for crystallography into buffers with minimal trace metals. Zn2+ levels were measured by atomic absorbance spectroscopy and were related to the protein levels as determined by the absorbance at 280 nm. The molar ratio was 0.56 for hTK1 and 0.51 for Uu-TK. No absorbance at visible wavelengths was detected with the two TK protein solutions.
Results and Discussion
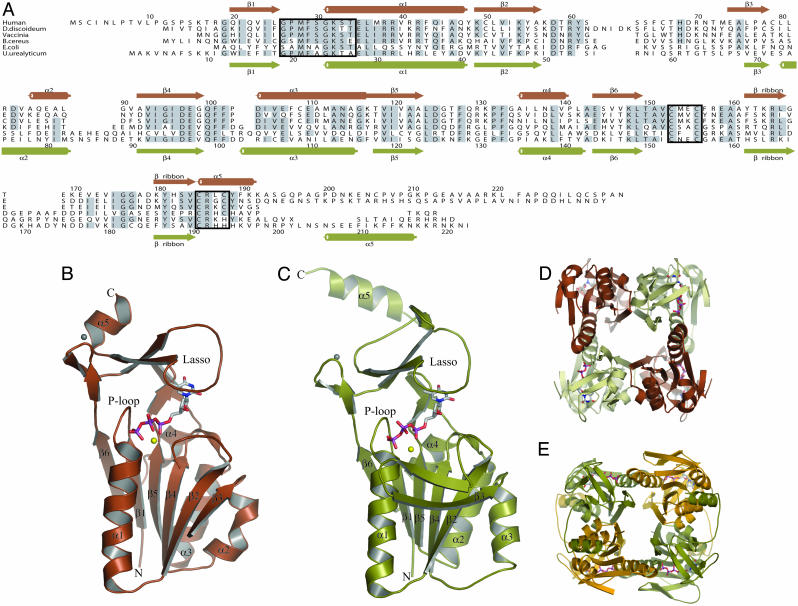
The Structure of TK1s. The difficulty in obtaining a TK1-type TK structure probably reflects that the enzyme has flexible parts. Because initial attempts to crystallize hTK1 were unsuccessful, we truncated the enzyme for the C-terminal 41 amino acids that previously have been shown to stabilize TK1 during the cell cycle without abolishing the specific activity (46). The enzymatic and oligomerization properties of the truncated hTK1 were virtually unchanged. For further stabilization of the enzyme, we used the feedback inhibitor dTTP and determined the 3D structures of C-terminally truncated hTK1 and the full-length Uu-TK in complex with dTTP. Both structures are tetramers, and each subunit consists of two domains, one α/β-domain and a small zinc-containing domain. The active site is buried between these domains (Fig. 1).
Fig. 1.
Structures of hTK1 and Uu-TK. (A) Structural alignment of the sequences of TKs from human (P04183), Dictyostelium discoideum (AAB03673.1), Vaccinia virus (AAB96503.1), B. cereus (ZP_00241105.1), E. coli (NP_287483.1), and U. urealyticum (U. parvum) (NP_078433). Secondary structure elements for hTK1 are shown above the alignment in brown, and those for Uu-TK are shown below in green. The P loop and the two zinc coordinating sequences are boxed. (B) Subunit structure of hTK1 with dTTP colored according to atom type. Mg2+ is shown in yellow, and Zn2+ is shown in gray. (C) Subunit structure of Uu-TK with dTTP, Mg2+, and Zn2+ shown in same colors as in B. hTK1 and Uu-TK are tetramers. (D and E) hTK1 (D) and Uu-TK (E) tetramers shown in different views. The C-terminal helix in Uu-TK interacts with a helix on the adjacent monomer.
The α/β-domain has a central, six-stranded parallel β-sheet, which is sandwiched between a long α-helix and the flexible loop region on one side and three shorter helices on the other side. The α/β-domain of TK1 is of the same type as ATP-binding domains in enzymes of the RecA-F1ATPase family and is present in several helicases and DNA repair proteins (47). The TK1 domain is generally smaller than those of the other members of the family. A search at the Dali server (48) reveals that the most similar structure in the database is CobA, which transfers the adenosyl moiety from ATP to corrinoid substrates (49). TK1 and CobA have all six parallel strands and the helices in common, and the CobA domain is roughly the same size as the α/β-domain of TK1. Ninety-one Cα atoms could be superimposed with an rms deviation of 1.53 Å. This superpositioning includes all strands and helices of the domains.
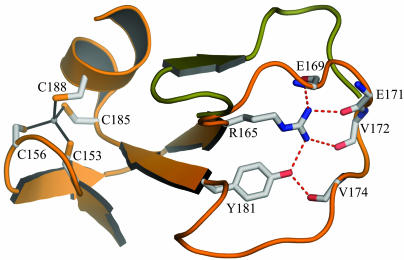
The small domain of 70–80 aa contains a long, lasso-shaped structure that covers the deoxynucleoside site in TK1 (Fig. 1). No similar structure was found in the database by using the Dali server. The domain contains two perpendicular β-ribbons connected by a metal that was interpreted as a zinc ion. Direct chemical analysis confirmed that the enzymes contained zinc. One of the β-ribbons opens up in a lasso-shaped loop that binds the base of the feedback inhibitor through main-chain hydrogen bonds. The lasso loop is kept in place by a conserved Arg–Tyr couple that is hydrogen-bonded and binds to main-chain atoms (Fig. 2). Arg and Tyr stack against the base of the nucleotide. The zinc is tetrahedrally coordinated by two cysteine pairs in the human enzyme as in structural zinc motifs (50, 51). The last of the cysteines is substituted by a His in the Ureaplasma enzyme as in other structural zinc motifs (52). The zinc atom is ≈20 Å from the active site.
Fig. 2.
Lasso domain with structural Zn2+. The coordination of Zn2+ is shown as present in hTK1. In Uu-TK, the coordinating amino acids are C153, C156, C191, and H194. The lasso is shown in orange for hTK1. In Uu-TK, this loop is slightly longer (shown in green). Additionally, the hydrogen bonding from the conserved Arg–Tyr couple is shown.
The hTK1 crystals contain two tetramers and Uu-TK contains one tetramer in the asymmetric unit; the tetramers are very similar to each other (Fig. 1). The monomers are organized with 222 symmetry, and the tetramer has a central channel lined mainly by polar and charged residues. There are two types of monomer–monomer interactions in the tetramers. One of these is dominated by a long helix from the two subunits forming an antiparallel pair. In the second interaction area, the β-sheet of one subunit is connected in an antiparallel manner by means of water molecules to the β-sheet in another subunit.
In the full-length Uu-TK enzyme, the C-terminal structure, with an extended arm-like loop and a terminal helix, interacts with a helix on the adjacent monomer. The missing C terminus of hTK1 contains a sequence motif (KEN) that serves as an ubiquination ligase signal involved in the mitotic degradation of TK1 (6). This part of the human enzyme is probably located in a similar, exposed subunit-contacting element.
The structures of the distantly related hTK1 and Uu-TK, with a sequence identity of 30%, are very similar. The subunits can be superimposed with an rms deviation of 1.2 Å for 147 Cα atoms. TK1-like sequences exist for a large number of species that all have a characteristic sequence pattern with many conserved residues, indicating that the members of the family have very similar structures.
The α/β-domain of TK1 differs from the α/β-domain of the other dNKs in strand order and positions of the helices. Particularly, the substrate-binding site that is formed by two helix pairs and the helical lid structure in the other dNKs are missing. The finding that TK1 uses a hitherto unknown Zn domain with a lasso-like loop to interact with the base and has an α/β-domain more similar to a domain in RecA proteins than to other dNK structures indicates a different evolutionary origin of the TK1 family.
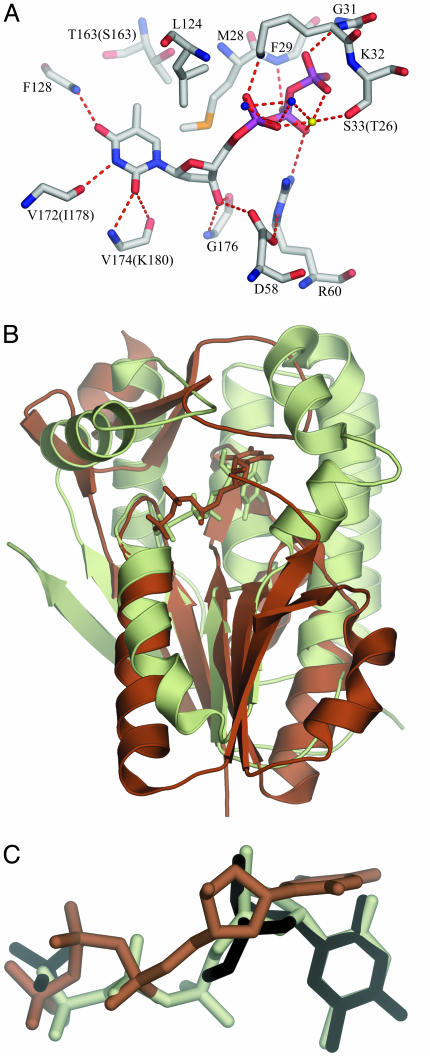
The Deoxyribonucleoside-Binding Site. dNKs catalyze the phosphoryl transfer from a phosphate donor, normally ATP, to the nucleoside substrate. The phosphate donor is bound at an exposed site containing the phosphate-binding P loop, whereas the deoxynucleoside is bound at a more buried site. dNKs are generally feedback-inhibited by the dNTP end product. Structural studies of the dNK from Drosophila (Dm-dNK) revealed a general mechanism for feedback-inhibition of dNKs (53). The end-product dNTP binds with its base and deoxyribose in the nucleoside-binding site like a deoxyribonucleoside substrate, and the triphosphates of the inhibitor bind backward to the phosphate-binding site at the P loop, i.e., as a bisubstrate inhibitor. The dTTP molecule in TK1s binds similarly as dTTP in Dm-dNK (Fig. 3) with the base of the dTTP buried within the protein; the triphosphates are more exposed.
Fig. 3.
Binding interactions of dTTP at the active site as found in hTK1. (A) The active-site amino acids are well conserved; only the residues that differ in Uu-TK are in brackets. The hydrogen-bond donors and acceptors of the base all form hydrogen bonds to main-chain atoms: O2 and N3 bond to main-chain atoms of the lasso loop, and O4 bonds to main-chain nitrogen of the α/β-domain. The methyl group of thymine is positioned in a hydrophobic pocket directed toward the β-carbon of a Ser in Uu-TK and a Thr in hTK1 surrounded by Met, Leu, and Tyr. The 3′-oxygen atom of the deoxyribose is hydrogen-bonded to the amino group of a conserved Gly in the lasso domain and to the side chain of the conserved Asp in the flexible loop of the α/β-domain. (B) Comparison of hTK1 and Dm-dNK with dTTP. hTK1 (brown) was superimposed with Dm-dNK bound with dTTP (light green). The P loop and the flanking regions were used in the superpositioning. (C) The thymidine base in hTK1 is bound perpendicularly compared with the way it is bound in Dm-dNK. dT and the sulfate ion bound to Dm-dNK are shown in black, dTTP bound to hTK1 is shown in brown, and dTTP bound to Dm-dNK is shown in light green.
Whereas the triphosphates bind to the α/β-domain, the base and the deoxyribose bind in a cleft between the α/β-domain and the lasso domain. The base stacks against the conserved Arg and Tyr on the lasso-domain side. The ring of the conserved Tyr is parallel to the base and covers most of this side. On the opposite side, the base stacks against two phenylalanines perpendicularly oriented relative to the base, with Leu and Val at its edges.
The hydrogen-bond donors and acceptors of the base all form hydrogen bonds to main-chain atoms (Fig. 3): O2 and N3 form bonds to main-chain atoms of the lasso-loop and O4 forms bonds to a main-chain nitrogen of the α/β-domain. The methyl group of thymine is positioned in a hydrophobic pocket directed toward the β-carbon of a Ser in Uu-TK and a Thr in hTK1 surrounded by Met, Leu, and Tyr.
The 3′-oxygen atom of the deoxyribose is hydrogen-bonded to the main-chain amino group of a conserved Gly in the lasso domain and to the side chain of the conserved Asp in the flexible loop of the α/β-domain. The 5′-oxygen of the deoxyribose is close to a Glu in the α/β domain. This Glu (Glu-98 in hTK1) may serve as the catalytic base abstracting a proton, thereby activating the 5′-oxygen to act as a nucleophile in the reaction. The other dNKs have a Glu in an equivalent position, implicating similar catalytic mechanisms (12). Beside a Mg ion bound to the phosphates, Arg-60 (hTK1 numbering) is in a position to stabilize the transition state of the reaction.
If the P loop, the preceding strand, and the subsequent helix of TK1 are superpositioned on the other dNKs, the base in TK1 is bound perpendicular to the others, with the tip displaced about 6 Å (Fig. 3). In the other dNKs, a Gln side chain binds at the edge of the base, and a Tyr–Glu couple binds to 3′-OH of the deoxyribose (12). These features are absent in TK1.
Substrate Specificity. The narrow substrate specificity of TK1 is consistent with a smaller binding site than in herpes virus 1–TK and other dNKs, with tight interactions of the protein and the base by main-chain hydrogen bonds to all polar groups of the base. At the 5-position of the base, the presence of a halogeno or ethyl group has no real effect on enzyme activity, consistent with 5-fluoro-2′-deoxyuridine being a good substrate. Bulkier or more polar groups, as amino and 2-bromovinyl, would be sterically hindered upon binding (54, 55). This property is consistent with the rather small hydrophobic pocket, where the methyl group fits snuggly.
It has been shown that TK1 accepts large substitutions at the 3-position of the ring (56). Although N3 is hydrogen-bonded to a main-chain carbonyl in this position, the 3-position of the ring points toward the contact area between the α/β-domain and the lasso domain. For large substituents at the 3-position, the lasso domain may not close down completely, but substituents at N3 may compensate the loss of a hydrogen bond to N3 by other interactions.
The high specificity for 2′-deoxyribonucleosides is consistent with the tight interactions at the 2′-carbon. Binding of ribonucleosides would cause steric interference among the 2′-OH and the carbonyl of the conserved Gly and the ring of the conserved Tyr in the lasso domain, because both residues are in van der Waals contact with the 2′-carbon. The 3′-OH interactions with the main-chain atoms of a conserved Gly in the lasso domain take part such that one side of the 3′-OH is exposed at the surface and thus tolerates substitutions. 3′-azido-3′-deoxythymidine is an example of such an analog modified in this position (11, 12).
TK1 and Drug Design. The high conservation of the dT-binding site with main-chain atoms makes selective drug development difficult, because there is little variation in the substrate cleft among different species. To create new prodrugs that could be selectively phosphorylated by Uu-TK but not hTK1, there are possibilities around the 5-methyl group of dT, where there is contact with the β-carbon of a Thr in hTK1 and of a Ser in Uu-TK. Otherwise, substitutions at N3 that have been show to be useful can be exploited further.
The environment of the deoxyribose is rather open around the 3′–4′ edge and may be used in the development of new prodrugs. An alternative is substitutions at O4′ of the ring, which is ≈4.5 Å from the conserved Gln-100 (human sequence numbering).
In gene therapy using viral TK, it was observed that some patients developed an immune response against the viral enzyme during the treatment (57). Gene therapy with hTK1 with modified specificity may circumvent the immunity problem. The two TK structures also will aid development of new immunological reagents for cell biology and cancer diagnostics.
Acknowledgments
We thank Marianne Lauridsen for excellent technical assistance and Alwyn Jones for help with new features of o. This work was supported by grants from the Swedish Research Council (to H.E. and S.E.), the Swedish Cancer Foundation (to H.E.), the Wallenberg Consortium North (to H.E.), the Swedish Research Council for Environment, Agricultural Sciences, and Spatial Planning (to L.W.), and the Danish Research Council (to B.M.-P.).
Author contributions: M.W., U.K., L.W., S.E., B.M.-P., and H.E. designed research; M.W., U.K., N.-E.M., C.C., C.Z., L.W., and H.E. performed research; M.W., U.K., and H.E. analyzed data; and M.W., U.K., S.E., B.M.-P., and H.E. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TK, thymidine kinase; hTK1, human TK 1; Uu-TK, TK from Ureaplasma urealyticum; dNK, deoxyribonucleoside kinase; Dm-dNK, dNK from Drosophila; dT, deoxythymidine; SeMet, selenomethionine.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 1XBT (hTK1) and 1XMR (Uu-TK)].
References
- 1.Jordan, A. & Reichard, P. (1998) Annu. Rev. Biochem. 67, 71–98. [DOI] [PubMed] [Google Scholar]
- 2.Arnér, E. S. J. & Eriksson, S. (1995) Pharmacol. Ther. 67, 155–186. [DOI] [PubMed] [Google Scholar]
- 3.Bello, L. J. (1974) Exp. Cell Res. 89, 263–274. [DOI] [PubMed] [Google Scholar]
- 4.Munch-Petersen, B. & Tyrsted, G. (1977) Biochim. Biophys. Acta 478, 364–375. [DOI] [PubMed] [Google Scholar]
- 5.Sherley, J. L. & Kelly, T. J. (1988) J. Biol. Chem. 263, 8350–8358. [PubMed] [Google Scholar]
- 6.Ke, P. Y. & Chang, Z. F. (2004) Mol. Cell. Biol. 24, 514–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kornberg, A., Lehman, I. R. & Simms, E. S. (1956) Fed. Proc. 15, 291–292. [Google Scholar]
- 8.Reichard, P. & Estborn, B. (1951) J. Biol. Chem. 188, 839–846. [PubMed] [Google Scholar]
- 9.Pontarin, G., Gallinaro, L., Ferraro, P., Reichard, P. & Bianchi, V. (2003) Proc. Natl. Acad. Sci. USA 100, 12159–12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobrovolsky, V. N., Bucci, T., Heflich, R. H., Desjardins, J. & Richardson, F. C. (2003) Mol. Genet. Metab. 78, 1–10. [DOI] [PubMed] [Google Scholar]
- 11.Furman, P. A., Fyfe, J. A., St. Clair, M. H., Weinhold, K., Rideout, J. L., Freeman, G. A., Lehrman, S. N., Bolognesi, D. P., Broder, S., Mitsuya, H., et al. (1986) Proc. Natl. Acad. Sci. USA 83, 8333–8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eriksson, S., Munch-Petersen, B., Johansson, K. & Eklund, H. (2002) Cell. Mol. Life Sci. 59, 1327–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gronowitz, J. S., Kallander, F. R., Diderholm, H., Hagberg, H. & Pettersson, U. (1984) Int. J. Cancer 33, 5–12. [DOI] [PubMed] [Google Scholar]
- 14.Wild, K., Bohner, T., Aubry, A., Folkers, G. & Schulz, G. E. (1995) FEBS Lett. 368, 289–292. [DOI] [PubMed] [Google Scholar]
- 15.Brown, D. G., Visse, R., Sandhu, G., Davies, A., Rizkallah, P. J., Melitz, C., Summers, W. C. & Sanderson, M. R. (1995) Nat. Struct. Biol. 2, 876–881. [DOI] [PubMed] [Google Scholar]
- 16.Sabini, E., Ort, S., Monnerjahn, C., Konrad, M. & Lavie, A. (2003) Nat. Struct. Biol. 10, 513–519. [DOI] [PubMed] [Google Scholar]
- 17.Pilger, B. D., Perozzo, R., Alber, F., Wurth, C., Folkers, G. & Scapozza, L. (1999) J. Biol. Chem. 274, 31967–31973. [DOI] [PubMed] [Google Scholar]
- 18.Bird, L. E., Ren, J., Wright, A., Leslie, K. D., Degreve, B., Balzarini, J. & Stammers, D. K. (2003) J. Biol. Chem. 278, 24680–24687. [DOI] [PubMed] [Google Scholar]
- 19.Knecht, W., Sandrini, M. P., Johansson, K., Eklund, H., Munch-Petersen, B. & Piskur, J. (2002) EMBO J. 21, 1873–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munch-Petersen, B., Tyrsted, G. & Cloos, L. (1993) J. Biol. Chem. 268, 15621–15625. [PubMed] [Google Scholar]
- 21.Carnrot, C., Wehelie, R., Eriksson, S., Bolske, G. & Wang, L. (2003) Mol. Microbiol. 50, 771–780. [DOI] [PubMed] [Google Scholar]
- 22.Glass, J. I., Lefkowitz, E. J., Glass, J. S., Heiner, C. R., Chen, E. Y. & Cassell, G. H. (2000) Nature 407, 757–762. [DOI] [PubMed] [Google Scholar]
- 23.Hudson, M. M. & Talbot, M. D. (1997) Int. J. STD AIDS 8, 546–551. [DOI] [PubMed] [Google Scholar]
- 24.Kristensen, T. (1996) Ph.D. thesis (Roskilde University, Roskilde, Denmark), pp. 45–50.
- 25.Berenstein, D., Christensen, J. F., Kristensen, T., Hofbauer, R. & Munch-Petersen, B. (2000) J. Biol. Chem. 275, 32187–32192. [DOI] [PubMed] [Google Scholar]
- 26.Bradshaw, H. D., Jr., & Deininger, P. L. (1984) Mol. Cell. Biol. 4, 2316–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frederiksen, H., Berenstein, D. & Munch-Petersen, B. (2004) Eur. J. Biochem. 271, 2248–2256. [DOI] [PubMed] [Google Scholar]
- 28.Gilles, S. I., Romain, S., Casellas, P., Ouafik, L., Fina, F., Combes, T., Vuaroquaux, V., Seitz, J. F., Bonnier, P., Galiegue, S., et al. (2003) Int. J. Biol. Markers 18, 1–6. [DOI] [PubMed] [Google Scholar]
- 29.Strausberg, R. L., Feingold, E. A., Grouse, L. H., Derge, J. G., Klausner, R. D., Collins, F. S., Wagner, L., Shenmen, C. M., Schuler, G. D., Altschul, S. F., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 16899–16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ducruix, A. & Giege, R. (1992) Crystallization of Nucleic Acids and Proteins: A Practical Approach (Oxford Univ. Press, London).
- 31.Stura, E. A. & Wilson, I. A. (1990) Methods 1, 38–49. [Google Scholar]
- 32.Bergfors, T. (2003) J. Struct. Biol. 142, 66–76. [DOI] [PubMed] [Google Scholar]
- 33.Collaborative Computational Project Number (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 34.Sheldrick, G. M., Hauptman, H. A., Weeks, C. M., Miller, M. & Usón, I. (2001) in International Tables for Crystallography, eds. Arnold, E. & Rossmann, M. (Kluwer, Dordrecht, The Netherlands), Vol. F, pp. 333–351. [Google Scholar]
- 35.La Fortelle, E. D. & Bricogne, G. (1997) Methods Enzymol. 276, 472–494. [DOI] [PubMed] [Google Scholar]
- 36.Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. (2003) Acta Crystallogr. D 59, 2023–2030. [DOI] [PubMed] [Google Scholar]
- 37.Cowtan, K. (1994) Joint CCP4 and ESF–EACBM Newsletter on Protein Crystallography 31, 34–38. [Google Scholar]
- 38.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 39.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski, Z. (1993) in Proceedings of the CCP4 Study Weekend, eds. Sawyer, L., Issacs, N. & Bailey, S. (Daresbury Laboratories, Warrington, U.K.), pp. 56–62.
- 41.Leslie, A. G. W. (1999) Acta Crystallogr. D 55, 1696–1702. [DOI] [PubMed] [Google Scholar]
- 42.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 43.Thompson, J. D., Higgins, D. G., Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barton, G. J. (1993) Protein Eng. 6, 37–40. [DOI] [PubMed] [Google Scholar]
- 45.DeLano, W. L. (2002) The pymol User's Manual (DeLano Scientific, San Carlos, CA).
- 46.Kauffman, M. G. & Kelly, T. J. (1991) Mol. Cell. Biol. 11, 2538–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sawaya, M. R., Guo, S., Tabor, S., Richardson, C. C. & Ellenberger, T. (1999) Cell 99, 167–177. [DOI] [PubMed] [Google Scholar]
- 48.Holm, L. & Sander, C. (1995) Trends Biochem. Sci. 20, 478–480. [DOI] [PubMed] [Google Scholar]
- 49.Bauer, C. B., Fonseca, M. V., Holden, H. M., Thoden, J. B., Thompson, T. B., Escalante-Semerena, J. C. & Rayment, I. (2001) Biochemistry 40, 361–374. [DOI] [PubMed] [Google Scholar]
- 50.Eklund, H., Nordström, B., Zeppezauer, E., Söderlund, G., Ohlsson, I., Boiwe, T., Söderberg, B. O., Tapia, O., Brändén, C. I. & Åkeson, Å. (1976) J. Mol. Biol. 102, 27–59. [DOI] [PubMed] [Google Scholar]
- 51.Luisi, B. F., Xu, W. X., Otwinowski, Z., Freedman, L. P., Yamamoto, K. R. & Sigler, P. B. (1991) Nature 352, 497–505. [DOI] [PubMed] [Google Scholar]
- 52.Cho, Y., Gorina, S., Jeffrey, P. D. & Pavletich, N. P. (1994) Science 265, 346–355. [DOI] [PubMed] [Google Scholar]
- 53.Mikkelsen, N. E., Johansson, K., Karlsson, A., Knecht, W., Andersen, G., Piskur, J., Munch-Petersen, B. & Eklund, H. (2003) Biochemistry 42, 5706–5712. [DOI] [PubMed] [Google Scholar]
- 54.Eriksson, S., Kierdaszuk, B., Munch-Petersen, B., Oberg, B. & Johansson, N. G. (1991) Biochem. Biophys. Res. Commun. 176, 586–592. [DOI] [PubMed] [Google Scholar]
- 55.Johansson, N. G. & Eriksson, S. (1996) Acta Biochim. Pol. 43, 143–160. [PubMed] [Google Scholar]
- 56.Al-Madhoun, A. S., Johnsamuel, J., Yan, J., Ji, W., Wang, J., Zhuo, J. C., Lunato, A. J., Woollard, J. E., Hawk, A. E., Cosquer, G. Y., et al. (2002) J. Med. Chem. 45, 4018–4028. [DOI] [PubMed] [Google Scholar]
- 57.Warren, P., Song, W., Holle, E., Holmes, L., Wei, Y., Li, J., Wagner, T. & Yu, X. (2002) Anticancer Res. 22, 599–604. [PubMed] [Google Scholar]