Abstract
Background
Despite being the most important cellulase producer, the cellulase-regulating carbon source signal transduction processes in Trichoderma reesei are largely unknown. Elucidating these processes is the key for unveiling how external carbon sources regulate cellulase formation, and ultimately for the improvement of cellulase production and biofuel production from lignocellulose.
Results
In this work, the role of the mitogen-activated protein kinase (MAPK) signal transduction pathways on cellulase formation was investigated. The deletion of yeast FUS3-like tmk1 in T. reesei leads to improved growth and significantly improved cellulase formation. However, tmk1 deletion has no effect on the transcription of cellulase-coding genes. The involvement of the cell wall integrity maintenance governing yeast Slt2-like Tmk2 in cellulase formation was investigated by overexpressing tmk3 in T. reesei Δtmk2 to restore cell wall integrity. Transcriptional analysis found little changes in cellulase-coding genes between T. reesei parent, Δtmk2, and Δtmk2::OEtmk3 strains. Cell wall integrity decreased in T. reesei Δtmk2 over the parent strain and restored in Δtmk2::OEtmk3. Meanwhile, cellulase formation is increased in T. reesei Δtmk2 and then decreased in T. reesei Δtmk2::OEtmk3.
Conclusions
These investigations elucidate the role of Tmk1 and Tmk2 on cellulase formation: they repress cellulase formation, respectively, by repressing growth and maintaining cell wall integrity, while neither MAPK regulates the transcription of cellulase-coding genes. This work, together with the previous investigations, suggests that all MAPKs are involved in cellulase formation, while Tmk3 is the only MAPK involved in signal transduction for the regulation of cellulase expression on the transcriptional level.
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-017-0789-x) contains supplementary material, which is available to authorized users.
Keywords: Trichoderma reesei, Cellulase, MAP kinase, Signal transduction, Cell wall integrity, Transcriptional analysis
Background
The economical and environment-friendly utilization of earth’s largest biomass reserve lignocellulose for the purpose of biofuel production requires the abundant production of cellulase, a multicomponent synergistic enzyme cocktail secreted by microorganisms [1]. Trichoderma reesei (syn. Hypocrea jecorina) is the most prominent industrial cellulase producer that was demonstrated capable of producing as much as 20 g L−1 cellulases [2]. This filamentous fungus has therefore been considered a model organism for cellulase formation and regulation mechanisms [3].
The formation of cellulases has been shown to be controlled on both transcriptional and secretion levels in filamentous fungi [4, 5]. Transcriptionally, cellulase-coding genes are co-regulated by external signals such as carbon signals (such as glucose, cellobiose, lactose, Avicel, and sophorose) and light signals [6–9]. Of these signals, glucose is a strong inhibitory signal, while Avicel, lactose, sophorose, cellobiose, and light signals are inducing signals for the expression of cellulase-coding genes. Transcription factors that govern the stimulation or inhibition of cellulase-coding genes have been investigated in great detail, leading to the identification of inhibitory transcription factors Cre1 and ACEI [10, 11], as well as activating transcription factors Xyr1 and ACEII [12, 13]. However, how signals are passed from extracellular space to transcription factors for the modulation of cellulase expression is largely unclear.
In the past decade, the most significant breakthrough on the understanding of cellulase-regulating signal transduction pathways is the elucidation of the light regulation pathway [9]. This signal transduction is executed by a complex system in which the second messenger cyclic AMP (cAMP) and protein kinase A (PKA) play a central role [14]. The understanding of the carbon signal transduction pathways, however, is still preliminary. Previous bioinformatic analysis suggested that cellulase-regulating transcription factors Cre1, Xyr1, ACEI, and ACEII all harbor phosphorylation sites for protein kinases such as casein kinase II (CKII) and mitogen-activated protein kinases (MAPKs) [15]. It was also shown that the disruption of CKII phosphorylation site on Cre1 leads to Cre1’s loss of phosphorylation and subsequently loss of glucose inhibition [16]. Combined with the findings that protein kinases such as Ime2, Sch9, and Yak1 regulate cellulase formation [17, 18], these results strongly suggest protein kinases may play an important role in relaying cellulase-regulating signals. Of these protein kinases, the most interesting protein kinases include MAPKs and CKII, primarily because bioinformatic and biochemical analysis suggested their direct involvement in the phosphorylation of cellulase-regulating transcription factors [15, 16, 19].
Mitogen-activated protein kinases are ubiquitous protein kinases in eukaryotes that play the role of signal transduction and amplification [20]. It is a key component of a MAPKKK–MAPKK–MAPK signal relay system. In this signal transduction cascade, MAPKKK (MAP kinase kinase kinase) is first phosphorylated upon activation, which phosphorylates and activates MAPKK (MAP kinase kinase), which in turn phosphorylates and activates MAPK for downstream functions [21]. Six major MAPK pathways are present in the model fungus Saccharomyces cerevisiae, the homologues of three of which have been widely found in filamentous fungi: Hog1-like MAPK pathway, Slt2-like MAPK pathway, and Fus3-like MAPK pathway [19, 22]. In S. cerevisiae, the Hog1 pathway responds to high osmolarity signals and functions in combating this environmental stress, the Slt2 pathway primarily functions in cell wall integrity maintenance, and the Fus3 pathway functions in the pheromone-induced mating type differentiation process [21, 23].
Mitogen-activated protein kinases are involved in a variety of physiological functions in filamentous fungi. Hog1 homologues in filamentous fungi function primarily in high osmolarity resistance similarly with S. cerevisiae [24–29]. Involvement of Hog1 homologues in other processes such as sporulation [24, 27], stress response [28, 30], and secondary metabolism [27] has also been reported. Like in S. cerevisiae, Slt2 homologues in filamentous fungi are also primarily involved in cell wall integrity maintenance with a few exceptions [31–42]. Previous reports also showed the involvement of Slt2 in a series of other processes such as virulence [31, 32, 41], sporulation [32, 36, 37, 42], female fertility, secondary metabolism [37, 40], hyphal polarity maintenance [37, 38, 43], surface hydrophobicity maintenance [39, 41], stress response [38] and circadian rhythm maintenance [44]. The function of Fus3 homologues in filamentous fungi is quite diverse, with reported participation in pathogenicity [45–50], sporulation [48, 51], hyphal development [52–55], secondary metabolism [55], and stress response [51]. In particular, Fus3 homologues in Trichoderma species were shown to participate in the production of mycoparasitism-related proteins such as chitinases and N-acetylglucosaminidases [49, 56–58], and in the production of cellulases [49]. This function raises curiosity on whether Fus3 homologues may function in regulating cellulase expression and secretion in T. reesei.
Three MAPKs have been identified from in silico analysis in T. reesei, namely, the Fus3-like Tmk1, the Slt2-like Tmk2, and the Hog1-like Tmk3 [19]. Previous research carried out in our laboratory has shown the physiological functions of Tmk2 and Tmk3 [15, 59]. Our results showed that both Tmk2 and Tmk3 are involved in cell wall integrity maintenance. Tmk3 also participates in high osmolarity resistance and biosynthesis, while Tmk2 also participates in sporulation. The deletion of tmk3 leads to a strong reduction of cellulase formation and transcription of cellulase-coding genes, suggesting the role of Tmk3 in positively regulating the expression of cellulase-coding genes [15]. On the contrary, the deletion of tmk2 has little effect on the transcriptional level of cellulase-coding genes but leads to a higher cellulase formation level [59]. The role of Tmk1 in T. reesei was still unknown.
In this work, we aim to generate a full picture on the role of MAPKs in cellulase formation in T. reesei. This was done by constructing and characterizing T. reesei Δtmk1, thereby finding out whether Tmk1 is involved in the regulation of cellulase formation, as well as the overexpression of tmk3 in a T. reesei Δtmk2 strain, which led to the identification of the mechanism on how Tmk2 influences cellulase formation. Based on this functional characterization, previously identified roles of Tmk2 and Tmk3 in T. reesei, as well as the mechanistic investigations carried out in this work on how MAPKs influence cellulase formation in T. reesei, this picture of the involvement of MAPKs in cellulase production can be obtained.
Results
Deletion of tmk1 in T. reesei leads to improved growth and cellulase formation
The deletion of tmk1 in T. reesei was successfully carried out with homologous recombination (see Additional file 1), generating two parallel tmk1 deletion strains, respectively, named T. reesei Δtmk1-1 and T. reesei Δtmk1-2. The parent T. reesei TU-6 strain and the tmk1 deletion strains (T. reesei Δtmk1-1 and T. reesei Δtmk1-2) were grown on potato dextrose agar (PDA) plates, minimal media plates containing glycerol, glucose, or lactose as the substrate, and Avicel double layer plates (Fig. 1a). It can be observed that the two tmk1 deletion strains form larger colonies in comparison with the parent strain. This observation is confirmed by measuring the colony diameters after 6 days of growth on Avicel double layer plates and 3 days of growth on all other plates (Fig. 1b). Without exception, both Δtmk1 strains grow significantly better than the parent strain. Interestingly, a larger clear zone was formed in the Avicel double layer plates on which either tmk1 deletion strain was grown in comparison with the T. reesei TU-6 parent strain (Fig. 1a). These results clearly lead to the suggestion that the deletion of tmk1 leads to improved growth and cellulase formation in T. reesei. No significant impact on cell wall integrity was observed after the deletion of tmk1 (see Additional file 2).
Fig. 1.
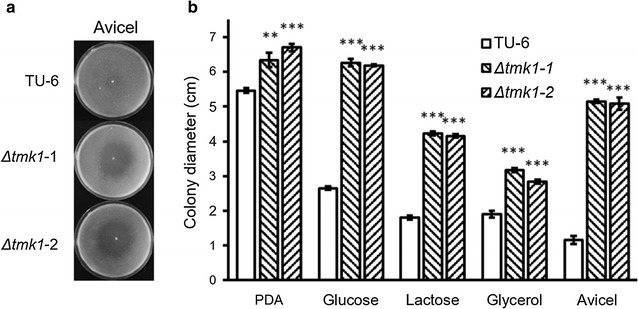
Growth of T. reesei TU-6 and tmk1 deletion strains on agar plates. a photographs of T. reesei colonies on double layer Avicel agar plates (Avicel); b comparison of colony diameters between T. reesei TU-6 and tmk1 deletion strains. TU-6, T. reesei TU-6; Δtmk1-1, T. reesei Δtmk1-1; Δtmk1-2, T. reesei Δtmk1-2. **p < 0.01; ***p < 0.001. Error bar standard deviation of three replicates
Growth and production analysis of T. reesei TU-6, T. reesei Δtmk1-1, and T. reesei Δtmk1-2 grown in 3 L fermenters showed the same phenomenon (Fig. 2). The overall filter paperase activities (FPase activity, FPA), CMCase activities that represent endoglucanase activities, pNPCase activities that represent cellobiohydrolase activities and total extracellular protein contents are all significantly improved in tmk1 deletion strains T. reesei Δtmk1-1 and T. reesei Δtmk1-2 in comparison with the parent strain (Fig. 2a–c, e). For determination of biomass of these strains, we used a previously reported method by which total intracellular A260 was used as a measure for biomass [15, 60]. This approach was validated by plotting intracellular A260 of different T. reesei strains grown under different growth conditions against respective dry mycelial weight (Fig. 3), confirming the use of total intracellular A260 as an applicable method for biomass determination. A significant improvement of total fungal biomass in the tmk1 deletion strains was observed with this method (Fig. 2d).
Fig. 2.
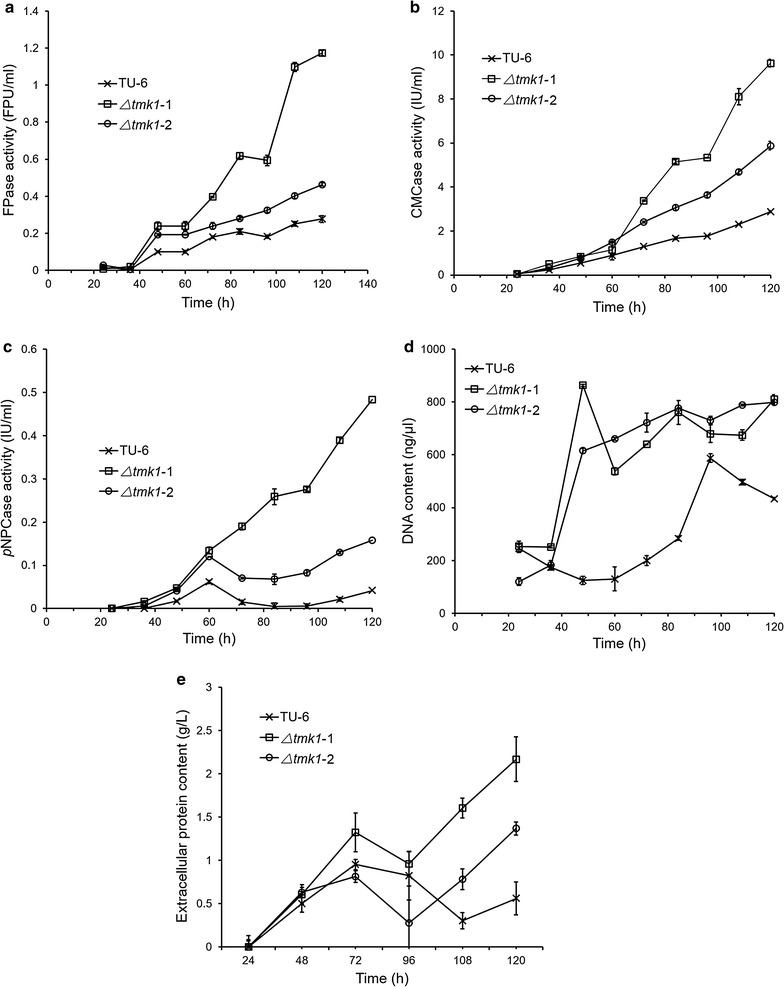
Production of cellulase activities by T. reesei TU-6, Δtmk1-1, and Δtmk1-2 in 3-L fermenters. a Production of filter paperase activity (FPase activity); b production of CMCase activity; c production of pNPCase activity; d biomass determination represented by A260; e total extracellular protein content. TU-6, T. reesei TU-6; Δtmk1-1, T. reesei Δtmk1-1; Δtmk1-2, T. reesei Δtmk1-2. Error bar standard deviation of three replicates
Fig. 3.

Correlation between A260 and dry mycelial weight
Tmk1 is not involved in regulation of cellulase transcription
Despite our observation that cellulase production is improved following tmk1 deletion, transcriptional analysis of major cellulase-coding genes cbh1, cbh2, egl1, and egl2 after 3 days of growth using Avicel and wheat bran as the carbon source showed little difference between the parent TU-6 strain and the two tmk1 deletion strains Δtmk1-1 and Δtmk1-2 (Fig. 4a). Change of transcriptional levels of the same genes in the first 24 h after the transfer from glycerol-containing media to Avicel- and wheat bran-containing media showed similar results (Fig. 4b–e). A suggestion can be made from these results that Tmk1 is not involved in the regulation of cellulase transcription in T. reesei. The reason for the improved cellulase formation in tmk1 deletion strains should therefore be the improved growth of T. reesei tmk1 deletion strains, rather than the specific regulation of Tmk1 on the expression of cellulase-coding genes. A further suggestion can be made from these results that Tmk1 is not involved in the signal transduction pathway transmitting cellulase induction cellulose signals.
Fig. 4.
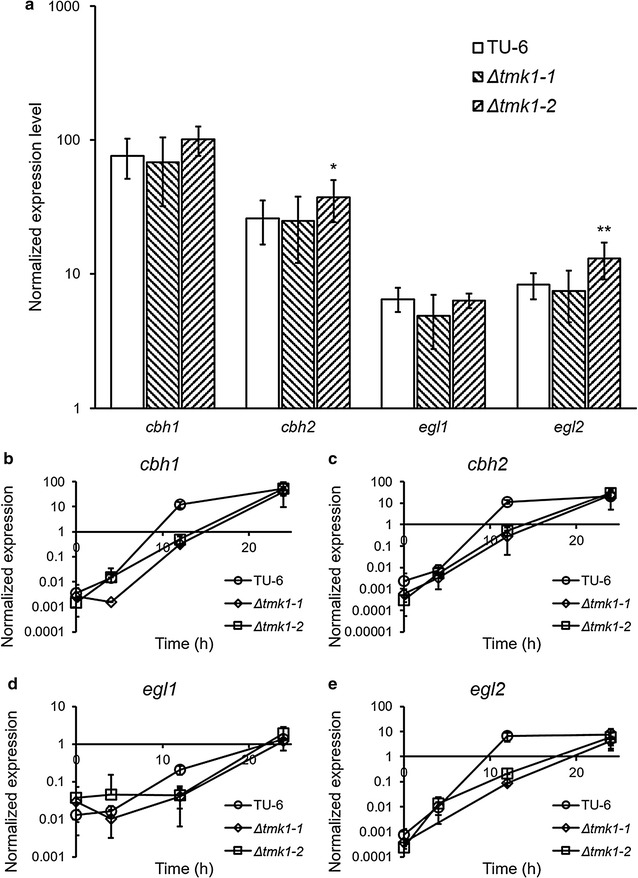
Transcriptional comparison of cellulase-coding genes between T. reesei TU-6, Δtmk1-1, and Δtmk1-2. a Expression levels 3 days after inoculation to Avicel- and wheat bran-containing media; b cbh1 expression levels after transfer to Avicel- and wheat bran-containing media; c cbh2 expression levels after transfer to Avicel- and wheat bran-containing media; d egl1 expression levels after transfer to Avicel- and wheat bran-containing media; e egl2 expression levels after transfer to Avicel- and wheat bran-containing media. Expression levels are normalized to the expression level of the actin-coding gene. TU-6, T. reesei TU-6; Δtmk1-1, T. reesei Δtmk1-1; Δtmk1-2, T. reesei Δtmk1-2. *p < 0.05; **p < 0.01. Error bar standard deviation of nine replicates
Damage of cell wall integrity by the deletion of tmk2 and reinforcing cell wall integrity by overexpressing tmk3
Both Tmk2 and Tmk3 were shown to participate in cell wall integrity maintenance, and the deletion of either tmk2 or tmk3 led to damaged cell wall integrity [15, 59]. They may therefore compensate each other’s function in cell wall integrity maintenance if one gene is deleted. In this work, we overexpressed tmk3 using the previously constructed T. reesei Δtmk2 strain [59] as the parent strain to improve cell wall integrity from T. reesei Δtmk2 (see Additional file 3). By replacing tmk3 promoter with a strong Pgpd promoter, we were able to increase tmk3 transcription by 11.80-folds (p = 1.82 × 10−10, n = 9). Sensitivity assays to CR and CFW were carried out for T. reesei TU-6, T. reesei Δtmk2, and T. reesei Δtmk2::OEtmk3 (Fig. 5). T. reesei TU-6 was able to tolerate 200 mg L−1 CR and 20 mg L−1 CFW, while T. reesei Δtmk2 cannot, suggesting that tolerance to CR and CFW is reduced in T. reesei Δtmk2 in comparison with the parent T. reesei TU-6 strain, in agreement with the previous report [59]. By overexpressing tmk3, this tolerance is regained, suggesting restoration of cell wall integrity in comparison with T. reesei Δtmk2 (Fig. 5).
Fig. 5.
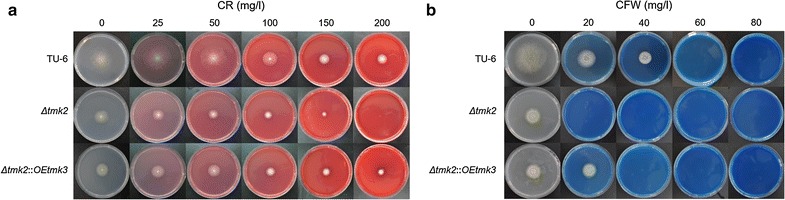
The sensitivity of T. reesei TU-6, Δtmk2 and Δtmk2::OEtmk3 to CR and CFW. a Growth of T. reesei TU-6, T. reesei Δtmk2 and T. reesei Δtmk2::OEtmk3 on CR-containing agar plates; b growth of T. reesei TU-6, T. reesei Δtmk2 and T. reesei Δtmk2::OEtmk3 on CFW-containing agar plates. TU-6, T. reesei TU-6; Δtmk2, T. reesei Δtmk2; Δtmk2::OEtmk3, T. reesei Δtmk2::OEtmk3
Cell wall integrity negatively impacts cellulase secretion in T. reesei TU-6, T. reesei Δtmk2, and T. reesei Δtmk2::OEtmk3
The transcription of cellulase-coding genes cbh1, cbh2, egl1, and egl2 was compared between T. reesei TU-6, T. reesei Δtmk2 and T. reesei Δtmk2::OEtmk3 grown using Avicel and wheat bran as the carbon source for 3 days (Fig. 6a). Although a slight difference can be seen between the three strains, the difference is too small to be meaningful. This finding is confirmed by the similar transcriptional levels of the same cellulase-coding genes in the three strains in the first 24 h after the carbon source changed from glycerol to Avicel and wheat bran (Fig. 6b–e). From these transcriptional analyses, suggestions can be made that the deletion of tmk2 and subsequent overexpression of tmk3 do not lead to changes in the transcriptional levels of cellulase-coding genes, in agreement with the previous reports [59].
Fig. 6.
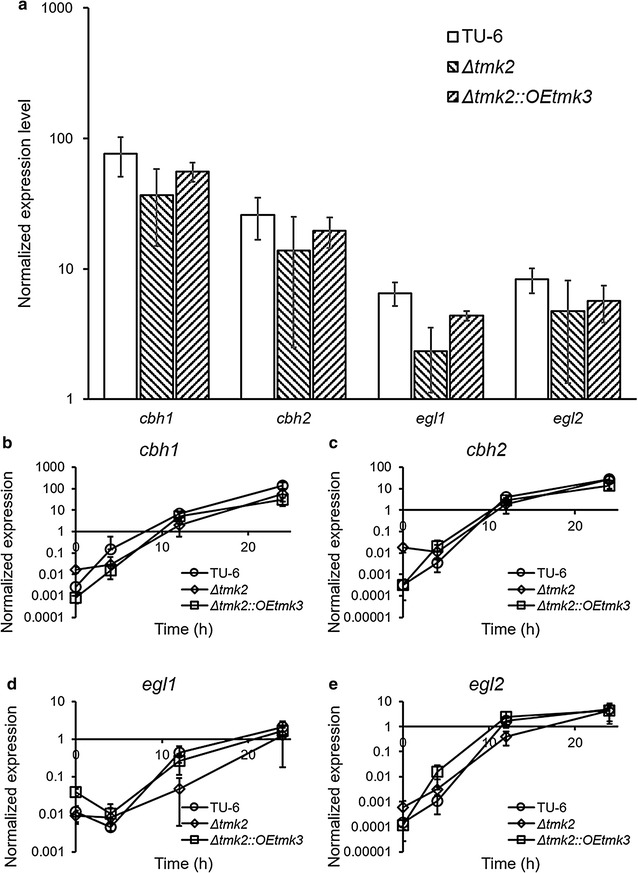
Transcriptional comparison of cellulase-coding genes between T. reesei TU-6, Δtmk2 and Δtmk2::OEtmk3. a Expression levels 3 days after inoculation to Avicel- and wheat bran-containing media; b cbh1 expression levels after transfer to Avicel- and wheat bran-containing media; c cbh2 expression levels after transfer to Avicel- and wheat bran-containing media; d egl1 expression levels after transfer to Avicel- and wheat bran-containing media; e egl2 expression levels after transfer to Avicel- and wheat bran-containing media. Expression levels are normalized to the expression level of the actin-coding gene. TU-6, T. reesei TU-6; Δtmk2, T. reesei Δtmk2; Δtmk2::OEtmk3, T. reesei Δtmk2::OEtmk3. Error bar standard deviation of nine replicates
Despite the lack of transcriptional changes in cellulase-coding genes, significant differences can be observed between cellulase production levels of the three strains in 3 L fermenters (Fig. 7). Although the biomass represented by volumetric A260 in the cultures decreased in T. reesei Δtmk2 in comparison with T. reesei TU-6 (Fig. 7e), the FPA, pNPCase, CMCase activities, and the extracellular protein content increased significantly in T. reesei Δtmk2 strain (Fig. 7a–d). This is in consistence with the previous reports that T. reesei Δtmk2 secrets more cellulases than the parent strain, despite no change in transcription of cellulase-coding genes is induced by tmk2 deletion [59]. Following the overexpression of tmk3, the total biomass remained largely similar in comparison with T. reesei Δtmk2 (Fig. 7e). The FPA, pNPCase, and CMCase activities, however, significantly decreased over T. reesei Δtmk2 (Fig. 7a, c, d). The same decrease is observed for extracellular protein content (Fig. 7b). A suggestion can be therefore made that the decrease of cell wall integrity by deleting tmk2 leads to improved cellulase production, while further increase of cell wall integrity by overexpressing tmk3 leads to decreased cellulase production.
Fig. 7.
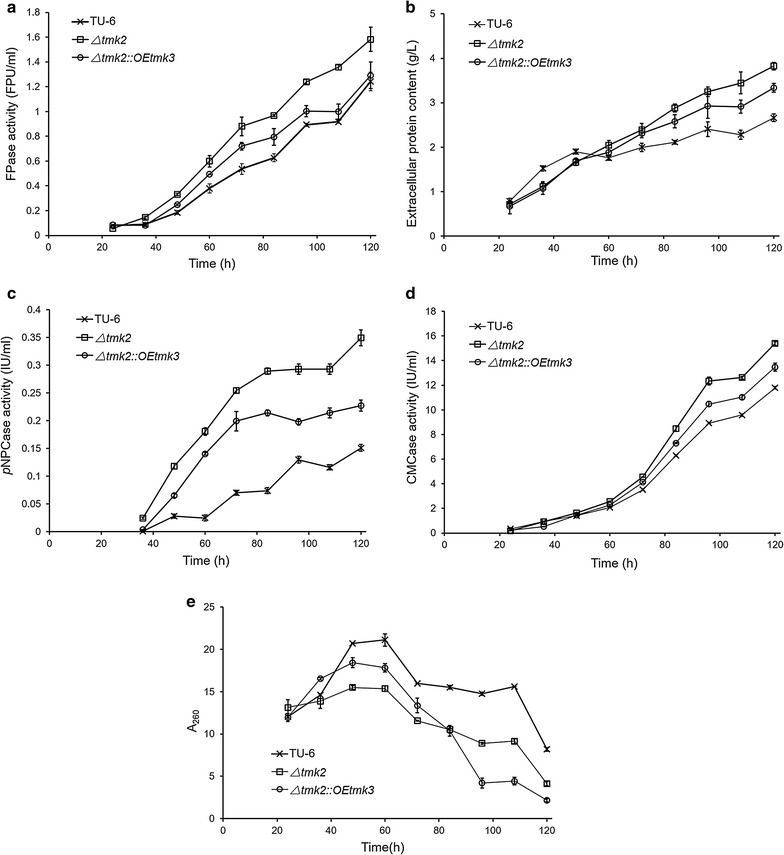
Production of cellulase by T. reesei TU-6, Δtmk2, and Δtmk2::OEtmk3 in 3-L fermenters. a Production of filter paperase activity (FPase activity); b production of extracellular proteins; c production of pNPCase activity; d production of CMCase activity; e biomass determination represented by A260. TU-6, T. reesei TU-6; Δtmk2, T. reesei Δtmk2; Δtmk2::OEtmk3, T. reesei Δtmk2::OEtmk3. Error bar standard deviation of three replicates
This observation of the relationship between cell wall integrity and cellulase secretion is confirmed by shake flask experiments assaying secretion of cellulase activities in T. reesei TU-6, T. reesei Δtmk2, and T. reesei Δtmk2::OEtmk3 (Fig. 8). T. reesei Δtmk2 that has a weaker cell wall secretes more cellulase activities than the parent T. reesei TU-6 strain, while decreased cellulase activities were observed for T. reesei Δtmk2::OEtmk3 whose cell wall integrity is partially restored.
Fig. 8.
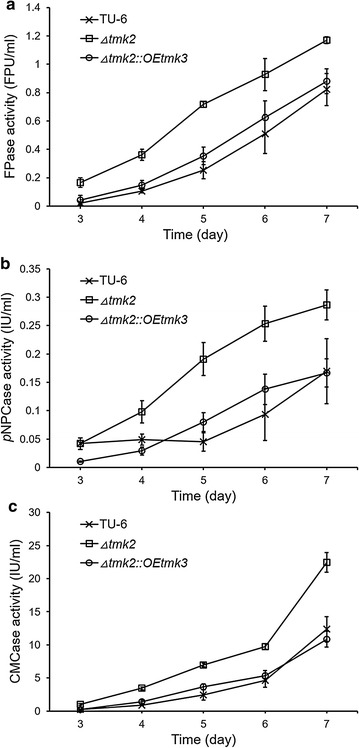
Production of cellulase by T. reesei TU-6, Δtmk2, and Δtmk2::OEtmk3 in shake flask experiments. a Production of filter paperase activity (FPase activity); b production of pNPCase activity; c production of CMCase activity. TU-6, T. reesei TU-6; Δtmk2, T. reesei Δtmk2; Δtmk2::OEtmk3, T. reesei Δtmk2::OEtmk3. Error bar standard deviation of three replicates
Discussion
The heterologous nature of T. reesei cells and the very complex mechanism of cellulase regulation have prevented us from fully elucidating the mechanisms for cellulase induction and repression by carbon sources. An example of the level of complexity of this regulation is the cross-regulation of transcription factors. As was reported previously with very delicate transcriptional analysis, full expression of the cellulase-inducing transcription factors Xyr1 and ACEII requires the presence of the cellulase repressing transcription factor Cre1 [61]. This seemingly aberrant phenomenon is actually a great testimony of how cellulase expression is fine-tuned by transcription factors in response to external signals. For this very reason, investigating the signal transduction pathways has the benefit of avoiding the complex cross-regulation between transcription factors, and can serve as an entry point for elucidating the complete cellulase induction and repression pathways, because the direct targets and upstream factors of a signal transduction pathway whose function is defined should participate in the same function.
Mitogen-activated protein kinase pathways are the best characterized signal transduction pathways in eukaryotes. Although the functions of many MAPK pathways are conserved in fungi, they also play specific roles for each species. Take Hog1-like pathways for instance, its role in high osmolarity resistance is universally conserved in fungi [24, 26], but the role in cellulase production is not reported in any fungal species except for T. reesei [15]. In Trichoderma species, the Fus3-like pathways are involved in mycoparasitism [49, 56–58]. The ability for mycoparasitism in T. reesei is, however, significantly reduced during evolution [62]. Since the function of mycoparasitism requires secretion of hydrolases such as chitinases and N-acetylglucosaminidases [49, 56, 57], we originally expected that during evolution, accompanying the reduction of mycoparasitism functions, T. reesei could use the Fus3-like Tmk1 to regulate the expression of cellulases which are also extracellular hydrolases. This, however, is not the case, and the role of Tmk1 on cellulase formation is limited to the growth rate-dependent production level, rather than signal transduction and transcription levels.
Both Tmk2 and Tmk3 pathways are involved in cell wall integrity maintenance in T. reesei [15, 59]. The primary cell wall integrity related MAPK conserved in fungi is the Slt2-like MAPK. The influence of Slt2-like Tmk2 on cellulase formation in T. reesei is found to be related to cell wall integrity in this work: Tmk2 maintains cell wall integrity, and negatively impacts cellulase secretion because of the intact cell wall. Indeed, the enhancement of protein secretion by damaging cell walls has been shown for other proteins [63], although reports on the relationship between cell wall integrity and cellulase secretion are rare. Suggestions can therefore be made that by hampering cell wall integrity, the secretion of cellulases is made easy because the enzymes now have an easier time penetrating cell wall, therefore leading to the promotion of cellulase formation.
Mechanistic investigations carried out in this and previous work lead to a suggestion that Tmk3 is the only MAPK that is involved in the transduction of cellulase-inducing signal [15, 59]. Recently, a phosphoproteomic investigation of T. reesei exposed to sophorose and distiller’s spent grain showed that Tmk3 is phosphorylated under these cellulase-inducing conditions [64]. What’s particularly interesting is that Tmk3 was also the only MAPK that is found to be phosphorylated when exposed to sophorose or distiller’s spent grain. These proteomic data are in full consistent with our finding, and supports our conclusion made in this work. We believe these findings could be a starting point on elucidating the transduction pathways of cellulase-inducing signals in T. reesei, with further work focused on finding out factors and pathways upstream and downstream of Tmk3.
Although Tmk1 and Tmk2 are not involved in signal transduction pathways leading to the induction or repression of cellulase expression, they can both potentially be used in engineering T. reesei for better cellulase titers because cellulase production is improved by deletion either gene even if growth was damaged for the case of tmk2. Conversely, although Tmk3 is involved in the transduction of cellulase induction signals, overexpressing this gene does not further improve transcription of cellulase-coding genes (Fig. 6), making it a bad target for engineering. This phenomenon gives Tmk1 and Tmk2 more practical/industrial values, although more focus should be given to Tmk3 on a microbial physiology perspective.
Conclusions
In conclusion, in this work, the role of MAPKs in T. reesei on cellulase formation was investigated. Two primary conclusions can be drawn from this work: Tmk1 is not involved in the regulation of cellulase-coding genes but represses cellulase formation by repressing cellular growth; Tmk2 is not involved in regulating the transcription of cellulase-coding genes either, but its role in maintaining cell wall integrity hampers cellulase secretion. The role of cell wall integrity in deterring cellulase formation, as strongly suggested by the hampered cellulase secretion in the cell wall integrity restored T. reesei Δtmk2::OEtmk3 strain, is a particularly interesting finding.
Results obtained from this and previous [15, 59] work lead to the answer to an important question: Which MAPK in T. reesei is involved in cellulase expression and production, and how? A full picture of the role of MAPKs on cellulase formation in T. reesei can be generated based on these investigations (Fig. 9). In this tentative model, Tmk3 stands out as the only MAPK involved in signal transduction pathways leading to the induction of cellulases, while Tmk1 and Tmk2 are both involved in repressing cellulase formation, although not by altering transcription of cellulase-coding genes. These investigations contribute to further understanding of the cellulase-regulating signal transduction processes in T. reesei, and contribute to the elucidation of cellulase regulation mechanisms in this key industrial cellulase-producing filamentous fungus. Furthermore and most importantly, findings from this work contribute to identifying appropriate targets for engineering T. reesei for better cellulase production, which is the key for biofuel production from lignocelluloics.
Fig. 9.
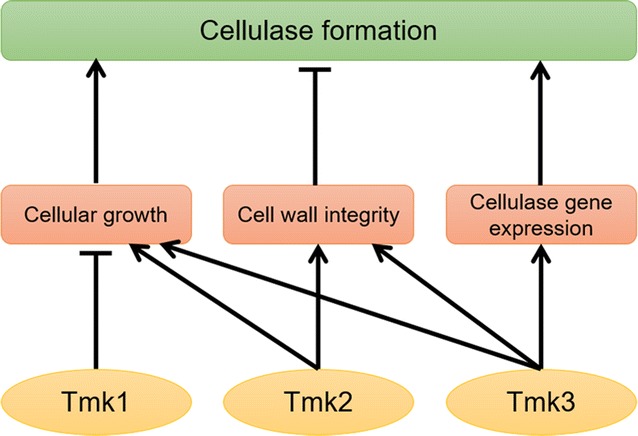
Involvement of T. reesei MAPKs in cellulase formation
Methods
Strains
The non-homologous end joining pathway-deficient, uridine auxotrophic defective T. reesei TU-6 (ATCC MYA-256) Δku70 strain was used as the parent strain in this work, and hereafter abbreviated as T. reesei TU-6 (or TU-6) strain. T. reesei Δtmk2 strain is the tmk2 deletion strain using T. reesei TU-6 as the parent strain [59].
Construction of T. reesei Δtmk1 and T. reesei Δtmk2::OEtmk3 strains
The construction of T. reesei Δtmk1 and T. reesei Δtmk2::OEtmk3 strains was carried out by homologous recombination essentially as previously described [65]. For construction of T. reesei Δtmk1, transformed DNA fragment contains 1.8 kb upstream homologous arm, pyr4 that restores uridine biosynthesis, and 1.8 kb downstream homologous arm (see Additional file 1). T. reesei TU-6 was used as the parent strain for tmk1 deletion. For construction of T. reesei Δtmk2::OEtmk3, transformed DNA fragment contains a 2 kb upstream homologous arm (3500 bp to 1500 bp upstream of tmk3), Pgpd from Aspergillus nidulans, ptrA gene from Aspergillus oryzae that confers pyrithiamine resistance and tmk3 sequence as the downstream homologous arm (2.2 kb) (see Additional file 3). T. reesei Δtmk2 was used as the parent strain for tmk3 overexpression. Two parallel tmk1 deletion strains T. reesei Δtmk1-1 and T. reesei Δtmk1-2 were generated.
Southern blotting analysis
Analysis of genotypes of T. reesei strains was carried out by Southern blotting following the previously published protocols [15]. For the analysis of T. reesei Δtmk1 genome, KpnI was used as the restriction digestion enzyme for genome digestion. For the analysis of T. reesei Δtmk2::OEtmk3 genome, HaeII was used as the restriction digestion enzyme for genome digestion.
Growth of T. reesei strains
Minimal media (MM) solution used for media preparation was prepared following the previously reported recipe [15]. For growth in shake flask in transcription abundance determination experiments, growth media contain MM solution, 2% Avicel, and 2% wheat bran. For growth in shake flask for genome extraction and Southern blotting, growth media contain MM solution and 2% glucose. For growth on plates, media contain MM solution, 2% agarose and 2% glucose, 2% lactose, or 2% glycerol. PDA plates contain 20% potato syrup, 2% agarose, and 2% sucrose. Double layer Avicel agar plates were prepared as previously described [15]. All plates and media contain 0.1% uridine. Congo Red (CR)- and Calcofluor White (CFW)-containing agar plates were prepared by adding CR or CFW to PDA plates. All cultures or plates were incubated at 30 °C for growth. Shake flask cultures were shaken at 200 rpm during cultivation. All parallel growth were carried out under the same illumination conditions.
For fermentation experiment, seed cultures were prepared by growing T. reesei strains in MM solution containing 2% Avicel and 2% wheat bran for 48 h (1 × 107 spore inoculation). Fermentation was carried out in 3-L fermenters (Model BLB10-3GJG, Bailun Bio-Technology Co., Ltd., Shanghai, China) containing 2 L of media (MM solution, 2% Avicel, 2% wheat bran). Two hundred milliliters of seed cultures were inoculated to initiate fermentation. Growth took place for 120 h at 30 °C. Agitation rate was adjusted to allow dissolved oxygen levels above 30%.
Phenotypic analysis
Phenotypic analysis of T. reesei Δtmk1 strains was carried out by inoculating 1 × 105 spores onto plates and grown for 3 days (6 days for double layer Avicel agar plates) prior to colony diameter determination. Three individual replicates were carried out.
Biochemical analysis
The analysis of A260, FPase (Filter Paperase) activities, pNPCase (p-nitrophenyl-β-d-cellobiosidase) activities, CMCase (carboxymethyl cellulase) activities, and protein concentration determination with the Lowry assay were carried out following the previously described protocols [15, 66, 67].
The correlation between A260 and dry mycelial weight was determined by first cultivating T. reesei TU-6 and T. reesei TU-6 Δtmk3 in potato dextrose broth (PDA minus agar) and MM media with glucose or lactose as the carbon sources in 3-L fermenters. For dry mycelial weight determination, approximately 11 mL of the cultures were extracted from fermenters, washed, filtered, and dried in an oven at 80 °C for over 12 h. The mycelia were subsequently weighed. The determination of A260 was carried out as previously described [15]. Four replicates were carried out.
CR and CFW resistance
For analysis of CR and CFW resistance, approximately 1 × 105 spores of T. reesei strains were inoculated on CR- and CFW-containing plates and were grown at 30 °C for 3 days prior to colony diameter determination. Three replicates were carried out.
qPCR analysis
For qPCR analysis, approximately 1 × 106 spores of T. reesei strains were inoculated to media containing 2% Avicel and 2% wheat bran and grown for 3 days prior to extraction of total RNA for transcription quantification. The qPCR procedures and data manipulation were carried out essentially as previously described [15]. Three biological replicates and three technical replicates for each biological replicate were carried out.
For determination of transcriptional abundance of cellulase-coding genes at different time points after inoculation to media containing Avicel + wheat bran, spores were first inoculated to MM media +2% glucose and grew for 2 days, after which mycelia were extracted, washed, and inoculated to MM media +2% glycerol for growth for 1 day. The mycelia were further washed and inoculated to MM media containing 2% wheat bran and 2% Avicel. Cells were extracted for transcriptional analysis upon transfer, as well as 4, 12, and 24 h after the transfer.
Statistics
To test if two sets of data are statistically different, two-tailed Student’s t-tests were carried out. p < 0.05 was considered statistically significant. A minimum of three replicates were carried out for each experiment.
Additional files
Additional file 1. Southern blotting analysis of T. reesei TU-6, Δtmk1-1 and Δtmk1-2. Panel A: Schematic drawing of strain construction and southern blotting analysis. Panel B: southern blotting of T. reesei strains. M: DNA marker; TU-6: T. reesei TU-6; Δtmk1-1: T. reesei Δtmk1-1; Δtmk1-2: T. reesei Δtmk1-2.
Additional file 2. The sensitivity of T. reesei TU-6, Δtmk1-1 and Δtmk1-2 to CR and CFW. Panel A: growth of T. reesei TU-6, T. reesei Δtmk1-1 and T. reesei Δtmk1-2 on CR-containing agar plates; Panel B: growth of T. reesei TU-6, T. reesei Δtmk1-1 and T. reesei Δtmk1-2 on CFW-containing agar plates. TU-6, T. reesei TU-6; Δtmk1-1, T. reesei Δtmk1-1; Δtmk1-2, T. reesei Δtmk1-2.
Additional file 3. Southern blotting analysis of T. reesei TU-6, Δtmk2 and Δtmk2::OEtmk3. Panel A: Schematic drawing of strain construction and southern blotting analysis. Panel B: southern blotting of T. reesei strains. M: DNA marker; Δtmk2: T. reesei Δtmk2; Δtmk2::OEtmk3: T. reesei Δtmk2::OEtmk3.
Authors’ contributions
MZ, LL, YD, YJ, KL, RZ, BJ, and KN performed the experiments. MW and XF wrote the manuscript. MW, MZ, LL, YD, and XF are involved in analysis and interpretation of experimental data. MW and XF conceived of the study. XF coordinated the project. All authors read and approved the final manuscript.
Acknowledgements
We thank Ms. Shaoli Hou for technical assistance during fermentation.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Funding
This work was supported by the National Key Technology R&D Program of China (No. 2014AA021903), Shandong Scientific and Technology Project (No. 2015ZDXX0403A01), National Natural Science Foundation (No. 31570040), Shandong Province Natural Science Foundation (No. ZR2013CM041), and the 111 Project (No. B16030).
The funders had no role in study design, data collection, analysis and interpretation, or writing of the manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mingyu Wang, Email: wangmingyu@sdu.edu.cn.
Meiling Zhang, Email: 893150816@qq.com.
Ling Li, Email: 893587738@qq.com.
Yanmei Dong, Email: 1379127157@qq.com.
Yi Jiang, Email: 250639559@qq.com.
Kuimei Liu, Email: 786335521@qq.com.
Ruiqin Zhang, Email: 2455645036@qq.com.
Baojie Jiang, Email: 313376858@qq.com.
Kangle Niu, Email: 271314114@qq.com.
Xu Fang, Email: fangxu@sdu.edu.cn.
References
- 1.Wang M, Li Z, Fang X, Wang L, Qu Y. Cellulolytic enzyme production and enzymatic hydrolysis for second-generation bioethanol production. Adv Biochem Eng Biotechnol. 2012;128:1–21. doi: 10.1007/10_2011_131. [DOI] [PubMed] [Google Scholar]
- 2.Peterson R, Nevalainen H. Trichoderma reesei RUT-C30 - thirty years of strain improvement. Microbiology. 2012;158:58–68. doi: 10.1099/mic.0.054031-0. [DOI] [PubMed] [Google Scholar]
- 3.Gusakov AV. Alternatives to Trichoderma reesei in biofuel production. Trends Biotechnol. 2011;29:419–425. doi: 10.1016/j.tibtech.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Kubicek CP. Systems biological approaches towards understanding cellulase production by Trichoderma reesei. J Biotechnol. 2013;163:133–142. doi: 10.1016/j.jbiotec.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang F, Liu K, Han L, Jiang B, Wang M, Fang X. Function of a p24 heterodimer in morphogenesis and protein transport in Penicillium oxalicum. Sci Rep. 2015;5:11875. doi: 10.1038/srep11875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sternberg D, Mandels GR. Induction of cellulolytic enzymes in Trichoderma reesei by sophorose. J Bacteriol. 1979;139:761–769. doi: 10.1128/jb.139.3.761-769.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Messner R, Kubicek CP. Carbon source control of cellobiohydrolase I and II formation by Trichoderma reesei. Appl Env Microbiol. 1991;57:630–635. doi: 10.1128/aem.57.3.630-635.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ilmén M, Saloheimo A, Onnela ML, Penttilä ME. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl Env Microbiol. 1997;63:1298–1306. doi: 10.1128/aem.63.4.1298-1306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tisch D, Schmoll M. Targets of light signalling in Trichoderma reesei. BMC Genom. 2013;14:657. doi: 10.1186/1471-2164-14-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aro N, Ilmén M, Saloheimo A, Penttilä M. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl Env Microbiol. 2003;69:56–65. doi: 10.1128/AEM.69.1.56-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antoniêto ACC, dos Santos Castro L, Silva-Rocha R, Persinoti GF, Silva RN. Defining the genome-wide role of CRE1 during carbon catabolite repression in Trichoderma reesei using RNA-Seq analysis. Fungal Genet Biol. 2014;73:93–103. doi: 10.1016/j.fgb.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Aro N, Saloheimo A, Ilmén M, Penttilä M. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J Biol Chem. 2001;276:24309–24314. doi: 10.1074/jbc.M003624200. [DOI] [PubMed] [Google Scholar]
- 13.Stricker AR, Grosstessner-Hain K, Würleitner E, Mach RL. Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and D-xylose metabolism in Hypocrea jecorina. Eukaryot Cell. 2006;5:2128–2137. doi: 10.1128/EC.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuster A, Tisch D, Seidl-Seiboth V, Kubicek CP, Schmoll M. Roles of protein kinase A and adenylate cyclase in light-modulated cellulase regulation in Trichoderma reesei. Appl Env Microbiol. 2012;78:2168–2178. doi: 10.1128/AEM.06959-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M, Zhao Q, Yang J, Jiang B, Wang F, Liu K, et al. A mitogen-activated protein kinase Tmk3 participates in high osmolarity resistance, cell wall integrity maintenance and cellulase production regulation in Trichoderma reesei. PLoS ONE. 2013;8:e72189. doi: 10.1371/journal.pone.0072189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cziferszky A, Mach RL, Kubicek CP. Phosphorylation positively regulates DNA bindign of the carbon catabolite repressor Cre1 of Hypocrea jecorina (Trichoderma reesei) J Biol Chem. 2002;277:14688–14694. doi: 10.1074/jbc.M200744200. [DOI] [PubMed] [Google Scholar]
- 17.Lv X, Zhang W, Chen G, Liu W. Trichoderma reesei Sch9 and Yak1 regulate vegetative growth, conidiation, and stress response and induced cellulase production. J Microbiol. 2015;53:236–242. doi: 10.1007/s12275-015-4639-x. [DOI] [PubMed] [Google Scholar]
- 18.Chen F, Chen XZ, Su XY, Qin LN, Huang ZB, Tao Y, et al. An Ime2-like mitogen-activated protein kinase is involved in cellulase expression in the filamentous fungus Trichoderma reesei. Biotechnol Lett. 2015;37:2055–2062. doi: 10.1007/s10529-015-1888-z. [DOI] [PubMed] [Google Scholar]
- 19.Schmoll M. The information highways of a biotechnological workhorse-signal transduction in Hypocrea jecorina. BMC Genom. 2008;9:430. doi: 10.1186/1471-2164-9-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gustin MC, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamel LP, Nicole MC, Duplessis S, Ellis BE. Mitogen-activated protein kinase signaling in plant-interacting fungi: distinct messages from conserved messengers. Plant Cell. 2012;24:1327–1351. doi: 10.1105/tpc.112.096156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Errede B, Vered L, Ford E, Pena MI, Elston TC. Pheromone-induced morphogenesis and gradient tracking are dependent on the MAPK Fus3 binding to Gα. Mol Biol Cell. 2015;26:3343–3358. doi: 10.1091/mbc.E15-03-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dixon KP, Xu JR, Smirnoff N, Talbot NJ. Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell. 1999;11:2045–2058. doi: 10.1105/tpc.11.10.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han KH, Prade RA. Osmotic stress-coupled maintenance of polar growth in Aspergillus nidulans. Mol Microbiol. 2002;43:1065–1078. doi: 10.1046/j.1365-2958.2002.02774.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Lamm R, Pillonel C, Lam S, Xu JR. Osmoregulation and fungicide resistance: the Neurospora crassa os-2 gene encodes a HOG1 mitogen-activated protein kinase homologue. Appl Environ Microbiol. 2002;68:532–538. doi: 10.1128/AEM.68.2.532-538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park SM, Choi ES, Kim MJ, Cha BJ, Yang MS, Kim DH. Characterization of HOG1 homologue, CpMK1, from Cryphonectria parasitica and evidence for hypovirus-mediated perturbation of its phosphorylation in response to hypertonic stress. Mol Microbiol. 2004;51:1267–1277. doi: 10.1111/j.1365-2958.2004.03919.x. [DOI] [PubMed] [Google Scholar]
- 28.Xue T, Nguyen CK, Romans A, May GS. A mitogen-activated protein kinase that senses nitrogen regulates conidial germination and growth in Aspergillus fumigatus. Eukaryot Cell. 2004;3:557–560. doi: 10.1128/EC.3.2.557-560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delgado-Jarana J, Sousa S, González F, Rey M, Llobell A. ThHog1 controls the hyperosmotic stress response in Trichoderma harzianum. Microbiology. 2006;152:1687–1700. doi: 10.1099/mic.0.28729-0. [DOI] [PubMed] [Google Scholar]
- 30.Ji Y, Yang F, Ma D, Zhang J, Wan Z, Liu W, et al. HOG-MAPK signaling regulates the adaptive responses of Aspergillus fumigatus to thermal stress and other related stress. Mycopathologia. 2012;174:273–282. doi: 10.1007/s11046-012-9557-4. [DOI] [PubMed] [Google Scholar]
- 31.Xu JR, Staiger CJ, Hamer JE. Inactivation of the mitogen-activated protein kianse Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc Natl Acad Sci USA. 1998;95:12713–12718. doi: 10.1073/pnas.95.21.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou Z, Xue C, Peng Y, Katan T, Kistler HC, Xu JR. A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol Plant Microbe Interact. 2002;15:1119–1127. doi: 10.1094/MPMI.2002.15.11.1119. [DOI] [PubMed] [Google Scholar]
- 33.Kojima K, Kikuchi T, Takano Y, Oshiro E, Okuno T. The mitogen-activated protein kinase gene MAF1 is essential for the early differentiation phase of appressorium formation in Collectotrichum lagenarium. Mol Plant Microbe Interact. 2002;15:1268–1276. doi: 10.1094/MPMI.2002.15.12.1268. [DOI] [PubMed] [Google Scholar]
- 34.Mey G, Held K, Scheffer J, Tenberge KB, Tudzynski P. CPMK2, an SLT2-homologous mitogen-activated protein (MAP) kinase, is essential for pathogenesis of Claviceps purpurea on rye: evidence for a second conserved pathogenesis-related MAP kinase cascade in phytopathogenic fungi. Mol Microbiol. 2002;46:305–318. doi: 10.1046/j.1365-2958.2002.03133.x. [DOI] [PubMed] [Google Scholar]
- 35.Mehrabi R, van der Lee T, Waalwijk C, Kema GHJ. MgSlt2, a cellular integrity MAP kinase gene of the fungal wheat pathogen Mycosphaerella graminicola, is dispensable for penetration but essential for invasive growth. Mol Plant Microbe Interact. 2006;19:389–398. doi: 10.1094/MPMI-19-0389. [DOI] [PubMed] [Google Scholar]
- 36.Rui O, Hahn M. The Slt2-type MAP kinase Bmp3 of Botrytis cinerea is required for normal saprotrophic growth, conidiation, plant surface sensing and host tissue colonization. Mol Plant Pathol. 2007;8:173–184. doi: 10.1111/j.1364-3703.2007.00383.x. [DOI] [PubMed] [Google Scholar]
- 37.Park G, Pan S, Borkovich KA. Mitogen-activated protein kinase cascade required for regulation of development and secondary metabolism in Neurospora crassa. Eukaryot Cell. 2008;7:2113–2122. doi: 10.1128/EC.00466-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valiante V, Heinekamp T, Jain R, Härtl A, Brakhage AA. The mitogen-activated protein kinase MpkA of Aspergillus fumigatus regulates cell wall signaling and oxidative stress response. Fungal Genet Biol. 2008;45:618–627. doi: 10.1016/j.fgb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 39.Kumar A, Scher K, Mukherjee M, Pardovitz-Kedmi E, Sible GV, Singh U, et al. Overlapping and distinct functions of two Trichoderma virens MAP kinases in cell-wall integrity, antagonistic properties and repression of conidiation. Biochem Biophys Res Commun. 2010;398:765–770. doi: 10.1016/j.bbrc.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 40.Jain R, Valiante V, Remme N, Docimo T, Heinekamp T, Hertweck C, et al. The MAP kinase MpkA controls cell wall integrity, oxidative stress response, gliotoxin production and iron adaptation in Aspergillus fumigatus. Mol Microbiol. 2011;82:39–53. doi: 10.1111/j.1365-2958.2011.07778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo X, Keyhani NO, Yu X, He Z, Luo Z, Pei Y, et al. The MAP kinase Bbslt2 controls growth, conidiation, cell wall integrity, and virulence in the insect pathogenic fungus Beauveria bassiana. Fungal Genet Biol. 2012;49:544–555. doi: 10.1016/j.fgb.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 42.Zeng F, Gong X, Hamid MI, Fu Y, Jiatao X, Cheng J, et al. A fungal cell wall integrity-associated MAP kinase cascade in Coniothyrium minitans is required for conidiation and mycoparasitism. Fungal Genet Biol. 2012;49:347–357. doi: 10.1016/j.fgb.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 43.Bussink HJ, Osmani SA. A mitogen-activated protein kinase (MPKA) is involved in polarized growth in the filamentous fungus, Aspergillus nidulans. FEMS Microbiol Lett. 1999;173:117–125. doi: 10.1111/j.1574-6968.1999.tb13492.x. [DOI] [PubMed] [Google Scholar]
- 44.Bennett LD, Beremand P, Thomas TL, Bell-Pedersen D. Circadian activation of the mitogen-activated protein kinase MAK-1 facilitates rhythms in clock-controlled genes in Neurospora crassa. Eukaryot Cell. 2013;12:59–69. doi: 10.1128/EC.00207-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu JR, Hamer JE. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Gene Dev. 1996;10:2696–2706. doi: 10.1101/gad.10.21.2696. [DOI] [PubMed] [Google Scholar]
- 46.Zheng L, Campbell M, Murphy J, Lam S, Xu JR. The BMP1 gene is essential for pathogenicity in the gray mold fungus Botrytis cinerea. Mol Plant Microbe Interact. 2000;13:724–732. doi: 10.1094/MPMI.2000.13.7.724. [DOI] [PubMed] [Google Scholar]
- 47.Pietro AD, García-Maceira FI, Méglecz E, Roncero MIG. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol Microbiol. 2001;39:1140–1152. doi: 10.1111/j.1365-2958.2001.02307.x. [DOI] [PubMed] [Google Scholar]
- 48.Ruiz-Roldán MC, Maier FJ, Schafer W. PTK1, a mitogen-activated-protein kinase gene, is required for conidiation, appressorium formation, and pathogenicity of Pyrenophora teres on barley. Mol Plant Microbe Interact. 2001;14:116–125. doi: 10.1094/MPMI.2001.14.2.116. [DOI] [PubMed] [Google Scholar]
- 49.Mukherjee PK, Latha J, Hadar R, Horwitz BA. TmkA, a mitogen-activated protein kinase of Trichoderma virens, is involved in biocontrol properties and repression of conidiation in the dark. Eukaryot Cell. 2003;2:446–455. doi: 10.1128/EC.2.3.446-455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin K, Han L, Xia Y. MaMk1, a FUS3/KSS1-type mitogen-activated protein kinase gene, is required for appressorium formation, and insect cuticle penetration of the entomopathogenic fungus Metarhizium acridum. J Invertebr Pathol. 2014;114:68–75. doi: 10.1016/j.jip.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 51.Leng Y, Zhong S. The role of mitogen-activated protein (MAP) kinase signaling components in the fungal development, stress response and virulence of the fungal cereal pathogen Bipolaris sorokiniana. PLoS ONE. 2015;10:e0128291. doi: 10.1371/journal.pone.0128291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pandey A, Roca MG, Read ND, Glass NL. Role of a mitogen-activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa. Eukaryot Cell. 2004;3:348–358. doi: 10.1128/EC.3.2.348-358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosales RCP, Pietro AD. Vegetative hyphal fusion is not essential for plant infection by Fusarium oxysporum. Eukaryot Cell. 2008;7:162–171. doi: 10.1128/EC.00258-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fleissner A, Leeder AC, Roca MG, Read ND, Glass NL. Oscillatory recruitment of signaling proteins to cell tips promotes coordinated behavior during cell fusion. Proc Natl Acad Sci USA. 2009;106:19387–19392. doi: 10.1073/pnas.0907039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Priegnitz BE, Brandt U, Pahirulzaman KA, Dickschat JS, Fleiβner A. The AngFus3 mitogen-activated protein kinase controls hyphal differentiation and secondary metabolism in Aspergillus niger. Eukaryot Cell. 2015;14:602–615. doi: 10.1128/EC.00018-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mendoza-Mendoza A, Pozo MJ, Grzegorski D, Martínez P, García JM, Olmedo-Monfil V, et al. Enhanced biocontrol activity of Trichoderma through inactivation of a mitogen-activated protein kinase. Proc Natl Acad Sci USA. 2003;100:15965–15970. doi: 10.1073/pnas.2136716100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reithner B, Schuhmacher R, Stoppacher N, Pucher M, Brunner K, Zeilinger S. Signaling via the Trichoderma atroviride mitogen-activated protein kinase Tmk1 differentially affects mycoparasitism and plant protection. Fungal Genet Biol. 2007;44:1123–1133. doi: 10.1016/j.fgb.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gruber S, Zeilinger S. The transcription factor Ste12 mediates the regulatory role of the Tmk1 MAP kinase in mycoparasitism and vegetative hyphal fusion in the filamentous fungus Trichoderma atroviride. PLoS ONE. 2014;9:e111636. doi: 10.1371/journal.pone.0111636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang M, Dong Y, Zhao Q, Wang F, Liu K, Jiang B, et al. Identification of the role of a MAP kinase Tmk2 in Hypocrea jecorina (Trichoderma reesei) Sci Rep. 2014;4:6732. doi: 10.1038/srep06732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morikawa Y, Kawamori M, Ado Y, Shinsha Y, Oda F, Takasawa S. Improvement of cellulase production in Trichoderma reesei. Agric Biol Chem. 1985;49:1869–1871. doi: 10.1080/00021369.1985.10866993. [DOI] [Google Scholar]
- 61.Portnoy T, Margeot A, Seidl-Seiboth V, Le Crom S, Ben Chaabane F, Linke R, et al. Differential regulation of the cellulase transcription factors XYR1, ACE2 and ACE1 in high and low cellulase producing strains of Trichoderma reesei. Eukaryot Cell. 2010;10:262–271. doi: 10.1128/EC.00208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kubicek CP, Herrera-Estrella A, Seidl-Seiboth V, Martinez DA, Druzhinina IS, Thon M, et al. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 2011;12:R40. doi: 10.1186/gb-2011-12-4-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang H, Wang S, Wang J, Song M, Xu M, Zhang M, et al. N-hypermannose glycosylation disruption enhances recombinant protein production by regulating secretory pathway and cell wall integrity in Saccharomyces cerevisiae. Sci Rep. 2016;6:25654. doi: 10.1038/srep25654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nguyen EV, Imanishi SY, Haapaniemi P, Yadav A, Saloheimo M, Corthals GL, et al. Quantitative site-specific phosphoproteomics of Trichoderma reesei signaling pathways upon induction of hydrolytic enzyme production. J Proteome Res. 2016;15:457–467. doi: 10.1021/acs.jproteome.5b00796. [DOI] [PubMed] [Google Scholar]
- 65.Zhang G, Hartl L, Schuster A, Polak S, Schmoll M, Wang T, et al. Gene targeting in a nonhomologous end joining deficient Hypocrea jecorina. J Biotechnol. 2009;139:146–151. doi: 10.1016/j.jbiotec.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 66.Wood TM, Bhat KM. Methods for measuring cellulase activities. Methods Enzym. 1988;160:87–112. doi: 10.1016/0076-6879(88)60109-1. [DOI] [Google Scholar]
- 67.Hideno A, Inoue H, Tsukahara K, Yano S, Fang X, Endo T, et al. Production and characterization of cellulases and hemicellulases by Acremonium cellulolyticus using rice straw subjected to various pretreatments as the carbon source. Enzyme Microb Technol. 2011;48:162–168. doi: 10.1016/j.enzmictec.2010.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Southern blotting analysis of T. reesei TU-6, Δtmk1-1 and Δtmk1-2. Panel A: Schematic drawing of strain construction and southern blotting analysis. Panel B: southern blotting of T. reesei strains. M: DNA marker; TU-6: T. reesei TU-6; Δtmk1-1: T. reesei Δtmk1-1; Δtmk1-2: T. reesei Δtmk1-2.
Additional file 2. The sensitivity of T. reesei TU-6, Δtmk1-1 and Δtmk1-2 to CR and CFW. Panel A: growth of T. reesei TU-6, T. reesei Δtmk1-1 and T. reesei Δtmk1-2 on CR-containing agar plates; Panel B: growth of T. reesei TU-6, T. reesei Δtmk1-1 and T. reesei Δtmk1-2 on CFW-containing agar plates. TU-6, T. reesei TU-6; Δtmk1-1, T. reesei Δtmk1-1; Δtmk1-2, T. reesei Δtmk1-2.
Additional file 3. Southern blotting analysis of T. reesei TU-6, Δtmk2 and Δtmk2::OEtmk3. Panel A: Schematic drawing of strain construction and southern blotting analysis. Panel B: southern blotting of T. reesei strains. M: DNA marker; Δtmk2: T. reesei Δtmk2; Δtmk2::OEtmk3: T. reesei Δtmk2::OEtmk3.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.


