Abstract
Background
Pancreatic ductal adenocarcinomas are among the most malignant neoplasms and have very poor prognosis. Our understanding of various cancers has recently improved the survival of patients with cancer, except for pancreatic cancers. Establishment of primary cancer cell lines of pancreatic ductal adenocarcinomas will be useful for understanding the molecular mechanisms of this disease.
Methods
Eighty-one surgically resected pancreatic ductal adenocarcinomas were collected. Six novel pancreatic cancer cell lines, AMCPAC01–06, were established and histogenetic characteristics were compared with their matched tissues. The clinicopathologic and molecular characteristics of the cell lines were investigated by KRAS and TP53 sequencing or SMAD4 and p53 immunohistochemistry. Xenografts using AMCPAC cell lines were established.
Results
From the 81 pancreatic ductal adenocarcinomas, six (7.4% success rate) patient-derived primary cell lines were established. The six AMCPAC cell lines showed various morphologies and exhibited a wide range of doubling times. AMCPAC cell lines contained mutant KRAS in codons 12, 13, or 61 and TP53 in exon 5 as well as showed aberrant p53 (5 overexpression and 1 total loss) or DPC4 (all 6 intact) expression. AMCPAC cell lines demonstrated homology for the KRAS mutation and p53 expression compared with matched primary cancer tissues, but showed heterogeneous DPC4 expression patterns.
Conclusions
The novel AMCPAC01–06 cell lines established in this study may contribute to the understanding of pancreatic ductal adenocarcinomas.
Trial registration Retrospectively registered
Electronic supplementary material
The online version of this article (doi:10.1186/s12935-017-0416-8) contains supplementary material, which is available to authorized users.
Keywords: Pancreas, Cancer, Ductal adenocarcinoma, Primary, Cell line
Background
Ductal adenocarcinomas are the most common malignant neoplasms of the pancreas and account for approximately 85–90% of malignant neoplasms arising from the pancreas (known as pancreatic cancers). Pancreatic cancers are the ninth most common cancer in Korea [1], and the 5-year survival rate of patients with this type of cancer is only 9% [1]. Surgical resection is the main stay for treating pancreatic cancer, but most patients are inoperable at the time of diagnosis, and only 30% of patients can undergo surgical resection [2]. Approximately 80% of patients show local recurrence or distant metastasis after surgical removal of tumors [2]. Other treatments, such as chemotherapies, are required to improve the survival time of patients with pancreatic cancer [3–6]. However, currently used chemotherapeutic regimens fail to significantly improve the survival time of these patients [7–9]. Therefore, development of new therapeutic modalities is important for improving the survival time of pancreatic cancer patients.
Recently, several trials were conducted to examine customized cancer treatment using patient-derived pancreatic cancer tissues [10–13]. However, using surgically resected pancreatic cancer tissue is difficult because of the limited amount of residual pancreatic cancer tissues after the submission of large amount of cancer tissues for pathologic examination for precise diagnosis and staging. To overcome this limitation, researchers have attempted to establish cancer cell lines from patient-derived cancer tissues and use various molecular pathologic studies with these cancer cell lines for tailored patient treatments. However, it is difficult to establish patient-derived primary cancer cell lines from pancreatic cancers because of specific histopathologic characteristics of pancreatic cancer such as low cancer cellularity of pancreatic cancer, and the occurrence of extensive desmoplastic reactions by overproduction of cancer-associated fibroblasts. Thus, the number of established patient-derived pancreatic cancer cell lines is currently much lower than that of cancers from other organs. At present, 21 pancreatic cancer cell lines have been established, including 11 cell lines from American Type Culture Collection and 3 cell lines form Korean Cell Line Bank [14–16].
In addition, some different clinicopathologic features were observed in pancreatic cancer patients of different ethnicities. Therefore, establishing novel primary pancreatic cancer cell lines derived from Korean pancreatic cancer patients and applying various chemotherapeutic regimens to the developed primary pancreatic cancer cell lines can help in the determination of highly sensitive chemotherapeutic regimens for individual pancreatic cancer patients, particularly when regional recurrence or distant metastasis develops.
Methods
Specimen collection
After approval (2015-0480) from the institutional review board, fresh tumor tissues measuring 0.5 × 0.5 × 0.2 cm3 in size were obtained from 81 surgically resected pancreatic ductal adenocarcinomas, immediately soaked in RPMI640 media (Sigma-Aldrich Corp., St. Louis, MO, USA), and transferred in a laminar flow biosafety cabinet.
In vitro cell culture
Pancreatic tumor tissues were rinsed with Hank’s balanced salt solution in clean bench, minced into <2 mm3 fragments, and digested with 0.1% (W/V) of collagenase type 1 (GIBCO, Grand Island, NY, USA) at 37 °C for 20 min. The resulting fragments were centrifuged at 200×g for 5 min, washed thrice with phosphate-buffered saline, plated onto RPMI1640 media (GIBCO) containing 10% fetal bovine serum (GIBCO) and 1% penicillin/streptomycin (GIBCO), and allowed to adhere. After incubation for several days, mixed growth of cancer cells and fibroblasts was observed in the tissue fragments. To overcome fibroblast overgrowth, periodic trypsinization was conducted by incubation with 0.005% trypsin/EDTA (GIBCO) at 37 °C for 3 min during 2–3 passages to remove fibroblasts, and unwanted fibroblasts were detached by pipetting. The primary cell culture was monitored with a phase-contrast microscope. Cancer cells were grown at 37 °C in a humidified atmosphere with 5% CO2.
Growth rate analysis of established cell lines
The cell growth rate was measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma-Aldrich) at 24-h intervals. After 1 × 104 cells were seeded into 96-well plates, 0.5 mg/mL MTT was added over consecutive days for violet pellet formation by living cells. The pellets were solubilized in 200 μL of dimethyl sulfoxide. The optical density of each sample was measured at 570 nm using a microplate reader (Sunrise Reader, Tecan, Männedorf, Switzerland). Growth rate was measured as a percentage of control growth. Cells from passage 15 were used to determine population doubling time, and all experiments were repeated twice in triplicate.
Characterization of cell lines
Construction cell microarray
After fixation of 5 × 106 cancer cells with a Cytorich Red fixative solution (BD Biosciences, Franklin Lakes, NJ, USA) for 48 h, the supernatant was removed after centrifugation. The pellets were additionally fixed with 95% ethanol for 60 min then embedded in paraffin. Each cancer cell block was selected as a donor, and the designated areas for each cell block were punched with a 5-mm diameter cylinder by a Manual Tissue Microarrayer (Uni TMA Co., Ltd., Seoul, Korea) and transferred to a recipient block, and cell microarrays (CMAs) were constructed.
Immunohistochemistry
Immunohistochemical labeling was performed by the immunohistochemical laboratory of the Department of Pathology, Asan Medical Center. Briefly, 4-μm tissue sections from the CMA and matched formalin-fixed paraffin-embedded (FFPE) primary cancer tissues of ductal adenocarcinomas were deparaffinized and hydrated in xylene and serially diluted with ethanol, respectively. Endogenous peroxidase was blocked by incubation in 3% H2O2 for 10 min, and heat-induced antigen retrieval was performed. Primary antibodies with Benchmark autostainer (Ventana Medical Systems, Tucson, AZ, USA) were used as per the manufacturer’s protocol. Primary antibodies for cytokeratin 19 (clone A53-B/A2.26; 1:200; Cell Marque, CA, USA), p53 (clone DO-7; 1:3000; DAKO, Glostrup, Denmark), and DPC4 (clone EP618Y, 1:100; GeneTex, Irvine, CA, USA) were incubated at room temperature for 32 min, and the sections were labeled with an automated immunostaining system with the I-View detection kit (Benchmark XT; Ventana Medical Systems). Immunostained sections were lightly counterstained with hematoxylin, dehydrated in ethanol, and cleared in xylene.
Detection of TP53 and KRAS mutations
The genomic DNA of the established cell lines was extracted using the QIAamp DNA Micro kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Polymerase chain reaction (PCR) amplification was performed with 10 ng of DNA covering exons 5–8 of the TP53 gene with intragenic primers flanking these exons as previously described [17]. PCR-amplified products were purified using a QIAquick column (Qiagen). TP53 gene sequencing was performed with BigDye 3.1 and a 3730xl DNA analyzer (Applied Biosystems, Foster City, CA, USA).
Similarly, pyrosequencing was performed to detect KRAS at codons 12, 13, and 61. Primer sequences of KRAS are summarized in Table 1. DNA (10 ng) was amplified using a biotin-labeled primer covering codons 12, 13, and 61 of KRAS. The biotin-labeled PCR products were immobilized on Streptavidin Sepharose HP beads (GE Healthcare, Little Chalfont, UK) and the immobilized PCR products were sequenced using a pyrosequencer, PyroMark Q96MD System (Qiagen) according to the manufacturer’s protocol. Primer sequences used to amplify TP53 and KRAS are shown in Table 1.
Table 1.
Primer sequences and PCR conditions of TP53 and KRAS
| Target | Forward primer | Reverse primer | AT (°C) | Size (bp) |
|---|---|---|---|---|
| TP53 exon 5 | 5′-CACTTGTGCCCTGACTTTCA-3′ | 5′-AACCAGCCCTGTCGTCTCT-3′ | 64 | 267 |
| TP53 exon 6 | 5′-CAGGCCTCTGATTCCTCACT-3′ | 5′-CTTAACCCCTCCTCCCAGAG-3′ | 64 | 185 |
| TP53 exon 7 | 5′-CCACAGGTCTCCCCAAGG-3′ | 5′-CCAGGTCAGGAGCCACTT-3′ | 64 | 179 |
| TP53 exon 8 | 5′-GCCTCTTGCTTCTCTTTTCC-3′ | 5′-TAACTGCACCCTTGGTCTCC-3′ | 62 | 217 |
| KRAS 12 and 13 | 5′-GGTGAGTTTGTATTAAAAGGTACTGG-3′ | 5′-Biotin-GCTGTATCGTCAAGGCACTCTT-3′ | 56 | 100 |
| KRAS 61 | 5′-TGGAGAAACCTGTCTCTTGGATAT-3′ | 5′-Biotin-TACTGGTCCCTCATTGCA CTGTA-3′ | 60 | 72 |
AT annealing temperature
In vivo tumorigenicity by tumor xenograft
Tumorigenicity in mice was confirmed by subcutaneous injection of approximately 2 × 106 cancer cells from culture bilaterally into each flank of male NSG (5 weeks old) mice. Xenograft tumor growth was monitored twice per week for 3 months. Tumor volume (TV) was calculated according to the formula: TV (mm3) = length × width2 × 0.5. When a tumor size of 100–200 mm3 was reached, tumors were explanted from the mice. The explanted tumor xenografts were used for reimplantation and FFPE to confirm the histology.
Statistical analysis
The significance of differences between experimental conditions was determined using the student’s t test and Mann–Whitney U test for unpaired observations.
Results
Establishment of pancreatic cancer cell lines
Fresh tissues from 81 patients diagnosed with pancreatic ductal adenocarcinomas were used to establish primary cancer cell lines. Among the 81 cases, 75 cases (93%) failed to establish cell lines because of fungal or mycoplasmal contamination in 30 cases (37%) and overgrowth of cancer-associated fibroblasts in 45 cases (56%). Six (7.4%) patient-derived pancreatic cancer cell lines were established and named, AMCPAC01–AMCPAC06. The first sub-culture was performed 7–15 days after initiating primary culture for attachment and spreading, and the AMCPAC cell lines were passed a minimum of 10 times.
Clinicopathologic characteristics of cancer cell lines
Clinicopathologic characteristics of the established cancer cell lines are summarized in Table 2. Briefly, cell lines were derived from five males and one female. The mean age of the patients was 52.0 ± 12.1 years (range 30–67 years). The pathologic diagnosis of all six cases was moderately differentiated ductal adenocarcinoma (Fig. 1). Tumor locations were as follows: tail of the pancreas in four cases, body in 1 case, and head in 1 case. The mean tumor size was 3.6 ± 1.8 cm (range 2.1–6.9 cm). Five cases showed peripancreatic soft tissue extension, while one case invaded the surrounding organs, spleen, and transverse colon (AMCPAC04). Lymphovascular invasions were identified in 3 cases (AMCPAC02, AMCPAC04, and AMCPAC05) and perineural invasions were observed in 5 cases, except for case #6. Lymph node metastases were detected in 3 cases (AMCPAC03, AMCPAC04, and AMCPAC06).
Table 2.
Clinicopathologic characteristics of established cancer cell lines
| Clinicopathologic factors | AMCPAC01 | AMCPAC02 | AMCPAC03 | AMCPAC04 | AMCPAC05 | AMCPAC06 |
|---|---|---|---|---|---|---|
| Age (years) | 30 | 67 | 56 | 53 | 52 | 54 |
| Sex | Male | Female | Male | Male | Male | Male |
| Operation name | DP | DP | DP | DP | PPPD | DP |
| Pathologic diagnosis | Ductal adenocarcinoma | Ductal adenocarcinoma | Ductal adenocarcinoma | Ductal adenocarcinoma | Ductal adenocarcinoma | Ductal adenocarcinoma |
| Differentiation | MD | MD | MD | MD | MD | MD |
| Location | Body | Tail | Tail | Tail | Head | Tail |
| Tumor size (cm) | 2.1 | 2.5 | 4.5 | 6.9 | 2.8 | 2.9 |
| pT classification | pT3 | pT3 | pT3 | pT4 | pT3 | pT3 |
| pN classification | pN0 | pN0 | pN1 | pN1 | pN0 | pN1 |
| Lymphovascular invasion | Absent | Present | Absent | Present | Present | Absent |
| Perineural invasion | Present | Present | Present | Present | Present | Absent |
DP distal pancreatectomy, PPPD pylorus preserving pancreatecticoduodenectomy, MD moderately differentiated
Fig. 1.
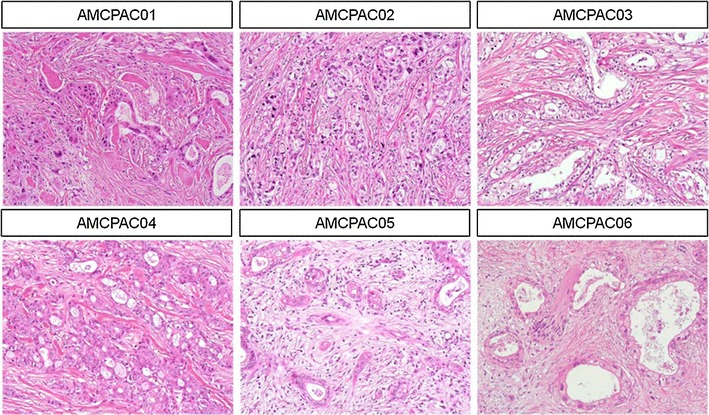
Representative hematoxylin and eosin staining images of FFPE pancreatic cancer tissue
Cytological characteristics of cancer cells
To more efficiently investigate cytological characteristics, CMAs were constructed with the AMCPAC cell lines. The CMA block was composed of 6 wells containing each AMCPAC cell line, and CMA sections were stained with hematoxylin and eosin; matched tissue samples of pancreatic ductal adenocarcinomas were included (Fig. 2b). Cytological evaluation revealed that all six cancer cell lines were adenocarcinomas with mucin production and monolayer growth on the culture dish surface (Fig. 2a). Cytologically, all 6 cancer cell lines showed round to oval (AMCPAC01, AMCPAC02, AMCPAC03, AMCPAC05, and AMCPAC06) or polygonal (AMCPAC04) shapes. The population doubling time of cancer cells was measured by MTT assay. The growth rates of AMCPAC04 and AMCPAC06 were rapid (doubling time of approximately 2 days), while those of AMCPAC02, AMCPAC03, and AMCPAC05 were relatively slow (approximately 3–4 days) based on 24-h growth after cell seeding (Table 3; Fig. 3).
Fig. 2.
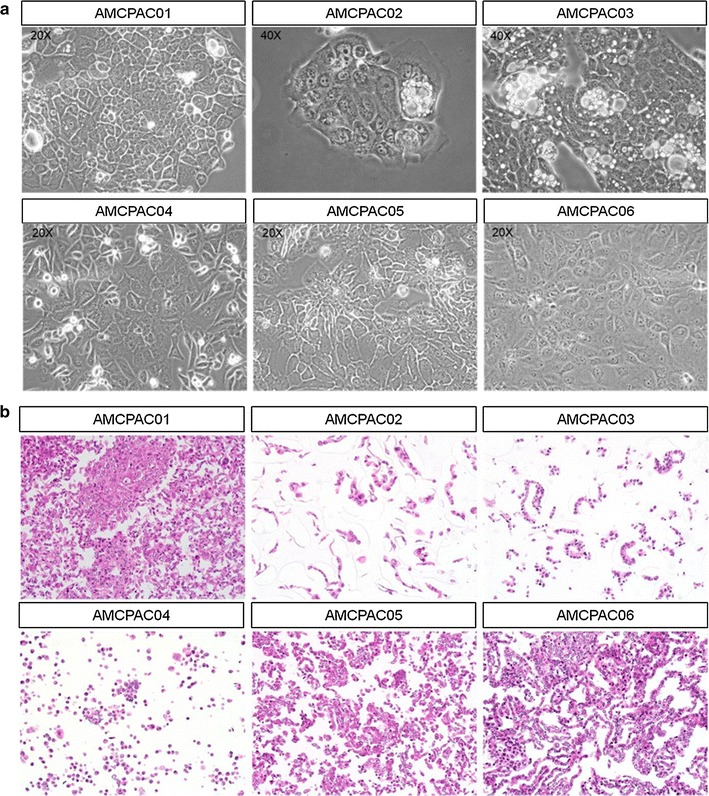
Representative AMCPAC cell line images. a Morphology of AMCPAC01–AMCPAC06 cell lines. b H&E staining images of AMCPAC01–AMCPAC06 (all, ×20)
Table 3.
Cytologic characteristics and growth rate of established cell lines
| Cell line | Growth characteristics | Cell morphology | Population doubling time (h) |
|---|---|---|---|
| AMCPAC01 | Adherent | Round/oval | 68 |
| AMCPAC02 | Adherent | Round/oval | 87 |
| AMCPAC03 | Adherent | Round/oval | 68 |
| AMCPAC04 | Adherent | Polygonal | 48 |
| AMCPAC05 | Adherent | Round/oval | 75 |
| AMCPAC06 | Adherent | Round/oval | 51 |
Fig. 3.
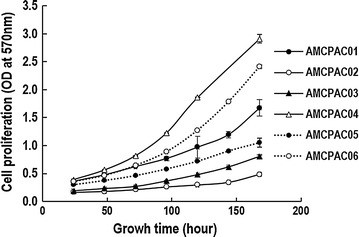
Growth curve of AMCPAC cell lines as determined by MTT assay. Cell proliferation represented by optical density (OD) at 570 nm was measured during seven days. Each value represents the mean ± SE of triplicate determinants
Mutational and immunohistochemical status of cancer cell lines
Mutant KRAS was detected in all AMCPAC cell lines. Mutations in KRAS codon 12 were detected in AMCPAC01, AMCPA02, AMCPA03, and AMCPA06, while a mutation in KRAS codon 13 was detected in AMCPAC04. And two mutations in KRAS codons 60 and 61 were detected in AMCPAC05 (Table 4). Representative images of KRAS mutations are depicted in Fig. 4.
Table 4.
KRAS mutation analysis in AMCPAC cell lines
| Cell line | c. Description | Codon number | Protein description | Mutation type |
|---|---|---|---|---|
| AMCPAC01 | c.35_36GT>TCa | 12 | p.Gly12Val(G12V) | Missense |
| AMCPAC02 | c.35G>Aa | 12 | p.Gly12Asp(G12D) | Missense |
| AMCPAC03 | c.35G>Aa | 12 | p.Gly12Asp(G12D) | Missense |
| AMCPAC04 | c.38G>Aa | 13 | p.Gly13Asp(G13D) | Missense |
| AMCPAC05 | c.180_181insCTA | 60, 61 | p.Gly60_Gln61insLeu | Insertion |
| AMCPAC05 | c.182A>Ta | 61 | p.Gln61Leu(Q61L) | Missense |
| AMCPAC06 | c.34G>Ca | 12 | p.Gly12Arg(G12R) | Missense |
NA not applicable
aPathogenic
Fig. 4.
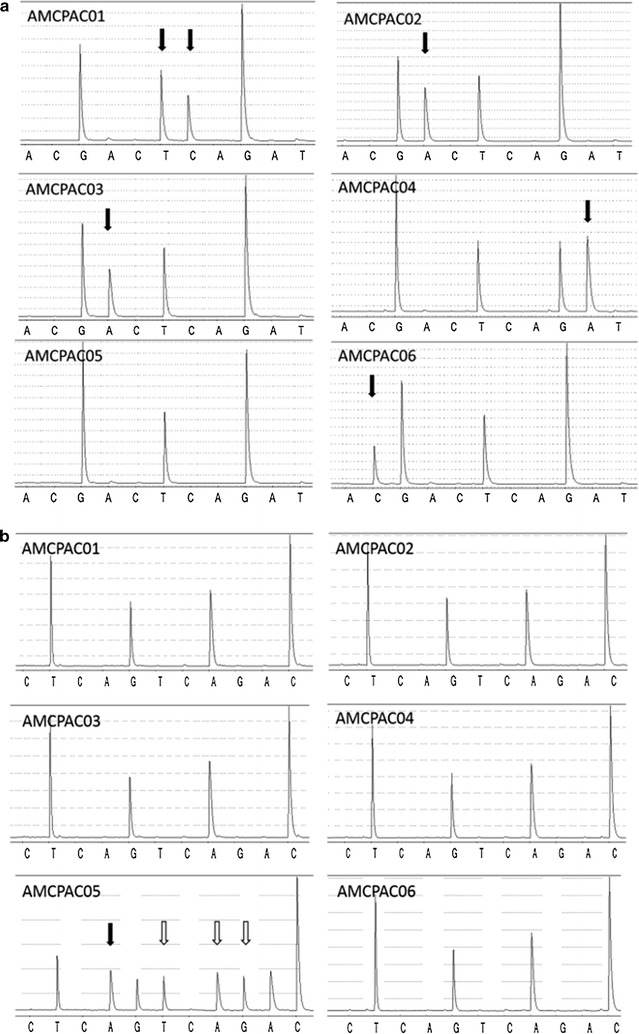
Representative pyrogram images of KRAS sequencing a of codons 12 and 13 and b codons 60 and 61. Different cell lines harbor different KRAS gene mutations. a Various missense mutations (black arrow) are noted in codons 12 and 13 of AMCPAC cell lines; G12V (GGT → GTC) in AMCPAC01, G12D (GGT → GAT) in AMCPAC02 and AMCPAC03, G13D (GGC → GAC) in AMCPAC04, and AMCPAC06 G12R (GGT → CGT). Only AMCPAC05 shows no mutation in codon 12 and 13 (GGTGGC), but codon 60 and 61 mutation of AMCPCA cell lines b. In codons 60–61, GGTCAA sequence assays in reverse orientation as TTGCAA. Only AMCPAC05 has 2 mutations; insertion (white arrow) and missense mutation (black arrow) (GGTCAA → GGTCTACTA). While other cell lines show wile type KRAS. Arrows indicate KRAS mutation site in each cell lines, and among them white arrow is newly founded mutated site in this study
Sequencing analysis of TP53 revealed the deletion of exon 5 in AMPCPAC01 and missense mutations in 3 lines (AMCPAC04, AMCPAC05, and AMCPAC06) based on Clinvar database analysis (Table 5, Additional file 1: Figure S1).
Table 5.
Exon 5 on TP53 mutation in AMCPAC cell lines
| Cell line | c. Description | Codon number | Protein description | Mutation type |
|---|---|---|---|---|
| AMCPAC01 | c.384_393del10 | 128–131 | N.D | Deletion |
| AMCPAC02 | N.A | N.A | N.A | Wild-type |
| AMCPAC03 | N.A | N.A | N.A | Wild-type |
| AMCPAC04 | c.380C>Ta | 127 | p.S127F | Missense |
| AMCPAC05 | c.398T>A | 133 | p.M133K | Missense |
| AMCPAC06 | c.451C>Tb | 151 | p.P151S | Missense |
NA not applicable, N.D not detectable
aLikely pathogenic
bPathogenic
Representative images of DPC4 and p53 immunolabeling are shown in Figs. 5 and 6 and summarized in Table 6. All 6 cancer cell lines showed intact DPC4 labeling (Fig. 5a). Overexpression of p53 protein was observed in 5 cancer cell lines (AMCPAC02–06; Fig. 6a), while 1 cell line showed a total loss of p53 expression (AMCPAC01). All 6 cases showed matched p53 expression patterns between pancreatic cancer tissue and primary cell lines (Fig. 6b). However, only AMCPAC04 showed matched DPC4 expression between the primary tumor and cancer cell line, while the other 5 tissues showed heterogeneous expression of DPC4 (Fig. 5b).
Fig. 5.
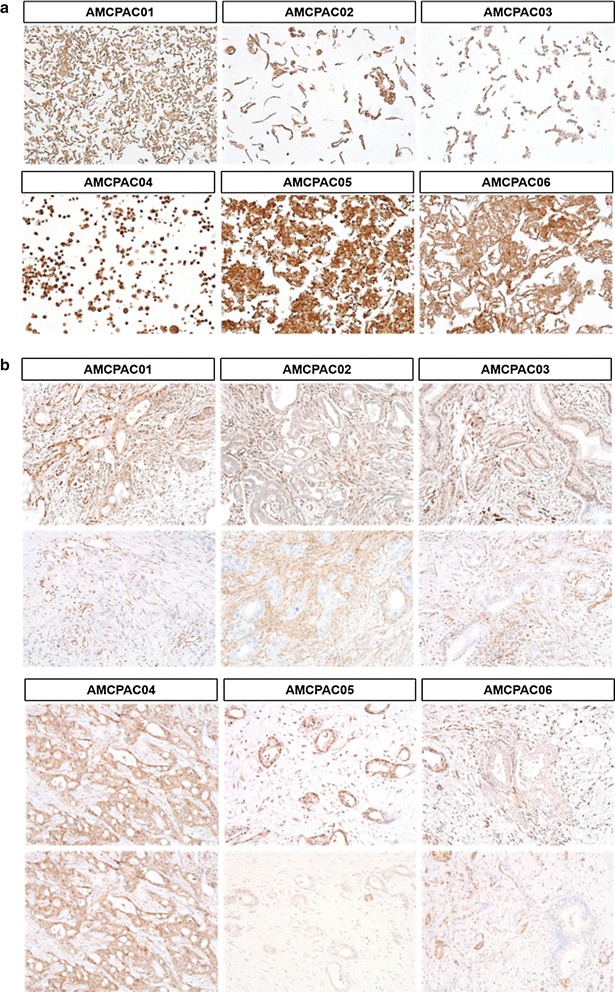
Heterogeneous DPC4 expression within original cancer tissues compared with AMCPAC cell line. a DPC4 expression in AMCPAC cell lines. b DPC4 expression in cancer tissues
Fig. 6.
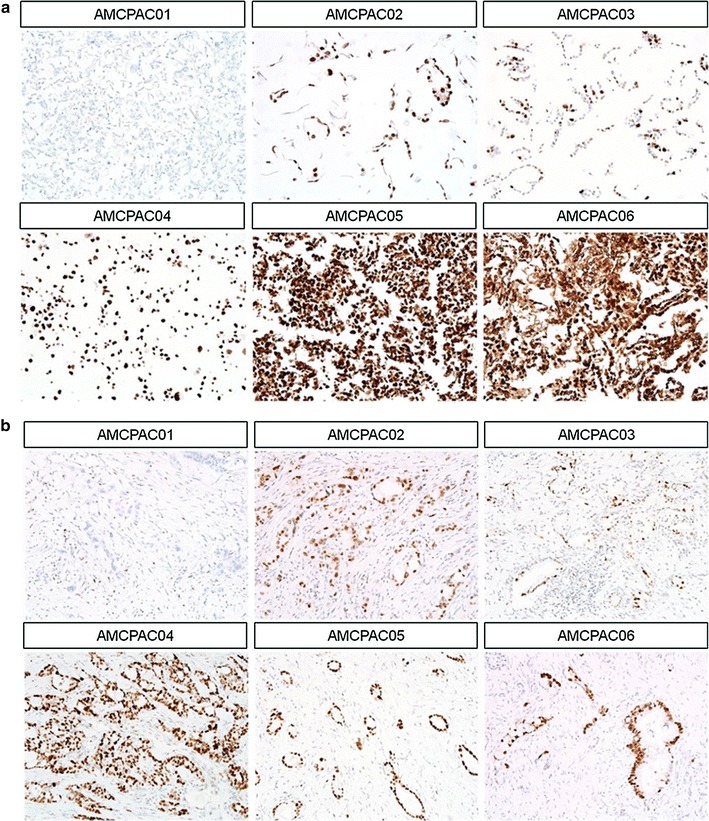
Representative p53 immunohistochemical staining. a p53 expression in AMCPAC cells lines. b p53 expression in cancer tissues
Table 6.
Summary of DPC4 and p53 immunohistochemistry staining in AMCPAC cell lines and matched cancer tissues
| Cell line | Primary cancer cell line | Cancer tissue | ||
|---|---|---|---|---|
| DPC4 | P53 | DPC4 | P53 | |
| AMCPAC01 | Intact | Total loss | Heterogeneous (intact/loss) | Total loss |
| AMCPAC02 | Intact | Overexpression | Heterogeneous (intact/loss) | Overexpression |
| AMCPAC03 | Intact | Overexpression | Heterogeneous (intact/loss) | Overexpression |
| AMCPAC04 | Intact | Overexpression | Intact | Overexpression |
| AMCPAC05 | Intact | Overexpression | Heterogeneous (intact/loss) | Overexpression |
| AMCPAC06 | Intact | Overexpression | Heterogeneous (intact/loss) | Overexpression |
Tumorigenicity of AMCPAC cell lines in immunodeficient mice
AMCPAC cell lines were implanted into 12 mice to test in vivo tumorigenicity. Three months after injection, the AMCPAC01, AMPCPAC04, and AMCPAC06 cell lines were developed, while AMCPAC02, AMPCPAC03, and AMPCPAC05 did not reach a sufficient tumor mass size for extraction from mice. AMCPAC01, AMCPAC04, and AMCPAC06 tumors collected from mice were 100–200 mm2 in diameter and used for construction of xenograft cell lines or xenograft FFPE tissue blocks for further analysis (Fig. 7).
Fig. 7.
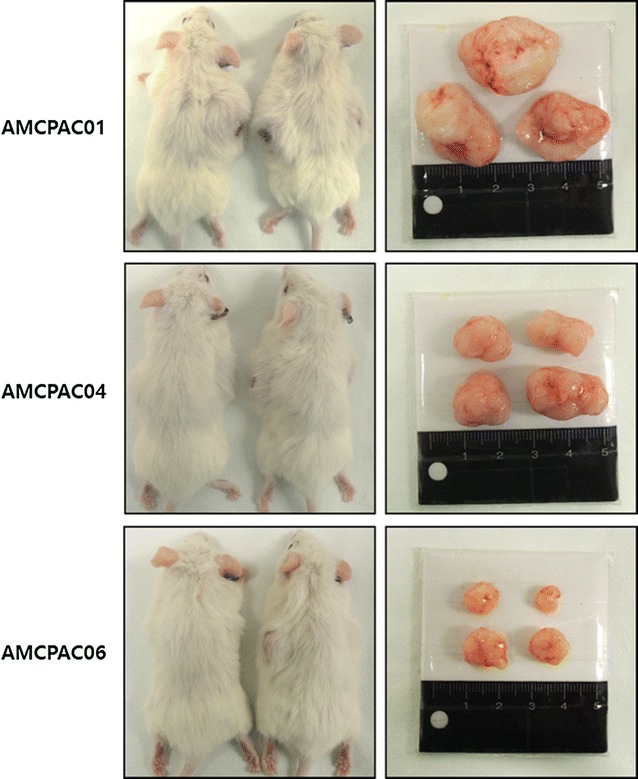
Construction of xenograft of AMCPAC cell lines
Discussion
In the present study, we established 6 novel patient-derived primary cancer cell lines (AMCPAC cell lines) from Korean patients with pancreatic ductal adenocarcinomas. The success rate of cancer cell line establishment was 7.4%. The main reason for failure was overgrowth of cancer-associated fibroblasts, accounting for 56% of samples. Ductal adenocarcinoma of the pancreas is characterized by extensive desmoplastic stromal reactions; therefore, the density of cancer cells in limited areas was very low. This low cellularity of cancer cells may prohibit the establishment of primary pancreatic cancer cell lines. To overcome these limitations, several primary cell culture methods have been developed such as patient-derived tumor xenografts or three-dimensional tumor organoid models [18–24]. In this study, we modified basic and rapid primary cell culture protocols based on the different reaction rates of trypsin-induced cell dissociation effects depending on cell type. This primary cell culture protocol did not show the expected success rate, but did select pure cancer cells.
KRAS is the most commonly activated oncogene in pancreatic cancer. More than 90% of pancreatic cancer patients show KRAS mutations, including a low grade of pancreatic intraepithelial neoplasm, a pancreatic cancer precursor [25, 26]. In Korean pancreatic cancer patients, 54% KRAS mutations were reported to commonly occur as G12D (GGT → GAT, 31%) and G12 V (GGT → GTT, 34%) on codon 12 by Sanger sequencing [27]. In the present study, KRAS mutations were evaluated by pyrosequencing, which is more sensitive compared to conventional Sanger sequencing. All 6 pancreatic cell lines contained various KRAS mutations, 4 of them had mutation in codon 12, one in codon 13, which was observed in previous whole exome sequencing studies [28–30]. The G12D, G12R, and G13D KRAS mutations detected in AMCPAC cell lines were reported in various cancers, such as pancreatic carcinoma, non-small cell lung cancer, stomach cancer, colorectal cancer, and leukemia [31–36]. Nucleotide exchanges (GGT → GTC) encoding the G12 V KRAS mutation in AMCPAC01 are rare, but have been previously reported in one pancreatic and one colorectal cancer patients [37, 38]. In AMCPAC05, two distinct KRAS mutations were detected throughout proximal region of codon 61. Q61L KRAS mutation is also rarely reported in pancreatic cancer or neoplasm [39, 40]. However, insertion of leucine between codon 60 and 61 (p.Gly60_Gln61insLeu) on KRAS has not reported any previous studies and variation databases, including ClinVar and COSMIC (catalogue of somatic mutations in cancer). Finally, AMCPAC cell lines harbor various and noble KRAS mutations and these are useful resources for pancreatic cancer research when associated with KRAS mutation type.
The mutational status of TP53 was analyzed because genetic alternations in KRAS, TP53, and SMAD4 play key roles in the tumorigenesis of pancreatic ductal adenocarcinoma [41–43]. TP53 mutations are frequently mutated in pancreatic cancer; approximately 38% of Korean pancreatic cancer patients contain TP53 alternations [27]. Four AMCPAC cell lines contained TP53 mutations only in exon 5, and AMCPAC02 and AMCPAC03 showed no TP53 gene alterations. TP53 mutations in AMCPAC04 and AMCPAC06 cells are known pathogenic mutations, while those in AMCPAC01 and AMCPAC05 were newly identified [44].
CMAs are novel multiplex array techniques for immunocytochemistry evaluation to detect the molecular composition and function of cell lines. The use of an agarose matrix or agarose mold for cell microarray construction has been reported. In this study, a cell block was used for cell microarray by directly paraffin embedding the cell pellet, resulting in the highest cell density in an array core compared to other methods [45].
It is known that KRAS, p16/DCKN2A, GNAS, and BRAF are mutated early in pancreatic cancer progression, while SMAD4/DPC4 and TP53 are mutated at later stages [25, 46]. To compare DPC4 and p53 expression levels between established cell lines and matched cancer tissues, immunocytochemistry was performed on CMA sections containing 6 AMCPAC cell lines and matched cancer tissues. Five cells lines, AMCPAC02–06, and matched cancer tissues showed p53 protein overexpression, while AMCPAC01 showed a total loss of p53 expression. A lack of p53 expression is associated with a nonsense mutation (or null mutation) in TP53 [47]. Our TP53 sequencing revealed a deletion (TGCCCTCAAC) on exon 5 of TP53 which was associated with the loss of p53 protein expression. Several studies reported worse prognosis of cancer patients with a lack of p53 expression compared to those with missense TP53 mutations in lung and ovarian cancers [48, 49].
Because DPC4 protein loss is correlated with SMAD4/DPC4 mutation, immunohistochemical staining for DPC4 protein is used as a diagnostic criteria of pancreatic cancer with lower cost than sequencing of SMAD4/DPC4 [50]. Loss of DPC4 expression is associated with distant metastasis, epithelial to mesenchymal transition, and treatment failure to local recurrence [51–53]. In the present study, all 6 AMCPAC cell lines showed homogeneously intact DPC4 expression. In contrast, matched cancer tissues showed heterogeneous DPC4 expression: one tumor area exhibited intact DPC4 expression, while another region showed loss of DPC4 protein expression (Fig. 8). Homogeneously intact DPC4 expression in the primary pancreatic cancer cell lines, which were obtained from cancer tissues with intratumoral DPC4 heterogeneity, can be explained by the positive selection of DPC4-expressing cancer cells during primary cell culture or most DPC4 expressing cancer cells presented in cancer tissues specimens at the time of primary cell culture (Fig. 8).
Fig. 8.
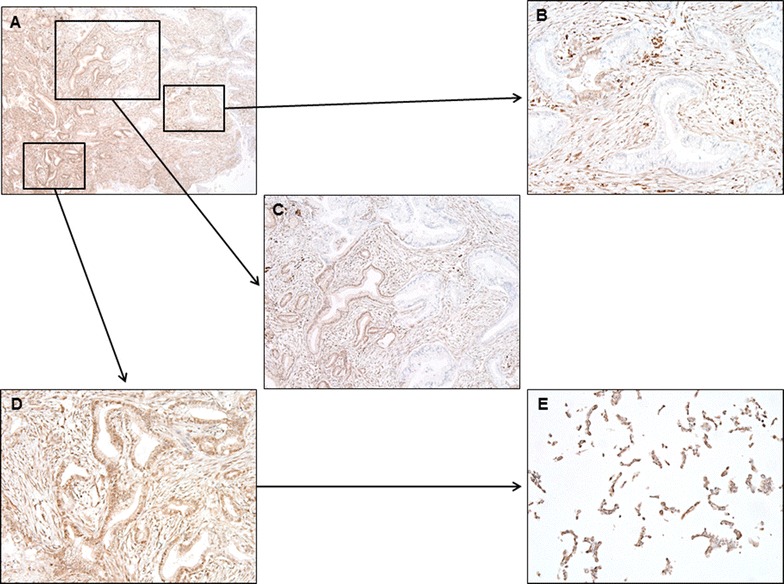
Original cancer tissue of AMCPAC04 showed heterogeneous DPC4 expression. A Low-power scanning view shows heterogeneous DPC4 expression (magnification, ×40). B Higher power view shows area of loss of DPC4 expression (×200). C Cancer cells on the left half show intact DPC labeling, while cancer cells on the right half show loss of DPC4 expression (×100). D Higher power view shows area of intact DPC4 expression (×200). E Cancer cells in AMCPAC04 cell line show intact DCP4 labeling (×200)
Genetic heterogeneity occurs in various situations, such as between separate cancers with the same histologic subtypes, primary and metastatic cancers from the same individuals, and separate regions from the same cancers [54–58]. Additionally, genetic heterogeneity has been observed in many solid tumors, including renal cell carcinomas, non-small cell lung cancers, and breast cancers [54, 55, 59–61]. The organoid model of pancreatic cancer was recently examined among preclinical models for predicting personalized tailored therapy [22–24]. This model is expected to conserve the tumor microenvironment, including ductal or acinar structures, and reflect tumor heterogeneity. Therefore, to understand interactions in tumor microenvironments or develop personalized medicine, application of the organoid model will be more effective than conventional primary cancer cell cultures. However, the traditional primary cancer cell culture model is more suitable for investigating the characteristics of pure cancer cells. Nevertheless, methods for establishing specific cell lines by modifying organic models and our culture protocols should be examined in future studies.
AMCPAC cell lines can provide a resource for pancreatic cancer studies, including the basic and translational research fields. For example, comparison of DPC4(+)/P53(−) (AMCPAC01) and DPC4(+)/P53(+) (AMCPAC02–06) cell lines will be helpful in understanding the interactions of TP53 and DPC4 signaling pathways. In total, 3 cell lines have been previously established from Korean pancreatic cancer patients, including SNU213, SNU324, and SNU410, which are available from the Korean Cell Line Bank. We have added 6 newly established pancreatic cancer cell lines from Korean patients. Different molecular pathologic characteristics of our new established AMCPAC cell lines may provide diversity and help in determining the molecular pathological basis of pancreatic cancer in different ethnicities from other previously established pancreatic cancer cell lines.
Conclusions
We established and characterized 6 novel pancreatic cancer cell lines (AMCPAC01–AMCPAC06). These novel cell lines may contribute to the understanding of pancreatic ductal adenocarcinomas, including the molecular basis of tumorigenesis, progression, and metastasis of pancreatic cancers.
Authors’ contributions
SJK, SYA, HSP, JHL, KBS, DWH, and SCK collected patient tumor tissue samples to establish cell lines and analyzed clinical information. MJK and MSK performed experimental work. JP implanted xenografts into mice. MJK, SYJ, KPK, SCK, and SMH conceived ideas and evaluated the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
The study protocol was approved by the institutional review board (IRB) of the Asan Medical Center (2015-0480). The animal care protocol for this study was approved by the International Animal Care and Use Committee (IACUC) of the Laboratory of Animal Research at the Asan Medical Center, Seoul, Korea.
Funding
This study was supported by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (Grant Number: HI14C2640).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- CMA
cell microarray
- FFPE
formalin-fixed paraffin-embedded
Additional file
Additional file 1: Figure S1. TP53 histogram of AMCPAC cell lines.
Contributor Information
Mi-Ju Kim, Email: kimmiju@nate.com.
Min-Sun Kim, Email: drusillaluv@gmail.com.
Sung Joo Kim, Email: ksj3641@naver.com.
Soyeon An, Email: soyan21@gmail.com.
Jin Park, Email: jin08@amc.seoul.kr.
Hosub Park, Email: parkhstm@gmail.com.
Jae Hoon Lee, Email: hbpsurgeon@gmail.com.
Ki-Byung Song, Email: mtsong21c@naver.com.
Dae Wook Hwang, Email: drdwhwang@gmail.com.
Suhwan Chang, Email: suhwan.chang@amc.seoul.kr.
Kyu-pyo Kim, Email: kkp1122@gmail.com.
Seong-Yun Jeong, Email: syj@amc.seoul.kr.
Song Cheol Kim, Phone: +82-3010-3936, Email: drksc@amc.seoul.kr.
Seung-Mo Hong, Phone: +82-2-3010-4558, Email: smhong28@gmail.com.
References
- 1.Jung KW, Won YJ, Kong HJ, Oh CM, Cho H, Lee DH, Lee KH. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2012. Cancer Res Treat. 2015;47(2):127–141. doi: 10.4143/crt.2015.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, Adenis A, Raoul JL, Gourgou-Bourgade S, de la Fouchardiere C, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364(19):1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 4.Philip PA, Goldman B, Ramanathan RK, Lenz HJ, Lowy AM, Whitehead RP, Wakatsuki T, Iqbal S, Gaur R, Benedetti JK, et al. Dual blockade of epidermal growth factor receptor and insulin-like growth factor receptor-1 signaling in metastatic pancreatic cancer: phase Ib and randomized phase II trial of gemcitabine, erlotinib, and cixutumumab versus gemcitabine plus erlotinib (SWOG S0727) Cancer. 2014;120(19):2980–2985. doi: 10.1002/cncr.28744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Z, Gabrielson A, Hwang JJ, Pishvaian MJ, Weiner LM, Zhuang T, Ley L, Marshall JL, He AR. Phase II study of lapatinib and capecitabine in second-line treatment for metastatic pancreatic cancer. Cancer Chemother Pharmacol. 2015;76(6):1309–1314. doi: 10.1007/s00280-015-2855-z. [DOI] [PubMed] [Google Scholar]
- 6.Wang-Gillam A, Li CP, Bodoky G, Dean A, Shan YS, Jameson G, Macarulla T, Lee KH, Cunningham D, Blanc JF, et al. Nanoliposomal irinotecan with fluorouracil and folinic acid in metastatic pancreatic cancer after previous gemcitabine-based therapy (NAPOLI-1): a global, randomised, open-label, phase 3 trial. Lancet. 2016;387(10018):545–557. doi: 10.1016/S0140-6736(15)00986-1. [DOI] [PubMed] [Google Scholar]
- 7.Fryer RA, Barlett B, Galustian C, Dalgleish AG. Mechanisms underlying gemcitabine resistance in pancreatic cancer and sensitisation by the iMiD lenalidomide. Anticancer Res. 2011;31(11):3747–3756. [PubMed] [Google Scholar]
- 8.Wang WB, Yang Y, Zhao YP, Zhang TP, Liao Q, Shu H. Recent studies of 5-fluorouracil resistance in pancreatic cancer. World J Gastroenterol. 2014;20(42):15682–15690. doi: 10.3748/wjg.v20.i42.15682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohkawa S, Okusaka T, Isayama H, Fukutomi A, Yamaguchi K, Ikeda M, Funakoshi A, Nagase M, Hamamoto Y, Nakamori S, et al. Randomised phase II trial of S-1 plus oxaliplatin vs S-1 in patients with gemcitabine-refractory pancreatic cancer. Br J Cancer. 2015;112(9):1428–1434. doi: 10.1038/bjc.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golan T, Atias D, Barshack I, Avivi C, Goldstein RS, Berger R. Ascites-derived pancreatic ductal adenocarcinoma primary cell cultures as a platform for personalised medicine. Br J Cancer. 2014;110(9):2269–2276. doi: 10.1038/bjc.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue A, Julovi SM, Hugh TJ, Samra JS, Wong MH, Gill AJ, Toon CW, Smith RC. A patient-derived subrenal capsule xenograft model can predict response to adjuvant therapy for cancers in the head of the pancreas. Pancreatology. 2015;15(4):397–404. doi: 10.1016/j.pan.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Logsdon CD, Arumugam T, Ramachandran V. Animal models of gastrointestinal and liver diseases. The difficulty of animal modeling of pancreatic cancer for preclinical evaluation of therapeutics. Am J Physiol Gastrointest Liver Physiol. 2015;309(5):G283–G291. doi: 10.1152/ajpgi.00169.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, Zhang C, Schnell C, Yang G, Zhang Y, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015;21(11):1318–1325. doi: 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]
- 14.Ku JL, Yoon KA, Kim WH, Jang Y, Suh KS, Kim SW, Park YH, Park JG. Establishment and characterization of four human pancreatic carcinoma cell lines. Genetic alterations in the TGFBR2 gene but not in the MADH4 gene. Cell Tissue Res. 2002;308(2):205–214. doi: 10.1007/s00441-001-0510-y. [DOI] [PubMed] [Google Scholar]
- 15.Ruckert F, Aust D, Bohme I, Werner K, Brandt A, Diamandis EP, Krautz C, Hering S, Saeger HD, Grutzmann R, et al. Five primary human pancreatic adenocarcinoma cell lines established by the outgrowth method. J Surg Res. 2012;172(1):29–39. doi: 10.1016/j.jss.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 16.Heller A, Angelova AL, Bauer S, Grekova SP, Aprahamian M, Rommelaere J, Volkmar M, Janssen JW, Bauer N, Herr I, et al. Establishment and characterization of a novel cell line, ASAN-PaCa, derived from human adenocarcinoma arising in intraductal papillary mucinous neoplasm of the pancreas. Pancreas. 2016;45(10):1452–1460. doi: 10.1097/MPA.0000000000000673. [DOI] [PubMed] [Google Scholar]
- 17.Kim SA, Kim MS, Kim MS, Kim SC, Choi J, Yu E, Hong SM. Pleomorphic solid pseudopapillary neoplasm of the pancreas: degenerative change rather than high-grade malignant potential. Hum Pathol. 2014;45(1):166–174. doi: 10.1016/j.humpath.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 18.Seol HS, Suh YA, Ryu YJ, Kim HJ, Chun SM, Na DC, Fukamachi H, Jeong SY, Choi EK, Jang SJ. A patient-derived xenograft mouse model generated from primary cultured cells recapitulates patient tumors phenotypically and genetically. J Cancer Res Clin Oncol. 2013;139(9):1471–1480. doi: 10.1007/s00432-013-1449-6. [DOI] [PubMed] [Google Scholar]
- 19.Inagawa Y, Yamada K, Yugawa T, Ohno S, Hiraoka N, Esaki M, Shibata T, Aoki K, Saya H, Kiyono T. A human cancer xenograft model utilizing normal pancreatic duct epithelial cells conditionally transformed with defined oncogenes. Carcinogenesis. 2014;35(8):1840–1846. doi: 10.1093/carcin/bgu112. [DOI] [PubMed] [Google Scholar]
- 20.Wennerstrom AB, Lothe IM, Sandhu V, Kure EH, Myklebost O, Munthe E. Generation and characterisation of novel pancreatic adenocarcinoma xenograft models and corresponding primary cell lines. PLoS ONE. 2014;9(8):e103873. doi: 10.1371/journal.pone.0103873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung J, Lee CH, Seol HS, Choi YS, Kim E, Lee EJ, Rhee JK, Singh SR, Jun ES, Han B, et al. Generation and molecular characterization of pancreatic cancer patient-derived xenografts reveals their heterologous nature. Oncotarget. 2016;7(38):62533–62546. doi: 10.18632/oncotarget.11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160(1–2):324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang L, Holtzinger A, Jagan I, BeGora M, Lohse I, Ngai N, Nostro C, Wang R, Muthuswamy LB, Crawford HC, et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat Med. 2015;21(11):1364–1371. doi: 10.1038/nm.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang HC, Kuo CJ. Personalizing pancreatic cancer organoids with hPSCs. Nat Med. 2015;21(11):1249–1251. doi: 10.1038/nm.3992. [DOI] [PubMed] [Google Scholar]
- 25.Kanda M, Matthaei H, Wu J, Hong SM, Yu J, Borges M, Hruban RH, Maitra A, Kinzler K, Vogelstein B, et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology. 2012;142(4):730–733 e739. doi: 10.1053/j.gastro.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi H, Kohno T, Ueno H, Hiraoka N, Kondo S, Saito M, Shimada Y, Ichikawa H, Kato M, Shibata T, et al. Utility of assessing the number of mutated KRAS, CDKN2A, TP53, and SMAD4 genes using a targeted deep sequencing assay as a prognostic biomarker for pancreatic cancer. Pancreas. 2017;46:335. doi: 10.1097/MPA.0000000000000760. [DOI] [PubMed] [Google Scholar]
- 27.Shin SH, Kim SC, Hong SM, Kim YH, Song KB, Park KM, Lee YJ. Genetic alterations of K-ras, p53, c-erbB-2, and DPC4 in pancreatic ductal adenocarcinoma and their correlation with patient survival. Pancreas. 2013;42(2):216–222. doi: 10.1097/MPA.0b013e31825b6ab0. [DOI] [PubMed] [Google Scholar]
- 28.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, Mollaee M, Wagner KU, Koduru P, Yopp A, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, Zakowski MF, Heelan RT, Kris MG, Varmus HE. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2(1):e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Motojima K, Urano T, Nagata Y, Shiku H, Tsurifune T, Kanematsu T. Detection of point mutations in the Kirsten-ras oncogene provides evidence for the multicentricity of pancreatic carcinoma. Ann Surg. 1993;217(2):138–143. doi: 10.1097/00000658-199302000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee KH, Lee JS, Suh C, Kim SW, Kim SB, Lee JH, Lee MS, Park MY, Sun HS, Kim SH. Clinicopathologic significance of the K-ras gene codon 12 point mutation in stomach cancer. An analysis of 140 cases. Cancer. 1995;75(12):2794–2801. doi: 10.1002/1097-0142(19950615)75:12<2794::AID-CNCR2820751203>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 34.Pinto P, Rocha P, Veiga I, Guedes J, Pinheiro M, Peixoto A, Pinto C, Fragoso M, Sanches E, Araujo A, et al. Comparison of methodologies for KRAS mutation detection in metastatic colorectal cancer. Cancer Genet. 2011;204(8):439–446. doi: 10.1016/j.cancergen.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Jun SY, Kim M, Jin GuM, Kyung Bae Y, Chang HK, Sun Jung E, Jang KT, Kim J, Yu E, Woon Eom D, et al. Clinicopathologic and prognostic associations of KRAS and BRAF mutations in small intestinal adenocarcinoma. Mod Pathol. 2016;29(4):402–415. doi: 10.1038/modpathol.2016.40. [DOI] [PubMed] [Google Scholar]
- 36.Matsuda K, Shimada A, Yoshida N, Ogawa A, Watanabe A, Yajima S, Iizuka S, Koike K, Yanai F, Kawasaki K, et al. Spontaneous improvement of hematologic abnormalities in patients having juvenile myelomonocytic leukemia with specific RAS mutations. Blood. 2007;109(12):5477–5480. doi: 10.1182/blood-2006-09-046649. [DOI] [PubMed] [Google Scholar]
- 37.Rachakonda PS, Bauer AS, Xie H, Campa D, Rizzato C, Canzian F, Beghelli S, Greenhalf W, Costello E, Schanne M, et al. Somatic mutations in exocrine pancreatic tumors: association with patient survival. PLoS ONE. 2013;8(4):e60870. doi: 10.1371/journal.pone.0060870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Solassol J, Ramos J, Crapez E, Saifi M, Mange A, Vianes E, Lamy PJ, Costes V, Maudelonde T. KRAS mutation detection in paired frozen and Formalin-Fixed Paraffin-Embedded (FFPE) colorectal cancer tissues. Int J Mol Sci. 2011;12(5):3191–3204. doi: 10.3390/ijms12053191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliveira-Cunha M, Hadfield KD, Siriwardena AK, Newman W. EGFR and KRAS mutational analysis and their correlation to survival in pancreatic and periampullary cancer. Pancreas. 2012;41(3):428–434. doi: 10.1097/MPA.0b013e3182327a03. [DOI] [PubMed] [Google Scholar]
- 40.Furukawa T, Kuboki Y, Tanji E, Yoshida S, Hatori T, Yamamoto M, Shibata N, Shimizu K, Kamatani N, Shiratori K. Whole-exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas. Sci Rep. 2011;1:161. doi: 10.1038/srep00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wood LD, Hruban RH. Pathology and molecular genetics of pancreatic neoplasms. Cancer J. 2012;18(6):492–501. doi: 10.1097/PPO.0b013e31827459b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu C, Hart SN, Bamlet WR, Moore RM, Nandakumar K, Eckloff BW, Lee YK, Petersen GM, McWilliams RR, Couch FJ. Prevalence of pathogenic mutations in cancer predisposition genes among pancreatic cancer patients. Cancer Epidemiol Biomarkers Prev. 2016;25(1):207–211. doi: 10.1158/1055-9965.EPI-15-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deer EL, Gonzalez-Hernandez J, Coursen JD, Shea JE, Ngatia J, Scaife CL, Firpo MA, Mulvihill SJ. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39(4):425–435. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen LC, Neubauer A, Kurisu W, Waldman FM, Ljung BM, Goodson W, 3rd, Goldman ES, Moore D, 2nd, Balazs M, Liu E, et al. Loss of heterozygosity on the short arm of chromosome 17 is associated with high proliferative capacity and DNA aneuploidy in primary human breast cancer. Proc Natl Acad Sci USA. 1991;88(9):3847–3851. doi: 10.1073/pnas.88.9.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moskaluk CA, Stoler MH. Agarose mold embedding of cultured cells for tissue microarrays. Diagn Mol Pathol. 2002;11(4):234–238. doi: 10.1097/00019606-200212000-00007. [DOI] [PubMed] [Google Scholar]
- 46.Wilentz RE, Iacobuzio-Donahue CA, Argani P, McCarthy DM, Parsons JL, Yeo CJ, Kern SE, Hruban RH. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res. 2000;60(7):2002–2006. [PubMed] [Google Scholar]
- 47.Yemelyanova A, Vang R, Kshirsagar M, Lu D, Marks MA, Shih Ie M, Kurman RJ. Immunohistochemical staining patterns of p53 can serve as a surrogate marker for TP53 mutations in ovarian carcinoma: an immunohistochemical and nucleotide sequencing analysis. Mod Pathol. 2011;24(9):1248–1253. doi: 10.1038/modpathol.2011.85. [DOI] [PubMed] [Google Scholar]
- 48.Shahin MS, Hughes JH, Sood AK, Buller RE. The prognostic significance of p53 tumor suppressor gene alterations in ovarian carcinoma. Cancer. 2000;89(9):2006–2017. doi: 10.1002/1097-0142(20001101)89:9<2006::AID-CNCR18>3.3.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 49.Hashimoto T, Tokuchi Y, Hayashi M, Kobayashi Y, Nishida K, Hayashi S, Ishikawa Y, Tsuchiya S, Nakagawa K, Hayashi J, et al. p53 null mutations undetected by immunohistochemical staining predict a poor outcome with early-stage non-small cell lung carcinomas. Cancer Res. 1999;59(21):5572–5577. [PubMed] [Google Scholar]
- 50.Tascilar M, Offerhaus GJ, Altink R, Argani P, Sohn TA, Yeo CJ, Cameron JL, Goggins M, Hruban RH, Wilentz RE. Immunohistochemical labeling for the Dpc4 gene product is a specific marker for adenocarcinoma in biopsy specimens of the pancreas and bile duct. Am J Clin Pathol. 2001;116(6):831–837. doi: 10.1309/WF03-NFCE-7BRH-7C26. [DOI] [PubMed] [Google Scholar]
- 51.Shin SH, Kim HJ, Hwang DW, Lee JH, Song KB, Jun E, Shim IK, Hong SM, Kim HJ, Park KM, et al. The DPC4/SMAD4 genetic status determines recurrence patterns and treatment outcomes in resected pancreatic ductal adenocarcinoma: A prospective cohort study. Oncotarget. 2017;8(11):17945–17959. doi: 10.18632/oncotarget.14901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iacobuzio-Donahue CA, Fu B, Yachida S, Luo M, Abe H, Henderson CM, Vilardell F, Wang Z, Keller JW, Banerjee P, et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol. 2009;27(11):1806–1813. doi: 10.1200/JCO.2008.17.7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamada S, Fujii T, Shimoyama Y, Kanda M, Nakayama G, Sugimoto H, Koike M, Nomoto S, Fujiwara M, Nakao A, et al. SMAD4 expression predicts local spread and treatment failure in resected pancreatic cancer. Pancreas. 2015;44(4):660–664. doi: 10.1097/MPA.0000000000000315. [DOI] [PubMed] [Google Scholar]
- 54.Cai W, Lin D, Wu C, Li X, Zhao C, Zheng L, Chuai S, Fei K, Zhou C, Hirsch FR. Intratumoral heterogeneity of ALK-rearranged and ALK/EGFR coaltered lung adenocarcinoma. J Clin Oncol. 2015;33(32):3701–3709. doi: 10.1200/JCO.2014.58.8293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chien BB, Burke RG, Hunter DJ. An extensive experience with postoperative pain relief using postoperative fentanyl infusion. Arch Surg. 1991;126(6):692–694. doi: 10.1001/archsurg.1991.01410300034004. [DOI] [PubMed] [Google Scholar]
- 57.Cargill S. Open learning. Snakes and ladders. Interview by Laura Swaffield. Nurs Times. 1991;87(4):42–43. [PubMed] [Google Scholar]
- 58.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geyer FC, Weigelt B, Natrajan R, Lambros MB, de Biase D, Vatcheva R, Savage K, Mackay A, Ashworth A, Reis-Filho JS. Molecular analysis reveals a genetic basis for the phenotypic diversity of metaplastic breast carcinomas. J Pathol. 2010;220(5):562–573. doi: 10.1002/path.2675. [DOI] [PubMed] [Google Scholar]
- 60.Wu X, Northcott PA, Dubuc A, Dupuy AJ, Shih DJ, Witt H, Croul S, Bouffet E, Fults DW, Eberhart CG, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482(7386):529–533. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bai H, Wang Z, Chen K, Zhao J, Lee JJ, Wang S, Zhou Q, Zhuo M, Mao L, An T, et al. Influence of chemotherapy on EGFR mutation status among patients with non-small-cell lung cancer. J Clin Oncol. 2012;30(25):3077–3083. doi: 10.1200/JCO.2011.39.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


