Conspectus
DNA is well-known as bearer of the genetic code. Since its structure elucidation nearly seven decades ago by Watson, Crick, Wilkins, and Franklin, much has been learned about its detailed structure, function, and genetic coding. The development of automated solid-phase synthesis, and with it the availability of synthetic DNA with any desired sequence in lengths of up to hundreds of bases in the best case, has contributed much to the advancement of the field of DNA research. In addition, classic organic synthesis has allowed introduction of a very large number of modifications in the DNA in a sequence specific manner, which have initially been targeted at altering the biological function of DNA. However, in recent years DNA has become a very attractive scaffold in supramolecular chemistry, where DNA is taken out of its biological role and serves as both stick and glue molecule to assemble novel functional structures with nanometer precision. The attachment of functionalities to DNA has led to the creation of supramolecular systems with applications in light harvesting, energy and electron transfer, sensing, and catalysis. Functional DNA is clearly having a significant impact in the field of bioinspired nanosystems.
Of particular interest is the use of porphyrins in supramolecular chemistry and bionanotechnology, because they are excellent functional groups due to their electronic properties that can be tailored through chemical modifications of the aromatic core or through insertion of almost any metal of the periodic table into the central cavity. The porphyrins can be attached either to the nucleobase, to the phosphate group, or to the ribose moiety. Additionally, noncovalent templating through Watson–Crick base pairing forms an alternative and attractive approach. With this, the combination of two seemingly simple molecules gives rise to a highly complex system with unprecedented possibilities for modulation of function, and with it applications, particularly when combined with other functional groups. Here, an overview is given on the developments of using porphyrin modified DNA for the construction of functional assemblies. Strategies for the synthesis and characterization are presented alongside selected applications where the porphyrin modification has proven to be particularly useful and superior to other modifiers but also has revealed its limitations. We also discuss implications on properties and behavior of the porphyrin–DNA, where similar issues could arise when using other hydrophobic and bulky substituents on DNA. This includes particularly problems regarding synthesis of the building blocks, DNA synthesis, yields, solubility, and intermolecular interactions.
1. Introduction
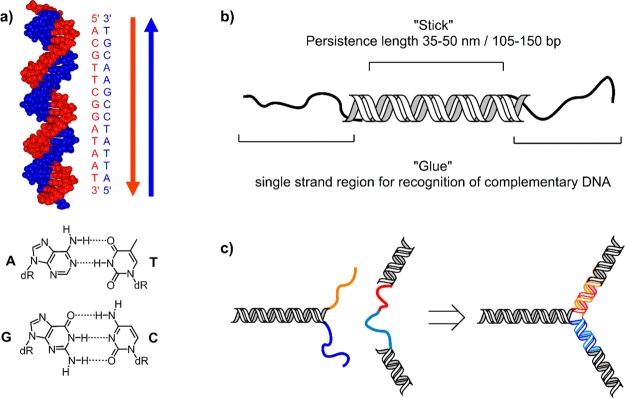
The construction of nanosized functional molecules by means of a bottom-up approach is best addressed using the concepts of supramolecular chemistry, which is defined as chemistry beyond the molecule. Large chemical constructs are made up of a discrete number of self-assembled molecular subunits. Biological systems can be regarded as the ultimate supramolecular assemblies, as they combine tailored structure and function to form living organisms, where smaller subunits are organized through noncovalent interactions. Therefore, biology provides us with ideas and templates that are ideal to draw upon. In this respect, DNA has shown to be an exciting construction material, owing to its distinct properties such as predictable three-dimensional structure in form of the double helix, its programmable nature, and synthetic availability. The basic principle of working with DNA is relatively straightforward: the molecule forms a well-understood duplex through complementary base pairing of two antiparallel DNA strands, where the recognition is based on the Watson–Crick (WC) base pairs (bp) of A–T and G–C (Figure 1).1−3 Yet there is far more to DNA than just this concept. DNA can act both as rigid stick (double strand, dsDNA) with a persistence length of about 40–50 nm (120–150 bp) and flexible glue (single strand, ssDNA), giving access to a Lego-like building block system to create architectures with nanometer precision. By taking DNA out of its biological context, new systems have emerged that are starting to play a major role in materials science, electronics, diagnostics, medicinal chemistry, and more.
Figure 1.
(a) Depiction of the DNA double helix (B-form), which is most commonly found in nature, with example of a complementary sequence for selective duplex formation via Watson–Crick base pairing of A=T and G≡C. (b) DNA as rigid stick and flexible glue molecule for the construction of DNA nanoarchitectures. (c) Concept of programmed self-assembly of DNA nanostructures: complementary ssDNA sequences (orange and red, light and dark blue) will hybridize to form predefined rigid constructs.
While most of the DNA based nanoconstructs are realized using unmodified DNA, it is intriguing to enhance the functionality of the nano-DNA systems by introducing functional groups not present in natural DNA. The various approaches showing the diversity of available functionalities have been reviewed independently,4−6 in particular using organic chromophores,7−10 and shall not be covered here in detail, but a focus is given on porphyrins as modifiers.11 The formation of porphyrin assemblies is fascinating from the point of view of creating new functional materials, and applications in the fields of energy or electron transfer, light harvesting, optics, catalysis, and many more have been reported numerously.12−16 The optical and electrochemical properties of the porphyrins are very diverse and can be tuned by either chemically modifying the porphyrin core or inserting metals in the central cavity, which is unique for porphyrins. Many different porphyrins and related compounds are available, though only a few have been used for attachment to DNA. Out of the many templates that have been studied for creating porphyrin assemblies,17−22 DNA is certainly among the most captivating of scaffolds as it allows for easier control over sequence and structure, though peptides have not yet been fully explored in this respect and could provide a complementary template. The use of covalent chemistry in the formation of porphyrin assemblies has the advantage of taking control over sequence using the very same chemistries that are applied to the synthesis of the DNA and starting from the same building blocks. Introduction of different porphyrins, potentially in combination with other functional entities, will give a well-defined array, where the porphyrins will be incorporated into a predetermined spatial arrangement. Noncovalent approaches where modified porphyrin units self-assemble on a ssDNA template through hydrogen bonding provide an alternative route,23,24 but this Account will focus on the formation, analysis, and application of covalently modified DNA strands.
2. Covalent Attachment of Porphyrins to DNA
2.1. Single End-of-DNA Porphyrin Attachment
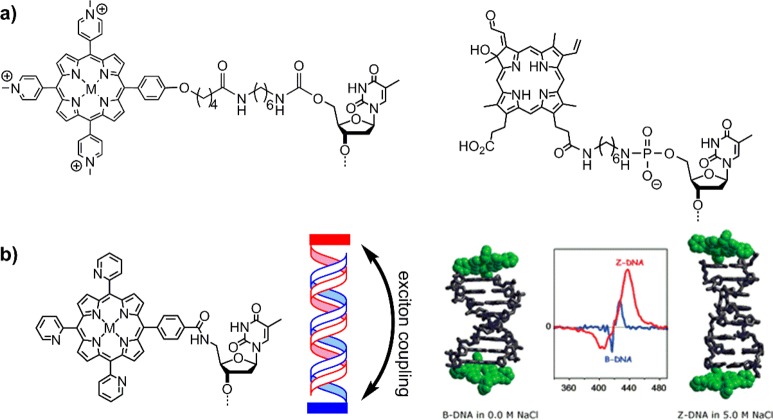
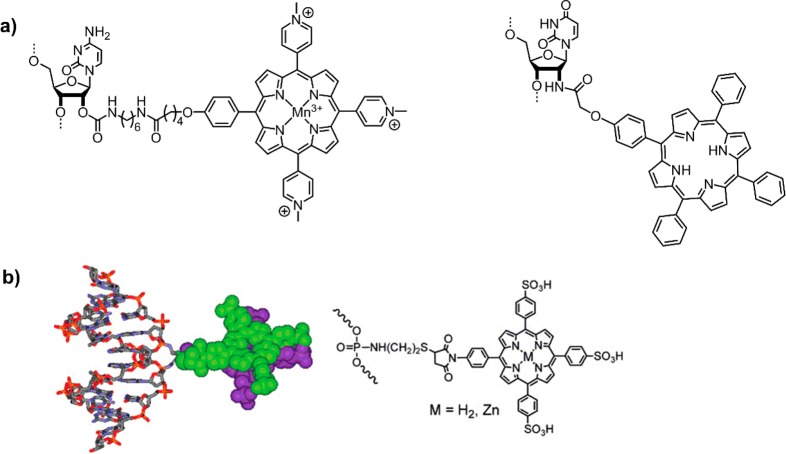
The first examples of porphyrins attached to DNA were reported by Meunier et al.25 and Hélène et al.,26 who created artificial nucleases with manganese porphyrins or chlorins, respectively (Figure 2a). Around the same time, Czuchajowski et al. used the H-phosphonate approach to add pyridinium-porphyrin to form a photoreactive antisense oligo-deoxynucleotide (ODN).27 This method was successful in adding one porphyrin modification to the DNA at its 5′-end. A similar approach was used by Berova and Balaz et al. to attach a porphyrin to the final phosphate group through monofunctional phosphoramidites, where it effectively acts as a cap on the blunt end of the DNA28,29 and can even stabilize non-Watson–Crick G–A base pairs.30 The groups showed that the porphyrins at the end of the DNA can act as a chiroptical marker for circular dichroism (CD) spectroscopy giving insight into structural aspects of the DNA and the environment of the porphyrin (Figure 2b), which is strongly dependent on the solvent (salt concentration) and central metal of the porphyrin (which influences hydrophobicity and sterics through potential axial water ligands).31−34 Since the porphyrin itself is achiral and produces no CD signal, its attachment to DNA invokes transfer of the chiral information on the DNA to the porphyrin. Porphyrins strongly absorb light around 420 nm (denoted Soret or B-band), thus the induced CD signal in this region provides an optimal handle to detect structural changes as it is well outside the window of the DNA absorbance. In this way, monitoring the structural change from B-DNA to Z-DNA with increasing salt concentration was successful.
Figure 2.
(a) First examples of end-of-DNA modification with porphyrinoids to create artificial sequence specific nucleases.25,26 (b) Structure (left) and schematic (middle) of porphyrins acting as caps (red and blue bars) for the blunt end of DNA; modeled DNA structure and induced Soret-band CD spectra of the porphyrins (right), showing DNA structure dependent exciton coupling. Reproduced from ref (28) with permission from the Royal Society of Chemistry.
Using the same methodology, Yamamura et al. developed a zinc porphyrin–DNA based on a p-tolylporphyrin.35 The zinc porphyrin–DNA did not exhibit long-range chromophore–chromophore exciton coupling at low salt concentration, but under high salt concentration strong interactions between porphyrins were observed. The system forms intermolecular stacks through interaction of the porphyrins, leading to the formation of insoluble aggregates. In fact, similar interstrand interactions were observed by Berova and Balaz,34 though here the systems remained soluble in aqueous solvents. Therefore, the nature of the porphyrin plays an important factor in how the modified DNA behaves. Overall, those examples demonstrate the versatility of porphyrins attached to DNA with variable functionality, that is, as enzyme mimic (artificial nuclease) or as chiroptical marker.
2.2. Porphyrins Embedded within the DNA
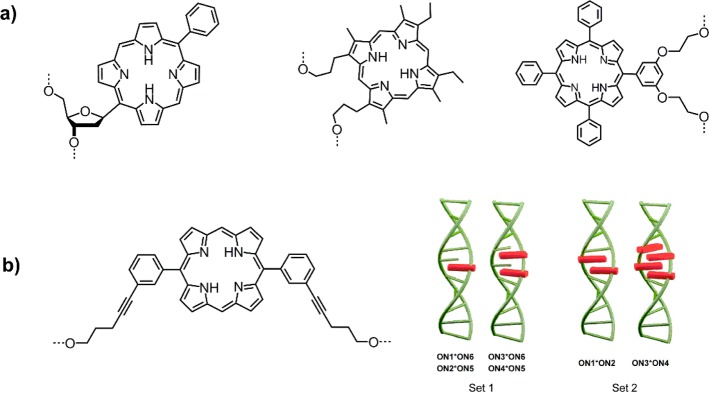
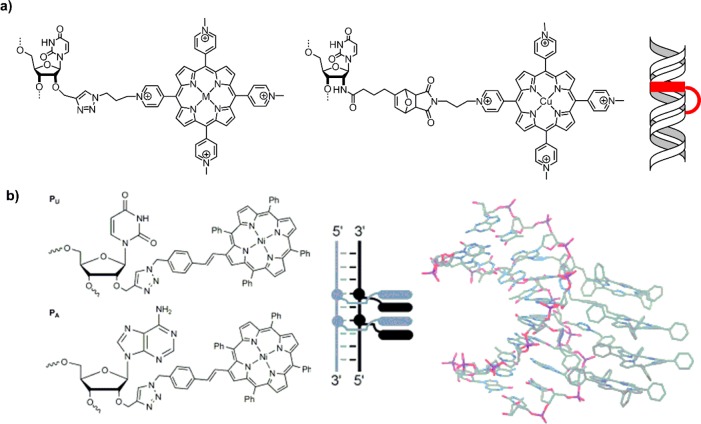
Very different approaches to porphyrin–DNA were explored by Kool et al.,36 Richert et al.,37 and Murashima and Sugimoto,38 where either the nucleobase or the entire nucleoside was replaced by a porphyrin (Figure 3a). The porphyrin here is actually positioned within the interior base-stacking region of the DNA. Recently, Häner et al.39 used this approach to create a four-porphyrin stack, where the complementary strands contain up to two porphyrins each; the interlocking nature of the array compensates the otherwise strongly destabilizing effect of the porphyrin modification (Figure 3b). Analysis of the constructs using CD, UV–vis, and fluorescence spectroscopy showed that in all these cases the DNA still forms a B-type duplex and that the porphyrins largely form an H-aggregate with concomitant exciton coupling; the porphyrin units also seem to stack very well with the neighboring base pairs.
Figure 3.
(a) Structures of base replacement36 and nucleoside surrogates37,38 for porphyrins embedded within the base stacking region of DNA. (b) DNA interior porphyrin stack creating stable H-aggregates through interlocked formation of the assembly from complementary porphyrin modified DNA strands. Reprinted with permission from ref (39). Copyright 2014 American Chemical Society.
Weaker hybridization (i.e., lower melting temperature, Tm) was seen in some of the systems, which depends on the nature of the modification, the nucleobase opposite to the porphyrin, and the number of adjacent porphyrins to create a stabilizing stacking effect. As rule of thumb, an abasic site opposite to the porphyrin can reduce steric clashes, and stacking of multiple porphyrins within the DNA stabilizes the duplex, though this has to be determined on a case by case basis.
2.3. Porphyrin Located on the Outside of the DNA
Several systems have been investigated to create porphyrin arrays with distinct composition, which are placed on the outside of the DNA, particularly within the minor or major groove. The attachment site of the porphyrin on the nucleoside will direct the porphyrin to the corresponding groove and is obviously crucial for the design of the array.
2.3.1. Porphyrin Arrays in the Minor Groove
Strategies that have been explored to place the porphyrins into the minor groove of the DNA include the attachment of the porphyrins to the 2′-position of the ribose moiety, or to the phosphate group. Generally the porphyrins are attached via cross-linking, including amide formation with either long tethers, as explored by Meunier et al. (Figure 4a),40 direct amidation of 2′-amino ribose with carboxy-porphyrin as in the system by Sitaula and Reed,41 or linking to a phosphoramidate through Michael addition as in the porphyrin–DNA of Majima et al. (Figure 4b)42 All provide alternative routes and represent a selection of the diversity of the chemistries that can be used. The study of Majima’s system, which included both free-base and zinc metalated porphyrins attached to complementary strands, showed that a B-type DNA duplex was retained upon hybridization, and the chromophores formed a face-to-face dimer near the minor groove of the ODN. The presence of the metal had a strong effect on the duplex formation and stability, where less stable dimers were obtained with two zinc-porphyrins. This confirms that the additional axial water ligands on the zinc make the zinc porphyrins less hydrophobic and can prevent efficient π–π stacking, analogous to the intermolecular interactions described above. Therefore, the choice of the central metal in the porphyrin not only influences the optical properties but also has an impact on the overall structure and stability through modulating interporphyrin stacking.
Figure 4.
(a) Attachment of the porphyrin to the 2′-position of the ribose through tethers (left)40 or direct amidation (right)41 will position the substituents in the minor groove of the DNA. (b) Conjugation to the phosphate backbone leads to external placement, and face-to-face stacked dimers can lead to a stabilizing effect in the duplex, which is dependent on the central metal and its potential additional axial ligands. Reprinted with permission from ref (42). Copyright 2008 American Chemical Society.
Postsynthetic modification with a TMPyP-type porphyrin was achieved in a straightforward manner using cycloadditions, such as copper catalyzed alkyne–azide click chemistry or Diels–Alder reaction, as shown by Wellner and Wagenknecht (Figure 5a).43 In this example, analysis of the Tm values and of the CD spectra revealed that the site of modification is not prevalent in the standard base-pairing and that the porphyrins intercalate into the DNA. This, however, is strongly dependent on the coupling chemistry and with it the nature of the linker used, where sterically less demanding triazole linkers favor intercalation. Analogously, Filichev et al.44 introduced the porphyrins via click chemistry but through the β-pyrrolic position, which provides a planar system between the porphyrin ring and substituent (Figure 5b). A convenient microwave assisted method to synthesize multiporphyrin-DNA arrays consisting of one to four porphyrin units in various locations within the ODN strand was developed. Attachment of four porphyrins in adjacent DNA strands lead to a significant stabilization of the DNA duplex through the formation of H-aggregates of the porphyrins in the minor groove of the DNA. It should be noted that the minor groove arrays have in general been little explored and thus contribute a field of research with great potential to grow.
Figure 5.
(a) Postsynthetic modification of DNA with porphyrins through click chemistry or Diels–Alder reaction; the flexibility of the linker allows the porphyrin to intercalate into the DNA.43 (b) Attachment of the porphyrins through the β-pyrrolic position reduces steric hindrances, leading to stable H-aggregates of the porphyrins in the minor groove. Reprinted by permission of John Wiley & Sons, Inc. from ref (44).
2.3.2. Porphyrin Arrays in the Major Groove
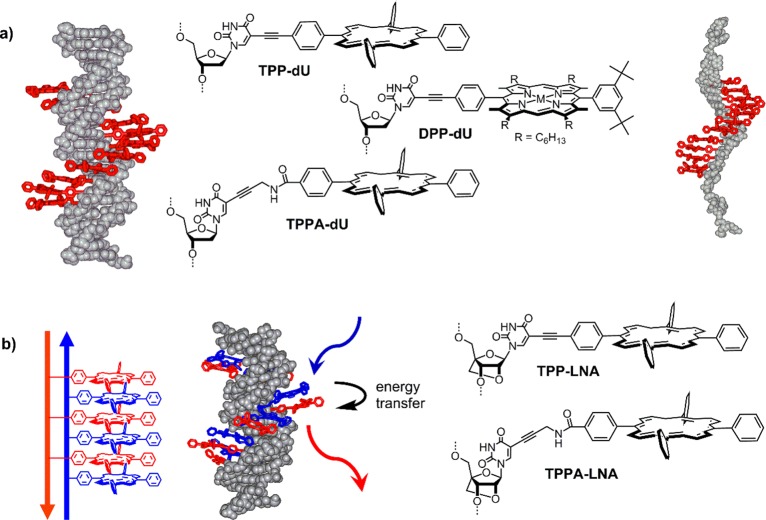
To create multiporphyrin arrays, where the porphyrins are held rigidly and in a predictable way on the DNA, new strategies had to be developed. We found that the use of Sonogashira coupling between 5-iodo-deoxyuridine (5-iodo-dU) and alkyne porphyrins is most versatile to synthesize building blocks for programmed insertion into DNA: the porphyrin is attached to the nucleobase and will protrude from the DNA into the major groove, leaving the Watson–Crick base recognition untouched.45 The use of metalated porphyrins is crucial to avoid copper metalation during the coupling; most conveniently zinc is used, which is subsequently lost in the DNA synthesis, yielding the free-base porphyrin–DNA. As alternative methods, amide coupling,46 click chemistry47 or maleimide–thiol conjugation48 can equally well be used. Structurally different modifiers such as diphenyl porphyrin (DPP-dU), tetraphenyl porphyrin (TPP-dU), or propargylamide-linked TPP (TPPA-dU) were thus synthesized (Figure 6a) Phosphitylation is straightforward, though the phosphoramidites are highly susceptible to oxidation due to the photosensitizing activity of the porphyrin. Silica gel column chromatography has thus to be performed under strict exclusion of light and oxygen, but precipitation from DCM-hexane is equally efficient for purification. The modifiers were successfully incorporated into DNA in variable numbers, ranging from one up to 12 porphyrins per DNA.49−51 This demonstrates that there is virtually no limitation in the number of large modifications that can be attached to DNA, and the programmable nature of DNA synthesis allows for easy design of the modified strand. Purification and solubility of highly modified DNA can be an issue, which strongly depends on the nature of the porphyrin and the overall length of the DNA. Purification is best performed by reverse phase HPLC using methanol and hexafluoro-isopropanol–triethylamine buffer.
Figure 6.
(a) First generation of porphyrin arrays with putative structure of the dsDNA array and induced helical stack in the ssDNA.49−51 Reprinted with permission from ref (49). Copyright 2007 American Chemical Society. (b) Second generation zipper-porphyrin array, where the porphyrins are attached to both complementary DNA strands; different metalation leads to a photonic wire showing efficient energy transfer from ZnTPP to 2HTPP.46,52,53 Reproduced from ref (46) with permission from the Royal Society of Chemistry.
The formation of DNA duplexes and the correct arrangement of the porphyrins in the major groove was confirmed by spectroscopy and molecular modeling, and the porphyrins form a nicely stacked helical chromophore array. While a single porphyrin shows unperturbed absorption and emission properties, the multiporphyrin array displays a significant broadening (TPP) or even splitting (DPP) of the porphyrin Soret band at 420 nm and quenched fluorescence. The analysis of the dipoles of the porphyrins indicate that they are coupled as a combination of H- and J-aggregates.51 We also observed that the porphyrins induce a stable helical arrangement in the ssDNA through stacking.49 This means that the duplex is actually not required to form a helical stack of chromophores, which can potentially act as electronic wires due to efficient coupling. But this will have to be judged on a case-by-case basis.
While this first generation of porphyrin–DNA was ideal for proof-of-concept, it also showed its limitations: the duplex stability is greatly reduced, which is strongly dependent on the nature of the porphyrin and the number of modifications. On average the thermodynamic destabilization is greater for DPP (ΔTm = −7 °C per porphyrin)51 compared to TPP (ΔTm = −3.5 °C per porphyrin),49 and most likely arises from local structural perturbation of the DNA. A way around this issue is to create interlocked arrays, called zipper arrays, where the porphyrins are attached to the complementary sequences in an alternate manner.46,52 This has several consequences: first the DNA duplex is stabilized by >40 °C in a 12-porphyrin array (ΔTm = +0.5 °C per porphyrin), and second the two porphyrin–DNA strands can be metalated separately with different metals. The duplex stability can further be increased by using the preorganized “locked nucleic acid” (LNA, Figure 6b) to give ΔTm of up to +1.7 °C per porphyrin.53 In terms of metalation, zinc, copper, or cobalt were inserted postsynthetically. With a mixed zinc–free-base porphyrin the first reversible photonic wire based on a DNA scaffolding approach was created,52 which shows efficient energy transfer in the annealed duplex state but not in the denatured single strand state.
The hydrophobic nature of the porphyrin has far reaching consequences on the structure and properties of the DNA. We found through spectroscopic studies (absorption and emission, CD, EPR, SAXS) that there are substantial intermolecular interactions through π-stacking of porphyrins, both in ssDNA and dsDNA,46,53,54 similar to what has been reported by other groups.34,35 Overall two to four DNA duplexes associate with an interstrand center-to-center distance of 6.5–8.9 Å of the porphyrins. Notably this does not lead to aggregation and precipitation, but to the formation of discrete bundles which remain soluble in water. These interactions are dominant at concentrations >5 μM DNA and >100 mM NaCl, but persist even in pure water. This so far prevents the analysis of pure intramolecular porphyrin interactions as there is always an intermolecular component present. On the other hand, this can lead to the stabilization of noncanonical DNA structures, such as GA-duplexes,30,55 i-motifs,47 and G-quadruplexes,56 in some cases leading to a change in the topology of those structures.57 This feature could very well be advantageous in the design of photonic systems, as it could allow organization of the chromophores in different orientations with respect to each other than would be observed from a DNA duplex.58 The intermolecular associations are also observed with small aromatic molecules, and might be a more general feature in modified DNA; this can be explored as molecular glue to add another level of interactions.59,60
3. Applications of Porphyrin–DNA
The chemistries and design approaches are now well set, and it is timely to consider applications of the porphyrin–DNA. The potential creation of energy transfer systems has been mentioned above; here we would like to introduce two other applications that we are pursuing and where porphyrins prove to be advantageous over other modifiers.
3.1. Porphyrins as Electrochemical Tags for DNA Sensing
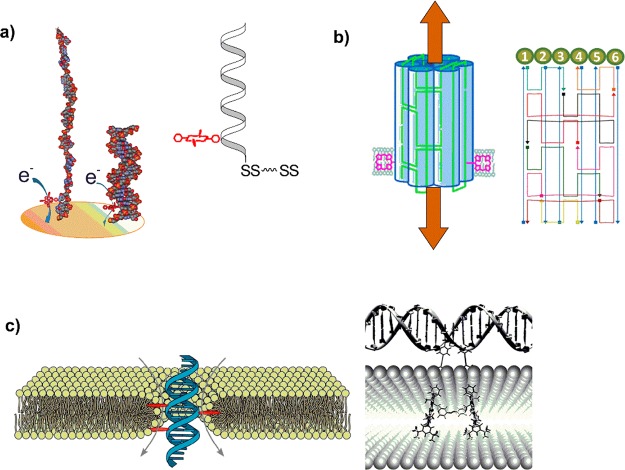
Porphyrins are not only optically active, but have a rich electrochemistry, which again depends strongly on the structure, central metal, and microenvironment. We used cobalt porphyrin–DNA to create highly sensitive geno-sensors (Figure 7a).61 The porphyrin is located close to the electrode surface, and we found that upon duplex formation the ionic current is greatly dimished. This is explained by placing the porphyrin into the hydrophobic major groove of the dsDNA and giving it limited access to the electrolyte, compared to the exposed single strand arrangement. The mechanism is distinctively different from other distance based “signal on–off” systems. The selective detection of complementary strands including single-nucleotide polymorphism was demonstrated, and it was calculated that as few as 1000 DNA molecules can be measured. In this way, an avian influenza virus (H5N1) based DNA sequence was detected at femtomolar levels from competing noncomplementary sequences.
Figure 7.
(a) Schematic of an electrochemical genosensor based on cobalt porphyrin with an efficient “signal-off” detection of the target sequence in the femtomolar range.61 (b) DNA bundle consisting of six DNA helices with two porphyrins as lipophilic anchors to create artificial nanopores with a 2 nm inner pore diameter, Reprinted by permission of John Wiley & Sons, Inc. from ref (63). (c) A minimal porphyrin–DNA pore where the current is induced through a flow of ions along the DNA backbone (left).64 This is compared to a two-porphyrin-DNA with longer linkers and elongated distance between the porphyrin attachment sites (right), embedding the porphyrins in the membrane while keeping the underlying DNA scaffold in the aqueous environment. Reproduced from ref (66) with permission from the Royal Society of Chemistry.
3.2. Porphyrin–DNA Based Lipid Bilayer Spanning Nanopores
Nanopores are currently under intense investigation; their facile insertion into membranes can be achieved on a single molecule level, and ionic current measurement through these nanopores gives rise to single molecule detectors. The research is focused on naturally occurring biological nanopores (e.g., α-hemolysin), solid-state nanopores, and hybrids of the two, with a strong focus on single molecule DNA sequencing. We have thus explored the formation of DNA origami based nanopores to create simple and tunable systems, because proteins are inherently tricky to modify at precise positions. Our pores consist of a bundle of six hexagonally arranged DNA duplexes, stabilized through crossover strands (Figure 7b).62 The DNA pore itself does not insert into the membrane due to the inherent energy mismatch to the hydrophobic environment of the membrane, therefore we initially added a hydrophobic belt consisting of ∼70 ethyl groups. Insertion was demonstrated by a steady ionic current through the DNA origami pore. The alkyl belt can be replaced by only two porphyrins, located on opposite sides of the pore, which showed equally efficient insertion.63 The porphyrin therefore clearly outweighs the alkyl groups by a large margin. The stable insertion of the DNA origami pores was confirmed by single molecule measurement of the ionic current and by fluorescence spectroscopy. We have further minimalized the system by using a simple, six porphyrin–DNA duplex (Figure 7c).64 We could demonstrate that this DNA duplex inserts stably into lipid bilayers, creating the smallest possible DNA based nanopore with maximum simplicity, which lacks a hollow central channel. By combining electrophysiology measurements with all-atom molecular dynamics simulations, we showed that ions flow at the DNA–lipid interface as the lipid head groups tilt toward the amphiphilic duplex, forming a toroidal pore filled with water and ions. The ionic current traces show well-defined insertion steps, closures, and gating, analogous to those observed for traditional protein channels or synthetic pores. In both larger and small pore, the porphyrin actually provides a very convenient handle with dual properties, namely, a hydrophobic anchor to efficiently embed the negatively charged DNA, which simultaneously acts as a chromophore to monitor the insertion and distribution of the DNA pore in the artificial membrane. It is interesting to note that the attachment site and number of porphyrins greatly influences the interaction with a lipid bilayer. While two porphyrins on opposite sides of DNA bundles direct larger constructs into the lipid bilayer, for simple dsDNA, this may not be sufficient. In our case, six porphyrins ensured complete transmembrane insertion, while single porphyrin modification of DNA retains the DNA in the aqueous environment. This has been used by Börjesson and Albinsson65−67 to arrange porphyrins within a lipid bilayer, and using the DNA as structural scaffold in the water phase where it shows normal duplex formation behavior, thereby creating a range of photoactive systems (Figure 7c). While this may sound counterintuitive, it can be explained by the overwhelming hydrophobic effect of the porphyrin, which compensates the energy barrier to insert the DNA only when in appropriate geometrical arrangement. Additionally, the linker length seems to be crucial as shorter linkers tend to embed the entire porphyrin–DNA, while longer linkers tend to only insert the porphyrin.
4. Conclusions
DNA certainly is a most versatile supramolecular scaffold, not only for porphyrins but for creating functional molecules in general, and for chromophore assemblies in particular. Porphyrins are ideal modifiers in many ways: their electronic properties can be fine-tuned and their lipophilicity can be used for anchoring DNA in hydrophobic environments, though this may lead to formation of intermolecular interactions and solubility issues. So far porphyrin–DNA has mainly been used for basic studies in terms of structure and optical properties. However, as outlined above many applications are emerging and may well be far-reaching in the fields of artificial photosynthesis or medicine. Where is the field heading? Future efforts should be directed to include other functionalities for applications in optoelectronics, which might be a major application;7 a large number of DNA modifiers are available, also commercially, and the chemistries are well laid out to create functional DNA where we can (more or less) reliably predict their properties. The programmable nature of DNA allows positioning the functional groups in well-defined spatial arrangements, for example, to alter the distance and avoid self-quenching through formation of H-aggregates. In addition, many other scaffolds for nanoarrays are compatible with DNA, such as MOFS, carbon nanotubes and -sheets, oligopeptides and proteins, nanoparticles, dendrimers, etc. Thus, the future of DNA nanoarchitectonics will certainly see more advances through the combination of a diverse set of scaffolds (including DNA origami) and modifiers, and it is timely to embed functional DNA into larger nanosystems to create truly tailored materials.
Biography
Eugen Stulz studied chemistry in Bern (Switzerland), from where he obtained his Ph.D. in 1998 in bioorganic chemistry, working on artificial nucleases. He received a SNF Fellowship to work as PDRA in Cambridge (U.K.), before starting his independent career in 2003 in Basel (Switzerland) as Treubel Fellow. In 2006, he was appointed lecturer in Southampton (U.K.), promoted to senior lecturer in 2010 and associate professor in 2014. His current research interests focus on the use of DNA as construction material for functional nanostructures, with applications in molecular electronics and biomedicinal chemistry.
Financial support by the BBSRC (BB/J001694/1), the European Union (FP7-PEOPLE-2012-IEF NANO-DNA), the EPSRC (EP/K039466/1), and Cancer Research UK (Pilot grant) is greatly acknowledged.
The author declares no competing financial interest.
References
- Wilkins M. H. F.; Stokes A. R.; Wilson H. R. Molecular Structure of Nucleic Acids: Molecular Structure of Deoxypentose Nucleic Acids. Nature 1953, 171, 738–740. 10.1038/171738a0. [DOI] [PubMed] [Google Scholar]
- Watson J. D.; Crick F. H. C. Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid. Nature 1953, 171, 737–738. 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Franklin R. E.; Gosling R. G. Molecular Configuration in Sodium Thymonucleate. Nature 1953, 171, 740–741. 10.1038/171740a0. [DOI] [PubMed] [Google Scholar]
- Stulz E. DNA Architectonics: towards the Next Generation of Bio-inspired Materials. Chem. - Eur. J. 2012, 18, 4456–4469. 10.1002/chem.201102908. [DOI] [PubMed] [Google Scholar]
- Bandy T. J.; Brewer A.; Burns J. R.; Marth G.; Nguyen T.; Stulz E. DNA as supramolecular scaffold for functional molecules: progress in DNA nanotechnology. Chem. Soc. Rev. 2011, 40, 138–148. 10.1039/B820255A. [DOI] [PubMed] [Google Scholar]
- Endo M.; Sugiyama H. Chemical Approaches to DNA Nanotechnology. ChemBioChem 2009, 10, 2420–2443. 10.1002/cbic.200900286. [DOI] [PubMed] [Google Scholar]
- Ensslen P.; Wagenknecht H. A. One-Dimensional Multichromophor Arrays Based on DNA: From Self-Assembly to Light-Harvesting. Acc. Chem. Res. 2015, 48, 2724–2733. 10.1021/acs.accounts.5b00314. [DOI] [PubMed] [Google Scholar]
- Malinovskii V. L.; Wenger D.; Häner R. Nucleic acid-guided assembly of aromatic chromophores. Chem. Soc. Rev. 2010, 39, 410–422. 10.1039/B910030J. [DOI] [PubMed] [Google Scholar]
- Teo Y. N.; Kool E. T. DNA-Multichromophore Systems. Chem. Rev. 2012, 112, 4221–4245. 10.1021/cr100351g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichev V. V.; Pedersen E. B.. Wiley Encyclopedia of Chemical Biology; John Wiley & Sons, Inc.: Hoboken N.J., 2009; Vol. 1; pp 493–524. [Google Scholar]
- Stulz E. Porphyrin-modified DNA as Construction Material in Supramolecular Chemistry and Nano-architectonics. Chimia 2015, 69, 678–683. 10.2533/chimia.2015.678. [DOI] [PubMed] [Google Scholar]
- Zhao L. Z.; Qu R.; Li A.; Ma R. J.; Shi L. Q. Cooperative self-assembly of porphyrins with polymers possessing bioactive functions. Chem. Commun. 2016, 52, 13543–13555. 10.1039/C6CC05449H. [DOI] [PubMed] [Google Scholar]
- Kc C. B.; D’Souza F. Design and photochemical study of supramolecular donor-acceptor systems assembled via metal-ligand axial coordination. Coord. Chem. Rev. 2016, 322, 104–141. 10.1016/j.ccr.2016.05.012. [DOI] [Google Scholar]
- Rioz-Martinez A.; Roelfes G. DNA-based hybrid catalysis. Curr. Opin. Chem. Biol. 2015, 25, 80–87. 10.1016/j.cbpa.2014.12.033. [DOI] [PubMed] [Google Scholar]
- Gust D. Supramolecular photochemistry applied to artificial photosynthesis and molecular logic devices. Faraday Discuss. 2015, 185, 9–35. 10.1039/C5FD00142K. [DOI] [PubMed] [Google Scholar]
- Day N. U.; Wamser C. C.; Walter M. G. Porphyrin polymers and organic frameworks. Polym. Int. 2015, 64, 833–857. 10.1002/pi.4908. [DOI] [Google Scholar]
- Hasobe T.; Kamat P. V.; Troiani V.; Solladie N.; Ahn T. K.; Kim S. K.; Kim D.; Kongkanand A.; Kuwabata S.; Fukuzumi S. Enhancement of light-energy conversion efficiency by multi-porphyrin arrays of porphyrin-peptide oligomers with fullerene clusters. J. Phys. Chem. B 2005, 109, 19–23. 10.1021/jp045246v. [DOI] [PubMed] [Google Scholar]
- Dunetz J. R.; Sandstrom C.; Young E. R.; Baker P.; Van Name S. A.; Cathopolous T.; Fairman R.; de Paula J. C.; Akerfeldt K. S. Self-assembling porphyrin-modified peptides. Org. Lett. 2005, 7, 2559–2561. 10.1021/ol050644h. [DOI] [PubMed] [Google Scholar]
- Sakamoto M.; Ueno A.; Mihara H. Multipeptide-metalloporphyrin assembly on a dendrimer template and photoinduced electron transfer based on the dendrimer structure. Chem. - Eur. J. 2001, 7, 2449–2458. . [DOI] [PubMed] [Google Scholar]
- Chen Y.; Zhao D.; Liu Y. Polysaccharide-porphyrin-fullerene supramolecular conjugates as photo-driven DNA cleavage reagents. Chem. Commun. 2015, 51, 12266–12269. 10.1039/C5CC04625D. [DOI] [PubMed] [Google Scholar]
- Clave G.; Chatelain G.; Filoramo A.; Gasparutto D.; Saint-Pierre C.; Le Cam E.; Pietrement O.; Guerineau V.; Campidelli S. Synthesis of a multibranched porphyrin-oligonucleotide scaffold for the construction of DNA-based nano-architectures. Org. Biomol. Chem. 2014, 12, 2778–2783. 10.1039/c4ob00202d. [DOI] [PubMed] [Google Scholar]
- D’Souza F.; Ito O. Supramolecular donor-acceptor hybrids of porphyrins/phthalocyanines with fullerenes/carbon nanotubes: electron transfer, sensing, switching, and catalytic applications. Chem. Commun. 2009, 4913–4928. 10.1039/b905753f. [DOI] [PubMed] [Google Scholar]
- DNA in Supramolecular Chemistry and Nanotechnology; Stulz E., Clever G. H., Eds.; John Wiley & Sons, Inc.: Chichester, U.K., 2015. [Google Scholar]
- Sargsyan G.; Leonard B. M.; Kubelka J.; Balaz M. Supramolecular ssDNA Templated Porphyrin and Metalloporphyrin Nanoassemblies with Tunable Helicity. Chem. - Eur. J. 2014, 20, 1878–1892. 10.1002/chem.201304153. [DOI] [PubMed] [Google Scholar]
- Casas C.; Lacey C. J.; Meunier B. Preparation of hybrid DNA cleaver oligonucleotide molecule based on a metallotris(methylpyridiniumyl)porphyrin motif. Bioconjugate Chem. 1993, 4, 366–371. 10.1021/bc00023a011. [DOI] [PubMed] [Google Scholar]
- Boutorine A. S.; Brault D.; Takasugi M.; Delgado O.; Hélène C. Chlorin-oligonucleotide conjugates: Synthesis, properties, and red light-induced photochemical sequence-specific DNA cleavage in duplexes and triplexes. J. Am. Chem. Soc. 1996, 118, 9469–9476. 10.1021/ja960062i. [DOI] [Google Scholar]
- Li H. D.; Fedorova O. S.; Trumble W. R.; Fletcher T. R.; Czuchajowski L. Site-specific photomodification of DNA by porphyrin-oligonucleotide conjugates synthesized via a solid phase h-phosphonate approach. Bioconjugate Chem. 1997, 8, 49–56. 10.1021/bc960074t. [DOI] [PubMed] [Google Scholar]
- Balaz M.; Li B. C.; Steinkruger J. D.; Ellestad G. A.; Nakanishi K.; Berova N. Porphyrins conjugated to DNA as CD reporters of the salt-induced B to Z-DNA transition. Org. Biomol. Chem. 2006, 4, 1865–1867. 10.1039/b603409h. [DOI] [PubMed] [Google Scholar]
- Balaz M.; Steinkruger J. D.; Ellestad G. A.; Berova N. 5′-Porphyrin-oligonucleotide conjugates: Neutral porphyrin-DNA interactions. Org. Lett. 2005, 7, 5613–5616. 10.1021/ol0522992. [DOI] [PubMed] [Google Scholar]
- Balaz M.; Li B. C.; Jockusch S.; Ellestad G. A.; Berova N. Tetraarylporphyrin as a selective molecular cap for non-Watson-Crick guanine-adenine base-pair sequences. Angew. Chem., Int. Ed. 2006, 45, 3530–3533. 10.1002/anie.200504431. [DOI] [PubMed] [Google Scholar]
- D’Urso A.; Holmes A. E.; Berova N.; Balaz M.; Purrello R. Z-DNA Recognition in B-Z-B Sequences by a Cationic Zinc Porphyrin. Chem. - Asian J. 2011, 6, 3104–3109. 10.1002/asia.201100161. [DOI] [PubMed] [Google Scholar]
- D’Urso A.; Mammana A.; Balaz M.; Holmes A. E.; Berova N.; Lauceri R.; Purrello R. Interactions of a Tetraanionic Porphyrin with DNA: from a Z-DNA Sensor to a Versatile Supramolecular Device. J. Am. Chem. Soc. 2009, 131, 2046–2047. 10.1021/ja808099u. [DOI] [PubMed] [Google Scholar]
- Pietropaolo A.; D’Urso A.; Purrello R.; Berova N. Effect of Different Z-Inducers on the Stabilization of Z Portion in BZ-DNA Sequence: Correlation Between Experimental and Simulation Data. Chirality 2015, 27, 773–778. 10.1002/chir.22502. [DOI] [PubMed] [Google Scholar]
- Mammana A.; Pescitelli G.; Asakawa T.; Jockusch S.; Petrovic A. G.; Monaco R. R.; Purrello R.; Turro N. J.; Nakanishi K.; Ellestad G. A.; Balaz M.; Berova N. Role of Environmental Factors on the Structure and Spectroscopic Response of 5′-DNA-Porphyrin Conjugates Caused by Changes in the Porphyrin-Porphyrin Interactions. Chem. - Eur. J. 2009, 15, 11853–11866. 10.1002/chem.200902029. [DOI] [PubMed] [Google Scholar]
- Onoda A.; Igarashi M.; Naganawa S.; Sasaki K.; Ariyasu S.; Yamamura T. Circular Dichroism of Neutral Zinc Porphyrin-Oligonucleotide Conjugates Modified with Flexible Linker. Bull. Chem. Soc. Jpn. 2009, 82, 1280–1286. 10.1246/bcsj.82.1280. [DOI] [Google Scholar]
- Morales-Rojas H.; Kool E. T. A porphyrin C-nucleoside incorporated into DNA. Org. Lett. 2002, 4, 4377–4380. 10.1021/ol0267376. [DOI] [PubMed] [Google Scholar]
- Berlin K.; Jain R. K.; Simon M. D.; Richert C. A porphyrin embedded in DNA. J. Org. Chem. 1998, 63, 1527–1535. 10.1021/jo9718051. [DOI] [Google Scholar]
- Murashima T.; Hayata K.; Saiki Y.; Matsui J.; Miyoshi D.; Yamada T.; Miyazawa T.; Sugimoto N. Synthesis, structure and thermal stability of fully hydrophobic porphyrin-DNA conjugates. Tetrahedron Lett. 2007, 48, 8514–8517. 10.1016/j.tetlet.2007.09.147. [DOI] [Google Scholar]
- Vybornyi M.; Nussbaumer A. L.; Langenegger S. M.; Häner R. Assembling Multiporphyrin Stacks Inside the DNA Double Helix. Bioconjugate Chem. 2014, 25, 1785–1793. 10.1021/bc500297e. [DOI] [PubMed] [Google Scholar]
- Dubey I.; Pratviel G.; Meunier B. Synthesis and DNA cleavage of 2′-O-amino-linked metalloporphyrin-oligonucleotide conjugates. J. Chem. Soc.-Perkin Trans. 1 2000, 3088–3095. 10.1039/b004431h. [DOI] [Google Scholar]
- Sitaula S.; Reed S. M. Porphyrin conjugated to DNA by a 2′-amido-2′-deoxyuridine linkage. Bioorg. Med. Chem. Lett. 2008, 18, 850–855. 10.1016/j.bmcl.2007.11.033. [DOI] [PubMed] [Google Scholar]
- Endo M.; Fujitsuka M.; Majima T. Diastereochemically controlled porphyrin dimer formation on a DNA duplex scaffold. J. Org. Chem. 2008, 73, 1106–1112. 10.1021/jo7025004. [DOI] [PubMed] [Google Scholar]
- Wellner C.; Wagenknecht H. A. Synthesis of DNA conjugates with metalated tetracationic porphyrins by postsynthetic cycloadditions. Org. Lett. 2014, 16, 1692–1695. 10.1021/ol500364j. [DOI] [PubMed] [Google Scholar]
- Stephenson A. W. I.; Bomholt N.; Partridge A. C.; Filichev V. V. Significantly Enhanced DNA Thermal Stability Resulting from Porphyrin H-Aggregate Formation in the Minor Groove of the Duplex. ChemBioChem 2010, 11, 1833–1839. 10.1002/cbic.201000326. [DOI] [PubMed] [Google Scholar]
- Bouamaied I.; Stulz E. Synthesis and spectroscopic properties of porphyrin-substituted uridine and deoxyuridine. Synlett 2004, 1579–1583. 10.1055/s-2004-829541. [DOI] [Google Scholar]
- Brewer A.; Siligardi G.; Neylon C.; Stulz E. Introducing structural flexibility into porphyrin-DNA zipper arrays. Org. Biomol. Chem. 2011, 9, 777–782. 10.1039/C0OB00535E. [DOI] [PubMed] [Google Scholar]
- Stephenson A. W. I.; Partridge A. C.; Filichev V. V. Synthesis of beta-Pyrrolic-Modified Porphyrins and Their Incorporation into DNA. Chem. - Eur. J. 2011, 17, 6227–6238. 10.1002/chem.201003200. [DOI] [PubMed] [Google Scholar]
- Endo M.; Fujitsuka M.; Majima T. Programmable conformational regulation of porphyrin dimers on geometric scaffold of duplex DNA. Tetrahedron 2008, 64, 1839–1846. 10.1016/j.tet.2007.11.096. [DOI] [Google Scholar]
- Fendt L. A.; Bouamaied I.; Thöni S.; Amiot N.; Stulz E. DNA as supramolecular scaffold for porphyrin arrays on the nanorneter scale. J. Am. Chem. Soc. 2007, 129, 15319–15329. 10.1021/ja075711c. [DOI] [PubMed] [Google Scholar]
- Bouamaied I.; Fendt L. A.; Häussinger D.; Wiesner M.; Thöni S.; Amiot N.; Stulz E. Porphyrin-DNA: A supramolecular scaffold for functional molecules on the nanometre scale. Nucleosides, Nucleotides Nucleic Acids 2007, 26, 1533–1538. 10.1080/15257770701544468. [DOI] [PubMed] [Google Scholar]
- Bouamaied I.; Nguyen T.; Rühl T.; Stulz E. Supramolecular helical porphyrin arrays using DNA as a scaffold. Org. Biomol. Chem. 2008, 6, 3888–3891. 10.1039/b813584c. [DOI] [PubMed] [Google Scholar]
- Nguyen T.; Brewer A.; Stulz E. Duplex Stabilization and Energy Transfer in Zipper Porphyrin-DNA. Angew. Chem., Int. Ed. 2009, 48, 1974–1977. 10.1002/anie.200805657. [DOI] [PubMed] [Google Scholar]
- Singleton D. G.; Hussain R.; Siligardi G.; Kumar P.; Hrdlicka P. J.; Berova N.; Stulz E. Increased duplex stabilization in porphyrin-LNA zipper arrays with structure dependent exciton coupling. Org. Biomol. Chem. 2016, 14, 149–157. 10.1039/C5OB01681A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.; Hakansson P.; Edge R.; Collison D.; Goodman B. A.; Burns J. R.; Stulz E. EPR based distance measurement in Cu-porphyrin-DNA. New J. Chem. 2014, 38, 5254–5259. 10.1039/C4NJ00673A. [DOI] [Google Scholar]
- Sargsyan G.; Balaz M. Porphyrin-DNA conjugates: porphyrin induced adenine-guanine homoduplex stabilization and interduplex assemblies. Org. Biomol. Chem. 2012, 10, 5533–5540. 10.1039/c2ob25710f. [DOI] [PubMed] [Google Scholar]
- Jayawickramarajah J.; Tagore D. M.; Tsou L. K.; Hamilton A. D. Allosteric control of self-assembly: Modulating the formation of guanine quadruplexes through orthogonal aromatic interactions. Angew. Chem., Int. Ed. 2007, 46, 7583–7586. 10.1002/anie.200701883. [DOI] [PubMed] [Google Scholar]
- Saha S.; Cai J. F.; Eiler D.; Hamilton A. D. Programing the formation of DNA and PNA quadruplexes by pi-pi-stacking interactions. Chem. Commun. 2010, 46, 1685–1687. 10.1039/b915955j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doluca O.; Withers J. M.; Loo T. S.; Edwards P. J. B.; Gonzalez C.; Filichev V. V. Interdependence of pyrene interactions and tetramolecular G4-DNA assembly. Org. Biomol. Chem. 2015, 13, 3742–3748. 10.1039/C4OB02499K. [DOI] [PubMed] [Google Scholar]
- Kashida H.; Hayashi T.; Fujii T.; Asanuma H. A Cationic Dye Triplet as a Unique “Glue” That Can Connect Fully Matched Termini of DNA Duplexes. Chem. - Eur. J. 2011, 17, 2614–2622. 10.1002/chem.201003059. [DOI] [PubMed] [Google Scholar]
- Baumstark D.; Wagenknecht H. A. Perylene bisimide dimers as fluorescent ″Glue″ for DNA and for base-mismatch detection. Angew. Chem., Int. Ed. 2008, 47, 2612–2614. 10.1002/anie.200705237. [DOI] [PubMed] [Google Scholar]
- Grabowska I.; Singleton D. G.; Stachyra A.; Gora-Sochacka A.; Sirko A.; Zagorski-Ostoja W.; Radecka H.; Stulz E.; Radecki J. A highly sensitive electrochemical genosensor based on Co-porphyrin-labelled DNA. Chem. Commun. 2014, 50, 4196–4199. 10.1039/c4cc00172a. [DOI] [PubMed] [Google Scholar]
- Burns J. R.; Stulz E.; Howorka S. Self-Assembled DNA Nanopores That Span Lipid Bilayers. Nano Lett. 2013, 13, 2351–2356. 10.1021/nl304147f. [DOI] [PubMed] [Google Scholar]
- Burns J. R.; Gopfrich K.; Wood J. W.; Thacker V. V.; Stulz E.; Keyser U. F.; Howorka S. Lipid-Bilayer-Spanning DNA Nanopores with a Bifunctional Porphyrin Anchor. Angew. Chem., Int. Ed. 2013, 52, 12069–12072. 10.1002/anie.201305765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göpfrich K.; Li C.-Y.; Mames I.; Bhamidimarri S. P.; Ricci M.; Yoo J.; Mames A.; Ohmann A.; Winterhalter M.; Stulz E.; Aksimentiev A.; Keyser U. F. Ion Channels Made from a Single Membrane-Spanning DNA Duplex. Nano Lett. 2016, 16, 4665–4669. 10.1021/acs.nanolett.6b02039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woller J. G.; Hannestad J. K.; Albinsson B. Self-Assembled Nanoscale DNA-Porphyrin Complex for Artificial Light Harvesting. J. Am. Chem. Soc. 2013, 135, 2759–2768. 10.1021/ja311828v. [DOI] [PubMed] [Google Scholar]
- Börjesson K.; Woller J. G.; Parsa E.; Martensson J.; Albinsson B. A bioinspired self assembled dimeric porphyrin pocket that binds electron accepting ligands. Chem. Commun. 2012, 48, 1793–1795. 10.1039/c2cc17434k. [DOI] [PubMed] [Google Scholar]
- Börjesson K.; Tumpane J.; Ljungdahl T.; Wilhelmsson L. M.; Norden B.; Brown T.; Martensson J.; Albinsson B. Membrane-Anchored DNA Assembly for Energy and Electron Transfer. J. Am. Chem. Soc. 2009, 131, 2831–2839. 10.1021/ja8038294. [DOI] [PubMed] [Google Scholar]