Conspectus
Cells are highly advanced microreactors that form the basis of all life. Their fascinating complexity has inspired scientists to create analogs from synthetic and natural components using a bottom-up approach. The ultimate goal here is to assemble a fully man-made cell that displays functionality and adaptivity as advanced as that found in nature, which will not only provide insight into the fundamental processes in natural cells but also pave the way for new applications of such artificial cells.
In this Account, we highlight our recent work and that of others on the construction of artificial cells. First, we will introduce the key features that characterize a living system; next, we will discuss how these have been imitated in artificial cells. First, compartmentalization is crucial to separate the inner chemical milieu from the external environment. Current state-of-the-art artificial cells comprise subcompartments to mimic the hierarchical architecture of eukaryotic cells and tissue. Furthermore, synthetic gene circuits have been used to encode genetic information that creates complex behavior like pulses or feedback. Additionally, artificial cells have to reproduce to maintain a population. Controlled growth and fission of synthetic compartments have been demonstrated, but the extensive regulation of cell division in nature is still unmatched.
Here, we also point out important challenges the field needs to overcome to realize its full potential. As artificial cells integrate increasing orders of functionality, maintaining a supporting metabolism that can regenerate key metabolites becomes crucial. Furthermore, life does not operate in isolation. Natural cells constantly sense their environment, exchange (chemical) signals, and can move toward a chemoattractant. Here, we specifically explore recent efforts to reproduce such adaptivity in artificial cells. For instance, synthetic compartments have been produced that can recruit proteins to the membrane upon an external stimulus or modulate their membrane composition and permeability to control their interaction with the environment. A next step would be the communication of artificial cells with either bacteria or another artificial cell. Indeed, examples of such primitive chemical signaling are presented. Finally, motility is important for many organisms and has, therefore, also been pursued in synthetic systems. Synthetic compartments that were designed to move in a directed, controlled manner have been assembled, and directed movement toward a chemical attractant is among one of the most life-like directions currently under research.
Although the bottom-up construction of an artificial cell that can be truly considered “alive” is still an ambitious goal, the recent work discussed in this Account shows that this is an active field with contributions from diverse disciplines like materials chemistry and biochemistry. Notably, research during the past decade has already provided valuable insights into complex synthetic systems with life-like properties. In the future, artificial cells are thought to contribute to an increased understanding of processes in natural cells and provide opportunities to create smart, autonomous, cell-like materials.
Introduction
Cells are regarded as the basic building blocks of life. The smallest entity generally considered to be living is a single cell, and all life forms are either uni- or multicellular organisms. Contemporary cells are a product of nature’s evolutionary sculpting. As such, they are highly complex and efficient microreactors, which have inspired scientists to construct synthetic equivalents.
Currently, we have a fair understanding of many processes that take place in a natural cell. The structure and function of individual components and even entire biochemical pathways have been elucidated. The interplay between all these factors, however, puts the complexity of a natural cell largely beyond the grasp of contemporary science. The field of synthetic biology tries to take the next step in understanding the integration of all these processes by creating a minimal cell either by genome-editing of a natural organism (top-down approach) or by constructing from individual building blocks a structure that mimics the essential aspects of a natural cell (bottom-up approach).1 The philosophy behind the bottom-up approach, which we will limit ourselves to in this Account, is that scientists can only truly understand a natural cell if they can make one from scratch.
Yet to make a living artificial cell, one first has to consider what the minimal criteria for life are. The chemoton model, developed by Tibor Gánti, is often used to describe minimal life.2 According to his model, an entity comprising (i) a chemical boundary system, (ii) a chemical information system, and (iii) a self-reproducing chemical motor (metabolism) can be considered “alive”. Additionally, (iv) growth and reproduction are needed for survival of the species. Finally, (v) adaptivity is paramount for life’s survival in a dynamic environment.
Integrating these characteristics in a single synthetic system is an ambitious yet daunting goal. In recent years, however, several groups have successfully recreated simplified characteristics of life in synthetic systems, in particular employing nano- or micrometer-sized self-assembled compartments that can encapsulate a wide variety of (macro)molecules.3,4 Such systems are usually termed artificial cells or semisynthetic minimal cells.
Here, we will discuss the design, assembly, and behavior of artificial cells with emphasis on strategies that integrate life-like characteristics and display complex and adaptive behavior. The five criteria of life defined above will be used to guide our discussion. We have limited ourselves to membrane-bound compartments; structures like water-in-oil droplets5 and coacervates6 are beyond the scope of this Account. Rather than being comprehensive, we aim to highlight the current state of the art, illustrate it with select examples, and discuss the direction in which this field is heading.
Synthetic Compartments
All living systems necessitate a semipermeable boundary to sustain life in a changing environment. Primarily, its permeability should be finely tuned to retain vital components while exchanging nutrients and waste with the environment. A natural cell carefully controls these processes using a semipermeable lipid membrane that contains channels, receptors, and carrier ionophores, among others. Over the years, a wide variety of synthetic compartments have been developed that also allow control over the permeability of their shell. Design factors that influence permeability include the chemical nature of the membrane building blocks, membrane thickness, the presence of pores and channels, and domain formation in heterogeneous membranes.3 Today, there are several established procedures for creating nano- and microcompartments that can facilitate reactions while exchanging reagents and products with the environment. These include lipid and polymeric vesicles (liposomes and polymersomes), hybrids of these, virus capsids, colloidosomes, and coacervates.3
While our control over the permeability, size, stimuli-responsiveness, and biodegradability of these compartments has greatly improved in recent years, they are still fairly basic mimics of the architecture of natural cells. Especially eukaryotic cells are characterized by an elaborate internal structure through which processes are separated via intracellular membranes. Recently, some designs have incorporated multiple compartments to mimic such natural structures.
Multicompartmentalized Vesicles
Prominent classes of multicompartmentalized systems that have been developed include liposomes within layer-by-layer capsules (capsosomes), liposome-in-liposome (vesosomes), and polymersome-in-polymersome architectures, and multisomes (Figure 1). Such structures have been extensively reviewed elsewhere;3,11 here we limit ourselves to highlighting some recently developed life-like geometries.
Figure 1.
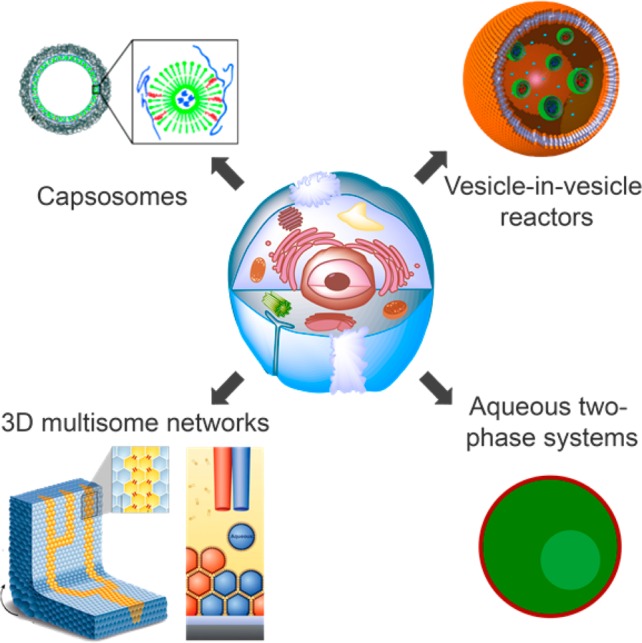
Prominent classes of multicompartmentalized vesicles. Their design is often inspired by the architecture of a eukaryotic cell (middle). Adapted with permission from refs (7−10). Copyrights 2009 and 2014 Wiley, 2013 American Association for the Advancement of Science, and 2010 American Chemical Society.
We and our collaborators have reported a multicompartmentalized vesicle whose architecture resembles a eukaryotic cell, as it contains organelles to create different chemical environments. In our system, the successive enzymes of an enzymatic cascade reaction were encapsulated in different polymeric nanoreactors within a large polymersome. The semipermeable nature of the nanoreactors facilitated the diffusion of reagents and products, while restricting the enzymes to their respective subcompartments. This design not only localized the successive reactions to specialized compartments, but also successfully separated incompatible enzymes into different subcompartments. Consequently, the cascade proceeded more efficiently in the multicompartmentalized vesicle than in bulk.8
Large networks of tightly connected aqueous compartments can be fabricated using the multisome-approach developed by Bayley’s group.12 These networks are formed when multiple lipid monolayer-covered water-in-oil droplets make contact and form droplet interface bilayers (DIBs). Recently, this approach was used to isolate enzymatic reactions in different compartments. Each enzymatic reaction produced the substrate for the following metabolic step in an adjacent compartment.13 Rather than mimicking the cellular architecture as closely as the above-described vesicle-in-vesicle approach, these multicompartment vesicle networks may provide a new tool to make tissue-like soft matter, although the number of bilayers separating these synthetic cells is in fact one too few. Nevertheless, impressive 3D networks of multisomes with defined geometries and conductivity have been created.9
There are subcompartments that are membraneless, both in nature and in synthetic systems. Macromolecular crowding in the cytosol leads to aqueous phase separation, which has been shown to play an important role in biological processes.5 Keating and co-workers created phase-separated subcompartments by encapsulating a dextran/poly(ethylene glycol) (PEG) aqueous two-phase system in a liposome. The resulting liposome contained two distinct aqueous compartments to which various macromolecules selectively partitioned.14 Such systems hold promise for the dynamic localization of macromolecules to distinct compartments of a synthetic cell, which can be used to study the effects of macromolecular crowding on essential biological functions.
For the next step in life-like multicompartmentalized vesicles, better control over the number, positioning, and membrane permeability of subcompartments is needed. Although for capsosomes the deposition of subcompartments is highly controllable, the current formation procedures for vesicle-in-vesicle reactors usually prohibit such control. Moreover, membrane transporters enable natural organelles to specifically uptake or excrete certain classes of molecules. Such discrimination is unreachable using current lipid or polymeric building blocks. Facile incorporation of selective membrane channels into synthetic vesicles would permit more diversified subcompartments with specific functions like natural organelles.
Genetically Programming Artificial Cells
Life requires not only some type of chemical boundary but also a chemical information system. Since all life forms use DNA to store information, it is also the information carrier of choice to construct a minimal cell. Yet, even the simplest organism contains hundreds of carefully regulated genes. Although regulation of synthetic networks of that size is yet unattainable, simpler synthetic gene circuits (SGCs) can already display complex behavior from temporal expression patterns generated by combining genetic modules. Such SGCs have been incorporated into living cells as well as in cell-free transcription–translation (TX-TL) extracts,15 but their implementation in cell-like compartments has been limited.
In 2004, the production of functional proteins using TX-TL extracts inside vesicles was demonstrated.16 The behavior of such bioreactors is relatively straightforward; contrarily, SGCs that regulate protein expression through feedback systems are much more relevant to constructing an artificial cell. A recently developed TX-TL system employing all seven regulatory Escherichia coli σ factors has enabled the construction of more extensive SGCs with complex behavior (Figure 2). For instance, in serial transcriptional activation cascades the expression of one σ factor activates the expression of a subsequent factor, ultimately producing a reporter protein (Figure 2A). Other circuits included AND gates, which require the simultaneous expression of two σ factors to produce the reporter, pulse circuits, in which a single factor first induced reporter expression and subsequently repressed it by a delayed repression pathway (Figure 2B), inducible transcriptional repression units that switched between two outputs depending on the inducer used (Figure 2C), positive feedback loops, and biosynthetic metabolite pathways. Some of these circuits have been constructed in vitro only, others in vesiculo as well.17,18
Figure 2.
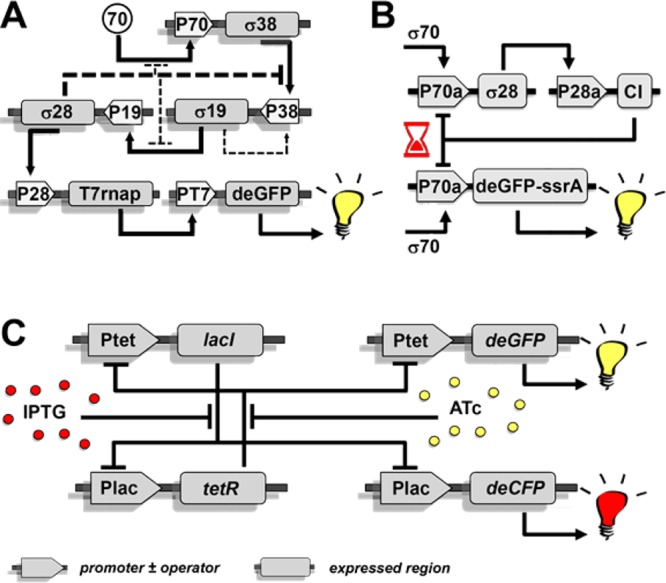
SGCs are examples of the complex behavior that arises when combining genetic elements. (A) A serial transcriptional activation cascade that produces deGFP. Each σ factor activates its successor by interacting with its promoter, as indicated by solid arrows. (B) This circuit generates a pulse in deGFP production due to two competing expression cascades. Addition of σ70 induces deGFP production by the stimulatory (lower) circuit, but the inhibitory (upper) circuit is triggered simultaneously and causes a delayed suppression. (C) An inducible transcriptional repression unit that can switch outputs. In the presence of IPTG, deCFP is produced; replacement by ATc represses deCFP production and stimulates deGFP expression. Adapted with permission from refs (17 and 18). Copyright 2012 and 2016 American Chemical Society.
These examples represent the most extensive SGCs currently realized in synthetic compartments. Far more complex behavior, however, like oscillations and pattern generation, has already been achieved in bulk systems.19 Such behavior usually occurs in a limited parameter space only and, therefore, requires tight control of the concentrations of all reagents. Unfortunately, most common techniques for vesicle formation yield a heterogeneous population of vesicles due to the stochastic nature of the encapsulation of dilute (macro-)molecules.20 This heterogeneity hinders the extent of control over reaction networks in compartments compared to bulk systems. Microfluidic platforms may provide a solution here, as they can construct synthetic compartments in a highly controlled way.
A very interesting next step in the programming of artificial cells would be the development of a replicating vesicle with strong genotype–phenotype linkages. That way, the genetic program of an artificial cell can alter vital biochemical properties, creating a selective pressure that can be used for directed evolution.21
Metabolism and Energy
Many processes constantly require a supply of building blocks and energy to maintain their activity. In natural cells, these are partially supplied by catabolic processes that recycle macromolecules and generate building blocks and energy-rich compounds like ATP. Most artificial cells reported to date, however, lack the mechanisms to maintain such a balance of resources. Therefore, their metabolism usually quickly grinds to a halt due to depletion of nutrients or accumulation of waste. Continuous feeding with fresh nutrients temporarily alleviates this problem,16 but the activity eventually decreases due to toxic byproducts or catalyst poisoning.
Only a few attempts to regenerate energy carriers, such as cofactors, in synthetic cells have been reported. For instance, we have shown that the natural cofactor NADPH could be regenerated in a polymersome using a set of enzymes and electron donors.22 In cell-free protein synthesis, ATP regeneration has received more attention, resulting in increased protein production times.23 Although lysate-based artificial cells in principle contain the machinery to regenerate nutrients, an efficient recycling of pivotal nutrients and removal of waste is still a distant goal. In the future, maintaining a supporting metabolism in artificial cells will become increasingly important when more complicated functions are pursued and maintaining homeostasis becomes pivotal for prolonged activity.
Analogous to photosynthesis, light would in this respect be a great energy source to generate energy-rich intermediates to fuel artificial cells. Light-powered ATP production using ATP synthase in conjunction with bacteriorhodopsin or a proton-pumping synthetic system has been demonstrated in polymersomes and liposomes, repectively.24,25 Their integration with an artificial cell system has, however, not been reported until now.
Growth and Division of Synthetic Compartments
Reproduction is essential to maintain a living population. For this, the artificial cell needs to copy all its vital components and divide these into daughter compartments. So far, most research has focused on replication of the (genetic) information carrier, as reviewed elsewhere.1 Recently, however, more groups have started to design artificial cells that produce new membranes and are capable of division.
Growth
Proliferation of cells requires the production of membrane components to prevent shrinkage of each subsequent generation. Generally, two approaches have been pursued to feed membranes with additional components. For dynamic membranes, like those based on fatty acids, externally added membrane components are spontaneously incorporated.21 Phospholipid membranes are less dynamic and require in situ production of membrane components for efficient incorporation. Polymersomes are generally even more stable and, therefore, used as nonreplicating model cells only.
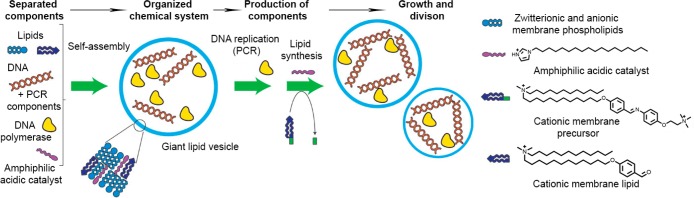
An often-adopted approach to generate membrane components inside an artificial cell comprises the encapsulation of a catalyst into the membrane or lumen, where it produces an amphiphilic molecule that is incorporated into the membrane. Typically, this membrane growth disturbs the compartment’s surface-to-volume ratio, which induces budding and fission (Figure 3).26 A problem in these systems, however, is the dilution of the catalyst after several rounds of growth and fission. To circumvent this problem, Devaraj and co-workers created a catalyst that can both generate new membrane components and undergo autocatalysis.27 Their system could perform 15 cycles of near-complete conversion of lipid precursors that produced many new vesicles.
Figure 3.
Self-reproduction of vesicles coupled to internal DNA amplification. A polymerase chain reaction (PCR) in the vesicle’s lumen amplifies the encapsulated DNA, and a catalyst in the membrane generates new membrane components from supplemented precursors. Importantly, the DNA accelerates membrane formation and induces budding and fission of the vesicle. Adapted with permission from refs (26 and 29). Copyright 2011 Macmillan Publishers Ltd.
To sustain a functional metabolism after division, however, the essential metabolic machinery should be replicated too. So far, little development has been realized in this area. Recently, however, an example of maintaining homeostasis upon growth of the compartment was demonstrated.28 Rather than replicating the encapsulated catalyst, it was activated due to dilution of its inhibitors upon growth of the compartment. Although such regulation was demonstrated for vesicle growth only, it is an interesting step toward maintaining homeostasis upon division. For future efforts, it is important that the replication of the information carrier and the compartment become coupled, to produce new generations with the same hereditary information. The work by Sugawara (Figure 3) has already provided an interesting approach to this challenge.26 Subsequently, the occurrence of small changes to the hereditary code and their subsequent propagation in the population can pave the way for evolution of such artificial life.
Division
Although the division of artificial cells is still far removed from the stringent control over this process found in nature, numerous studies have reported basic methods of division (fission or budding) in artificial cells. The divisions were instigated by external mechanical shearing,30 membrane growth,26,27 volume reduction,31 or phase separation.31 Frequently, an increased surface-to-volume ratio due to membrane growth makes spontaneous budding and fission thermodynamically favorable.32 Indeed, sustained membrane growth can fuel repetitive growth/division cycles of artificial cells, albeit with limited control over the divisions.27,30 Nevertheless, recent studies have demonstrated that macromolecules can provide some control over division by interacting with the growing membrane.26,32
To gain more spatiotemporal control over synthetic division cycles, several groups have used regulatory elements from nature, like proteins that play a pivotal role in the division of simple bacteria, to divide artificial cells. Most notably, the FtsZ and Min proteins from E. coli have been reconstituted into artificial cells to achieve controlled division.33 Recently, some evidence was presented that a contractile Z-ring formed by FtsZ and FtsA could completely pinch off and divide an artificial cell.34 Although such strategies could provide much better spatiotemporal control over the division process, their drawback is that the regulatory function of these protein networks is difficult to reproduce ex cellulo. The need to move toward more complicated division machinery is, however, obvious, since the field still faces major challenges here, like equal distribution of genetic information upon division, and implementing adequate checkpoints in the cell cycle.
Adaptivity
Many of the artificial cells developed over the past years can be used to study processes in isolation and under controlled conditions. In the previous sections, we reviewed such systems in our discussion of the first four characteristics of life we introduced earlier: (i) compartment, (ii) information carrier, (iii) metabolism, and (iv) reproduction. To survive in a dynamic environment, however, cells have to continuously sense and respond to changes in their milieu. Here, we examine efforts to mimic such adaptivity in artificial cells that dynamically respond to their environment or each other and discuss our recent work on nanomotors that use fuel from their surroundings to move.
Sensing the Environment
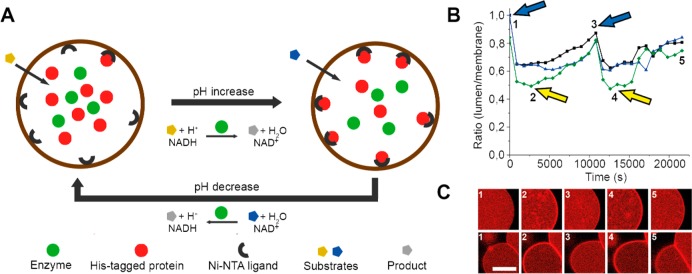
We recently developed a giant vesicle in which protein–ligand interactions were reversibly controlled by the addition of external small molecule triggers.35 The interaction of a His-tagged protein with its ligand in the membrane of giant unilamellar vesicles (GUVs) was regulated by the activity of a pH-modifying enzyme, alcohol dehydrogenase (ADH) (Figure 4). The pH inside the GUV depended on the redox equilibrium of the natural cofactor NAD(H), which was reduced or oxidized by ADH in the presence of isopropanol or acetone, respectively. Thus, addition of either substrate could change the pH inside the GUVs, altering the association of protein to the membrane. Importantly, this process could be reversed by replacement of the original substrate by its antagonist. Hence, this artificial cell provides a platform to drive the dynamic membrane association and dissociation of any His-tagged biomolecule. Additionally, the covalent addition of targeted proteins to lipid membranes with spatiotemporal control has recently been reported as well.36 Such platforms are especially useful for studying proteins that change conformation or that form functional complexes upon membrane association.
Figure 4.
Reversible assembly of a His-tagged protein on the membrane of a GUV. The assembly is driven by the catalytic activity of ADH, which changes the pH of the vesicle’s lumen. (A) Schematic of the reversible assembly. (B) Alternating addition of two substrates (indicated by arrows) drives membrane assembly and disassembly of the His-tagged protein. (C) Fluorescence microscopy images of GUVs corresponding to the time points in panel B. Adapted with permission from ref (35). Copyright 2015 Wiley.
Whereas natural cells can alter their membrane composition to regulate signaling or binding, synthetic membranes are usually static. Devaraj et al. addressed this discrepancy using a reversible native chemical ligation strategy to create lipid analogs that can easily exchange their acyl chains and hydrophilic head groups. The lipids self-assembled into GUVs, after which they could exchange their tails and head groups by addition of reactive precursors to the GUV solution. This way, the GUV’s membrane composition was remodeled to induce the formation of lipid domains, as well as to recruit a protein through electrostatic interactions with a newly introduced lipid headgroup.37
Not only the composition but also the shape of natural cells is dynamic; their morphology can change upon external or internal triggers. In an effort to translate this property to synthetic compartments, polymersomes and colloidosomes were shown to undergo oscillatory shape deformations in response to temperature variations and an internal oscillatory reaction (Figure 5).38,39
Figure 5.
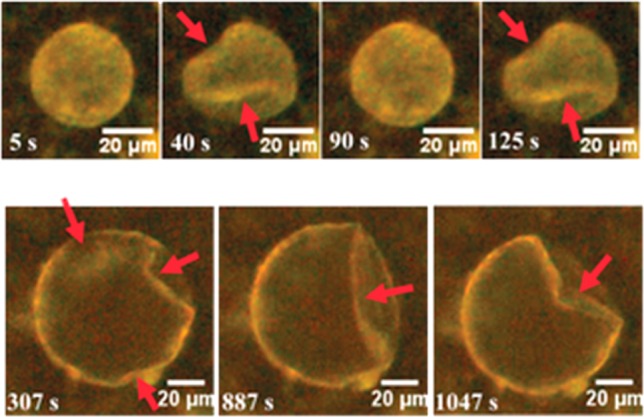
Oscillating buckling patterns observed for two colloidosomes in response to temperature variations and an internal oscillating reaction. Reproduced with permission from ref (39). Copyright 2016 Wiley.
These examples show the range of possibilities for creating adaptive artificial cells. Currently, the efforts seem to lack a clear main objective, and groups are just exploring some curious designs. An important goal would be the modulation of membrane permeability upon an external or internal stimulus. The Mann group already showed they could influence the membrane permeability of a colloidosome sufficiently to regulate an internal reaction.40 Preferably though, the uptake should be more selective than just based on size or charge. For this, activatable membrane channels could be a very interesting option. Additionally, sensing signals from other cells and converting a chemical fuel into motion are very interesting cases of adaptivity that will be discussed below.
Communication between Cells
Cells actively communicate to coordinate their actions not only in multicellular but also in unicellular organisms. Efforts to mimic this behavior comprise artificial communication between both natural and artificial cells. Here we limit ourselves to systems that involve artificial cells.
The first artificial cell capable of communicating with a natural cell used formaldehyde and boric acid as a simple feedstock to generate a variety of sugar derivatives.41 Due to the similarity of these borates to an inducer of bioluminescence in the bacterium Vibrio harveyi, the artificial cells could signal the bacterium to produce light. More recently, protocell-to-cell signaling was also engineered such that artificial cells translated a chemical compound that E. coli cannot sense to a signaling molecule that triggered a cellular response in the bacteria.42
Few systems comprising signaling between two artificial cells have been reported.43,44 In one example, the Mann group engineered one-way signaling from a hydrogen peroxide-producing colloidosome to a secondary colloidosome. The hydrogen peroxide triggered formation of an outer shell of thermoresponsive PNIPAM on the secondary colloidosome. This new shell altered the colloidosome’s permeability, which influenced the rate of an enclosed enzymatic reaction (Figure 6).43 The same group also engineered interesting predatory behavior in communities of artificial cells. Coacervate droplets could seek and attack protein-based vesicles and, after lysing them, hijack their contents.45
Figure 6.
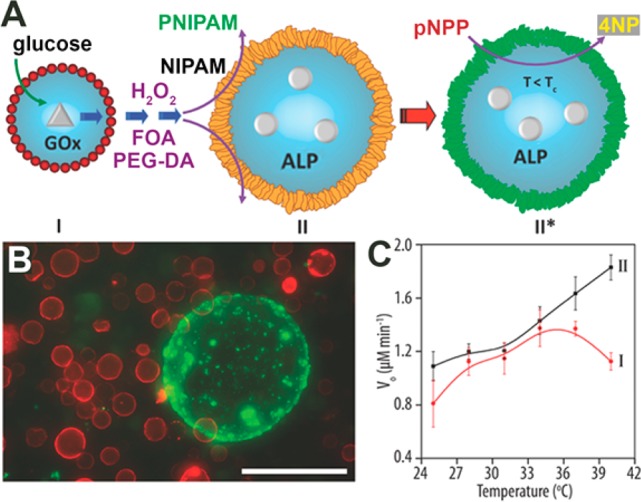
Colloidosome signaling. (A) A glucose oxidase (GOx)-filled colloidosome produces hydrogen peroxide, which induces polymerization of the NIPAM shell of a secondary colloidosome. Consequently, the permeability of the PNIPAM-colloidosome becomes thermoresponsive, influencing the kinetics of an internal reaction. (B) Fluorescence microcopy image of red fluorescent GOx-colloidosomes that induced the polymerization of a green fluorescent shell around a PNIPAM-colloidosome. (C) Kinetics of an enzymatic reaction inside the PNIPAM-colloidosomes before (black) and after (red) polymerization of the NIPAM shell. Adapted with permission from ref (43). Copyright 2016 Wiley.
Despite these recent examples, interactions between artificial cells remain a relatively unexplored part of the field. As such, the current designs are mere primitive versions of the complex communication that exists between natural cells. Reciprocal communication between two artificial cells, for example, has not yet been demonstrated. Such mutual regulation between artificial cells could form the basis for the design of communities that can coordinate their actions, for instance, to maintain homeostasis. As such, the behavior of communities of artificial cells merits further research because it represents the first steps to the bottom-up synthesis of tissue-like organizations.
Motility and Chemotaxis
Nature offers plenty examples of the importance of motility for natural cells. Many of these trajectories follow a chemical gradient, in a process known as chemotaxis. So far, however, in synthetic cell-like compartments active and directed motility has seldom been produced. Such systems would not only yield insight in the movement of natural cells but also pose interesting platforms for drug delivery as smart materials that can migrate to specific chemical milieus.
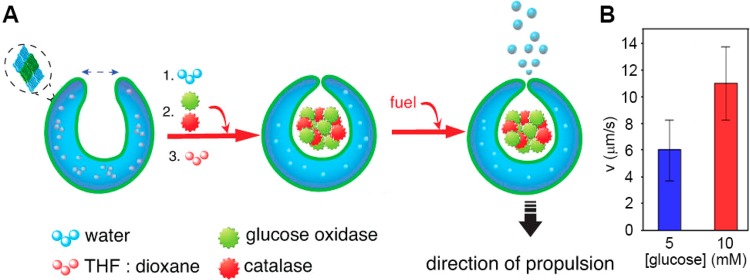
Synthetic compartments can be propelled by a magnetic, acoustic, or electric field, light, or a chemical fuel. Using a chemical fuel has the advantage that no external force is needed to drive propulsion. However, the examples of fuel-driven propulsion of micro- and nanomotors that have been reported are mostly very different from cell-like structures.46 Our group has used bowl-shaped polymeric vesicles, termed stomatocytes, that are propelled by an encapsulated hydrogen peroxide-consuming catalyst (Figure 7).47 These nanomotors are obtained by controlled deformation of polymersomes, during which catalytic species such as enzymes are encapsulated in their cavity to catalyze the decomposition of fuel molecules like glucose and hydrogen peroxide.48 The nanomotors were shown to move in a directed manner in the presence of fuel, which is thought to be caused by a combination of diffusiophoresis and oxygen bubble formation.47
Figure 7.
Glucose-fueled propulsion of enzyme-loaded stomatocytes. (A) The nanomotor’s cavity is loaded with enzymes during self-assembly. Fuel can diffuse in, but the enzymes remain trapped, producing thrust. (B) The nanomotors’ speed varies with glucose concentration. Adapted with permission from ref (48). Copyright 2016 American Chemical Society.
To demonstrate life-like behavior, synthetic motor systems should also show chemotactic characteristics. Until now, however, the number of motors demonstrating guided movement to an attractant is limited.49 Mainly, they are based on bimetallic particles or metal-coated microspheres. We recently created a more cell-like chemotactic system based on stomatocytes. The nanomotors moved along gradients of hydrogen peroxide, which could even direct them toward hydrogen peroxide-producing neutrophils.50 This chemotactic behavior is thought to result from the longer distances traveled at high fuel concentrations due to a fuel concentration-dependent increase in speed. Currently, we are investigating whether the speed, directionality, and temporal behavior of such chemotactic nanomotors can be further controlled. For instance, we have created nanomotors that use an enzymatic reaction network with several feedforward loops to maintain a constant speed independent of fuel concentration.51
Conclusions and Outlook
During the past decade, the bottom-up construction of artificial cells has started to pick up steam. Prominent advances in their functionality include the construction of gene circuits that display increasingly complex behavior and of self-reproducing compartments with improved control over their division. An important recent development is the construction of artificial cells that are adaptive. Their dynamic nature and ability to interact with the environment are significant next steps in the realization of fully autonomous artificial cells.
For future developments, it is important to ensure that life-like systems are provided with an effective metabolism to sustain the biomimetic processes performed within the compartment. Furthermore, strategies should be designed that allow the replication of not only the genetic information or the membrane components, but also the functional units that execute the biomimetic processes. A first step in this direction would be integrating advanced functional elements while maintaining their mutual compatibility. In this regard, accommodating the relatively static multicompartmentalized vesicle platforms currently developed with the functionality and adaptivity realized in other systems seems a promising strategy. Additionally, communication between artificial cells would open up interesting avenues to collective behavior inspired by bacterial colonies or multicellular organisms. Taking adaptivity a step further, artificial cells would also provide an interesting platform to study the principles of genetic evolution.
Finally, the bottom-up construction of artificial cells will not only enhance our understanding of fundamental physical and chemical processes in living systems but may also find applications in biomedicine and environmental science via the development of smart, autonomous microreactors that can monitor their environment and intervene if necessary.
Biographies
Bastiaan Buddingh’ received his M.Sc. in molecular life sciences from Radboud University (The Netherlands) in 2014. During his master’s, he worked on both polymeric nanoreactors for enzyme therapy in the bio-organic chemistry group at Radboud University, and new bioorthogonal reactions in the group of Prof. Carolyn Bertozzi at UC Berkeley (USA). Currently, he is a Ph.D. candidate working on new functions for artificial cells in the group of Prof. Jan van Hest.
Jan van Hest obtained his Ph.D. in 1996 from Eindhoven University of Technology (The Netherlands) under supervision of Prof. Bert Meijer. In 2000, Radboud University appointed him as full professor in bio-organic chemistry. Recently, he relocated to Eindhoven University of Technology to become professor at the Departments of Chemical Engineering and Chemistry and Biomedical Engineering. His group develops bioinspired materials and processes by combining the functionality of biological systems with the flexibility and robustness of synthetic structures.
The Dutch Ministry of Education, Culture and Science (Gravitation program 024.001.035) is acknowledged for funding.
The authors declare no competing financial interest.
References
- Blain J. C.; Szostak J. W. Progress toward Synthetic Cells. Annu. Rev. Biochem. 2014, 83, 615–640. 10.1146/annurev-biochem-080411-124036. [DOI] [PubMed] [Google Scholar]
- Gánti T.; Griesemer J.; Szathmáry E.. The Principles of Life, 1st ed.; Oxford University Press: Oxford, 2003. [Google Scholar]
- Schoonen L.; van Hest J. C. M. Compartmentalization Approaches in Soft Matter Science: From Nanoreactor Development to Organelle Mimics. Adv. Mater. 2016, 28 (6), 1109–1128. 10.1002/adma.201502389. [DOI] [PubMed] [Google Scholar]
- Tu Y.; Peng F.; Adawy A.; Men Y.; Abdelmohsen L. K. E. A.; Wilson D. A. Mimicking the Cell: Bio-Inspired Functions of Supramolecular Assemblies. Chem. Rev. 2016, 116 (4), 2023–2078. 10.1021/acs.chemrev.5b00344. [DOI] [PubMed] [Google Scholar]
- Sokolova E.; Spruijt E.; Hansen M. M. K.; Dubuc E.; Groen J.; Chokkalingam V.; Piruska A.; Heus H. A.; Huck W. T. S. Enhanced Transcription Rates in Membrane-Free Protocells Formed by Coacervation of Cell Lysate. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (29), 11692–11697. 10.1073/pnas.1222321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga S.; Williams D. S.; Perriman A. W.; Mann S. Peptide-Nucleotide Microdroplets as a Step towards a Membrane-Free Protocell Model. Nat. Chem. 2011, 3 (9), 720–724. 10.1038/nchem.1110. [DOI] [PubMed] [Google Scholar]
- Städler B.; Chandrawati R.; Price A. D.; Chong S.-F.; Breheney K.; Postma A.; Connal L. A.; Zelikin A. N.; Caruso F. A Microreactor with Thousands of Subcompartments: Enzyme-Loaded Liposomes within Polymer Capsules. Angew. Chem., Int. Ed. 2009, 48 (24), 4359–4362. 10.1002/anie.200900386. [DOI] [PubMed] [Google Scholar]
- Peters R. J. R. W.; Marguet M.; Marais S.; Fraaije M. W.; van Hest J. C. M.; Lecommandoux S. Cascade Reactions in Multicompartmentalized Polymersomes. Angew. Chem., Int. Ed. 2014, 53 (1), 146–150. 10.1002/anie.201308141. [DOI] [PubMed] [Google Scholar]
- Villar G.; Graham A. D.; Bayley H. A Tissue-Like Printed Material. Science 2013, 340 (6128), 48–52. 10.1126/science.1229495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominak L. M.; Gundermann E. L.; Keating C. D. Microcompartmentation in Artificial Cells: pH-Induced Conformational Changes Alter Protein Localization. Langmuir 2010, 26 (8), 5697–5705. 10.1021/la903800e. [DOI] [PubMed] [Google Scholar]
- de Hoog H.-P. M.; Nallani M.; Tomczak N. Self-Assembled Architectures with Multiple Aqueous Compartments. Soft Matter 2012, 8 (17), 4552. 10.1039/c2sm06934b. [DOI] [Google Scholar]
- Villar G.; Heron A. J.; Bayley H. Formation of Droplet Networks That Function in Aqueous Environments. Nat. Nanotechnol. 2011, 6 (12), 803–808. 10.1038/nnano.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elani Y.; Law R. V.; Ces O. Vesicle-Based Artificial Cells as Chemical Microreactors with Spatially Segregated Reaction Pathways. Nat. Commun. 2014, 5, 5305. 10.1038/ncomms6305. [DOI] [PubMed] [Google Scholar]
- Keating C. D. Aqueous Phase Separation as a Possible Route to Compartmentalization of Biological Molecules. Acc. Chem. Res. 2012, 45 (12), 2114–2124. 10.1021/ar200294y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson E. D.; Gan R.; Hodgman C. E.; Jewett M. C. Cell-Free Protein Synthesis: Applications Come of Age. Biotechnol. Adv. 2012, 30 (5), 1185–1194. 10.1016/j.biotechadv.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noireaux V.; Libchaber A. A Vesicle Bioreactor as a Step toward an Artificial Cell Assembly. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (51), 17669–17674. 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.; Noireaux V. An E. Coli Cell-Free Expression Toolbox: Application to Synthetic Gene Circuits and Artificial Cells. ACS Synth. Biol. 2012, 1 (1), 29–41. 10.1021/sb200016s. [DOI] [PubMed] [Google Scholar]
- Garamella J.; Marshall R.; Rustad M.; Noireaux V. The All E. Coli TX-TL Toolbox 2.0: A Platform for Cell-Free Synthetic Biology. ACS Synth. Biol. 2016, 5 (4), 344–355. 10.1021/acssynbio.5b00296. [DOI] [PubMed] [Google Scholar]
- van Roekel H. W. H.; Rosier B. J. H. M.; Meijer L. H. H.; Hilbers P. A. J.; Markvoort A. J.; Huck W. T. S.; de Greef T. F. A. Programmable Chemical Reaction Networks: Emulating Regulatory Functions in Living Cells Using a Bottom-up Approach. Chem. Soc. Rev. 2015, 44 (21), 7465–7483. 10.1039/C5CS00361J. [DOI] [PubMed] [Google Scholar]
- Nishimura K.; Tsuru S.; Suzuki H.; Yomo T. Stochasticity in Gene Expression in a Cell-Sized Compartment. ACS Synth. Biol. 2015, 4 (5), 566–576. 10.1021/sb500249g. [DOI] [PubMed] [Google Scholar]
- Adamala K. P.; Szostak J. W. Competition between Model Protocells Driven by an Encapsulated Catalyst. Nat. Chem. 2013, 5 (6), 495–501. 10.1038/nchem.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeuwissen S. A.; Rioz-Martínez A.; de Gonzalo G.; Fraaije M. W.; Gotor V.; van Hest J. C. M. Cofactor Regeneration in Polymersome Nanoreactors: Enzymatically Catalysed Baeyer-Villiger Reactions. J. Mater. Chem. 2011, 21 (47), 18923. 10.1039/c1jm12407b. [DOI] [Google Scholar]
- Jewett M. C.; Swartz J. R. Mimicking the Escherichia Coli Cytoplasmic Environment Activates Long-Lived and Efficient Cell-Free Protein Synthesis. Biotechnol. Bioeng. 2004, 86 (1), 19–26. 10.1002/bit.20026. [DOI] [PubMed] [Google Scholar]
- Steinberg-Yfrach G.; Rigaud J. L.; Durantini E. N.; Moore A. L.; Gust D.; Moore T. A. Light-Driven Production of ATP Catalysed by F0F1-ATP Synthase in an Artificial Photosynthetic Membrane. Nature 1998, 392 (6675), 479–482. 10.1038/33116. [DOI] [PubMed] [Google Scholar]
- Choi H. J.; Montemagno C. D. Artificial Organelle: ATP Synthesis from Cellular Mimetic Polymersomes. Nano Lett. 2005, 5 (12), 2538–2542. 10.1021/nl051896e. [DOI] [PubMed] [Google Scholar]
- Kurihara K.; Tamura M.; Shohda K.; Toyota T.; Suzuki K.; Sugawara T. Self-Reproduction of Supramolecular Giant Vesicles Combined with the Amplification of Encapsulated DNA. Nat. Chem. 2011, 3 (10), 775–781. 10.1038/nchem.1127. [DOI] [PubMed] [Google Scholar]
- Hardy M. D.; Yang J.; Selimkhanov J.; Cole C. M.; Tsimring L. S.; Devaraj N. K. Self-Reproducing Catalyst Drives Repeated Phospholipid Synthesis and Membrane Growth. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (27), 8187–8192. 10.1073/pnas.1506704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhart A. E.; Adamala K. P.; Szostak J. W. A Simple Physical Mechanism Enables Homeostasis in Primitive Cells. Nat. Chem. 2016, 8 (5), 448–453. 10.1038/nchem.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisi P. L.; Stano P. Synthetic Biology: Minimal Cell Mimicry. Nat. Chem. 2011, 3 (10), 755–756. 10.1038/nchem.1156. [DOI] [PubMed] [Google Scholar]
- Zhu T. F.; Szostak J. W. Coupled Growth and Division of Model Protocell Membranes. J. Am. Chem. Soc. 2009, 131 (15), 5705–5713. 10.1021/ja900919c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andes-Koback M.; Keating C. D. Complete Budding and Asymmetric Division of Primitive Model Cells to Produce Daughter Vesicles with Different Interior and Membrane Compositions. J. Am. Chem. Soc. 2011, 133 (24), 9545–9555. 10.1021/ja202406v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasawa H.; Nishimura K.; Suzuki H.; Matsuura T.; Yomo T. Coupling of the Fusion and Budding of Giant Phospholipid Vesicles Containing Macromolecules. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (16), 5942–5947. 10.1073/pnas.1120327109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martos A.; Jiménez M.; Rivas G.; Schwille P. Towards a Bottom-up Reconstitution of Bacterial Cell Division. Trends Cell Biol. 2012, 22 (12), 634–643. 10.1016/j.tcb.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Osawa M.; Erickson H. P. Liposome Division by a Simple Bacterial Division Machinery. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (27), 11000–11004. 10.1073/pnas.1222254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters R. J. R. W.; Nijemeisland M.; van Hest J. C. M. Reversibly Triggered Protein-Ligand Assemblies in Giant Vesicles. Angew. Chem., Int. Ed. 2015, 54 (33), 9614–9617. 10.1002/anie.201502920. [DOI] [PubMed] [Google Scholar]
- Rudd A. K.; Valls Cuevas J. M.; Devaraj N. K. SNAP-Tag-Reactive Lipid Anchors Enable Targeted and Spatiotemporally Controlled Localization of Proteins to Phospholipid Membranes. J. Am. Chem. Soc. 2015, 137 (15), 4884–4887. 10.1021/jacs.5b00040. [DOI] [PubMed] [Google Scholar]
- Brea R. J.; Rudd A. K.; Devaraj N. K. Nonenzymatic Biomimetic Remodeling of Phospholipids in Synthetic Liposomes. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (31), 8589–8594. 10.1073/pnas.1605541113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamate R.; Ueki T.; Yoshida R. Self-Beating Artificial Cells: Design of Cross-Linked Polymersomes Showing Self-Oscillating Motion. Adv. Mater. 2015, 27 (5), 837–842. 10.1002/adma.201404757. [DOI] [PubMed] [Google Scholar]
- Tamate R.; Ueki T.; Yoshida R. Evolved Colloidosomes Undergoing Cell-like Autonomous Shape Oscillations with Buckling. Angew. Chem., Int. Ed. 2016, 55 (17), 5179–5183. 10.1002/anie.201511871. [DOI] [PubMed] [Google Scholar]
- Li M.; Harbron R. L.; Weaver J. V. M; Binks B. P.; Mann S. Electrostatically Gated Membrane Permeability in Inorganic Protocells. Nat. Chem. 2013, 5 (6), 529–536. 10.1038/nchem.1644. [DOI] [PubMed] [Google Scholar]
- Gardner P. M.; Winzer K.; Davis B. G. Sugar Synthesis in a Protocellular Model Leads to a Cell Signalling Response in Bacteria. Nat. Chem. 2009, 1 (5), 377–383. 10.1038/nchem.296. [DOI] [PubMed] [Google Scholar]
- Lentini R.; Santero S. P.; Chizzolini F.; Cecchi D.; Fontana J.; Marchioretto M.; Del Bianco C.; Terrell J. L.; Spencer A. C.; Martini L.; Forlin M.; Assfalg M.; Serra M. D.; Bentley W. E.; Mansy S. S. Integrating Artificial with Natural Cells to Translate Chemical Messages That Direct E. Coli Behaviour. Nat. Commun. 2014, 5, 4012. 10.1038/ncomms5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.; Li M.; Dong F.; Wang S.; Tian L.; Mann S. Chemical Signaling and Functional Activation in Colloidosome-Based Protocells. Small 2016, 12 (14), 1920–1927. 10.1002/smll.201600243. [DOI] [PubMed] [Google Scholar]
- Adamala K. P.; Martin-Alarcon D. A.; Guthrie-Honea K. R.; Boyden E. S. Engineering Genetic Circuit Interactions within and between Synthetic Minimal Cells. Nat. Chem. 2016, 10.1038/nchem.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y.; Li M.; Booth R.; Mann S. Predatory Behaviour in Synthetic Protocell Communities. Nat. Chem. 2016, 10.1038/nchem.2617. [DOI] [PubMed] [Google Scholar]
- Wu Z.; Lin X.; Si T.; He Q. Recent Progress on Bioinspired Self-Propelled Micro/Nanomotors via Controlled Molecular Self-Assembly. Small 2016, 12 (23), 3080–3093. 10.1002/smll.201503969. [DOI] [PubMed] [Google Scholar]
- Wilson D. A.; Nolte R. J. M.; van Hest J. C. M. Autonomous Movement of Platinum-Loaded Stomatocytes. Nat. Chem. 2012, 4 (4), 268–274. 10.1038/nchem.1281. [DOI] [PubMed] [Google Scholar]
- Abdelmohsen L. K. E. A.; Nijemeisland M.; Pawar G. M.; Janssen G.-J. A.; Nolte R. J. M.; van Hest J. C. M.; Wilson D. A. Dynamic Loading and Unloading of Proteins in Polymeric Stomatocytes: Formation of an Enzyme-Loaded Supramolecular Nanomotor. ACS Nano 2016, 10 (2), 2652–2660. 10.1021/acsnano.5b07689. [DOI] [PubMed] [Google Scholar]
- Yadav V.; Duan W.; Butler P. J.; Sen A. Anatomy of Nanoscale Propulsion. Annu. Rev. Biophys. 2015, 44 (1), 77–100. 10.1146/annurev-biophys-060414-034216. [DOI] [PubMed] [Google Scholar]
- Peng F.; Tu Y.; van Hest J. C. M.; Wilson D. A. Self-Guided Supramolecular Cargo-Loaded Nanomotors with Chemotactic Behavior towards Cells. Angew. Chem., Int. Ed. 2015, 54 (40), 11662–11665. 10.1002/anie.201504186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijemeisland M.; Abdelmohsen L. K. E. A.; Huck W. T. S.; Wilson D. A.; van Hest J. C. M. A Compartmentalized Out-of-Equilibrium Enzymatic Reaction Network for Sustained Autonomous Movement. ACS Cent. Sci. 2016, 2 (11), 843–849. 10.1021/acscentsci.6b00254. [DOI] [PMC free article] [PubMed] [Google Scholar]