Abstract
The eukaryotic translation initiation factor eIF4E is deregulated in many human cancers, and its overexpression in cells leads to malignant transformation. Oncogenic properties of eIF4E are directly linked to its ability to bind 7-methyl guanosine of the 5′ mRNA. Here, we observe that the antiviral guanosine analogue ribavirin binds to eIF4E with micromolar affinity at the functional site used by 7-methyl guanosine mRNA cap, competes with eIF4E:mRNA binding, and, at low micromolar concentrations, selectively disrupts eIF4E subcellular organization and transport and translation of mRNAs posttranscriptionally regulated by eIF4E, thereby reducing levels of oncogenes such as cyclin D1. Ribavirin potently suppresses eIF4E-mediated oncogenic transformation of murine cells in vitro, of tumor growth of a mouse model of eIF4E-dependent human squamous cell carcinoma in vivo, and of colony formation of eIF4E-dependent acute myelogenous leukemia cells derived from human patients. These findings describe a specific, potent, and unforeseen mechanism of action of ribavirin. Quantum mechanical and NMR structural studies offer directions for the development of derivatives with improved cytostatic and antiviral properties. In all, ribavirin's association with eIF4E may provide a pharmacologic means for the interruption of posttranscriptional networks of oncogenes that maintain and enhance neoplasia and malignancy in human cancer.
Keywords: cancer, drug, virus, oncogenic network
Whereas the precise causes of neoplasia and malignancy are known only for a few human cancers, deregulated tumor suppressors and oncogenes that maintain and enhance the malignant phenotype are becoming relatively well described (1, 2). Among these molecules are tumor suppressors like p53, Rb, and APC and oncogenes such as myc, cyclin D1, and eIF4E. Their interactions constitute a network of self-reinforcing feedback loops, whereby inactivation of principal elements can lead to reversal and at times even sustained loss of neoplastic phenotype (3, 4).
eIF4E is overexpressed in a wide variety of malignant cell lines and primary human tumors such as carcinomas of the breast (5), colon (6), and head and neck (7), non-Hodgkin's lymphomas (8), and chronic and acute M4/M5 myelogenous leukemias (9). Consistently, even moderate overexpression of eIF4E in rodent cells leads to deregulated proliferation and malignant transformation (10). eIF4E is essential for growth and survival of eukaryotes by acting at a critical step of cap-dependent translation and recruiting transcripts to the ribosome as a result of a specific interaction with the 5′ 7-methyl guanosine (m7G) mRNA cap (11). It is important to stress that, although the interaction of eIF4E with the 5′ mRNA cap is required for initiation of translation of cap-dependent mRNAs, up-regulation of eIF4E does not increase translation of all cap-dependent transcripts, but only of a specific subset of eIF4E-sensitive transcripts, as described below.
As much as 70% of eIF4E is present in the nuclei of mammalian cells, where it is organized in nuclear bodies (12–16). Here, eIF4E promotes transport of mRNAs of a specific set of transcripts such as cyclin D1 but not of housekeeping genes such as β-actin and GAPDH (12–16). Posttranscriptional regulation of gene expression at the levels of eIF4E-mediated mRNA transport and translation exhibits different gene specificities, with some genes being regulated at the level of transport (e.g., cyclin D1), some at the level of translation (e.g., VEGF), others at the level of both [e.g., ornithine decarboxylase (ODC)], and yet others at neither (e.g., GAPDH) (17). Binding to the m7G cap is required both for mRNA transport and translation by eIF4E, both of which contribute to its ability to transform cells (17, 18).
A guanosine ribonucleoside analogue (19), 1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide (ribavirin, also known as Virazole) is currently used for the treatment of infections with Lassa fever virus, respiratory syncytial virus, hepatitis C virus (HCV), and severe acute respiratory syndrome coronavirus (20). Recently, Cameron and colleagues (21, 22) showed that ribavirin can be misincorporated into mRNA by viral RNA-dependent RNA polymerases because of its chemical similarity to guanosine, and this outcome leads to lethal mutagenesis of genomes of polio and HCV. Ribavirin triphosphate (RTP) binds the HCV polymerase with an observed dissociation constant (Kd) of 0.58 mM (23), which is consistent with micromolar concentrations necessary to achieve a therapeutic effect against HCV clinically (24). In addition to effects at millimolar concentrations, ribavirin is also active at micromolar concentrations. For example, ribavirin inhibits growth of human lymphocytes with an EC50 of 2 μM (25). The mechanism underlying these more potent effects is not understood.
Here, we observe direct binding of ribavirin to eIF4E with micromolar affinity in vitro, efficient competition with eIF4E:m7G mRNA cap binding in vitro and in cells, and specific disruption of eIF4E:m7G functions in the transport and translation of eIF4E-regulated genes at low micromolar concentrations in cells, with consequent down-regulation of oncogenic proteins, cell-cycle arrest, and suppression of eIF4E-mediated transformation in vitro and in vivo. Evaluation of physical properties of guanosine analogues by using quantum mechanical calculations and their molecular recognition by eIF4E using NMR spectroscopy leads us to conclude that ribavirin is a physical mimic of m7G and offers specific directions for the design of derivatives with improved cytostatic and antiviral properties.
Methods
eIF4E protein was purified from BL21 cells, and fluorescence studies were performed in 0.3 M NaCl/10 mM Na2PO4, pH 7.5/1 μM zinc in a 0.3 × 0.3 cm2 fluorescence cuvette (Hellma, Forest Hills, NY) by using an eIF4E concentration of 2 μM (26). Standard methods for cell culture and cell fractionation were used as described in ref. 9. Western analysis was carried out by using SuperSignal reagent (West Pico), and Northern analysis was performed with the Ambion kit as in ref. 9. For immunofluorescence, cells were fixed in methanol, and results were visualized by confocal microscopy as described in ref. 9. Cells were transfected by using GeneJammer reagent and selected by using G418, and foci were counted manually from three independent experiments as in ref. 14. Primary human leukemia and normal bone marrow cells were obtained from patients and processed as in ref. 9. Mouse xenograft studies were performed as described (27) by using female 5- to 7-week old athymic NCr-nu/nu mice from Taconic Farms that were injected with FaDu cells (American Type Culture Collection). NMR spectroscopy was performed by using a 500 MHz Bruker DRX spectrometer (28) in 0.1 M NaCl/50 mM Na2PO4, pH 6.5/5mM DTT/5% (vol/vol) 2H2O, at 288 and 298 K, using protein concentrations of 0.8 mM. For NMR, the G4E eIF4E was purified from BL21 cells by m7G cap affinity chromatography as in ref. 29. For treatments, drugs were dissolved in PBS (pH 7.4) and filter-sterilized. Untreated cells and animals received filter-sterilized PBS. Detailed descriptions of procedures are provided in Supporting Methods, which is published as supporting information on the PNAS web site.
Results and Discussion
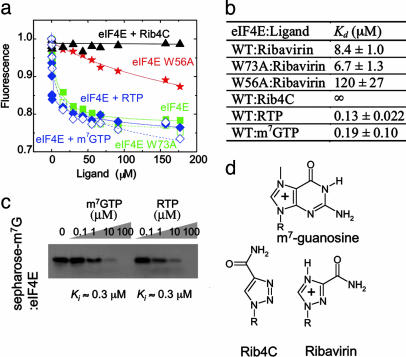
High-affinity binding of the m7G mRNA cap to mammalian eIF4E occurs by way of specific recognition of the methylated and consequently positively charged quaternary amine m7G base by two conserved tryptophans, W56 and W102, which form an aromatic stack as a result of cation–π and π–π interactions (30, 31). Binding of the uncharged tertiary amine guanosine to eIF4E is >5,000-fold weaker (31). Because the pKa values of 1,2,4-triazoles are >12 (32), making them protonated and thus positively charged at physiological pH, we investigated whether eIF4E binds to the putatively cationic 1,2,4-triazole-3-carboxamide of ribavirin (Fig. 1). The affinity of eIF4E for its nucleoside ligands in vitro can be measured by using tryptophan fluorescence emission spectroscopy, whereby binding of ligand quenches fluorescence of tryptophans that stack with it (26). Ribavirin binds to eIF4E with an apparent Kd of 8.4 μM, similar to that of m7G nucleoside (Fig. 1 a and b). Mutation of one of the tryptophans, W56A, in the cap-binding site reduces affinity by 14-fold, whereas mutation of W73A on the dorsal surface of eIF4E away from the cap-binding site has no significant effect on ribavirin affinity (Fig. 1 a and b). Similar results are obtained by using m7G (26). Furthermore, the ribavirin analogue 1-β-d-ribofuranosyl-1,2,3-triazole-4-carboxamide (Rib4C), which exhibits reduced antiviral and cellular effects (32) and contains an uncharged 1,2,3-triazole with a reduced pKa (32), fails to bind eIF4E (Fig. 1). Because ribavirin is nearly completely converted to RTP in cells (34), we measured the affinity of eIF4E for RTP. eIF4E binds RTP and m7GTP with equal apparent dissociation constants of ≈0.1 μM (Fig. 1 a and b). By using m7G-Sepharose affinity chromatography, we observe that RTP competes with eIF4E:m7G binding with an apparent inhibition constant (Ki) of ≈0.3 μM, nearly indistinguishable from m7GTP itself (Fig. 1c). In all, these results indicate that ribavirin binds eIF4E with high affinity, at the functional site used by 5′ m7G mRNA cap, as a result of cationic interaction with the cap-binding tryptophans, and suggest that ribavirin competes with m7G 5′ mRNA cap binding to eIF4E in cells. More detailed NMR structural studies are presented below.
Fig. 1.
Ribavirin, but not Rib4C, binds to the functional m7G cap-binding site of eIF4E with the same affinity as m7G mRNA cap. (a) Normalized corrected tryptophan fluorescence intensity quenching and their fits for binding to ribavirin to eIF4E wild-type (▪), W73A (□), W56A (stars), Rib4C to wild-type eIF4E (▴), RTP to wild-type eIF4E (♦), and m7GTP to wild-type eIF4E (⋄). (b) Apparent dissociation constants in micromolar for nucleoside/nucleotide:eIF4E binding. (c) Western blot of eIF4E remaining bound to m7G-Sepharose upon competition with various concentrations of m7GTP or RTP. Both m7GTP and RTP lead to 50% reduction of binding at a concentration of ≈1 μM. (d) Chemical structures of the keto forms of m7G, ribavirin, and Rib4C nucleosides. +, positive charge; R, ribose.
In mammalian cells, functions of eIF4E depend on its subcellular organization. In the cytoplasm, eIF4E associates with ribosomes and functions in m7G cap-dependent mRNA translation (11). Up-regulation of eIF4E increases translation of only a specific set of sensitive mRNAs, those that are posttranscriptionally regulated by eIF4E at the level of mRNA translation (17). In the nucleus, eIF4E forms multiprotein structures, termed eIF4E nuclear bodies, and plays a role in nucleocytoplasmic mRNA transport of a specific set of mRNA transcripts (18). The formation and function of these structures are linked with eIF4E's mRNA cap binding because treatment of permeabilized cells with excess m7G cap analogue disrupts eIF4E nuclear bodies but not other subnuclear structures (14, 35). Consistently, disruption of eIF4E bodies impedes nucleocytoplasmic eIF4E-dependent mRNA transport (9). Because ribavirin binds eIF4E with high affinity and competes with eIF4E:m7G binding in vitro, we examined whether it affects subcellular organization of eIF4E in cells. Thus, we treated NIH 3T3 fibroblasts with varying concentrations of ribavirin for 48 h and monitored their subcellular organization by using immunofluorescence in conjunction with confocal microscopy. Ribavirin treatment has no apparent effects on chromatin structure (DAPI), organization of nucleoli and Cajal bodies (nucleolar protein Nopp140), structure of splicing speckles (Sc35 domains), and cellular morphology (Fig. 5a, which is published as supporting information on the PNAS web site). In contrast, ribavirin treatment disrupts eIF4E nuclear bodies, with this effect evident at 1 μM and nearly complete at 10 μM (Fig. 5a).
To confirm this effect, we fractionated cells and examined relative protein abundance in nuclear and cytoplasmic fractions by using Western blotting methods. In agreement with the above microscopy studies, ribavirin treatment leads to redistribution of eIF4E without affecting the distributions of predominantly nuclear Sc35 and cytoplasmic β-actin (Fig. 5b). Importantly, ribavirin treatment does not alter total protein levels of eIF4E but, rather, relocalizes the majority of the protein to the cytoplasm. Thus, ribavirin may interfere with mRNA transport and translation of genes posttranscriptionally regulated by eIF4E.
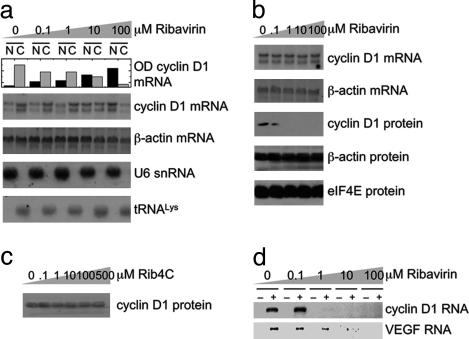
To test this possibility directly, we fractionated cells treated with ribavirin and assessed effects on nucleocytoplasmic mRNA transport by monitoring cyclin D1 mRNA levels of nuclear and cytoplasmic fractions by using subcellular fractionation and Northern methods or independently, using quantitative PCR. Ribavirin treatment impedes nucleocytoplasmic transport of cyclin D1 mRNA with an apparent EC50 of ≈1 μM, with nearly complete nuclear retention at 100 μM (Fig. 2a and Fig. 6, which is published as supporting information on the PNAS web site). On the other hand, nucleocytoplasmic transport of β-actin and VEGF mRNAs is not affected even at 100 μM (Figs. 2a and 6b), which is consistent with insensitivity of their transport to eIF4E activity (13, 15). Ribavirin treatment does not appear to affect splicing and 5′ capping of pre-mRNAs because cotranscriptional capping is required for pre-mRNA splicing, and both cyclin D1 and β-actin mRNAs are correctly spliced (Fig. 2a). Moreover, ribavirin does not appear to affect expression or localization of nuclear RNAs with methylphosphate cap structures such as U small nuclear RNAs, because the levels and distribution of U6 small nuclear RNA are not affected (Fig. 2a). Similarly, ribavirin treatment has no effect on mRNA transcription and stability, because the total steady-state levels of cyclin D1, VEGF, and β-actin mRNAs are not affected (Figs. 2b and 6).
Fig. 2.
Ribavirin specifically inhibits eIF4E:mRNA binding, inhibits nucleocytoplasmic mRNA transport, and depletes levels of transport-regulated proteins. (a) Northern blots of RNA extracts of nuclear and cytoplasmic fractions of ribavirin-treated NIH 3T3 cells, which were probed as indicated. U6 small nuclear RNA and tRNALys serve as controls for quality of the fractionation. Ribavirin inhibits nucleocytoplasmic mRNA transport of cyclin D1, but not β-actin, with an apparent EC50 of ≈1 μM, as judged from the bar graph quantification (top row). N, nuclear; C, cytoplasmic. This effect was confirmed by using quantitative real-time PCR (Fig. 6b). (b) Northern and Western blots of total extracts of ribavirin-treated cells, exhibiting depletion of cyclin D1, without affecting transcription, mRNA stability, and protein synthesis. (c) Western blot of total protein extract of Rib4C-treated cells that were probed for cyclin D1. (d) Semiquantitative RT-PCR of cyclin D1 mRNA contained in eIF4E purified from the nuclei of ribavirin-treated cells. Control samples were purified by using IgG antibody (–) instead of antibody specific for eIF4E (+). Semiquantitative PCR of VEGF from cytoplasmic extracts was immunopurified as above.
We extended our studies to examine the effects of ribavirin on mRNA translation in the cytoplasm by monitoring polysomal loading of mRNAs translationally regulated by eIF4E. Polysomal fractions were prepared, and mRNA content was assessed by using real-time PCR. Ribavirin treatment has no significant effect on the polysomal loading profile of cyclin D1 mRNA (Fig. 6a), which is consistent with lack of regulation by eIF4E of cyclin D1 levels at the level of translation (13). In contrast, ribavirin treatment leads to a shift of VEGF and ODC mRNAs from heavier polysomal to lighter monosomal fractions, which have decreased translational efficiency. The decrease of polysomal loading is >1,000-fold (Fig. 6a), in agreement with translational regulation of VEGF and ODC levels by eIF4E (17). Thus, the apparent sensitivity of genes to ribavirin parallels their sensitivity to regulation by eIF4E, in terms of which genes are affected and the level(s) of regulation.
Because many genes are posttranscriptionally regulated by eIF4E, we focused on cyclin D1 as a model transcript because eIF4E's ability to modulate its mRNA transport is well characterized (13–16). Ribavirin treatment reduces levels of cyclin D1 protein with an apparent EC50 of 0.1–1 μM (Fig. 2b), which is consistent with its inhibition of nucleocytoplasmic cyclin D1 mRNA transport with an EC50 of ≈1 μM (Figs. 2a and 6). In contrast, treatment with Rib4C, which does not bind to eIF4E in vitro (Fig. 1), cannot repress cyclin D1 protein production in cells (Fig. 2c). Furthermore, levels of β-actin and eIF4E proteins, which are not posttranscriptionally regulated by eIF4E (Fig. 2a), are not reduced by ribavirin treatment (Fig. 2b). Thus, ribavirin's specific interaction with eIF4E is required for ribavirin's ability to suppress eIF4E-dependent mRNA transport of cyclin D1.
We tested the ability of ribavirin to directly alter the ability of eIF4E to form ribonucleoproteins with transcripts sensitive to eIF4E-dependent mRNA transport (e.g., cyclin D1) and at the translation level (e.g., VEGF). Thus, we immunopurified eIF4E from nuclei of ribavirin-treated cells and assessed its mRNA content by using semiquantitative RT-PCR (Fig. 2d). Ribavirin treatment leads to inhibition of eIF4E binding to cyclin D1 mRNA in cells with an apparent EC50 of ≈1 μM (Fig. 2d), similar to the Kd for binding of its triphosphate to eIF4E in vitro (Fig. 1) and to the EC50 for inhibition of nucleocytoplasmic cyclin D1 mRNA transport and depletion of cyclin D1 protein in cells (Fig. 2). Similarly, cytoplasmic eIF4E:VEGF mRNA complexes are partly abrogated, even by 1 μM ribavirin (Fig. 2d), which is consistent with the observed alterations in polysomal loading by ribavirin (Fig. 6). Importantly, cytoplasmic eIF4E:actin mRNA complexes are not disrupted, even at 100 μM ribavirin (data not shown), which is consistent with the insensitivity of actin protein levels to ribavirin. Ribavirin's effects are likely not limited to eIF4E-mediated regulation of cyclin D1 mRNA transport and VEGF mRNA translation and include other genes regulated posttranscriptionally by eIF4E (36, 37).
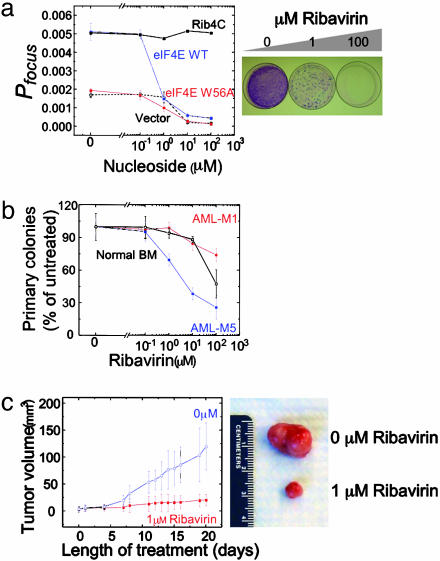
eIF4E causes malignant transformation of cells when overexpressed (10). Mutagenesis studies indicate that its oncogenic properties are due, at least in part, to deregulated transport of mRNAs of oncogenes and growth regulatory genes such as cyclin D1 (9, 14–16). Thus, we examined whether ribavirin treatment and its inhibition of eIF4E-dependent mRNA transport and translation suppress eIF4E-mediated oncogenic transformation. We overexpressed eIF4E in NIH 3T3 cells and assayed transformation by monitoring foci formation as a result of loss of contact growth inhibition. eIF4E levels in transfected cells are 10-fold greater than endogenous levels in control cells (Fig. 7a, which is published as supporting information on the PNAS web site), leading to transformation and a significant increase in foci formation (Fig. 3a). Overexpression of eIF4E W56A cap-binding mutant fails to transform cells (Fig. 3a), in agreement with earlier studies, even though it is expressed to similar levels as wild-type eIF4E (14). Ribavirin suppresses eIF4E-mediated transformation with an apparent EC50 of 0.1–1 μM (Fig. 3 a and b). In contrast, addition of Rib4C fails to reduce the number of foci formed, even at 100 μM (Fig. 3a), which is consistent with its inability to bind eIF4E in vitro and inhibit eIF4E-mediated regulation of mRNA transport and translation in cells (Figs. 1 and 2c). Observed suppression of transformation is not due to nonspecific effects such as metabolic toxicity or cell death (Fig. 7b). Furthermore, low micromolar concentrations of ribavirin induce G1 cell-cycle arrest (Fig. 7c), which is consistent with ribavirin's down-regulation of cyclin D1 (Fig. 2b) and with earlier studies (25).
Fig. 3.
Ribavirin suppresses eIF4E-mediated oncogenic transformation. (a) (Left) Anchorage-dependent foci formation of NIH 3T3 cells treated with ribavirin and transfected with empty vector (black dashed line), eIF4E WT (blue line), eIF4E W56A (red line), and cells treated with Rib4C and transfected with eIF4E WT (black solid line). Error bars represent ± 1σ of three independent experiments. Probability of focus formation (Pfocus) is defined as the number of foci formed divided by the number of cells plated. (Right) Photograph of Giemsa-stained dishes of ribavirin-treated cells transformed by eIF4E. (b) Colony formation of primary human CD34+ myeloid progenitors isolated from patients with AML (M1, solid circles; M5, solid squares) and normal bone marrow (BM, open squares), as a function of ribavirin concentration. Ribavirin reduces colony formation of eIF4E-dependent AML-M5 with an apparent IC50 of ≈1μM, and with no effect on M1 and normal bone marrow myeloid progenitor cells at this concentration. Note that data are internally normalized and that absolute colony formation efficiencies of AML myeloid progenitors are greater than that of BM (data not shown). Error bars represent ± 1σ of four independent experiments. (c)(Left) Mean tumor volume in nude mice engrafted with cells derived from a hypopharyngeal eIF4E-dependent tumor, as a function of treatment with daily 1 μM ribavirin orally at a dose of 40μg per kg per day (solid squares). Error bars represent ± 1σ of 10 mice. (Right) Photograph of tumors resected after 20 days of treatment.
To examine the effect of ribavirin on tumor growth in vivo, we obtained specimens of primary myeloid progenitor cells from patients with acute myelogenous leukemias (AMLs) and comparable cells from normal bone marrow (9). Previous studies (9) indicated that a subset of AMLs have very high levels of nuclear eIF4E and that cyclin D1 mRNA transport is substantially upregulated in these cells. Reduction of nuclear eIF4E levels led to a decrease in cyclin D1 mRNA transport to normal levels (9). Thus, we examined whether ribavirin specifically alters growth of this subset of AMLs. Isolated CD34+ progenitor cells were resuspended in methylcellulose medium and cultured in the presence of various concentrations of ribavirin for 14 days to assess their ability to form colonies. Ribavirin potently repressed colony formation of primary AML-M5 (French-American-British classification) progenitor cells with an apparent IC50 of ≈1 μM (Fig. 3b), which is consistent with their overexpression and dysregulation of eIF4E (9). In contrast, similar concentrations of ribavirin failed to repress colony formation of AML-M1 progenitor cells (Fig. 3b), which is consistent with non-up-regulated eIF4E levels and nondysregulated cyclin D1 mRNA transport in these cells (9). This tumor-suppressive effect of ribavirin at micromolar concentrations is distinct from its cellular toxicity at millimolar concentrations (Fig. 7b), as is evident from the lack of an effect on colony formation of normal bone marrow myeloid progenitors at micromolar concentrations (Fig. 3b).
Treatment with ribavirin caused a marked suppression of tumor growth in a mouse model of human squamous cell carcinoma (Fig. 3c). We used FaDu cells derived from a hypopharyngeal squamous cell carcinoma because they overexpress eIF4E, and form tumors in nude mice, as compared with nonmalignant epithelial cells (27, 38). Importantly, when levels of eIF4E are reduced to nonmalignant levels by using antisense RNA, these cells are markedly less tumorigenic (27). Thus, nude mice were engrafted by using sub-cutaneous injection of eIF4E-dependent FaDu cells and treated with 40 μg/kg ribavirin orally each day, yielding a mean body concentration of ≈1 μM. After 20 days of ribavirin treatment, mean tumor volume of animals in the treatment group was 6-fold less than those in the untreated control group (P = 0.023, n = 10; Fig. 3c). At this low concentration, ribavirin was apparently well tolerated and minimally toxic, as suggested by the absence of treatment-associated mortality and of effect on body weight (data not shown). Thus, ribavirin's inhibition of eIF4E at low micromolar concentrations is correlated with inhibition of eIF4E-mediated oncogenic transformation and tumor suppression in vitro and in vivo.
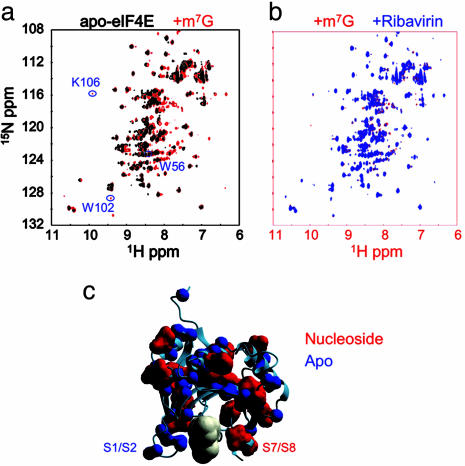
Binding of m7G mRNA cap by eIF4E is required for its nucleocytoplasmic mRNA transport, cytoplasmic translation, and oncogenic transformation. High-affinity binding of m7G cap by eIF4E is accomplished as a result of specific recognition of the cationic methylated base. Because ribavirin, but not its neutral analogue, Rib4C, binds to eIF4E in vitro with the same apparent affinity as the m7G cap and inhibits eIF4E's ability to bind mRNA and function in mRNA transport and translation in cells, we assessed the extent of similarity and molecular recognition by eIF4E of ribavirin and m7G mRNA cap. Thus, we performed 1H, 15N heteronuclear single-quantum correlation NMR spectroscopy (HSQC NMR) titrations of eIF4E with m7G and ribavirin. 1H, 15N HSQC NMR spectroscopy reports on the chemical environment of the individual 15NH amides of the polypeptide backbone, thereby providing a sensitive probe of ligand binding and accompanying conformational rearrangements. In solution, eIF4E exists in low- and high-affinity conformations, the interconversion of which is regulated by binding of partner proteins such as PML and ligands such as m7G mRNA cap, as observed by using CD spectroscopy titrations (26, 36, 39, 40). Here, we observe a similar phenomenon upon the conversion from apo- to m7G-bound eIF4E by using HSQC NMR titrations, with the structuring or reorganization of 19 of 64 assigned residues of 217 residues in eIF4E (Fig. 4a), distributed throughout the structure (Fig. 4c), in agreement with CD measurements (26). These residues include the S7/S8 loop with W102, which stacks with the m7G base, and K106, which coordinates the ribose (Fig. 4c). On the other hand, the S1/S2 loop is preorganized in apo-eIF4E in the high-affinity conformation, with W56 showing no significant changes in resonance intensity or chemical shift upon cap binding (Fig. 4a). Strikingly, conversion of apo-eIF4E to ribavirin-bound eIF4E involves an almost identical conformational rearrangement, with little perturbation of the S1/S2 loop and W56 and significant structuring of the S7/S8 loop and W102 (Fig. 4b), as indicated by nearly exact overlay of cap- and ribavirin-bound spectra of eIF4E (Fig. 4b). These data are consistent with the reduced ribavirin affinity of W56A mutant and ribavirin's ability to efficiently compete with m7G for binding to eIF4E (Fig. 1). Double 15N-edited, filtered 1H, 1H NOESY spectra of nucleoside-saturated eIF4E, which specifically identify 15NH groups of eIF4E in close proximity (<5 Å) to nucleoside as a result of intermolecular NOE transfer, are consistent with the binding sites of ribavirin and m7G overlapping (data not shown). Thus, eIF4E binds and recognizes ribavirin in a manner similar to m7G cap, which is consistent with their similar binding activities (Fig. 1).
Fig. 4.
Ribavirin and m7G mRNA cap are recognized similarly by eIF4E. (a) 1H, 15N HSQC NMR spectra of eIF4E in the absence (black) and presence (red) of saturating concentrations of m7G nucleoside. Note that of the 273 residues of the construct, 207 resonances are observed. (b) 1H, 15N HSQC NMR spectra of eIF4E in the presence of saturating concentrations of m7G (red) and ribavirin nucleosides (blue). (c) eIF4E backbone residues that exhibit (red) and do not exhibit (blue) 1H, 15N HSQC NMR chemical shift perturbation upon binding of ribavirin and m7G mRNA cap. The difference between conformational rearrangements upon cap binding of mouse eIF4E observed here and those reported for yeast eIF4E (29) may be because of differences between mouse and yeast proteins as well as micelle binding to yeast eIF4E (29).
To assess the physical origin of ribavirin's mimicry of m7G, we calculated electrostatic properties of guanosine and ribavirin analogues by using ab initio quantum mechanical and continuum electrostatic methods. Only m7G and ribavirin exhibit significant electropositive character in their aromatic rings (Fig. 8, which is published as supporting information on the PNAS web site). Other nucleoside bases exhibit various degrees and patterns of electronegativity, including the inactive ribavirin analogue Rib4C, which is not protonated and uncharged at neutral pH due to its 1,2,3-triazole, inactive ribavirin metabolite ICN3297, which is neutral because of its oxidized carboxamide (data not shown), guanosine analogue and inosine monophosphate dehydrogenase inhibitor tiazofurin, which is neutral due to its thiazole, and uncharged guanosine itself (Fig. 4). Thus, ribavirin is a physical mimic of m7G.
There are two major cap-binding proteins in the cell, eIF4E and the cap-binding complex (CBC). Although both proteins intercalate m7G between two aromatic residues, the affinity of the CBC for m7GpppG cap is substantially higher (Kd ≈ 10 nM), as compared with eIF4E (Kd ≈ 200 nM), because of more extensive interactions of the CBC with the methylated base, as well as with the adjacent pyrophosphate nucleotide as compared with eIF4E (41). Because ribavirin's triazole ring would be missing many of these additional contacts with the CBC and is missing the adjacent base, ribavirin interferes only with the functions of eIF4E, and not those of the CBC, as observed here (Fig. 2).
Conclusions
Although widely studied, mechanisms of cellular action of ribavirin and origins of its antiviral effects remain enigmatic. Because of its similarity to guanosine, ribavirin is suggested to inhibit 5′ mRNA capping by competing with guanosine for guanylyl transferase, to inhibit guanosine biogenesis by mimicking guanosine for interaction with inosine monophosphate dehydrogenase, and to be a mutagen by competing with guanosine for mRNA incorporation by RNA polymerases (42). Indeed, at millimolar concentrations, such effects occur, leading to lethal mutagenesis of poliovirus (EC50 ≈ 0.2 mM) and depletion of cellular guanosine pools (EC50 ≈ 0.1 mM), for example (21, 43). Importantly, at low micromolar concentrations, ribavirin does not appear to participate in guanosine metabolism, likely because of structural and energetic differences in m7G and/or guanosine-binding sites of involved proteins. Ribavirin does not appear to cause physiologic depletion of guanosine pools, as suggested by lack of metabolic toxicity (Fig. 7), and is not apparently mutagenic, as suggested by lack of cell death and unaffected synthesis and stability of produced proteins (Figs. 2 and 7). Thus, the origin of more specific, growth-suppressive effects of ribavirin at low micromolar concentrations has remained poorly understood. Here, we observe that ribavirin inhibits the ability of eIF4E to promote mRNA transport and translation of eIF4E-sensitive transcripts by antagonizing eIF4E:m7G mRNA cap binding and disrupting subcellular eIF4E organization.
eIF4E overexpression does not increase protein synthesis globally but, rather, affects the expression of a subset of transcripts defined as eIF4E-sensitive, including those studied here, such as cyclin D1, ODC, and VEGF (13, 36, 37, 44). Although the major point of this work was to elucidate a mechanism of action of ribavirin and to characterize its potential anti-cancer activities, our findings have implications for mRNA translation as well. We show that selectivity of ribavirin's inhibition of eIF4E stems from the selectivity of eIF4E's activity itself in terms of eIF4E's posttranscriptional regulation of a specific set of eIF4E-sensitive transcripts. Thus, just as eIF4E overexpression does not globally increase protein translation (11, 36), ribavirin is not a global inhibitor. Sensitivity to eIF4E appears to be inversely related to the complexity of UTRs of corresponding transcripts. Hence, ribavirin-induced inhibition of eIF4E specifically reduces translation of the transcripts that contain long and highly structured 5′ UTRs, including a number of protooncogenic mRNAs, e.g., VEGF, c-myc, and ODC. Conversely, ribavirin does not affect translational rates of housekeeping mRNAs, such as GAPDH, that bear short, unstructured 5′ UTRs.
Electrostatic properties of guanosine-related nucleosides correlate directly with their point of action in cellular guanosine metabolism. For example, tiazofurin, despite having the same molecular geometry as ribavirin, is electronically similar to guanosine (Fig. 8) and, consequently, is a potent inosine monophosphate dehydrogenase inhibitor, binding to the guanosine allosteric effector site on inosine monophosphate dehydrogenase (45). Similarly, Rib4C is neutral (Fig. 8) and neither binds nor inhibits eIF4E (Figs. 1, 2, and 4). On the other hand, ribavirin is positively charged at physiological pH (32) because of its electronic structure (Fig. 8) and, as a result, antagonizes m7G mRNA cap binding by eIF4E (Figs. 1 and 3).
This specific and potent activity of ribavirin is unlikely to account for its entire spectrum of antiviral effects because ribavirin exhibits effects against viruses such as HCV and poliovirus that bypass cellular m7G cap-dependent mRNA processing (46–48). However, ribavirin's inhibition of eIF4E may contribute to its effects against viruses that coopt cellular eIF4E, such as Lassa fever virus (49), or those that use m7G, such as severe acute respiratory syndrome coronavirus (50). Because ribavirin has the widest spectrum of activity of all antiviral drugs to date, our findings offer specific directions (Figs. 4 and 8) for the design of improved derivatives, with increased efficacy against very deadly viral infections such as Lassa fever, and against emergent pathogens such as severe acute respiratory syndrome coronavirus.
Ribavirin and its derivatives offer a pharmacologic means to interrupt networks of tumor suppressors and oncogenes that maintain and enhance neoplasia and malignancy. For instance, deregulation of eIF4E leads to deregulation of oncogenes such as cyclin D1 and myc, which, in turn, leads to further deregulation of eIF4E (51). eIF4E is a target of mitogenic stimulation and a direct transcriptional target of myc (11). Consistent with such self-reinforcing behavior, inactivation of myc leads to differentiation and sustained regression of tumors in a transgenic mouse model of osteogenic sarcoma (4). Similarly, antisense cyclin D1 reverts the phenotype of human carcinoma cells toward normal and prevents tumor formation in mice (3). Complementarily, rapamycin suppresses chemoresistance in a mouse lymphoma model, and this effect is reversed by dysregulation of eIF4E (52). Here, we demonstrate that a similar effect can be accomplished pharmacologically by inhibiting eIF4E-dependent nucleocytoplasmic mRNA transport and translation.
It is becoming increasingly evident that posttranscriptional regulation of gene expression plays a paramount role in regulation of growth and development in eukaryotes, and disruption of this level of regulation contributes to a variety of human cancers. Our findings indicate that ribavirin acts in a previously unsuspected manner, at the level of posttranscriptional, eIF4E-mediated regulation of growth regulatory genes. It is likely that the apparent potency of ribavirin's suppression of eIF4E-mediated oncogenic transformation in vitro and in vivo involves down-regulation of a combination of oncogenes, with cyclin D1 being a model transcript examined here. Further characterization of this unforeseen mechanism of ribavirin action and development of derivatives with improved antiviral and cytostatic properties are important directions for future work.
Supplementary Material
Acknowledgments
We thank Maria Salvato, Roman Osman, Allan Capili, and Jonathan Licht for helpful discussions, Lei Zeng for assistance with NMR data collection, Zhi Hong (Valeant, Costa Mesa, CA) and Gerhard Wagner (Harvard University, Cambridge, MA) for kind gifts of Rib4C and eIF4E, respectively, and Monica Guzman and Craig Jordan (University of Rochester, Rochester, NY) for leukemia specimens. This work was supported by the National Institutes of Health Medical Scientist Training Program (to A.K. and L.S.). K.L.B.B. is a Scholar of the Leukemia and Lymphoma Society and holds a Canada Research Chair. Funding was provided by National Institutes of Health Grants CA 88991, CA 98571, and PO1 AI44236-01.
Author contributions: A.K. and K.L.B.B. designed research; A.K., I.T., B.C., and L.S. performed research; A.K. and K.L.B.B. analyzed data; and A.K. and K.L.B.B. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: m7G, 7-methyl guanosine; ODC, ornithine decarboxylase; HCV, hepatitis C virus; ribavirin, 1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide; Rib4C, 1-β-d-ribofuranosyl-1,2,3-triazole-4-carboxamide; RTP, ribavirin triphosphate; AML, acute myelogenous leukemia; HSQC NMR, heteronuclear single-quantum correlation NMR spectroscopy; CBC, cap-binding complex.
References
- 1.Weinstein, I. B. (2000) Carcinogenesis 21, 857–864. [DOI] [PubMed] [Google Scholar]
- 2.Pomeroy, S. L., Tamayo, P., Gaasenbeek, M., Sturla, L. M., Angelo, M., McLaughlin, M. E., Kim, J. Y., Goumnerova, L. C., Black, P. M., Lau, C., et al. (2002) Nature 415, 436–442. [DOI] [PubMed] [Google Scholar]
- 3.Colomer, R., Lupu, R., Bacus, S. S. & Gelmann, E. P. (1994) Br. J. Cancer 70, 819–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jain, M., Arvanitis, C., Chu, K., Dewey, W., Leonhardt, E., Trinh, M., Sundberg, C. D., Bishop, J. M. & Felsher, D. W. (2002) Science 297, 102–104. [DOI] [PubMed] [Google Scholar]
- 5.Kerekatte, V., Smiley, K., Hu, B., Smith, A., Gelder, F. & De Benedetti, A. (1995) Int. J. Cancer 64, 27–31. [DOI] [PubMed] [Google Scholar]
- 6.Rosenwald, I. B., Chen, J. J., Wang, S., Savas, L., London, I. M. & Pullman, J. (1999) Oncogene 18, 2507–2517. [DOI] [PubMed] [Google Scholar]
- 7.Nathan, C. A., Carter, P., Liu, L., Li, B. D., Abreo, F., Tudor, A., Zimmer, S. G. & De Benedetti, A. (1997) Oncogene 15, 1087–1094. [DOI] [PubMed] [Google Scholar]
- 8.Wang, S., Rosenwald, I. B., Hutzler, M. J., Pihan, G. A., Savas, L., Chen, J. J. & Woda, B. A. (1999) Am. J. Pathol. 155, 247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Topisirovic, I., Guzman, M. L., McConnell, M. J., Licht, J. D., Culjkovic, B., Neering, S. J., Jordan, C. T. & Borden, K. L. (2003) Mol. Cell. Biol. 23, 8992–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazaris-Karatzas, A., Montine, K. S. & Sonenberg, N. (1990) Nature 345, 544–547. [DOI] [PubMed] [Google Scholar]
- 11.Gingras, A. C., Raught, B. & Sonenberg, N. (1999) Annu. Rev. Biochem. 68, 913–963. [DOI] [PubMed] [Google Scholar]
- 12.Iborra, F. J., Jackson, D. A. & Cook, P. R. (2001) Science 293, 1139–1142. [DOI] [PubMed] [Google Scholar]
- 13.Rousseau, D., Kaspar, R., Rosenwald, I., Gehrke, L. & Sonenberg, N. (1996) Proc. Natl. Acad. Sci. USA 93, 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen, N., Sharma, M., Kentsis, A., Perez, J. M., Strudwick, S. & Borden, K. L. (2001) EMBO J. 20, 4547–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topisirovic, I., Culjkovic, B., Cohen, N., Perez, J. M., Skrabanek, L. & Borden, K. L. (2003) EMBO J. 22, 689–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Topisirovic, I., Capili, A. D. & Borden, K. L. (2002) Mol. Cell. Biol. 22, 6183–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Benedetti, A. & Harris, A. L. (1999) Int. J. Biochem. Cell Biol. 31, 59–72. [DOI] [PubMed] [Google Scholar]
- 18.Strudwick, S. & Borden, K. L. (2002) Differentiation 70, 10–22. [DOI] [PubMed] [Google Scholar]
- 19.Sidwell, R. W., Huffman, J. H., Khare, G. P., Allen, L. B., Witkowski, J. T. & Robins, R. K. (1972) Science 177, 705–706. [DOI] [PubMed] [Google Scholar]
- 20.Tam, R. C., Lau, J. Y. & Hong, Z. (2001) Antiviral Chem. Chemother. 12, 261–272. [DOI] [PubMed] [Google Scholar]
- 21.Crotty, S., Maag, D., Arnold, J. J., Zhong, W., Lau, J. Y., Hong, Z., Andino, R. & Cameron, C. E. (2000) Nat. Med. 6, 1375–1379. [DOI] [PubMed] [Google Scholar]
- 22.Crotty, S., Cameron, C. E. & Andino, R. (2001) Proc. Natl. Acad. Sci. USA 98, 6895–6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maag, D., Castro, C., Hong, Z. & Cameron, C. E. (2001) J. Biol. Chem. 276, 46094–46098. [DOI] [PubMed] [Google Scholar]
- 24.Zoulim, F., Haem, J., Ahmed, S. S., Chossegros, P., Habersetzer, F., Chevallier, M., Bailly, F. & Trepo, C. (1998) J. Viral Hepat. 5, 193–198. [DOI] [PubMed] [Google Scholar]
- 25.Drach, J. C., Thomas, M. A., Barnett, J. W., Smith, S. H. & Shipman, C., Jr. (1981) Science 212, 549–551. [DOI] [PubMed] [Google Scholar]
- 26.Kentsis, A., Dwyer, E. C., Perez, J. M., Sharma, M., Chen, A., Pan, Z. Q. & Borden, K. L. (2001) J. Mol. Biol. 312, 609–623. [DOI] [PubMed] [Google Scholar]
- 27.DeFatta, R. J., Nathan, C. A. & De Benedetti, A. (2000) Laryngoscope 110, 928–933. [DOI] [PubMed] [Google Scholar]
- 28.Capili, A. D., Edghill, E. L., Wu, K. & Borden, K. L. (2004) J. Mol. Biol. 340, 1117–1129. [DOI] [PubMed] [Google Scholar]
- 29.McGuire, A. M., Matsuo, H. & Wagner, G. (1998) J. Biomol. NMR 12, 73–88. [DOI] [PubMed] [Google Scholar]
- 30.Marcotrigiano, J., Gingras, A. C., Sonenberg, N. & Burley, S. K. (1997) Cell 89, 951–961. [DOI] [PubMed] [Google Scholar]
- 31.Niedzwiecka, A., Marcotrigiano, J., Stepinski, J., Jankowska-Anyszka, M., Wyslouch-Cieszynska, A., Dadlez, M., Gingras, A. C., Mak, P., Darzynkiewicz, E., Sonenberg, N., et al. (2002) J. Mol. Biol. 319, 615–635. [DOI] [PubMed] [Google Scholar]
- 32.O'Neil, M. J., Smith, A., Heckelman, P. E. & Obenchain, J. R., Jr. (2001) The Merck Index (Merck, Whitehouse Station, NJ), 13th Ed.
- 33.Smith, R. A., Knight, V. & Smith, J. A. D. (1984) Clinical Applications of Ribavirin (Academic, Orlando, FL).
- 34.Page, T. & Conner, J. D. (1990) Int. J. Biochem. 22, 379–383. [DOI] [PubMed] [Google Scholar]
- 35.Dostie, J., Lejbkowicz, F. & Sonenberg, N. (2000) J. Cell Biol. 148, 239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von der Haar, T., Gross, J. D., Wagner, G. & McCarthy, J. E. (2004) Nat. Struct. Mol. Biol. 11, 503–511. [DOI] [PubMed] [Google Scholar]
- 37.Graff, J. R. & Zimmer, S. G. (2003) Clin. Exp. Metastasis 20, 265–273. [DOI] [PubMed] [Google Scholar]
- 38.Sorrells, D. L., Ghali, G. E., Meschonat, C., DeFatta, R. J., Black, D., Liu, L., De Benedetti, A., Nathan, C. A. & Li, B. D. (1999) Head Neck 21, 60–65. [DOI] [PubMed] [Google Scholar]
- 39.Wang, Y., Sha, M., Ren, W. Y., van Heerden, A., Browning, K. S. & Goss, D. J. (1996) Biochim. Biophys. Acta 1297, 207–213. [DOI] [PubMed] [Google Scholar]
- 40.Kentsis, A., Gordon, R. E. & Borden, K. L. (2002) Proc. Natl. Acad. Sci. USA 99, 15404–15409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calero, G., Wilson, K. F., Ly, T., Rios-Steiner, J. L., Clardy, J. C. & Cerione, R. A. (2002) Nat. Struct. Biol. 9, 912–917. [DOI] [PubMed] [Google Scholar]
- 42.Hong, Z. & Cameron, C. E. (2002) Prog. Drug Res. 59, 41–69. [DOI] [PubMed] [Google Scholar]
- 43.Fouchier, F., Forget, P., Bellan, C., Marvaldi, J., Champion, S. & Pichon, J. (1996) J. Recept. Signal Transduct. Res. 16, 39–58. [DOI] [PubMed] [Google Scholar]
- 44.Graff, J. R., De Benedetti, A., Olson, J. W., Tamez, P., Casero, R. A., Jr., & Zimmer, S. G. (1997) Biochem. Biophys. Res. Commun. 240, 15–20. [DOI] [PubMed] [Google Scholar]
- 45.Colby, T. D., Vanderveen, K., Strickler, M. D., Markham, G. D. & Goldstein, B. M. (1999) Proc. Natl. Acad. Sci. USA 96, 3531–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rijnbrand, R. C. & Lemon, S. M. (2000) Curr. Top Microbiol. Immunol. 242, 85–116. [DOI] [PubMed] [Google Scholar]
- 47.Sonenberg, N. & Pelletier, J. (1989) BioEssays 11, 128–132. [DOI] [PubMed] [Google Scholar]
- 48.Crotty, S., Cameron, C. & Andino, R. (2002) J. Mol. Med. 80, 86–95. [DOI] [PubMed] [Google Scholar]
- 49.Campbell Dwyer, E. J., Lai, H., MacDonald, R. C., Salvato, M. S. & Borden, K. L. (2000) J. Virol. 74, 3293–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Grotthuss, M., Wyrwicz, L. S. & Rychlewski, L. (2003) Cell 113, 701–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zimmer, S. G., DeBenedetti, A. & Graff, J. R. (2000) Anticancer Res. 20, 1343–1351. [PubMed] [Google Scholar]
- 52.Wendel, H. G., De Stanchina, E., Fridman, J. S., Malina, A., Ray, S., Kogan, S., Cordon-Cardo, C., Pelletier, J. & Lowe, S. W. (2004) Nature 428, 332–337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.