Abstract
The identification of recurring patterns within a sequence of events is an important task in behavior research. In this paper, we consider a general probabilistic framework for identifying such patterns, by distinguishing between events that belong to a pattern and events that occur as part of background processes. The event processes, both for background events and events that are part of recurring patterns, are modeled as competing renewal processes. Using this framework, we develop an inference procedure to detect the sequences present in observed data. Our method is compared to a current approach used within the ethology literature on both simulated data and data collected to study the impact of fragmented and unpredictable maternal behavior on cognitive development of children.
1 Introduction
The study of behavior plays an important role in a variety of fields, such as psychology, neuroscience, sociology, and zoology. However, most behavior studies are qualitative in nature, as constructs of behavior have proven difficult to characterize quantitatively. In particular, a great deal of subjectivity is applied when determining what does or does not constitute an interesting or scientifically relevant behavior, and when attempting to differentiate between various types of behaviors. Researchers often want both to predict when an actor will exhibit various behaviors, and to use past behavior to predict other characteristics. Thus a quantitative construct of behavior would be beneficial to help drive these fields when investigating behavior.
One possible construct that has proven popular are recurring behavioral patterns, which are relatively simple to identify and have proven useful for characterizing temporal streams of behavior [Eibl-Eibesfeldt, 1970]. However, most analyses fix patterns of interest a priori, and attempt to identify those specific patterns. Approaches to automatically detect such patterns are in their infancy. In this paper, we present a probabilistic model which serves as an attempt to model some of these intricacies of behavior, which allows the extraction of patterns for further analysis.
We describe a behavioral sequence as a series of events performed by an actor or actors, with each event occurring at some point in continuous time. The data we consider can be represented as a sequence of time-event pairs, including the type of event and the time that it occurred. Event types are defined from a fixed set of possible behaviors identified by subject matter experts. Our model identifies repeated patterns within these sequences of events, which we posit describe an element of the behavior of an individual or individuals. These types of repeated behavioral patterns have proven beneficial to behavior research in the past.
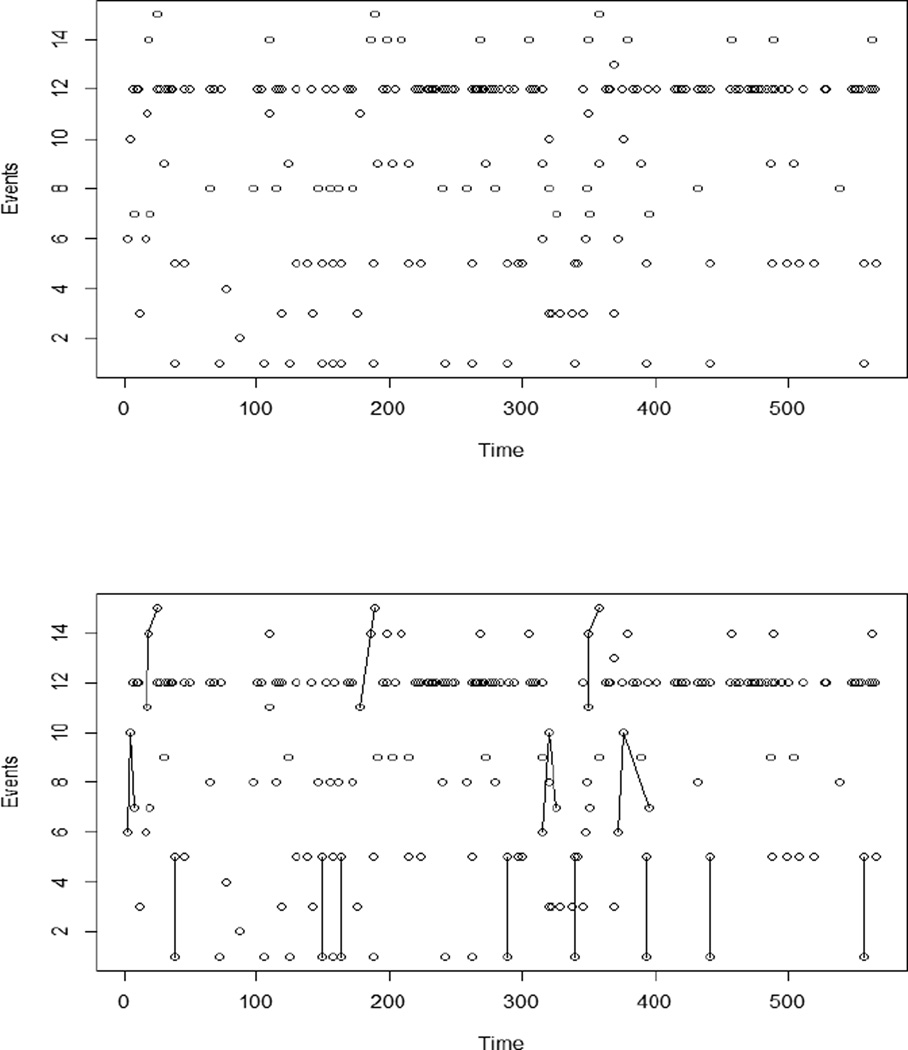
Often, when patterns are not known a priori, they are identified by visual inspection. However, it can be very difficult to identify patterns with visual inspection alone, see Fig 1. Thus most analysts resort to guessing which patterns they expect to exist, and then noting how often the expected patterns occur.
Figure 1.
Graphs of behavior data. The 15 events types, represented as integers, are on the vertical axis. Horizontal axis gives time in seconds. Each dot represents a recorded event. Top and bottom panels are the same, with patterns identified in the lower plot.
The aim of this paper is to introduce a general purpose model that can automatically extract these repeated patterns in sequences of time-indexed events. The existence of such patterns, and their varied numbers in different individuals, often carry scientific significance, such as in behavior studies.
In Section 2, we provide a brief background on existing approaches to event sequence modeling. In Section 3, we describe the data generating process for our model in detail. This is followed by a discussion of our approach to inference follows in Section 4, experiments and application in Section 5, and a discussion of future directions in Section 6.
2 Background
There are several techniques for identifying patterns in discrete sequences of observations. For instance, a rich literature exists on pattern finding within DNA sequences where repeated patterns, known as motifs, are important for understanding gene expression [Kellis et al., 2004, Liu et al., 1995]. A motif finding algorithm typically operates by first establishing a background model and then searching for any sequences that occur more often than expected by the background model.
Several compression methods can be applied to discrete time series to identify recurring sequences, such as the Lempel-Ziv algorithm and its derivatives [Welch, 1984, Ziv and Lempel, 1978]. These lossless compression methods focus on creating a dictionary of recurring patterns in a series of events, allowing us to represent the original data space with fewer bits.
The study that motivates our work attempts to search for patterns in sequences of events measured in continuous time. Time series pattern finding for continuous time data (e.g., looking for change points) usually occurs in the context of continuous measurements made at regular time points. This allows the use of standard time series models as the foundation for a model. However, our data are categorical measurements occurring at irregular time points. This type of data could instead be modeled as a series of states, such as a Markov process, but this would be insufficient for addressing our scientific questions. Instead, we would like to model each action as an instantaneous event, and then identify temporal relationships between several events.
In cases where continuous time points exist, discrete time series methods could be implemented by simply removing the time element, and assuming that all events are equally spaced. The times series could also be discretized into equally sized time bins, with several ’no action’ events assigned to bins where no events are recorded. The former method could run into issues when the equal time spacing assumption is incorrect, and different scales of discretization could have significant effects on the latter.
The software package Theme is one existing method to extract sequences from continuous data [Magnusson, 2000]. Theme begins by assuming that all events occur uniformly within the observation period, and uses exhaustive search methods to determine if any sequence of events occurs more often than expected under a uniformly distributed model. However there are some aspects of Theme that we have found difficult to navigate. The program, using fairly standard settings, identifies a large number of potential patterns. The assessment of which of these are genuine and which are false positives is not explicit in Theme’s output. We discuss this further below.
At present, we are not aware of any alternative models that address these concerns. With those concerns in mind, we have developed a probabilistic model that explicitly incorporates continuous time.
3 Data Generating Process
It is easiest to describe our model and our approach to inference by focusing on the model’s data generating process. Events are assumed to be generated by competing renewal processes, which we classify as either background or sequence processes. The model assumes most observed events occur independently of other recent events; these are assumed to belong to the background processes. Other events are assumed to occur as part of a sequence; these events are generated by sequence processes.
We consider data comprised of a set of n events that occur over some period of time. Events are observed as time-event pairs {Ei, Ti} with occurrence times Ti ∈ ℝ+ and event types Ei ∈ Q, where Q = {1, 2, …, J} is a set of possible event types. Occurrence times are strictly increasing, so Ti > Ti−1, and we define event interarrival times as ti = Ti − Ti−1. While events are observed as {Ei, Ti}, our model is most easily described with the equivalent interarrival time-event pairs {Ei, ti}.
3.1 Background Processes
To begin, we describe the background process corresponding to a single event type. For the event type j, we define its background process as a continuous time renewal process with independent and identically distributed interarrival times from some arbitrary strictly-positive distribution ti ~ Fj (ti). For example, if Fj (·) is defined as an exponential distribution, the resulting process would be a Poisson process.
Our data are assumed to include J possible event types. For each event type, we assume the events belong to an independent background process, each with its own unique interarrival time distribution. Thus we consider a multi-event model with several independent processes, each one corresponding to an event in Q. We will distinguish the interarrival times of each of these processes by a superscript, so for event type j ∈ Q we have .
We consider a competing risks framework to generate the event sequence {Ei, ti}i=1, …, n from the independent renewal processes. In this scenario, each of the possible actions in Q is a competing event, where the first to occur will be labeled as the next event to occur in our observed sequence. The competing risks framework will prove especially useful when we incorporate sequence processes in the next subsection.
At the initial time T0, we want to know the next pair {E1, t1} to occur in the sequence. The data generating mechanism generates interarrival times from the J event processes, denoted . The events are sorted into a schedule according to increasing times, and we observe the first scheduled event next. Thus the time until the next event can be modeled as the minimum over the interarrival times and the corresponding event type is labeled the next event .
Now suppose we are at time Ti and have just observed the pair {Ei, ti}. Recall that the next event in each background process is scheduled to occur at for j ≠ Ei. We keep these other events that were already scheduled, , to determine the next event in sequence. Two additional steps are required before determining the next time-event pair. First we need to account for the fact that these events did not occur between Ti−1 and Ti. We update the corresponding interarrival times by decrementing each time by the value of ti, for j ≠ Ei. Second, we need to include a new event corresponding to Ei, so we draw a new interarrival time and append it to the set of interarrival times. The new set of active interarrival times can now be denoted as .
As a consequence of the decrement, is no longer distributed according to Fj (ti+1). We can derive the correct distribution most easily by introducing the survival function. The survival function for the original interarrival time for event type j, , is defined as . Then, for event types j ≠ Ei, the conditional survival function of the decremented interarrival time, , is easily derived as
| (1) |
We refer to (1) as the event specific conditional survival functions S̄j (t), and refer to the survival function for the next event as the multi-event survival function S (t). Because we assume that the J processes are independent, the multi-event survival function can be defined as the product of the relevant conditional survival functions,
| (2) |
To perform inference for our model (see Section 4), we need to derive the density of the interarrival times from the survival function. The hazard function λ (t), the instantaneous rate of events conditional on survival to time t, can be defined via the survival function, . The density of the interarrival times, f (t), is the product of the survival and hazard functions, f (t) = λ (t) S (t). Following the notation above, we use the superscript j with the hazard function λj (t) to denote the hazard function for the process of the jth event type, and let λ (t) denote the hazard function for the multi-event background process. We can use the relationship of the hazard and survival functions to establish that the multi-event hazard is , and thus is the density of the interarrival times.
The previous paragraph defines the density function for the interarrival time. We are interested in the joint density of the event type and the interarrival time, or the event-time pair {Ei, ti}. Because the probability density for the interarrival times marginalizes the joint density over all possible events j, it follows that the background process time-event pairs have the following density:
| (3) |
where S (t) is as defined in (2).
3.2 Sequence Processes
In addition to background events, we also want to consider the possibility of recurring sequences, where certain events tend to occur more often after some other set of events has occurred. We model these sequences of events as part of sequence processes, which are assumed independent of the background processes. Sequence processes are identified when events occur in sequence more often than expected if they belonged to the background processes. We require all preceding events must have occurred to observe an event in a sequence process.
For notational convenience, let s index a specific sequence, and let s {ℓ} be the event index corresponding to the ℓth event in that sequence. Thus for the sequence of behaviors A → B → C, we say Es{1} = A, Es{2} = B, and Es{3} = C. We require notation of this form because it is not necessary that the events in a recurring sequence be consecutive observations. It is possible that an unrelated event may occur in the midst of a sequence by chance. We refer to these events as noise events, intervening events that occur during a sequence but do not belong to that sequence. These events can occur as part of either background processes or other sequence processes. Furthermore, our model allows a single event to belong to multiple sequence processes.
We introduce a parameter that governs the maximum time allowed between events in a sequence. We included this parameter both because it is well accepted that events occurring well in the past have much less direct influence on current behavior, and for computational convenience. However, no limit is placed on either the number of events in a sequence or its overall duration, so it remains possible that events well in the past can have indirect influence on current events.
We denote the time parameter as τ, a positive number that represents the maximum amount of time that can elapse between events Es{ℓ−1} and Es{ℓ} for each ℓ. Hence we require that the ℓth event in the sequence s {·} must occur within the time window [Ts{ℓ−1}, Ts{ℓ−1} + τ]. If Es{l} does not occur by the end of this window, then the sequence does not occur. Given events Es{1}, …, Es{ℓ−1} have occurred, there is positive probability that event Es{ℓ} does not occur, and thus neither does the sequence s {·}. This probability is equivalent to the probability that the interarrival time is greater than τ,
| (4) |
Here we introduce Ṡs{ℓ} (τ) as notation for the survival function of the next event. The censoring at τ complicates the calculation of the survival distribution associated with the sequence event s {ℓ}. The survival function is right censored at τ, with a nonzero probability that the event does not occur. We define a new censored survival function S̃s{ℓ} (t) as
| (5) |
The latter half of the sum in (5) refers to a point mass giving the probability that the event does not occur. Let λ̇ (t) denote the hazard function associated with Ṡ (t). Then by the properties of the density function described earlier, the probability density of a sequence event can be derived as
| (6) |
To model the noise events defined earlier, assume the ℓth event in the sequence occurs at Ti, so s {ℓ} = i. The previous event in the sequence, Es{ℓ−1}, occurs at some time Ts{ℓ−1} which is less than Ti−1 if intervening events are present, but equal to Ti−1 if not. Thus, the time of the intervening event can be represented as Ti−1 = Ts{ℓ−1} + r where r ≥ 0 is the time between the previous event in sequence and the noise event. If no noise event is present, then r = 0, but if a noise event is present, then s {ℓ − 1} < i − 1 and r > 0. The interarrival time for the sequence can then be represented as Ts{ℓ} − Ts{ℓ−1} = t + r, where t is the time between Ts{ℓ} and Ti−1 and r is the time between Ts{ℓ−1} and Ti−1. Here we considered the noise event occurring directly before Es{ℓ}, but any number of noise events may occur between events Es{ℓ−1} and Es{ℓ}.
The hazard function λ̇ (t) remains unchanged, so the new density for event s {ℓ} can be expressed as
| (7) |
This defines the density for a single event in a particular sequence process. We now want to consider multiple sequence processes, in addition to the background processes that were described earlier.
3.3 Combining Background and Sequence Processes
Our model assumes that background and sequence events occur independently, which allows us to combine them in a relatively straightforward manner. We assume that all sequence events are initialized by a background event as their initial event, thus s {1} effectively belongs to both a background and a sequence process.
As before, assume the current time is Ti and we observe {Ei, ti}. Now the pool of possible next events is comprised both of all background events as well as events belonging to all live sequences. We define a live sequence as any process such that s {1} through s {ℓ − 1} has occurred previously and Ts{ℓ−1} < Ti < Ts{ℓ−1} + τ. Let ℬ = {1, …, J} denote all background processes, and 𝒮 = {s1, …, sK} denote all K live sequence processes at time Ti, so the next event must occur from either ℬ or S.
From our independence assumption, and . Furthermore, from the definition of the sequence process, we have
| (8) |
where .
Finally, we get the following density, inserting the modification from (7) as needed:
| (9) |
where Λ̇e (t) = ∑s∈𝒮 for which s{ℓ}=e λ̇s (t)
Finally, we can calculate the likelihood of the model by taking the product of the densities for all time-event pairs: , where Θ denotes the set of all model parameters.
Though not the focus of our paper, we note that our probabilistic model also has a built in mechanism for prediction. The equation in (9) provides the density of a given time-event pair. One could simply calculate the likelihood for each possible event type, and then normalize to obtain the predictive probability that the next event is a particular event type.
4 Inference
To this point, we have not specified the functional form for the survival and hazard functions of the renewal processes. Fortunately, our inference approach is general, and can handle a wide variety of functional forms. To describe our inference approach, we assume each background and sequence process has its own parameters θ, which includes the maximum time parameter τ for sequence processes. We denote the entire parameter set as Θ. We take a Bayesian approach to learning the parameters in our model. For our experiments, we place independent Cauchy priors on all parameters, truncated to the support of each parameter. Parameters will be sampled via a Markov Chain Monte Carlo (MCMC) procedure, described below.
Our model allows for a potentially unlimited number of sequence processes to exist, so we need to determine which sequences actually exist in the observed data. Including all possible sequences is impractical; when we consider sequences of increasing maximum length, the number of possible sequences grows exponentially. Instead, we only include a small subset of all possible sequences in our model at any given point in the sampler, and use a birth-death MCMC technique to vary the subset of sequences.
Sequences that are very unlikely to occur will have a hazard rate near zero, and thus a survival function constantly near one. When λ (t) ≈ 0 and S (t) ≈ 1 for all t, from (9) we see that the sequence is effectively not included in the model. In this case, we fix the sequence process parameters θ such that the hazard rate is fixed at zero.
Our complete approach to inference uses the No-U-Turns Sampler (NUTS) for sampling the parameters, with occasional birth-death steps incorporated for model selection. First, we describe NUTS, implemented with the Stan software [Hoffman and Gelman, 2012, Stan Development Team, 2013], and then describe the birth-death steps.
NUTS is a form of Hamiltonian Monte Carlo (HMC), an MCMC algorithm that avoids the random walk behavior of several other popular MCMC methods. However, standard HMC is sensitive to user provided parameters: the number of leap frog steps and the step size. NUTS improves upon the standard HMC implementation by eliminating the need to set both parameters, though our implementation does still require that we set the step size, as described below. Furthermore, the Stan implementation uses reverse-mode algorithmic differentiation to calculate the gradient, eliminating the need for us to calculate it by hand.
While NUTS is very effective for sampling from the posterior distribution, it does not address model selection. To determine which sequence processes belong in the model, we apply the birth-death process of Stephens for model selection, an alternative to reversible jump steps [Stephens, 2000]. Stephen’s birth-death MCMC views the parameters of the model as a point process, which allows the number of components to vary by allowing new ones to be ’born’ and existing ones to ’die.’ At various times within the overall NUTS chain, we perform a birth-death step by constructing an easily simulated process that changes the number of sequences in the model. The births and deaths both occur according to Poisson processes. Births occur at a constant rate, while deaths occur at a low rate for sequences critical for explaining the data, and a high rate for sequences that do not help explain the data.
When a birth-death step occurs, we fix some simulation time W and run through a series of birth or death steps until the simulated time exceeds W. Birth steps occur according to a Poisson process with rate δB. Each sequence s currently in the model receives an independent death process with rate , described in the next paragraph. The overall rate of death steps is . The time to the next step is exponentially distributed with rate δB + δD. Times are generated until they pass time W, with a birth or death step occurring at each time with probability proportional to the rate for the birth/death.
To ensure that important sequences are retained within the model, the death rate for a given sequence s will be equal to the birth rate multiplied by the ratio of the time-event likelihoods with and without sequence s included. Thus sequences that have a profound positive effect on the likelihood when included, and thus for which the data suggests they belong in the model, will have very low death rates.
For the birth step, we choose a sequence not currently in the model, and draw parameters from an appropriate distribution, e.g. the prior distribution for it. To choose the new sequence, we pool all existing sequences in the model, randomly sample two of them and combine them to form a new sequence. For these purposes, we include all single events as sequences in addition to the multi-event sequences in the model. For example, assume the model has background events A, B, and C, as well as the C→A pattern. We sample, with replacement, two of those four possible events, and combine them to form the new pattern. Possible new patterns include A→B, C→A→B, or C→A→C→A, among others.
A death step simply chooses an existing sequence process, and fixes its parameters such that the hazard function is zero for all possible times t.
The simulated birth-death process proceeds as follows:
Algorithm 1.
Birth-Death Process
| Fix δB, W ; // Birth Rate, BirthDeath Sim Time |
| w ← 0; |
| while w < W: |
| ; // Sequence Death Rate |
| ; // Overall Death Rate |
| Sample u ~ unif (0, 1); |
| if : |
| Birth Step |
| else: |
| Death Step |
| Sample ; // Next Jump Time |
| w ← w + t; |
Here Θ − θs denotes the parameter set Θ excluding parameters related to the sequence s. The birth-death process helps the model selection process traverse the posterior distribution more efficiently than reversible jump steps, as well as improve sampling efficiency by never rejecting a proposed change in model.
Finally, we describe how our model selection technique is integrated within the NUTS algorithm. The birth-death process is run once every m iterations of the sampler, allowing NUTS to explore the state space for every model. The standard implementation of NUTS within Stan determines the step size needed for HMC during the warmup phase. To ensure ergodicity, we estimate the step size for the initial model (typically without sequence processes included), and then fix the step size at this value when running NUTS.
5 Results
For this paper, all experiments assume sequences have a constant hazard function, and therefore the interarrival times are exponentially distributed. However, our model can accommodate more general hazard rates. We have implemented our model to include renewal processes with Weibull increments, and have plans to consider other alternative distributions.
5.1 Simulation
We first consider the results for simulated data under three different simulation scenarios. The first simulation is a simple scenario, with only three possible event types which we label events 1, 2, and 3. We include only one sequence process, a simple 2-event sequence: 3 → 1. We fix the background exponential process parameters λ1 = 0.16, λ2 = 0.57, and λ3 = 0.40, the sequence exponential process parameter λ3→1 = 0.25 and the corresponding time window parameter as τ3→1 = 1.12. Using these parameters, we sample M = 100 data sets of length n = 500 events, and fit the model to each data set.
Our other two simulations consider data which are more consistent with possible applications. Both simulations are comprised of 20 different possible event types. The first has three 2-event sequences, representing an actor with very few repeated patterns. The second simulates data with both more patterns (six) and more complex patterns (up to 4-events in one pattern). Again, we sample M = 100 data sets of length n = 500 events for both of the more complex scenarios.
For each simulated data set in each scenario, we run our inference procedure treating all parameters as unknown. We calculate a 95% posterior credible interval for each parameter. Table 1 shows the coverage probabilities for the posterior intervals for the parameters in the first scenario. All are 0.95 or higher. For the second and third scenarios, there are considerably more parameters; all have coverage probabilities above 0.90.
Table 1.
Coverage probability estimates for each parameter for M = 100 simulated data sets
| λ1 | λ2 | λ3 | λ3→1 | τ3→1 |
|---|---|---|---|---|
| 0.97 | 0.95 | 0.97 | 0.97 | 0.95 |
We also record which sequences were included by the model selection process for each data set in each scenario. We record the proportion of trials in which the correct sequence was identified (power), the number of null sequences incorrectly identified as being present (false positives), and the proportion of declared sequences that are true sequences (precision). Our Renewal Process Model’s results for the first scenario are in the final column of Table 2.
Table 2.
Number of sequences for M = 100 simulated data sets
| Theme | Renewal Process Model |
|
|---|---|---|
| # of Correct Sequences |
94/100 | 99/100 |
| # of False Positives | 384 | 57 |
| # of False Negatives | 6 | 1 |
| Power/Recall | 0.94 | 0.99 |
| Precision | 0.20 | 0.64 |
We also ran Theme with its default settings on each of the simulated data sets for the first scenario, and recorded the same summaries. As described earlier, assessing significance in Theme is not always straightforward, and not all patterns are assumed real. After the patterns are identified, a permutation procedure can be used, which compares the results with the results using randomized data. However, it does not indicate which patterns are most important. Here we just summarize the patterns identified recognizing that practiced Theme users would not consider all of these as real. These results are also included in Table 2. Theme identified the correct sequence about as often as our model, though it often identified several other sequences that do not actually exist.
For the second simulation scenario, all 3 patterns were found in 91% of the simulations using the RPM, with an average of 0.87 false positive patterns per simulation. In the final scenario, all 6 patterns were found in 87% of all simulations with an average of 1.6 false positives per simulation. Thus our method performs similarly well for more complex data. Furthermore, these simulations were designed to be consistent with our application (see below), and it does not appear that the complexity of the model, including the larger parameter space, has any negative effects on our model. For applications where more complexity is expected (more event types, more patterns, longer patterns), further simulation studies would be recommended.
5.2 Mother-Child Data
Previous experimental research has identified fragmented and unpredictable maternal behavior in rodents as a risk factor for outcomes that are analogous to emotional and cognitive disorders in humans [Baram et al., 2012]. However, how to best characterize fragmented and unpredictable behavior in humans in clinically meaningful ways remains an open research question. One proposed method evaluates maternal behaviors and defines fragmented and unpredictable behavior using the number and length of recurring sequences.
We define consistent behavior as long repeated sequences of actions, whereas fragmented behavior has few, if any, long repeated sequences. Unpredictable behavior refers to the inability to determine the mother’s next action after considering the previous actions performed by her and the child. For the purposes of this paper, the degree to which a mother’s behavior is deemed fragmented or unpredictable will be determined via the length of the longest repeated sequence and the number of different sequences identified. Note that the definition of fragmented and unpredictable behavior does not consider whether the actions are positive or negative, but rather only considers the structure of the behavior.
As part of a longitudinal study to examine the impact of early interactions between a mother and her child on the child’s development through adolescence, our collaborators have recorded several instances of mothers playing with their children in the research lab. The data consists of short videos, about 10 minutes long, which have been annotated by the researchers to include the type of each event and the time when it occurs. Event types include both changes in the mother’s expression or emotional state (smiling, content, bored) as well as physical interactions (hug, play with toy, pick up child, speak). The number of events in any particular session ranges from about 100 to 400 events, with about 20 unique event types per session. An example of a mother-child session can be seen in Figure 1, where time is on the horizontal axis, and events (with numerically assigned labels) are on the vertical axis.
We ran both our model and Theme for five different mothers. Each mother has one 10 minute session, and we consider both models independently across the five mothers. Results are summarized in Table 3. As before, Theme detects considerably more patterns than our model, as well as longer sequences.
Table 3.
Number of Patterns and Longest Pattern in Maternal Behavior Data for Theme and the Renewal Process Model (RPM)
| Mother | One | Two | Three | Four | Five |
|---|---|---|---|---|---|
| Seq. Length | 313 | 160 | 213 | 273 | 210 |
| Number of Patterns Identified | |||||
| Theme | 39 | 27 | 73 | 80 | 64 |
| RPM | 5 | 3 | 4 | 3 | 4 |
| Longest Pattern Identified | |||||
| Theme | 9 | 6 | 10 | 11 | 9 |
| RPM | 2 | 3 | 2 | 2 | 3 |
While our model detects considerably fewer sequences, the sequences that are detected are largely quite sensible. Examples of detected patterns, with the mothers demonstrating each pattern in parentheses, include:
New Toy → Manipulating Toy (1, 2, 3, 4, 5)
Smiling → Laughing (1, 2, 3)
Carrying Child → Setting Child Down (5)
Speaking → Child in Lap → Hug/Kiss (2)
Sing → Smile → Silent (5)
The first two patterns are shared across several mothers, and it does seem likely that certain patterns will be common across different mothers. We return to this point below.
6 Conclusion
In this paper, we proposed a generative model, the Renewal Process Model, and an inference procedure for identifying recurring patterns from a stream of events in continuous time. Our experiments show that our model correctly identifies patterns in simulated data, performs admirably with increasing complexity, and finds considerably fewer false positives than the current state of the art algorithm. Furthermore our model identifies both interesting and intuitive patterns in real data. The Renewal Process Model appears to provide several advantages, including a rigorous probability structure that is easily generalized and a marked improvement on the number of false positives that are generated.
Behavior data, such as the maternal care behavior we examined, can be expensive to collect, and time consuming to annotate. This means that we have relatively little data per individual, which can make it difficult to identify patterns. As noted above, it is reasonable to expect similar patterns across the population of mothers. This suggests a hierarchical model in which each actor has her own model parameters, but the parameters are assumed to come from a common population distribution. Such a hierarchical model would improve our inference by borrowing strength across actors, which can help us to identify sequences that may not be significant in just one observation period. This is an area of current research.
Furthermore, our model can easily be adapted to incorporate different probability distributions for the interarrival times. In our experiments, we only consider sequence processes with constant hazards, but including more complicated hazard or survival functions would allow for more complex modeling. In particular, we are working to expand the model to include several families of distributions, as well as to develop a version that can accommodate nonparametric interarrival time distributions.
Acknowledgments
This work was supported by NIH grant MH096889.
Footnotes
Appearing in Proceedings of the 17th International Conference on Artificial Intelligence and Statistics (AISTATS) 2014, Reykjavik, Iceland.
Contributor Information
Kevin Heins, Department of Statistics, University of California, Irvine.
Hal Stern, Department of Statistics, University of California, Irvine.
References
- Baram TZ, Davis E, Obenaus A, Sandman C, Small S, Solodkin A, Stern H. Fragmentation and unpredictability of early-life experience in mental disorders. Am J Psychiatry. 2012;169(9):907–915. doi: 10.1176/appi.ajp.2012.11091347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibl-Eibesfeldt I. Ethology: The Biology of Behavior. Holt, Rinehart and Winston: 1970. [Google Scholar]
- Hoffman MD, Gelman A. The no-U-turn sampler: Adaptively setting path lengths in Hamiltonian Monte Carlo. Journal of Machine Learning Research. 2012 [Google Scholar]
- Kellis M, Patterson N, Birren B, Berger B, Lander ES. Methods in comparative genomics: genome correspondence, gene identification and regulatory motif discovery. Journal of Computational Biology. 2004;11(2–3):319–55. doi: 10.1089/1066527041410319. [DOI] [PubMed] [Google Scholar]
- Liu JS, Neuwald AF, Lawrence CE. Bayesian models for multiple local sequence alignment and gibbs sampling strategies. Journal of the American Statistical Association. 1995;90(432):1156–1170. [Google Scholar]
- Magnusson MS. Discovering hidden time patterns in behavior: T-patterns and their detection. Behavior Research Methods, Instruments, and Computers: a Journal of the Psychonomic Society, Inc. 2000;32(1):93–110. doi: 10.3758/bf03200792. [DOI] [PubMed] [Google Scholar]
- Stan Development Team. Stan: A c++ library for probability and sampling, version 1.3. 2013 URL http://mc-stan.org/ [Google Scholar]
- Stephens M. Bayesian analysis of mixture models with an unknown number of components- an alternative to reversible jump methods. The Annals of Statistics. 2000;28(1):40–74. [Google Scholar]
- Welch TA. A technique for high-performance data compression. Computer. 1984;17(6):8, 19. [Google Scholar]
- Ziv J, Lempel A. Compression of individual sequences via variable-rate coding. IEEE Trans. Inf. Theor. 1978;24(5):530–536. [Google Scholar]