Abstract
Allopolyploidy is a significant evolutionary process, resulting in new species with diploid or greater chromosome complements derived from two or more progenitor species. We examined the chromosomal consequences of genomic merger in Arabidopsis suecica, the allotetraploid hybrid of Arabidopsis thaliana and Arabidopsis arenosa. Fluorescence in situ hybridization with centromere, nucleolus organizer region (NOR), and 5S rRNA gene probes reveals the expected numbers of progenitor chromosomes in natural A. suecica, but one pair of A. thaliana NORs and one pair of A. arenosa-derived 5S gene loci are missing. Similarly, in newly formed synthetic A. suecica-like allotetraploids, pairs of A. thaliana NORs are gained de novo, lost, and/or transposed to A. arenosa chromosomes, with genotypic differences apparent between F3 siblings of the same F2 parent and between independent lines. Likewise, pairs of A. arenosa 5S genes are lost and novel linkages between 5S loci and NORs arise in synthetic allotetraploids. By contrast, the expected numbers of A. arenosa-derived NORs and A. thaliana-derived 5S loci are found in both natural and synthetic A. suecica. Collectively, these observations suggest that some, but not all, loci are unstable in newly formed A. suecica allotetraploids and can participate in a variety of alternative rearrangements, some of which resemble chromosomal changes found in nature.
Keywords: plant, polyploidy, speciation, translocation
Polyploidy is a widespread mechanism for speciation in eukaryotes, having occurred in almost all plants and vertebrates (1–7). Polyploid species can arise through duplication of a single genome (autopolyploidy) or by combining two or more distinct genomes through hybridization (allopolyploidy). Many species that appear to be diploids, including maize, Arabidopsis thaliana, and Saccharomyces cerevisiae, are actually ancient polyploids (6, 8, 9) that have undergone “diploidization,” a variety of chromosomal and genic changes that result in diploid-like chromosome pairing, segregation, and gene function (10). Although many wild and cultivated allopolyploid plants are fertile and genetically stable, there is often evidence that changes in chromosome structure, heterochromatin content, and gene content have occurred during their evolutionary history (refs. 11–17; for reviews see refs. 4 and 18–21). Little is known about when such polyploidy-associated chromosomal changes occur. Amplified fragment-length polymorphism and DNA blot analyses of newly formed (neo) wheat allopolyploids have suggested that DNA sequence elimination is an immediate response to allopolyploid formation (22–25). However, a similar amplified fragment-length polymorphism analysis of cotton neoallopolyploids failed to detect evidence for sequence elimination (26), making it unclear whether allopolyploidy-induced genomic changes are a common occurrence.
In this study, using Arabidopsis suecica as a model system, we provide visual cytogenetic evidence for chromosomal changes in a neoallopolyploid. A. suecica is an allotetraploid species formed by combining a diploid A. thaliana genome (2n = 2x = 10, where n = the number of chromosomes in gametes and x = the fundamental chromosome number) with a 2x chromosome complement from the autotetraploid, Arabidopsis arenosa (2n = 4x = 32) (27, 28). By using FISH with centromere, 45S rRNA gene [nucleolus organizer region (NOR)], and 5S rRNA gene (5S gene) probes, we show that genome restructuring occurs in both natural and newly synthesized A. suecica. Unexpected locus numbers are evident for both NORs and 5S gene clusters, as are novel linkages among these loci. In synthetic A. suecica, multiple chromosomal changes occur and can differ between F3 siblings of the same F2 parent and in independent lines derived from different F1 hybrids. Our data suggest that a combination of predictable trends and stochastic outcomes contribute to restructuring the neoallopolyploid genome.
Materials and Methods
Plant Material. Diploid A. thaliana (ecotype Nossen, No-0); autotetraploid A. thaliana [2n = 4x = 20; ecotype Landsberg erecta (Ler); Arabidopsis Biological Resource Center stock CS3900], A. arenosa (strain 3651, also known as Care-1 in the L.C. laboratory), A. suecica (2n = 4x = 26; natural strain LC1, also known as Sue-1 in the L.C. laboratory), and synthetic A. suecica lines were grown in a greenhouse (20-h photoperiod at 25 ± 2°C). The synthetic lines resulted from crossing autotetraploid A. thaliana (which arose spontaneously during regeneration in tissue culture) with A. arenosa Care-1 as described (29, 30).
FISH. Species-specific centromere FISH probes were made from plasmids pARR20–1 or pAaCEN containing 180 bp of A. thaliana (31) or ≈250 bp of A. arenosa (32) pericentromeric repeats, respectively. Species-specific A. thaliana or A. arenosa NOR probes were made from pAt2 (33) or pCaIGS (GenBank accession no. AF177417), respectively. A 5S gene FISH probe was made from pCT4.1, containing an ≈500-bp A. thaliana 5S gene repeat (34). NOR probes were labeled by nick translation using biotin-dUTP or digoxigenin-dUTP (Roche, Gipf-Oberfrick, Switzerland). The A. thaliana centromere probe was amplified by PCR by using primers 5′-ATCCTCTAGAGTCGACCTGCA-3′ and 5′-TTCCCAGTCACGACGTTGTAA-3′, a-4 min denaturation step at 94°C, and 35 cycles of 45 s at 94°C, 45 s at 56°C, and 45 s at 72°C. The pAaCEN and pCT4.1 probes were amplified by using M13 universal primers. All PCR products were labeled with digoxigenin-dUTP or biotin-dUTP.
Flower buds and roots for cytological analysis were prepared according to refs. 35 and 36, respectively. FISH was carried out according to ref. 37. Preparations were counterstained with DAPI in CitiFluor antifade buffer (AF1, Agar Scientific, Essex, U.K.). Chromosomes were examined by using epifluorescence microscopy (Zeiss Axioskop2) and recorded with a Zeiss Axio-Cam digital camera. Figures were composed by using photoshop 6.0 software (Adobe Systems, San Jose, CA).
Results
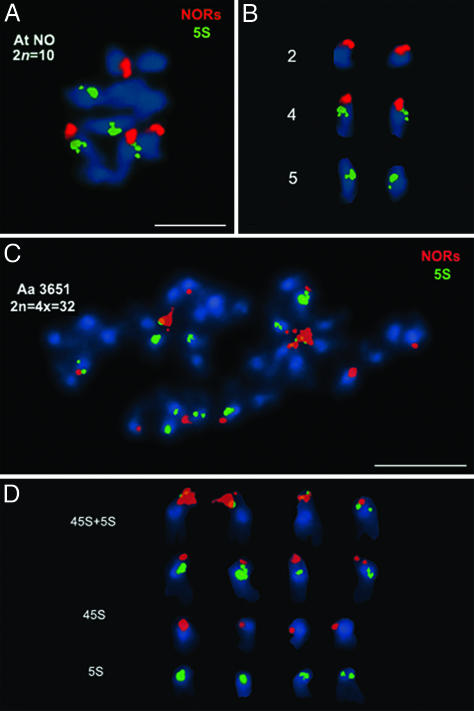
We analyzed A. thaliana (ecotype No-0), A. arenosa (strain 3651), and natural A. suecica (strain LC1/Sue-1) genomes by using FISH probes to visualize centromeres, 5S rRNA gene loci, or 45S rRNA gene clusters (NORs). A. thaliana ecotype No-0 was chosen because it shares with A. suecica a class of transposable elements that is missing in most A. thaliana ecotypes, suggesting a possible relationship (38). Hybridization of 45S rRNA and 5S gene probes to A. thaliana No-0 metaphase chromosomes revealed four NORs (Fig. 1A, red signals) and four 5S gene clusters (Fig. 1 A, green signals). Both pairs of NORs are similar in size and are terminally located on the short arms of acrocentric chromosomes (Fig. 1 A and B, red signals) as in other ecotypes (39–41), consistent with the known locations of NOR2 and NOR4 on chromosomes 2 and 4, respectively (42–44). Furthermore, one pair of NOR-bearing chromosomes bears a 5S gene cluster (Fig. 1B, middle pair), which is a conserved characteristic of chromosome 4 (39, 45–47). No-0 also has an NOR-bearing chromosome pair that lacks 5S genes (Fig. 1B, top pair), a conserved characteristic of chromosome 2 (39, 46). A chromosome pair that lacks NORs has 5S loci located in or near pericentromeric heterochromatin (Fig. 1B, bottom pair), a conserved characteristic of chromosome 5.
Fig. 1.
Metaphase chromosomes of A. thaliana (At) ecotype No-0 and A. arenosa (Aa) hybridized with 5S gene and species-specific NOR probes. (A and B) A. thaliana No-0 (2n = 2x = 10) hybridized with an A. thaliana-specific NOR probe (red) and a 5S gene probe (green). Chromosomes were counterstained with DAPI (blue). (C and D) A. arenosa strain 3651 (2n = 4x = 32) hybridized with an A. arenosa-specific NOR probe (red) and 5S gene probe (green). NO, ecotype Nossen, No-0. (Scale bars, 5 μm.)
FISH analysis of A. arenosa revealed 12 NORs, all on acrocentric chromosomes (Fig. 1C, red signals), and 12 5S gene loci (Fig. 1C, green signals). A. arenosa is an apparent autotetraploid (2n = 4x = 32), suggesting that three NORs and three 5S gene loci are present in each 1x complement of eight chromosomes. If so, one would predict that there should be four chromosomes displaying any given pattern of NOR and/or 5S loci. This prediction is borne out, as shown in Fig. 1D. Four chromosomes bear NORs and 5S rRNA gene loci in close proximity within their short arms (Fig. 1D, top row). Four chromosomes have subterminal NORs and pericentromeric 5S clusters (Fig. 1D, second row from top). Four chromosomes have NORs but lack 5S gene loci (Fig. 1D, third row from top). Finally, four chromosomes have 5S loci in the pericentromeric heterochromatic regions but lack NORs (Fig. 1D, bottom row). Despite the occurrence of four chromosomes for each of the patterns described above, the most similar chromosomes, based on FISH signal location and intensity, occur in pairs (evident in Fig. 1D), which is indicative of diploidization.
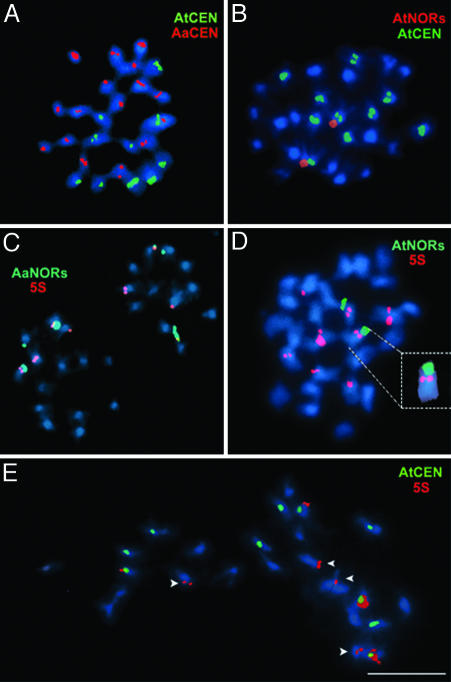
In A. suecica strain LC1, A. thaliana- and A. arenosa-derived chromosomes can be discriminated by FISH using species-specific centromere probes. The A. thaliana probe hybridized to 10 A. suecica chromosomes (Fig. 2A, green signals), whereas the A. arenosa probe hybridized to 16 chromosomes (Fig. 2 A, red signals), as expected (48). Despite the presence of the 10 expected A. thaliana chromosomes in A. suecica, only two NORs of A. thaliana origin were detected rather than the expected diploid number of four (Fig. 2B, red signals; compare with Fig. 1 A). We conclude that one pair of A. thaliana NORs was lost during the evolutionary history of natural A. suecica strain LC1. One pair of A. thaliana NORs is also missing in natural A. suecica strain 9502 (data not shown; see ref 36). By contrast, FISH with an A. arenosa-specific 45S gene probe revealed six A. arenosa-derived NORs in A. suecica strains LC1 (Fig. 2C, green) and 9502 (see ref 36), which is the expected 2x number. Dual FISH with A. thaliana NOR and 5S probes (Fig. 2D, green and red signals, respectively) revealed pericentromeric 5S gene signals on both chromosomes that bear A. thaliana-derived NORs (Fig. 2 D, D Inset, and E), suggesting that it is NOR4 that has been retained in A. suecica and NOR2 that has been lost. Molecular-marker data are consistent with this deduction (see Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 2.
Centromeres, NORs, and 5S gene loci at metaphase in natural A. suecica, the allotetraploid hybrid of A. thaliana (At), and A. arenosa (Aa). (A) Dual FISH with A. thaliana (green) and A. arenosa (red) centromere (CEN) probes. Chromosomes were counterstained with DAPI (in blue). (B) A. suecica metaphase chromosomes hybridized with A. thaliana-specific centromere (in green) and NOR probes (red). (C) Dual FISH with the A. arenosa-specific NOR probe (green) and the 5S gene probe (red). (D) Dual FISH with A. thaliana-specific NOR (green) and 5S gene probes (red), revealing 5S gene loci on the NOR-bearing A. thaliana chromosomes (see Inset). (E) A. suecica strain LC1 has the expected four A. thaliana-derived 5S gene loci but only four of the expected six A. arenosa-derived 5S gene loci. The A. thaliana-specific centromere repeat probe (green) was used to discriminate the parental genomes. All 5S gene signals are red. (Scale bar, 5 μm.)
As a molecular test of the deduction that the pair of A. thaliana NORs retained in natural A. suecica correspond to NOR4, we looked for the persistence of gene At4g00030, a linked protein-coding gene located only ≈12 kb from the centromere-proximal border of NOR4. Using a restriction endonuclease site polymorphism that allows A. arenosa and A. thaliana At4g00030 homologs to be discriminated after PCR amplification and HhaI digestion (Fig. 7, lanes 1 and 3), it is clear that both parental alleles are retained in A. suecica strain LC1 (Fig. 1, lane 2).
Ten or 12 5S loci are expected in A. suecica, six derived from A. arenosa and either four or six 5S loci derived from A. thaliana, depending on the ecotype (see below). However, only eight 5S gene clusters are observed (Fig. 2 C–E). Dual FISH localized four of these 5S loci to A. thaliana-derived chromosomes (Fig. 2E). Therefore, we deduce that only four of the six expected A. arenosa-derived 5S loci are retained in A. suecica [those located on NOR-bearing chromosomes (Fig. 2C)], and two have been lost. It is conceivable that two 5S loci may also have been lost from A. thaliana chromosomes if an ecotype with six 5S loci (as in Ler; see below) was the progenitor of A. suecica. However, the simplest explanation presumes that the A. thaliana progenitor had only four 5S loci per diploid genome, as in ecotype No-0 (see Fig. 1 A and B) and that all these loci are retained.
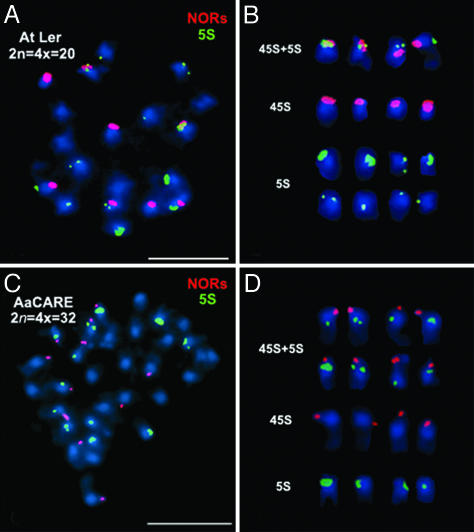
A. suecica is found in regions of northern Europe that were covered by glaciers during the last ice age, suggesting an origin since the glacial retreat (27, 49–51). Nonetheless, the species may have had thousands of years in which to lose NORs and 5S gene loci. To determine whether NOR and 5S locus instabilities occur early after formation of the allopolyploid genome, we examined synthetic A. suecica-like allotetraploids that were generated by crossing autotetraploid A. thaliana (ecotype Ler) with A. arenosa (29, 30). Ler has NORs on chromosomes 2 and 4 and 5S loci on chromosomes 3, 4, and 5. The autotetraploid A. thaliana thus has 8 NORs and 12 5S loci, as expected, indicating that chromosome doubling did not induce changes in NOR or 5S locus numbers (Fig. 3 A and B). Furthermore, four of the NORs (NOR4 loci) are present on chromosomes with 5S loci, as expected, suggesting that no rearrangements of NORs and 5S loci had occurred. This observation contrasts with an autotetraploid line of the ecotype Wilna in which NOR4 was found to be dissociated from its linked 5S locus on one pair of chromosomes after cultivation for 20–30 generations (41). The A. arenosa parent of the synthetic A. suecica-like allotetraploids (strain Care-1) has 12 NORs and 12 5S gene loci, as expected (Fig. 3 C and D).
Fig. 3.
NORs and 5S loci of metaphase chromosomes in autotetraploid A. thaliana (At) Ler and A. arenosa (Aa) progenitors of synthetic A. suecica. (A and C) Dual FISH detection of NORs (red) and 5S gene loci (green) in autotetraploid A. thaliana (A) or A. arenosa strain Care-1 (C). (B and D) Karyotyping of NOR and 5S loci-bearing chromosomes in autotetraploid A. thaliana (B) or A. arenosa (D). Chromosomes were counterstained with DAPI (blue). (Scale bars, 5 μm.)
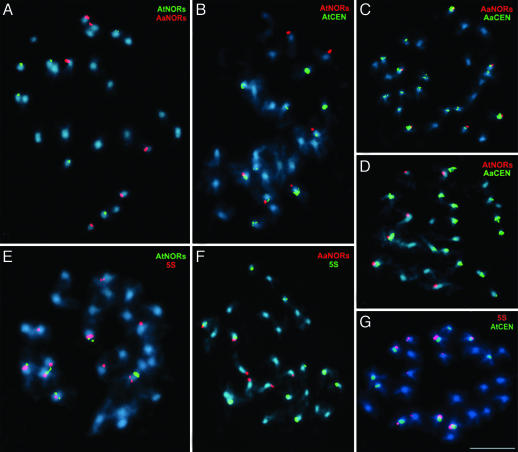
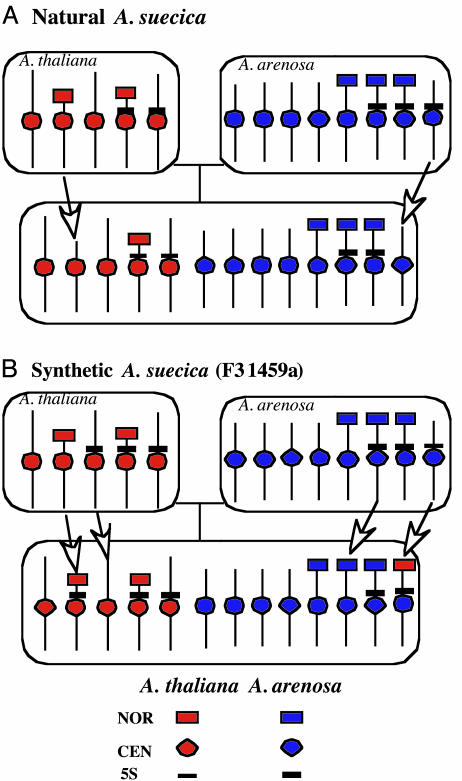
The tetraploid progenitors analyzed in Fig. 3 were crossed by using A. thaliana as the maternal parent, the only direction in which the cross is successful in the laboratory (29) or is detected in nature (52). F1 hybrid plants are obtained at low frequency and set relatively few F2 seeds. Seeds of two F2 A. suecica-like synthetic allotetraploids, each descended from a unique F1 plant, comprise the F3 families 1459 and 1460. Multiple siblings of both the 1459 and 1460 families were found to possess the expected 26 chromosomes, 10 derived from A. thaliana and 16 inherited from A. arenosa. Fig. 4 details the analysis of one individual from the F3 family 1459 (individual 1459a). The most striking change in this individual is that six NORs of A. thaliana origin are detected rather than the expected four (Fig. 4A, green signals). Six A. arenosa-derived NORs (the expected number) are also present (Fig. 4A, red signals). Dual FISH with species-specific rRNA gene and centromere probes shows that four of the A. thaliana NORs are located on chromosomes with A. thaliana centromeres (Fig. 4B), but two (presumably the novel pair) are located on A. arenosa chromosomes (Fig. 4D). All six A. arenosa NORs are present on chromosomes with A. arenosa centromeres, as expected (Fig. 4C). Only 10 5S gene loci are present in the synthetic allotetraploid (Fig. 4 E–G), instead of the expected 12 (6 from each progenitor; see Fig. 3). Six of the 5S loci are on chromosomes with A. thaliana centromeres (Fig. 4G), which is the expected number, indicating that the two missing 5S loci were eliminated from A. arenosa chromosomes, as in natural A. suecica strain LC1 (refer to Fig. 2). It is interesting that only two A. arenosa-derived NORs in the synthetic F3 A. suecica line are present on chromosomes with 5S gene loci (Fig. 4F), whereas four are expected (see Fig. 3D). By contrast, the four A. thaliana NORs carried on A. thaliana chromosomes in the synthetic allotetraploid (as well as the two novel A. thaliana NORs located on A. arenosa chromosomes) are present on chromosomes that also have 5S loci (Fig. 4E), whereas only two such chromosomes are expected (see Fig. 3B), namely the chromosome 4 pair. Collectively, the dual-FISH analyses shown in Fig. 4 reveal altered NOR and 5S gene locus numbers, positions, and linkages in F3 neoallotetraploid individual 1459a, as summarized in Fig. 5.
Fig. 4.
Gain, loss, and reorganization of NORs and 5S gene loci in a synthetic A. suecica-like F3 allotetraploid (individual 1459a). Shown are metaphase chromosomes subjected to dual FISH with pairs of probes that are indicated in the upper-right corners. The color of the lettering (red or green) corresponds to the color of the FISH signal in each case. Chromosomes were counterstained with DAPI (blue). At, A. thaliana; Aa, A. arenosa; CEN, centromere. (Scale bar, 5 μm.)
Fig. 5.
Summary of chromosome changes observed in natural and synthetic A. suecica. Depicted are diagrams of 1x chromosome complements of A. thaliana, A. arenosa, or A. suecica, highlighting NORs, centromeres (CEN), and 5S loci. Arrows point out chromosomal changes detected in natural A. suecica strain LC1 or synthetic A. suecica-like allotetraploid F3 individual 1459a. For simplicity, the A. thaliana progenitor of natural A. suecica is depicted with only two 5S loci, as in ecotype No-0 (and many other ecotypes), but may have had three, as in Ler. Note that the precise chromosomes involved in the various gains, losses, or rearrangements of NORs and 5S loci are not known; thus, the arrows reflect only one of several possible explanations for the changes observed.
To determine whether the same chromosomal changes recur in newly synthesized A. suecica, we examined a sibling of the individual analyzed in Fig. 4 and two individuals (1460a and 1460b) of an independently derived F3 family (Fig. 6). Unlike the 1459a individual with six A. thaliana NORs (Fig. 4 B and D), its sibling (1459b) had only four A. thaliana-derived NORs, the expected 2x number (Fig. 6A). Nonetheless, a shared feature of 1459a and 1459b is that one pair of A. thaliana NORs is present on A. arenosa chromosomes (compare Figs. 4D and 6A). Presumably, the F2 parent possessed one pair of A. thaliana NORs transposed onto a pair of A. arenosa chromosomes such that both siblings inherited the resulting chromosomes. An additional pair of A. thaliana NORs then apparently arose de novo in 1459a or arose de novo in the F1 or F2 generation and was retained in 1459a but lost in 1459b. Examination of the siblings of F3 family 1460 revealed additional A. thaliana NOR genotypes including one individual (1460a) with only two A. thaliana NORs, both on chromosomes with A. thaliana centromeres (Fig. 6B), thereby resembling natural A. suecica strain LC1 (see Fig. 2B). A sibling of this plant, 1460b, possessed four A. thaliana-derived NORs, all on A. thaliana chromosomes (Fig. 6C), as expected if no deletions, amplifications, or transpositions occurred. Collectively, these results suggest that multiple alternative chromosomal changes can occur in the early generations of neoallotetraploid A. suecica.
Fig. 6.
Variability in A. thaliana (At) NOR number and/or chromosomal position in F3 individuals of two independent A. suecica-like allotetraploid lines. The individual shown in A (1459b) and the individual analyzed in Fig. 4 (1459a) are siblings descended from the same F2 parent. F3 individuals 1460a and 1460b in B and C, respectively, are siblings descended from a different F1 hybrid. The color of the lettering (red or green) reflects the color of the FISH probe. Note that 1459b has four A. thaliana-derived NORs, whereas its sibling, 1459a, had six A. thaliana-derived NORs. In both 1459a and 1459b, two of these NORs are located on chromosomes with A. arenosa (Aa) centromeres (CEN) (compare A with Fig. 4D). Individuals 1460a and 1460b have either two or four A. thaliana-derived NORs, respectively, all located on chromosomes with A. thaliana centromeres. (Scale bar, 5 μm.)
It is important to note that all F3 individuals examined from both the 1459 and 1460 families had the expected six A. arenosa-derived NORs (data not shown), as in natural A. suecica strains LC1 and 9502, indicating that only A. thaliana NORs are unstable and hypervariable. Likewise, the expected six A. thaliana-derived 5S gene loci were found in both F3 families, but at least one pair (sometimes two) of A. arenosa-derived 5S loci was missing (data not shown), as in natural A. suecica strain LC1 (see Fig. 2 C–E). Therefore, NOR instability in A. suecica seems to be a characteristic of A. thaliana-derived loci, and 5S gene locus instability seems to be a characteristic of A. arenosa-derived loci.
Discussion
Our study shows that loss, gain, and/or rearrangement of NORs and 5S loci occur in the early generations after formation of the allotetraploid A. suecica genome and can give rise to alternative chromosomal changes, some of which resemble those found in nature. Our observations suggest that chromosomal alterations in natural and synthetic A. suecica follow certain trends, with specific loci tending to be unstable. However, the consequences of this instability are unpredictable, as demonstrated by the variability in A. thaliana-derived NOR numbers and chromosomal positions in neoallotetraploids. It is possible that rapid losses or de novo generation of NORs might play a significant role in the homogenization of rRNA genes in natural allotetraploids such as cotton (12) in addition to (or rather than) gene conversion or recurrent unequal crossing-over events that are likely to require longer time spans. Likewise, unequal losses and/or de novo generation of NORs from the different progenitors might be an explanation for DNA blot and DNA sequencing evidence suggesting rapid changes in rRNA gene allele frequency in glycine (soybean) neoallopolyploids (16).
Several mechanisms could mediate the chromosome-restructuring events we observe. For instance, novel linkages among NORs and 5S loci could result from homologous or homeologous recombination, the latter being possible between homeologous chromosomes or within homeologous regions of nonhomeologous chromosomes. Although analysis of meiosis in synthetic A. suecica-like allotetraploids at the F4 generation reveals primarily homologous pairing, homeologous pairing and recombination cannot be ruled out (A.M. and L.C., unpublished data). Chromosome breakage followed by loss or translocation of broken chromosomal segments is also a possible mechanism to explain changes in NOR number and/or position. In synthetic A. suecica-like allotetraploid lines at the F4 generation, abnormal meioses can be observed in which chromosome breaks are evident (L.C., unpublished data). Such breaks may occur because of misdivision of unpaired chromosomes at anaphase or as a consequence of recombination events that result in dicentric chromosomes.
In natural A. suecica, loss of one pair of A. thaliana NORs might have been relatively easy to accomplish because of the fact that NOR2 and NOR4 are located at the very tips of chromosomes 2 and 4, respectively, at which they are capped by telomeres TEL2N and TEL4N (42, 43). Because of their terminal positions, a double-strand break in an NOR-bearing chromosome could remove a large portion, or the entirety, of an NOR without affecting nonredundant essential genes that flank the NOR. The de novo generation of a novel pair of A. thaliana-derived NORs in synthetic A. suecica (see Fig. 4) could have resulted from breakage midway within an A. thaliana NOR and subsequent ligation of the distal fragment, with its attached telomere, to the ends of a pair of A. arenosa chromosomes. The broken end of the donor chromosome then might be healed by de novo telomere addition to the end of the truncated NOR. Evidence that de novo telomere addition can occur is that in A. thaliana, consensus telomere repeats (T3AG3) directly abut rRNA gene sequences, having been added to an rRNA gene sequence motif that resembles an incomplete telomere repeat (42).
A third mechanism that might contribute to loss or translocation of NORs is transposable element-mediated chromosome breakage (53). In regard to the loss of one pair of A. thaliana NORs in natural A. suecica, which we deduce to be NOR2, it is interesting that NOR2 is flanked by a complex jumble of transposable element sequences spanning ≈50 kb in the A. thaliana ecotype Columbia (8). By contrast, NOR4 has only two putative nonautonomous retrotransposons in the analogous region (8, 54). Thus, the relative instability of A. thaliana NOR2 in A. suecica could be related to the density of transposable elements flanking this NOR. Likewise, the loss of 5S gene clusters also might be related to the fact that 5S loci are embedded in pericentromeric heterochromatin that is typically rich in transposable elements. Furthermore, because genes of transposable elements have been shown to be released from suppression in newly formed polyploids (23, 55), including newly formed synthetic A. suecica (A.M. and L.C., unpublished work) we suspect that transposon-mediated rearrangements might play an important role in the generation of novel linkages between NORs and 5S gene clusters. The genomic resources available for Arabidopsis, including the known identities of its various classes of transposable elements, make A. suecica an attractive model system for testing this hypothesis.
Supplementary Material
Acknowledgments
We thank Eric Richards and Pat Heslop-Harrison for centromere clones, Paul Fransz for the 5S probe, Richard Lawrence for nucleolus organizer region probes, and Augusta Barão for technical assistance. C.S.P. laboratory research was supported by National Institutes of Health Grant R01-GM60380. Research in the W.V. laboratory was supported by Fundação para a Ciência e Tecnologia project Programa Operacional “Ciência, Tecnologia, Inovação”/BCI/38557/2001 (to N.N.). Publication costs were defrayed in part by a research prize from the Jean Lowenhaupt Botany fund (to O.P.).
Author contributions: O.P., M.S., and M.S.L. performed research; O.P., N.N., M.S., A.M., L.C., W.V., and C.S.P. analyzed data; N.N., W.V., and C.S.P. designed research; A.M. and L.C. contributed new reagents/analytic tools; and C.S.P. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NOR, nucleolus organizer region; Ler, Landsberg erecta.
Note. After completion of our cytogenetic studies, a test for five genetic markers that can discriminate alleles of natural A. suecica from alleles of our A. thaliana or A. arenosa lab strains found three of the five A. suecica alleles in one individual of synthetic allotetraploid F3 family 1459 (A.M., unpublished data). No natural A. suecica alleles were identified in two 1460 F3 individuals tested. We note that the chromosomal rearrangements in NORs and 5S loci that we describe in this article cannot be explained by the presence of natural A. suecica alleles. However, natural alleles may be segregating among the 1459 F3 family; researchers wanting to use this material should be aware of that possibility.
References
- 1.Masterson, J. (1994) Science 264, 421–424. [DOI] [PubMed] [Google Scholar]
- 2.Leitch, L. J. & Bennett, M. D. (1997) Trends Plant Sci. 2, 470–476. [Google Scholar]
- 3.Soltis, D. E. & Soltis, P. S. (1999) Trends Ecol. Evol. 14, 348–352. [DOI] [PubMed] [Google Scholar]
- 4.Comai, L. (2000) Plant Mol. Biol. 43, 387–399. [DOI] [PubMed] [Google Scholar]
- 5.Ohno, S., Wolf, U. & Atkin, N. B. (1968) Hereditas 59, 169–187. [DOI] [PubMed] [Google Scholar]
- 6.Wolfe, K. H. & Shields, D. C. (1997) Nature 387, 708–713. [DOI] [PubMed] [Google Scholar]
- 7.Spring, J. (1997) FEBS Lett. 400, 2–8. [DOI] [PubMed] [Google Scholar]
- 8.The Arabidopsis Genome Initiative (2000) Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 9.Gaut, B. S., Le Thierry d'Ennequin, M., Peek, A. S. & Sawkins, M. C. (2000) Proc. Natl. Acad. Sci. USA 97, 7008–7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfe, K. H. (2001) Nat. Rev. Genet. 2, 333–341. [DOI] [PubMed] [Google Scholar]
- 11.Volkov, R. A., Borisjuk, N. V., Panchuk, I. I., Schweizer, D. & Hemleben, V. (1999) Mol. Biol. Evol. 16, 311–320. [DOI] [PubMed] [Google Scholar]
- 12.Wendel, J. F., Schnabel, A. & Seelanan, T. (1995) Proc. Natl. Acad. Sci. USA 92, 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao, X. P., Si, Y., Hanson, R. E., Crane, C. F., Price, H. J., Stelly, D. M., Wendel, J. F. & Paterson, A. H. (1998) Genome Res. 8, 479–492. [DOI] [PubMed] [Google Scholar]
- 14.McGrath, J. M., Quiros, C. F., Harada, J. J. & Landry, B. S. (1990) Mol. Gen. Genet. 223, 198–204. [DOI] [PubMed] [Google Scholar]
- 15.Song, K., Lu, P., Tang, K. & Osborn, T. C. (1995) Proc. Natl. Acad. Sci. USA 92, 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joly, S., Rauscher, J. T., Sherman-Broyles, S. L., Brown, A. H. & Doyle, J. J. (2004) Mol. Biol. Evol. 21, 1409–1421. [DOI] [PubMed] [Google Scholar]
- 17.Vaughan, H. E., Jamilena, M., Ruiz, R. C., Parker, J. S. & Garrido, R. M. A. (1993) Heredity 71, 574–580. [Google Scholar]
- 18.Pikaard, C. S. (2001) Trends Genet. 17, 675–677. [DOI] [PubMed] [Google Scholar]
- 19.Osborn, T. C., Chris Pires, J., Birchler, J. A., Auger, D. L., Jeffery Chen, Z., Lee, H. S., Comai, L., Madlung, A., Doerge, R. W., Colot, V., et al. (2003) Trends Genet. 19, 141–147. [DOI] [PubMed] [Google Scholar]
- 20.Liu, B. & Wendel, J. F. (2002) Curr. Genomics 3, 489–505. [Google Scholar]
- 21.Rieseberg, L. H. (2001) Curr. Biol. 11, R925–R928. [DOI] [PubMed] [Google Scholar]
- 22.Ozkan, H., Levy, A. A. & Feldman, M. (2001) Plant Cell 13, 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaked, H., Kashkush, K., Ozkan, H., Feldman, M. & Levy, A. A. (2001) Plant Cell 13, 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feldman, M., Liu, B., Segal, G., Abbo, S., Levy, A. A. & Vega, J. M. (1997) Genetics 147, 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kashkush, K., Feldman, M. & Levy, A. A. (2002) Genetics 160, 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, B., Brubaker, C. L., Mergeai, G., Cronn, R. C. & Wendel, J. F. (2001) Genome 44, 321–330. [PubMed] [Google Scholar]
- 27.Al-Shehbaz, I. A. & O'Kane, S. L. (2002) in The Arabidopsis Book, eds. Somerville, C. R. & Meyerowitz, E. M. (Am. Soc. Plant Biologists, Rockville, MD), 10.1199/tab.0009. Available at www.aspb.org/publications/arabidopsis.
- 28.Redei, G. P. (1974) Arabidopsis Info. Serv. 11, 5. [Google Scholar]
- 29.Comai, L., Tyagi, A. P., Winter, K., Holmes-Davis, R., Reynolds, S. H., Stevens, Y. & Byers, B. (2000) Plant Cell 12, 1551–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madlung, A., Masuelli, R. W., Watson, B., Reynolds, S. H., Davison, J. & Comai, L. (2002) Plant Physiol. 129, 733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vongs, A., Kakutani, T., Martienssen, R. A. & Richards, E. J. (1993) Science 260, 1926–1928. [DOI] [PubMed] [Google Scholar]
- 32.Kamm, A., Galasso, I., Schmidt, T. & Heslop-Harrison, J. S. (1995) Plant Mol. Biol. 27, 853–862. [DOI] [PubMed] [Google Scholar]
- 33.Doelling, J. H., Gaudino, R. J. & Pikaard, C. S. (1993) Proc. Natl. Acad. Sci. USA 90, 7528–7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campell, B. R., Song, Y., Posch, T. E., Cullis, C. A. & Town, C. D. (1992) Gene 112, 225–228. [DOI] [PubMed] [Google Scholar]
- 35.Schwarzacher, H. G. & Mosgoeller, W. (2000) Cytogenet. Cell Genet. 91, 243–252. [DOI] [PubMed] [Google Scholar]
- 36.Pontes, O., Lawrence, R. J., Neves, N., Silva, M., Lee, J. H., Chen, Z. J., Viegas, W. & Pikaard, C. S. (2003) Proc. Natl. Acad. Sci. USA 100, 11418–11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones, G. H. & Heslop-Harrison, J. S. (2000) in Arabidopsis, a Practical Approach, ed. Wilson, Z. A. (Oxford Univ. Press, Oxford), pp. 105–124.
- 38.Henikoff, S. & Comai, L. (1998) Genetics 149, 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fransz, P., Armstrong, S., Alonso-Blanco, C., Fischer, T. C., Torres-Ruiz, R. A. & Jones, G. (1998) Plant J. 13, 867–876. [DOI] [PubMed] [Google Scholar]
- 40.Maluszynska, J. & Heslop-Harrison, J. S. (1991) Plant J. 1, 159–166. [Google Scholar]
- 41.Weiss, H. & Maluszynska, J. (2000) Hereditas 133, 255–261. [DOI] [PubMed] [Google Scholar]
- 42.Copenhaver, G. P. & Pikaard, C. S. (1996) Plant J. 9, 259–272. [DOI] [PubMed] [Google Scholar]
- 43.Copenhaver, G. P., Doelling, J. H., Gens, J. S. & Pikaard, C. S. (1995) Plant J. 7, 273–286. [DOI] [PubMed] [Google Scholar]
- 44.Haberer, G., Fischer, T. C. & Torres-Ruiz, R. A. (1996) Mol. Gen. Genet. 250, 123–128. [DOI] [PubMed] [Google Scholar]
- 45.Fransz, P. F., Alonso-Blanco, C., Liharska, T. B., Peeters, A. J., Zabel, P. & de Jong, J. H. (1996) Plant J. 9, 421–430. [DOI] [PubMed] [Google Scholar]
- 46.Fransz, P. F., Armstrong, S., de Jong, J. H., Parnell, L. D., van Drunen, C., Dean, C., Zabel, P., Bisseling, T. & Jones, G. H. (2000) Cell 100, 367–376. [DOI] [PubMed] [Google Scholar]
- 47.Murata, M., Heslop-Harrison, J. S. & Motoyoshi, F. (1997) Plant J. 12, 31–37. [DOI] [PubMed] [Google Scholar]
- 48.Comai, L., Tyagi, A. P. & Lysak, M. A. (2003) Chromosome Res. 11, 217–226. [DOI] [PubMed] [Google Scholar]
- 49.O'Kane, S., Schaal, B. & Al-Shehbaz, I. (1995) Syst. Bot. 21, 559–566. [Google Scholar]
- 50.O'Kane, S. L. (1997) Novon 7, 323–327. [Google Scholar]
- 51.Lind-Hallden, C., Hallden, C. & Sall, T. (2002) Hereditas 136, 45–50. [DOI] [PubMed] [Google Scholar]
- 52.Sall, T., Jakobsson, M., Lind-Hallden, C. & Hallden, C. (2003) J. Evol. Biol. 16, 1019–1029. [DOI] [PubMed] [Google Scholar]
- 53.McClintock, B. (1952) Cold Spring Harbor Symp. Quant. Biol. 16, 13–47. [DOI] [PubMed] [Google Scholar]
- 54.Lewis, M. S. & Pikaard, C. S. (2001) Proc. Natl. Acad. Sci. USA 98, 14536–14540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu, B. & Wendel, J. F. (2000) Genome 43, 874–880. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.