Abstract
Expression profiles of breast carcinomas are difficult to interpret when they are obtained from tissue in toto, which may contain a large proportion of non-cancer cells. To avoid this problem, we microscopically isolated cells from a primary invasive ductal carcinoma of the breast and from an axillary node harboring a metastatic breast carcinoma, to obtain pure populations of carcinoma cells (≈500) and used them for serial analysis of gene expression. The expression profiles generated from both populations of cells were compared with the profile of a disease-free mammary epithelium. We showed that the expression profiles obtained are exclusive of carcinoma cells with no contribution of non-epithelial cells. From a total of 16,939 unique tags analyzed, we detected 559 statistically significant changes in gene expression; some of these genes have not been previously associated with breast cancer. We observed that many of the down-regulated genes are the same in both cancers, whereas the up-regulated genes are completely different, suggesting that the down-regulation of a set of genes may be the basic mechanism of cancer formation, while the up-regulation may characterize and possibly control the state of evolution of individual cancers. The results obtained may help in characterizing the neoplastic process of breast cancer.
Keywords: breast cancer, serial analysis of gene expression, cell microdissection, carcinoma
Breast cancer progresses through a series of stages, starting as atypical duct hyperplasia, to ductal carcinoma in situ, invasive ductal carcinoma, and finally, metastatic disease. Global expression profiling has been extensively used to classify the disease and to predict its clinical outcome. Most of these studies used array-based platforms, and therefore, were limited to the analysis of known but most likely incomplete selection of genes and ESTs. More recently, global changes of gene expression have been determined by using serial analysis of gene expression (SAGE), which does not have this limitation (1, 2). Both microarray and SAGE data suggest that there is considerable diversity among breast tumor profiles (1–5). However, these data were obtained primarily from tumor tissue, rather than from a homogenous population of epithelial cancer cells. The problem that arises is that, if in the context of a complex tissue, only a small proportion of cells corresponds to the cells of interest, many important regulatory genes, often expressed at low levels, but essential for determining the pathological cell phenotype, will be undetected. In fact, purified and unpurified samples were shown to produce different expression profiles (2), because contaminating nontumoral cells may have been present in different amounts. For instance, in tumor samples cancer cells may be present in proportions ranging from 5% to 50% of the total cell mass (6, 7). For these reasons, it is necessary to determine the expression profile on a pure population of carcinoma cells.
This work is a pilot study carried out by following this concept. We performed SAGE on highly homogeneous populations of cells microscopically isolated from a primary invasive ductal carcinoma of the breast and from an axillary node harboring a metastatic breast carcinoma. A useful feature of the SAGE technology is that databases can be compared directly with each other. The purity of the cell population is shown in our SAGE libraries by the absence or minimal expression of genes that are markers of nonepithelial cells such as endothelial and stromal cells, adipose cells, B and T lymphocytes, and macrophages.
Materials and Methods
Clinical Information and Cell Microdissection. Samples from both a primary invasive ductal carcinoma of the breast and a nodal metastasis were obtained at the Istituto Nazionale Tumori (Milan) after patient consent. For the invasive library construction, ≈500 cancer cells (99% pure) were microscopically isolated from a primary estrogen- and progesterone-receptor-positive invasive ductal carcinoma. For the metastatic library, the same amount of cells were also microscopically isolated from a metastatic lymph node derived from an estrogen- and progesterone-receptor-negative breast-invasive ductal carcinoma. Cancer cells were microscopically dissected from methylene blue-stained 20-μm frozen sections, kept at low temperature during the entire manipulation, by using microneedle aspiration (8); cancer cells are less attached to the connective tissue stroma and are preferentially released by mechanical force.
cDNA Preparation, Library Construction, and Sequencing. The microdissected SAGE libraries were generated by following a protocol described (9), with modifications made necessary due to the small number of cells used as starting material. Total RNA was obtained by using the PicoPure RNA isolation kit (Arcturus). A pre-SAGE linear amplification step was performed with T7 RNA polymerase by using the RiboAmp kit (Arcturus).
Tools for SAGE. Tags were analyzed by using sage2000 (www.sagenet.org/index.htm) and esage software (10). The National Center for Biotechnology Information SAGEmap (www.ncbi.nlm.nih.gov/SAGE) and the CGAP SAGEgenie (http://cgap.nci.nih.gov/SAGE/AnatomicViewer) databases were also used. The Gene Expression Level Probability Function was obtained by using the discrete Pareto-like probability function (11). The assignment of molecular function of proteins and chromosomal location of individual genes was based on the LocusLink database (www.ncbi.nlm.nih.gov/projects/LocusLink/).
In Situ Hybridization. In situ hybridization was performed on human breast tissues as described (12). Sense and antisense riboprobes were generated by in vitro transcription using T7 or Sp6 polymerase primers, from the cloned region of 762 nt, between primers CXCL6F/CXCL6R (5′-TCATAAAATTGCCCAGTCTTC-3′ and 5′-TGTTTTTGGGCTTCTTCATCT-3′) of the human CXCL6 mRNA sequence NM_002993.
Supporting Information. Tables 3–8 are published as supporting information on the PNAS web site.
Results
Generation of SAGE Libraries from Microdissected Cells. Two SAGE libraries were obtained: one from cancer epithelial cells isolated by microscopic dissection from a primary invasive (INV) breast ductal carcinoma (the INV library, 17,306 tags), and the other from cancer cells microscopically isolated from a lymph node harboring a metastatic (MET) breast carcinoma (the MET library, 10,363 tags). In both cases, the purified cells were obtained in very limited amounts (≈500 cells) and, due to the very limited amount of RNA obtained, amplification was required. To verify that the RNA amplification preserved the original mRNA abundance, an amplification diagnosis test was carried out by performing hybridization comparison of the RNA before and after amplification, in a dot-blot analysis using radioactive probes for 10 different genes, expressed at different levels of abundance in normal and tumoral mammary gland epithelial cells. This control showed that the RNA amplification did not induce any preferential amplification (data not shown). The purity of the cell population is shown in our SAGE libraries by the absence or minimal expression of genes that are markers of nonepithelial cells, such as endothelial and stromal cells, adipose cells, B and T lymphocytes, and macrophages (data not shown).
SAGE Library Comparison. Our INV library was matched against the tag list of the SAGE library Br_N (library name: “N2”), obtained by Porter et al. (1), from a disease-free mammary epithelium through immunomagnetic purification, using an anti-Ber-Ep4 antibody coupled to magnetic beads (1). We compared the tag abundance distribution of our INV library with that of the Br_N SAGE library and observed that the abundance distribution of distinct SAGE tags in the two libraries was similar (data not shown). However, the Br_N library is different in size in respect to our INV library, and this fact may affect the results of the statistical comparison of the gene expression levels in the libraries (13). To avoid this problem, we constructed similar size sublibraries of 10,082 tags each, by choosing tags randomly without replacement from the Br_N library and from our INV library.
The comparison of the two sublibraries showed a greater abundance of rare transcripts in our INV sublibrary with respect to the normal Br_N sublibrary. Also, in our INV sublibrary, the fraction of rarely expressed transcripts is statistically higher (P < 0.001, data not shown). Thus, we could expect to observe not only a more specific gene expression profile but also novel genes in our INV library. We in fact detected 29 tags matching to ESTs or mRNAs encoding for hypothetical proteins, and 11 no-matching tags that had not been detected previously (see Tables 6 and 8).
TAG Analysis. From a total of 37,751 tags analyzed, 16,939 unique tags were detected. By using a P value of <0.05 and a ratio of difference in expression of >1.5-fold, 559 tags were identified as differentially expressed in a significant way in the INV and MET libraries, when compared with the Br_N library. Of these 559 tags, 392 (70.1%) correspond to known genes or ESTs, and of the other 167 tags (29.9%), 152 match to multiple genes, and 15 do not match to any genes. A selection of the 50 most down-regulated genes is reported in Table 1 and a selection of the 50 most up-regulated genes is reported in Table 2. Tables 3–5 list tags down-regulated in the INV and MET libraries, as follows: 221 tags matching to known genes, including 40 ribosomal genes are listed in Table 3, 76 multiple-gene-matching tags are reported in Table 4, and 3 no-matching tags are reported in Table 5. Tables 6–8 list tags up-regulated in the INV and MET libraries, as follows: 171 tags matching to known genes, including 30 ribosomal genes, are listed in Table 6, 76 multiple-gene-matching tags are reported in Table 7, and 12 no-matching tags are reported in Table 8.
Table 1. Selection of the 50 genes most down-regulated in the INV and/or MET libraries with respect to the Br_N library from normal mammary epithelium.
| Gene symbol | Br_N | INV | MET | Unigene ID | Genetic map |
|---|---|---|---|---|---|
| Transcription factors/chromatin/nuclear proteins | |||||
| NFKBLA | 20 | 0 | 1 | 81328 | 14q13 |
| LMNA | 9 | 1 | 1 | 436441 | 1q21.2-q21.3 |
| JUNB | 8 | 0 | 0 | 400124 | 19p13.2 |
| HMGN1 | 7 | 1 | 1 | 356285 | 21q22.3 |
| PNRC1 | 6 | 0 | 0 | 75969 | 6q16.1 |
| JUND | 5 | 0 | 0 | 2780 | 19p13.2 |
| DDB1 | 5 | 0 | 1 | 290758 | 11q12-q13 |
| DDX5 | 12 | 4 | 1 | 279806 | 17q21 |
| BAP1 | 4 | 0 | 1* | 106674 | 3p21.31-p21.2 |
| Cell cycle/apoptosis/cell growth | |||||
| MSF | 6 | 0 | 0 | 288094 | 17q25 |
| GADD45B | 6 | 1 | 0 | 110571 | 19p13.3 |
| PPM1G | 6 | 1 | 1 | 17883 | 2p23.3 |
| AREG | 5 | 0 | 0 | 270833 | 4q13-q21 |
| LGALS3 | 5 | 0 | 0 | 411701 | 14q21-q22 |
| DOC-1R | 4 | 0 | 0 | 379039 | 11q13 |
| PDCD5 | 4 | 0 | 0 | 443831 | 19q12-q13.1 |
| Membrane proteins/antigens/receptors/adhesion | |||||
| TM4SF1 | 16 | 0 | 0 | 351316 | 3q21-q25 |
| ANXA1 | 9 | 1 | 1 | 287558 | 9q12-q21.2 |
| ZYX | 7 | 0 | 0 | 75873 | 7q32 |
| CLDN4 | 4 | 0 | 0 | 5372 | 7q11.23 |
| Signal transduction | |||||
| TACSTD2 | 12 | 1 | 0 | 23582 | 1p32-p31 |
| ARHA | 8 | 0 | 0 | 77273 | 3p21.3 |
| STRN4 | 5 | 0 | 0 | 406918 | 19q13.2 |
| RASD1 | 4 | 0 | 0 | 25829 | 17p11.2 |
| YWHAE | 7 | 2 | 1 | 79474 | 17p13.3 |
| Secreted and ECM proteins | |||||
| LOC118430 | 60 | 1 | 0 | 348419 | 12q |
| CXCL1 | 45 | 0 | 0 | 789 | 4q21 |
| IL8 | 44 | 0 | 0 | 624 | 4q13-q21 |
| CXCL2 | 22 | 0 | 0 | 75765 | 4q21 |
| LIF | 10 | 0 | 0 | 2250 | 22q12.2 |
| SCGB1D2 | 7 | 0 | 0 | 204096 | 11q13 |
| CXCL3 | 5 | 0 | 0 | 89690 | 4q21 |
| CCL20 | 4 | 0 | 0 | 75498 | 2q33-q37 |
| CXCL6 | 4 | 0 | 0 | 164021 | 4q21 |
| IL6 | 4 | 0 | 0 | 512234 | 7p21 |
| SEMA3B | 4 | 0 | 0 | 82222 | 3p21.3 |
| CST3 | 6 | 0 | 2* | 304682 | 20p11.21 |
| Cytoskeleton | |||||
| ACTG1 | 21 | 1 | 0 | 14376 | 17q25 |
| KRT6B | 8 | 0 | 0 | 432677 | 12q12-q13 |
| PDLIM1 | 8 | 0 | 0 | 75807 | 10q22-q26.3 |
| KRT7 | 14 | 2 | 2 | 23881 | 12q12-q13 |
| PFN1 | 20 | 6 | 4 | 408943 | 17p13.3 |
| Protein synthesis, transport, and degradation | |||||
| EIF4A1 | 11 | 0 | 2 | 129673 | 17p13 |
| SEC61B | 8 | 0 | 0 | 191887 | 9q22.32-q31.3 |
| SUI1 | 10 | 2 | 3 | 150580 | 17q21.31 |
| EIF3S4 | 4 | 0 | 0 | 28081 | 19p13.2 |
| PICALM | 4 | 0 | 0 | 39252 | 11q14 |
| ST13 | 4 | 0 | 1* | 377199 | 22q13.2 |
| Metabolism | |||||
| SOD2 | 26 | 0 | 9 | 384944 | 6q25.3 |
| SAT | 10 | 0 | 0 | 28491 | Xp22.1 |
The frequency for each tag is given for a 10,000-tag library. Asterisks refer to tags whose P value is not statistically significant in one of the libraries. The complete data are published in Tables 3-8.
Table 2. Selection of the 50 genes most up-regulated in the INV and/or MET libraries with respect to the Br_N library from normal mammary epithelium.
| Gene symbol | Br_N | INV | MET | Unigene ID | Genetic map |
|---|---|---|---|---|---|
| Transcription factors/chromatin/nuclear proteins | |||||
| HNRPC | 1 | 35 | 20 | 476302 | 14q11.2 |
| HIF1A | 0 | 0* | 31 | 412416 | 14q21-q24 |
| HMGA1 | 1 | 1* | 26 | 57301 | 6p21 |
| RMP | 0 | 23 | 0* | 7943 | 19q12 |
| H2AFZ | 0 | 1* | 23 | 119192 | 4q24 |
| SRCAP | 1 | 19 | 0* | 136227 | 16p11.2 |
| DHX9 | 0 | 0* | 17 | 374524 | 1q25 |
| PTMA | 1 | 1* | 15 | 459927 | 2q35-q36 |
| ZNF9 | 0 | 0* | 12 | 2110 | 3q21 |
| HMGB1 | 2 | 7* | 22 | 434102 | 13q12 |
| SFRS6 | 1 | 0* | 9 | 6891 | 20q12-q13.1 |
| NME1 | 0 | 1* | 8 | 118638 | 17q21.3 |
| ADNP | 0 | 7 | 0* | 448540 | 20q13.13 |
| Cell cycle/apoptosis/cell growth | |||||
| CEB1 | 0 | 1* | 20 | 26663 | 4q22.1-q23 |
| TGFA | 0 | 15 | 0* | 170009 | 2p13 |
| CTGF | 0 | 13 | 0* | 410037 | 6q23.1 |
| CCNA2 | 0 | 5 | 1* | 85137 | 4q25-q31 |
| SPY1 | 3 | 11 | 0* | 511956 | 2p23.3 |
| Membrane proteins/antigens/receptors/adhesion | |||||
| GAGED2 | 0 | 0* | 36 | 112208 | Xp11.22-p11.21 |
| IFITM3 | 0 | 1* | 27 | 374650 | 11p15.5 |
| STEAP | 0 | 0* | 9 | 61635 | 7q21 |
| Signal transduction | |||||
| DUSP23 | 1 | 1* | 22 | 425801 | 1q23.1 |
| GNG11 | 0 | 0* | 19 | 83381 | 7q31-q32 |
| PTPN1 | 0 | 19 | 0* | 418004 | 20q13.1-q13.2 |
| YWHAZ | 0 | 1* | 15 | 386834 | 8q23.1 |
| GNB2L1 | 3 | 1* | 30 | 5662 | 5q35.3 |
| DGKQ | 0 | 8 | 0* | 99932 | 4p16.3 |
| Secreted and ECM proteins | |||||
| IGFBP4 | 1 | 1* | 12 | 1516 | 17q12-q21.1 |
| FDC-SP | 0 | 9 | 0* | 320147 | 4q13 |
| EDN2 | 1 | 7 | 0* | 1407 | 1p34 |
| FN1 | 0 | 1* | 6 | 418138 | 2q34 |
| Cytoskeleton | |||||
| CYFIP1 | 0 | 14 | 4 | 26704 | 15q11 |
| FSCN1 | 0 | 1* | 23 | 118400 | 7p22 |
| CAP1 | 1 | 0* | 11 | 104125 | 1p34.2 |
| Protein synthesis, transport, and degradation | |||||
| SAE1 | 1 | 0* | 18 | 32748 | 19q13.33 |
| CCT5 | 1 | 2* | 17 | 1600 | 5p15.31 |
| UBA52 | 2 | 3* | 31 | 5308 | 19p13.1-p12 |
| TRAP1 | 0 | 0* | 14 | 183803 | 16p13.3 |
| RPN2 | 0 | 0* | 11 | 406532 | 20q12-q13.1 |
| PPIA | 17 | 27* | 159 | 356331 | 7p13-p11.2 |
| YKT6 | 1 | 8 | 0* | 296244 | 7p15.1 |
| HSPA8 | 7 | 0 | 31 | 180414 | 11q24.1 |
| TPT1 | 48 | 6 | 90 | 374596 | 13q12-q14 |
| Metabolism | |||||
| LDHA | 3 | 1* | 68 | 2795 | 11p15.4 |
| NNMT | 1 | 0* | 16 | 364345 | 11q23.1 |
| ENO1 | 0 | 0* | 11 | 433455 | 1p36.3-p36.2 |
| AHCY | 1 | 1* | 9 | 388004 | 20cen-q13.1 |
| Others | |||||
| SNCAIP | 0 | 23 | 0* | 24948 | 5q23.1-q23.3 |
| ANKRD10 | 0 | 19 | 0* | 164969 | 13q34 |
| OBTP | 1 | 1* | 12 | 525899 | 6p21.31 |
The frequency for each tag is given for a 10,000-tag library. Asterisks refer to tags whose P value is not statistically significant in one of the libraries. Values in italics indicate a P value that is statistically significant, but with expression in the opposing direction in one of the libraries. The complete data are published in Tables 3-8.
Genes Down-Regulated in INV and MET Carcinoma Libraries. Our findings are in part similar and in part different in relation to previously published work (1, 2). In agreement with the earlier work, we observed that the most dramatic difference in gene expression, between cancer and normal cells, involves genes down-regulated in cancer. Of 221 down-regulated genes (Table 3), 134 genes are down-regulated in both the INV and MET libraries, 77 genes are down-regulated only in the INV library, and 10 genes are down-regulated only in the MET library. A large fraction of these genes are of unknown function or their function has not been associated to breast cancer. In previous work by Polyak and coworkers (1, 2), 32 genes were found to be down-regulated in all breast cancer samples analyzed in their study. Nineteen of these genes are also down-regulated in our INV and MET libraries in a significant way, and they are as follows: PNRC1, CEBPD, TM4SF1, ANXA1, TNFRSF10B, RASD1, CXCL1, IL8, TFF1, SCGB3A1, LIF, LAMC2, CXCL3, CCL20, CXCL6, KRT6B, SOD2, SAT, and STC2; the remaining 13 are also down-regulated in our INV and MET libraries, but are not reported in Tables 1 and 3 because, according to our parameters, they are not down-regulated significantly (P > 0.05). In agreement with Porter et al.'s results (1, 2), a large fraction of genes down-regulated in the INV and MET libraries encodes secreted proteins such as chemokines CXCL1, CXCL2, CXCL3, CXCL6 and CCL20; cytokines IL6, IL8, and LIF; and the protein LOC118430 (small breast epithelial mucin).
Additional genes down-regulated in Table 1 are not associated to breast cancer but are described to be important in the genesis of other tumor types. They include the following: the tumor suppressor genes: MSF, DOC1R (a potential human tumor suppressor gene), SUI1 (a putative translation initiation factor), and ST13 (also called SNC6) (14–17); genes involved in the apoptotic pathway are as follows: GADD45B and PDCD5 (18, 19); genes controlling cell proliferation are: LIF, which regulates the growth of normal human breast epithelial cells, and SEMA3B, a mediator of p53 tumor-suppressor activity (20–23).
Other genes in Table 1 include PD-LIM1, which has a role in cytoskeletal organization, cell structure and shape, cell migration, cell polarity, and cytokinesis (24); PFN1, that, when overexpressed, reduces the migration of invasive breast cancer cells (25); CLDN4 and KRT7, whose expression was detected in normal estrogen-responsive cells (26), suggesting that they may encode factors important in the autocrine/paracrine estrogen-signaling pathway regulating normal mammary epithelium; SCGB1D2 and CST3, which are both of unknown function. We also found down-regulation of mitochondrial genes, such as SOD2 (1, 2), and down-regulation of the ribosomal genes S18, S29, L13, L35, L36, and L36a (see Tables 1 and 3). It is reported that these ribosomal genes may act as tumor suppressors, because loss-of-function mutations in these genes result in tumor formation at high frequency in zebrafish cell lines (27). We observed that many of the genes, the expression of which was found down-regulated in our SAGE libraries, play important roles in the regulation of cell growth, differentiation, and morphogenesis of the normal mammary epithelium (2). This finding suggests that loss of specific functions, including the ability to differentiate, or the loss of the epithelial phenotype, may have an essential role in tumorigenesis.
Genes Up-Regulated in INV and MET Libraries. The genes up-regulated in the INV and MET libraries are listed in Table 6. From this list, 57 genes are up-regulated in the INV and 110 genes are up-regulated in the MET libraries, with moderate-to-high increase of expression in respect to the normal epithelium library; we found few genes up-regulated in both libraries. This gene list includes some key regulatory genes that play crucial roles in cell proliferation, invasion, and metastasis in breast cancer or in other tumor types; genes not yet associated to the genesis of cancer and several ESTs of unknown function.
The list of genes highly expressed in the INV library (Table 2) includes some genes already known to be linked to cancer, such as ADNP, implicated in maintaining cell survival (28); TGFA, which has a role in breast tumor growth and progression (29, 30); CTGF, which stimulates angiogenesis (31); CCNA2, an oncogene overexpressed in liver tumors (32); SPY1, which induces cell-cycle progression (33); PTPN1, an activator of the cSrc and RAS signaling pathway, amplified in ovarian cancer (34); and YKT6 (also called SNARE), which is associated with invasive and metastatic phenotypes in breast cancer (35). The list of genes highly expressed in the INV library also includes genes not previously associated with the genesis of cancer, such as CYFIP1, SNCAIP, and ANKRD10; 11 no-matching tags and 29 tags matching to ESTs or mRNAs encoding for hypothetical proteins.
Most of the up-regulated genes were found in the MET library. Many of these were already recognized to be associated with cancer, such as DHX9, which inhibits BRCA1 (36); PTMA, which prevents apoptosis (37); CEB1, which is elevated when the function of p53 and RB are compromised (38); GAGED2, which is overexpressed in a variety of tumors (39); STEAP, which is associated with prostate cancer and tumor progression (40); GNB2L1/RACK1, which inhibits apoptosis (41); PPIA, which is associated with prostate cancer (42); and TPT1, which is overexpressed in colon cancer (43). Some other genes, like OBTP, have no known function. The activity of some of the up-regulated genes (see Table 2) correlates with increased cell proliferation: four of these genes are up-regulated in the INV library (ADNP, TGFA, SPY1 and PTPN1), and three are up-regulated in the MET library (PTMA, CEB1 and GNB2L1).
Some of the genes from Table 2, such as NME1 and FN1, have been already described through different expression-profiling methods to be up-regulated in breast carcinoma (44), and the genes IGFBP4 and CCNA2 were found differentially regulated in estrogen-responsive breast cancer cells (26). Several up-regulated genes of the MET library, such as PTMA, NME1, CEB1, GNB2L1, TRAP1, HSPA8, TPT1, LDHA, RPS5, RPS7, RPL8, RPL32, and RPL34 are known to be induced by ectopic expression of c-MYC (45). Two genes (EDN2 and NNMT) were found up-regulated in human mammary epithelial cells expressing high levels of ERBB2 (46).
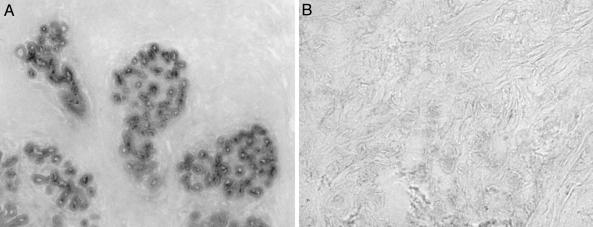
Confirming Gene Expression by RNA in Situ Hybridization. In situ hybridization, performed on normal (Fig. 1A) and tumor samples of breast cells (Fig. 1B), confirmed the expression of CXCL6 in the normal mammary tissue and its epithelial localization.
Fig. 1.
CXCL6 expression in human breast tissues. Frozen sections of normal breast tissue (A) and breast-invasive ductal carcinoma (B) were hybridized with a digitonin-labeled CXCL6 antisense riboprobe. Strong expression of CXCL6 was detected in normal breast samples (A; ×10 magnification) but not in the invasive breast cancer samples (B; ×40 magnification). The sense probe gives no signal (data not shown).
Conclusions
In this paper, we report our initial work on the profiles of gene expression in human breast carcinoma cells compared with normal epithelial cells, by using the SAGE approach. We used very pure populations of cancer cells obtained by microscopic isolation from frozen sections of a primary invasive breast carcinoma (N.0) and from a nodal metastasis of a breast carcinoma. By using this approach, we collected ≈500 cells from each sample, and the amount of mRNA obtained from these number of cells was very small; therefore the mRNA was amplified. We show that this procedure can be performed without altering the original mRNA abundance. This approach is shown to be feasible, and the analysis of the two libraries documents the presence of changes in the expression of many genes, in agreement with what has been already shown by others, but there are differences in the genes affected. As previously shown, we also found that several classes of genes are down-regulated or up-regulated in both libraries. A more general observation can be made: many of the down-regulated genes are the same in both libraries, whereas the up-regulated genes are almost completely different in the two libraries. The difference in the up-regulated genes may be due to the fact that our two libraries were obtained from different patients, who had cancers with different characteristics, especially in estrogen receptor expression, and could therefore have arisen by different mechanisms. However, there is a more interesting explanation: the down-regulation of a set of genes may be the basic mechanism of cancer formation, whereas the up-regulation may characterize and possibly control the state of evolution of individual cancers, but further work is required to verify this hypothesis. This information should be useful for clarifying the mechanism involved in the formation and progression of breast cancer.
This work describes, in an accurate and objective way, the gene expression profiles of two human breast carcinomas uncontaminated by accidental intrusions from non-cancer cells, and not biased by arbitrary gene selection. This approach has the potential to be used in other systems where the cells of interest are very few and the contaminating cells can be a problem.
Supplementary Material
Acknowledgments
We thank Drs. N. A. Datson, V. Velculescu, and S. M. Wang for their advice on the SAGE protocol; Drs. S. Bertuzzi and P. Taglialatela for their advice on the in situ hybridization protocol; Dr. G. Bertalot for assistance and suggestions; and L. Susani and B. Vergani for their technical assistance. This work was supported by Associazione Italiana per la Ricerca sul Cancro Grant 115/2003 (to I.Z.); Italy-USA Project on Cancer Pharmacogenomics Grant N.527/B-B7 (to I.Z.); Ministero dell'Istruzione, dell'Università e della Ricerca/Fondo per gli Investimenti della Ricerca di Base Grant RBME019J9W (to P.V. and I.Z.); a Compagnia di San Paolo Grant (to P.V.); and Progetto Strategico: Genomica Funzionale, Consiglio Nazionale delle Ricerche Grant 449/97 (to P.V.). This manuscript is no. 80 of the Genoma 2000/Istituto Tecnologie Biomediche Avanzate Project funded by the Cariplo Foundation.
Author contributions: I.Z. designed research; I.Z., E.M., M.S., V.V., and E.V. performed research; V.A.K., S.P., R.R., P.V., and A.A. contributed new reagents/analytic tools; E.M. analyzed data; I.Z. and R.D. wrote the paper; V.A.K. performed library comparison; S.P. performed all microdissection experiments; B.S. and G.V. performed sequencing; and R.D. contributed to the main original concepts and outline of all of the experiments.
Abbreviations: SAGE, serial analysis of gene expression; INV, invasive; MET, metastatic.
References
- 1.Porter, D. A., Krop, I. E., Nasser, S., Sgroi, D., Kaelin, C. M., Marks, J. R., Riggins, G. & Polyak, K. (2001) Cancer Res. 61, 5697–5702. [PubMed] [Google Scholar]
- 2.Porter, D., Lahti-Domenici, J., Keshaviah, A., Bae, Y. K., Argani, P., Marks, J., Richardson, A., Cooper, A., Strauberg, R. & Riggins, G. J. (2003) Mol. Cancer Res. 1, 362–375. [PubMed] [Google Scholar]
- 3.Perou, C. M., Sorlie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., Rees, C. A., Pollack, J. R., Ross, D. T., Johnsen, H., Akslen, L. A., et al. (2000) Nature 406, 747–752. [DOI] [PubMed] [Google Scholar]
- 4.van't Veer, L. J., Dai, H., van de Vijver, M. J., He, Y. D., Hart, A. A., Mao, M., Peterse, H. L., van der Kooy, K., Marton, M. J., Witteveen, A. T., et al. (2002) Nature 415, 530–536. [DOI] [PubMed] [Google Scholar]
- 5.Sorlie, T., Tibshirani, R., Parker, J., Hastie, T., Marron, J. S., Nobel, A., Deng, S., Johnsen, H., Pesich, R., Geisler, S., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 8418–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iacobuzio-Donahue, C. A., Argani, P., Hempen, P. M., Jones, J. & Kern, S. E. (2002) Cancer Res. 62, 5351–5357. [PubMed] [Google Scholar]
- 7.Ronnov-Jessen, L., Peterson, O. W. & Bissel, M. (1996) Phisyol. Rev. 76, 69–125. [DOI] [PubMed] [Google Scholar]
- 8.Franzen, B., Linder, S., Okuzawa, K., Kato, H. & Auer G. (1999) Electrophoresis 14, 1045–1053. [DOI] [PubMed] [Google Scholar]
- 9.Velculescu, V. E., Zhang, L., Vogelstein, B. & Kinzler, K. W. (1995) Science 270, 484–487. [DOI] [PubMed] [Google Scholar]
- 10.Margulies, E. H. & Innis, J. W. (2000) Bioinformatics 16, 650–651. [DOI] [PubMed] [Google Scholar]
- 11.Kuznetsov, V. A., Knott, G. D. & Bonner, R. F. (2002) Genetics 161, 1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook, J. & Russel, D. W. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 13.Kuznetsov, V. A. (2001) EURASIP J. on Applied Signal Processing 4, 285–296. [Google Scholar]
- 14.Russell, S. E. H., McIlhatton, M. A., Burrows, J. F., Donaghy, P. G., Chanduloy, S., Petty, E. M., Kalikin, L. M., Church, S. W., McIlroy, S., Harkin, D. P., et al. (2000) Cancer Res. 60, 4729–4734. [PubMed] [Google Scholar]
- 15.Todd, R., McBride, J., Tsuji, T., Donoff, R. B., Nagai, M., Chou, M. Y., Chiang, T. & Wong, D. T. (1995) FASEB J. 9, 1362–1370. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka, K., Tamura, H., Tanaka, H., Katoh, M., Futamata, Y., Seki, N., Nishimune, Y. & Hara, T. (2002) Dev. Biol. 246, 466–479. [DOI] [PubMed] [Google Scholar]
- 17.Zhang, Y., Cai, X., Schlegelberger, B. & Zheng, S. (1998) Cytogenet. Cell Genet. 83, 56–57. [DOI] [PubMed] [Google Scholar]
- 18.Yoo, J., Ghiassi, M., Jirmanova, L., Balliet, A. G., Hoffman, B., Fornace, A. J., Jr., Liebermann, D. A., Bottinger, E. P. & Roberts, A. B. (2003) J. Biol. Chem. 278, 43001–43007. [DOI] [PubMed] [Google Scholar]
- 19.Chen, Y., Sun, R., Han, W., Zhang, Y., Song, Q., Di, C. & Ma, D. (2001) FEBS Lett. 509, 191–196. [DOI] [PubMed] [Google Scholar]
- 20.Grant, S. L., Douglas, A. M., Goss, G. A. & Begley, C. G. (2001) Growth Factors 19, 153–162. [DOI] [PubMed] [Google Scholar]
- 21.Ochi, K., Mori, T., Toyama, Y., Nakamura, Y. & Arakawa, H. (2002) Neoplasia 4, 82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuroki, T., Trapasso, F., Yendamuri, S., Matsuyama, A., Alder, H., Williams, N. N., Kaiser, L. R. & Croce, C. M. (2003) Cancer Res. 63, 3352–3355. [PubMed] [Google Scholar]
- 23.Lerman, M. I. & Minna, J. D. (2000) Cancer Res. 60, 6116–6133. [PubMed] [Google Scholar]
- 24.Vallenius, T. & Makela, T. P. (2002) J. Cell Sci. 115, 2067–2073. [DOI] [PubMed] [Google Scholar]
- 25.Roy, P. & Jacobson, K. (2004) Cell Motil. Cytoskeleton 57, 84–95. [DOI] [PubMed] [Google Scholar]
- 26.Vendrell, J. A., Magnino, F., Danis, E., Duchesne, M. J., Pinloche, S., Pons, M., Birnbaum, D., Nguyen, C., Theillet, C. & Cohen, P. A. (2004) J. Mol. Endocrinol. 32, 397–414. [DOI] [PubMed] [Google Scholar]
- 27.Amsterdam, A., Sadler, K. C., Lai, K., Farrington, S., Bronson, R. T., Lees, J. A. & Hopkins, N. (2004) PLoS Biol. 2, 690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zamostiano, R., Pinhasov, A., Gelber, E., Steingart, R. A., Seroussi, E., Giladi, E., Bassan, M., Wollman, Y., Eyre, H. J., Mulley, J. C., et al. (2001) J. Biol. Chem. 276, 708–714. [DOI] [PubMed] [Google Scholar]
- 29.Jiang, D., Yang, H., Willson, J. K., Liang, J., Humphrey, L. E., Zborowska, E., Wang, D., Foster, J., Fan, R. & Brattain, M. G. (1998) J. Biol. Chem. 273, 31471–31479. [DOI] [PubMed] [Google Scholar]
- 30.Umekita, Y., Ohi, Y., Sagara, Y. & Yoshida, H. (2000) Int. J. Cancer. 89, 484–487. [DOI] [PubMed] [Google Scholar]
- 31.Shimo, T., Kubota, S., Kondo, S., Nakanishi, T., Sasaki, A., Mese, H., Matsumura, T. & Takigawa, M. (2001) Cancer Lett. 174, 57–64. [DOI] [PubMed] [Google Scholar]
- 32.Haddad, R., Morrow, A. D., Plass, C. & Held, W. A. (2000) Biochem. Biophys. Res. Commun. 274, 188–196. [DOI] [PubMed] [Google Scholar]
- 33.Porter, L. A., Dellinger, R. W., Tynan, J. A., Barnes, E. A., Kong, M., Lenormand, J. L. & Donoghu, D. J. (2002) J. Cell Biol. 157, 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dube, N., Cheng, A. & Tremblay, M. L. (2004) Proc. Natl. Acad. Sci. USA 101, 1834–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kluger, H. M., Kluger, Y., Gilmore-Hebert, M., DiVito, K., Chang, J. T., Rodov, S., Mironenko, O., Kacinski, B. M., Perkins, A. S. & Sapi, E. (2004) Lab. Invest. 84, 320–331. [DOI] [PubMed] [Google Scholar]
- 36.Schlegel, B. P., Starita, L. M. & Parvin, J. D. (2003) Oncogene 22, 983–991. [DOI] [PubMed] [Google Scholar]
- 37.Jiang, X., Kim, H. E., Shu, H., Zhao, Y., Zhang, H., Kofron, J., Donnelly, J., Burns, D., Ng, S. C., Rosenberg, S., et al. (2003) Science 299, 223–226. [DOI] [PubMed] [Google Scholar]
- 38.Mitsui, K., Nakanishi, M., Ohtsuka, S., Norwood, T. H., Okabayashi, K., Miyamoto, C., Tanaka, K., Yoshimura, A. & Ohtsubo, M. (1999) Biochem. Biophys. Res. Commun. 266, 115–122. [DOI] [PubMed] [Google Scholar]
- 39.Egland, K. A., Kumar, V., Duray, P. & Pastan, I. (2002) Mol. Cancer Ther. 1, 441–450. [PubMed] [Google Scholar]
- 40.Hubert, R. S., Vivanco, I., Chen, E., Rastegar, S., Leong, K., Mitchell, S. C., Madraswala, R., Zhou, Y., Kuo, J., Raitano, A. B., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 14523–14528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiely, P. A., Sant, A. & O'Connor, R. (2002) J. Biol. Chem. 277, 22581–22589. [DOI] [PubMed] [Google Scholar]
- 42.Cande, C., Vahsen, N., Kouranti, I., Schmitt, E., Daugas, E., Spahr, C., Luban, J., Kroemer, R. T., Giordanetto, F., Garrido, C., et al. (2004) Oncogene 23, 1514–1521. [DOI] [PubMed] [Google Scholar]
- 43.Chung, S., Kim, M., Choi, W., Chung, J. & Lee, K. (2000) Cancer Lett. 156, 185–190. [DOI] [PubMed] [Google Scholar]
- 44.Amatschek, S., Koenig, U., Auer, H., Steinlein, P., Pacher, M., Gruenfelder, A., Dekan, G., Vogl, S., Kubista, E., Heider, K. H., et al. (2004) Cancer Res. 64, 844–856. [DOI] [PubMed] [Google Scholar]
- 45.Menssen, A. & Hermeking, H. (2002) Proc. Natl. Acad. Sci. USA 99, 6274–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mackay, A., Jones, C., Dexter, T., Silva, R. L., Bulmer, K., Jones, A., Simpson, P., Harris, R. A., Jat, P. S., Neville, A. M., et al. (2003) Oncogene 22, 2680–2688. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.