Abstract
The UL41 protein of herpes simplex virus 1 has been reported to mediate the degradation of both viral and cellular mRNAs. Extensive studies on β-actin and some viral mRNAs were consonant with this conclusion. In earlier studies, we reported that the UL41-dependent degradation of cellular mRNAs up-regulated after infection was selective. One class of the up-regulated mRNAs, exemplified by the stress-inducible immediate-early 1 mRNA, is deadenylated, 3′ to 5′ degraded and is not translated. Another class of up-regulated mRNAs, exemplified by GADD45β, does not undergo this pattern of degradation and is translated. A puzzling feature of the earlier results is that the amounts of up-regulated mRNAs accumulating in the cytoplasm of ΔUL41 mutant virus-infected cells was lower than in WT virus-infected cells, a contradiction, inasmuch as if the rates of accumulation were identical and degradation of the mRNAs were higher in WT virus-infected cells, the steady-state levels should have been higher in ΔUL41 mutant virus-infected cells. In this report, we show that in ΔUL41 mutant virus-infected cells, the rates of degradation of the stress-inducible immediate-early response gene 1 and other up-regulated mRNAs are approximately the same as those observed in mock-infected cells and are faster than in WT virus-infected cells. This is contrary to the observed UL41-dependent degradation of β-actin and other mRNAs. The UL41 protein thus mediates two functions, i.e., it mediates rapid degradation of some mRNAs exemplified by β-actin and stabilizes or delays the degradation of other mRNAs exemplified by GADD45β, tristetraprolin, etc. A model unifying both activities of the UL41 protein is presented.
Keywords: AU-rich elements, nucleases, virion host shutoff
It has been known for many decades that herpes simplex viruses (HSVs) shut off host macromolecular metabolism primarily for two reasons: (i) to preclude host responses to infection and (ii) to divert the resources of the cells to viral macromolecular synthesis (1). The key protagonist in this process is a protein encoded by UL41 ORF, also known as the virion host shutoff protein, or vhs (2–4). This protein is incorporated into the virion and is released into the cytoplasm after the entry of the virus into the newly infected cells. The functions mediated by this protein are readily demonstrated by the rapid decrease in incorporation of amino acids into cellular proteins and degradation of both viral and cellular mRNAs. The view maintained over two decades is that UL41 protein mediates the degradation of both viral and cellular mRNAs in an indiscriminate fashion, and that viral mRNAs prevail because the rate of transcription of viral ORFs exceeds the rate of degradation of the mRNAs, and finally, that upon synthesis of late viral proteins, RNase-mediated function of UL41 protein is neutralized by association with the protein encoded by the UL48 ORF (reviewed in ref. 5). In earlier publications (6, 7), we reported that the degradation of cellular mRNAs up-regulated after infection is selective. Specifically, a class of mRNAs, exemplified by the stress-inducible immediate-early response gene 1 (IEX-1) and containing AU-rich elements (AREs) in their 3′-UTRs, were deadenylated, subjected to endonucleolytic cleavage and 3′ to 5′ degradation, and were not translated. Moreover, truncated fragments consisting of the 5′ termini of the mRNAs tended to persist in the cytoplasm for many hours. In contrast, the mRNAs of tristetraprolin (TTP) and growth arrest and DNA damage-inducible gene 45β (GADD45β) did not appear to be subjected to similar degradation and were translated. The processive degradation of IEX-1 including the lingering of the 5′ portion of the mRNAs and the stability and translation of TTP, e.g., were mediated by UL41 inasmuch as they did not take place in ΔUL41 mutant virus-infected cell.
The genesis of the studies reported here stemmed from the observation that the amounts of mRNAs accumulating in ΔUL41 mutant virus-infected cells were lower than in WT virus-infected cells. Because the degradation of mRNA appeared to be ΔUL41-dependent, the observation made sense only if the rates of degradation of the up-regulated mRNAs were faster in ΔUL41-infected cells as compared with those of WT virus-infected cells. In this report, we show that this is indeed the case. We also show that associated with the functions of UL41 protein are two pathways of degradation of mRNAs. Thus, the mRNAs of GAPDH and β-actin are rapidly degraded in a UL41-dependent manner. In contrast, the up-regulated mRNAs, exemplified by IEX-1, GADD45β, and TTP mRNAs, are more rapidly degraded in ΔUL41 mutant-virus infected cells than in WT virus-infected cells. Last, we propose a hypothesis that unifies these two apparently divergent functions of the UL41 protein.
Materials and Methods
Cells and Viruses. HeLa cells obtained from American Type Culture Collection were propagated in DMEM supplemented with 5% newborn calf serum. HSV-1(F) is the prototype HSV-1 strain used in this laboratory (8). The ΔUL41 mutant virus R2621, as well as the repaired virus, R2626 (ΔUL41R), were reported elsewhere (9).
Cell Infection and Treatment. HeLa cell monolayers were either mock-infected or exposed for 1 h to 10 plaque-forming units (pfu) of WT or mutant virus per cell at 37°C. At 3 h after infection, the cultures were mock-treated or incubated with medium containing actinomycin D (Sigma, 5 μg/ml). Because the stock solution of actinomycin D was made in DMSO (Sigma), all control cultures were incubated in medium containing 0.5% DMSO.
Isolation of Total and Cytoplasmic RNA. Total RNA was extracted with the aid of TRIzol reagent (Life Technologies, Rockville, MD) according to the manufacturer's instructions. DNase treatment (Life Technologies), phenol-chloroform extraction, and ethanol precipitation (Fisher Scientific) were carried out to remove possible DNA contamination. Cytoplasmic RNA was isolated with the aid of RNeasy mini kit according to the protocol furnished by the manufacturer (Qiagen, Valencia, CA).
Northern Blot Analyses. For these analyses, 12 μg of total RNA or 8 μg of cytoplasmic RNA was loaded onto denaturing formaldehyde gel and probed with random hexanucleotide-primed 32P-labeled specific probe after transfer onto a nylon membrane. IEX-1, TTP, and GADD45β probe templates were generated as described (6, 7). For β-tubulin mRNA detection, a fragment containing part of the β-tubulin-coding sequence was amplified by RT-PCR of total RNA purified from HSV-1-infected human-foreskin fibroblasts by using the following pairs of primers: forward, 5′-TTCCAGCTGACCCACTCTCT-3′; reverse, 5′-TTGGAGTCGAACATCTGCTG-3′. Probe templates for GAPDH and β-actin were purchased from Ambion (Austin, TX). Prehybridization and hybridization were performed with the ULTRAhyb buffer (Ambion) supplemented with 200 μg of denatured salmon sperm DNA per ml (Stratagene). The membranes were prehybridized for 2 h at 42°C and then overnight after the addition of the 32P-labeled probe. The membranes were rinsed as suggested by the manufacturer of ULTRAhyb and exposed to film for signal detection.
Real-Time PCR. Real-time PCR was carried out as described (10). The following primers were used: IEX-1 forward, 5′-CCGTCCTCGAGCCCTTTAA-3′, and reverse 5′-TGCTGAGGTCCAGAGCGTAGT-3′; and TTP forward, 5′-CGCGCTACAAGACTGAGCTATG-3′, and reverse 5′-CATGGGCAAACTGGCACTT-3′. Both sets of primers were designed inside the coding sequence of the two transcripts. cDNA quantities were normalized to 18S rRNA quantities (primers from Ambion) obtained from the same plate. The percentage of remaining mRNAs (IEX-1 or TTP) for each time point was calculated by using as 100% the mRNA level at time 3 for each condition [mock, HSV-1(F), or ΔUL41].
Results
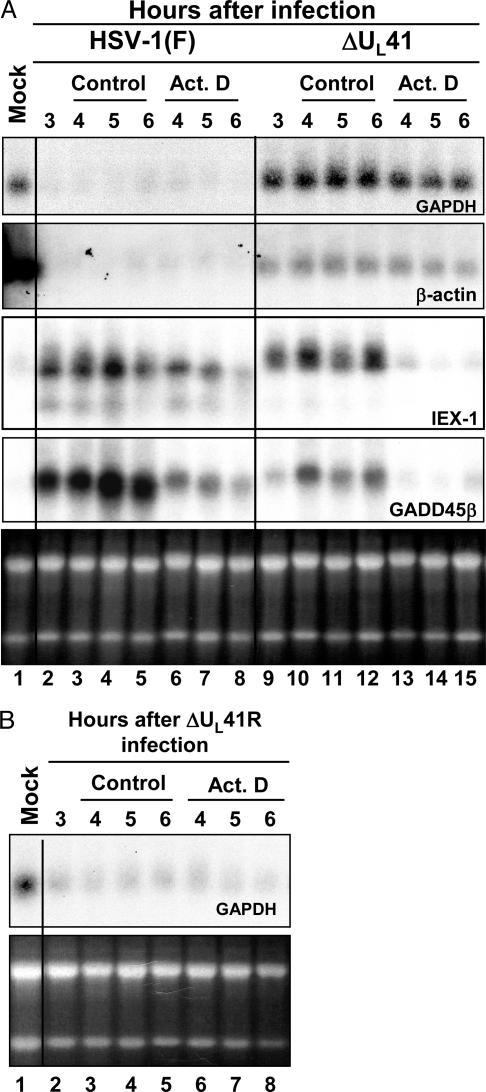
Constitutively expressed RNAs are rapidly degraded after HSV-1 infection in a UL41-dependent manner. To analyze the stability of cellular mRNAs after HSV-1 infection, cells were exposed to actinomycin D to block transcription. Confluent HeLa cell monolayers were mock-infected or exposed to 10 pfu of HSV-1(F) or ΔUL41 mutant viruses per cell at 37°C. After 1 h, the inoculum was replaced with fresh medium and the cells were incubated at 37°C for an additional 3 h (time 0). Actinomycin D was then added to the cell cultures, and total RNA was extracted 1, 2, or 3 h after exposure to the drug. The times of harvest corresponded to 4, 5, or 6 h after infection. As control, total RNA was also purified from replicate cultures of infected cells processed as above but incubated in medium containing 0.5% DMSO, the solvent used to prepare the actinomycin D stock solution. In this series of experiments, we have selected for analysis GAPDH and β-actin mRNAs. Both mRNAs are constitutively expressed in uninfected cells and neither is up-regulated after infection (10). The accumulation of the GAPDH and β-actin transcripts was analyzed by Northern blot hybridization, and the results are shown in Fig. 1A. Both transcripts were rapidly degraded upon HSV-1(F) infection (lanes 2–8) compared to mock-infected cells (lane 1). The infection with the mutant virus lacking UL41 (ΔUL41) did not result in degradation of either GAPDH or β-actin mRNAs (lanes 9–12). Moreover, the persistence of the mRNA even in the presence of actinomycin D (lanes 13–15) suggested that the half-lives of these mRNAs extended beyond the 3 h of observation and could not be measured in this experiment.
Fig. 1.
Total RNA accumulations in HeLa cells infected with WT and ΔUL41 mutant virus. (A) HeLa cells were either mock-infected or infected with 10 pfu of HSV-1(F) or ΔUL41 mutant viruses per cell for 3 h. Transcription was then stopped by addition of 5 μg of actinomycin D (Act. D) per ml. In parallel, an identical series was incubated in medium containing 0.5% DMSO (control). The cells were harvested at the time intervals shown; total mRNA was extracted, and amounts of GAPDH, β-actin, IEX-1, and GADD45β transcripts were determined by Northern blotting. The ethidium bromide staining of total RNA samples is shown as a loading control. (B) HeLa cells were either mock-infected or exposed to 10 pfu of ΔUL41 repaired virus (ΔUL41R) per cell and incubated for 3 h. The cells were treated as described above, and total RNA was analyzed for GAPDH mRNA. The ethidium bromide staining of total RNA samples is shown as a loading control. The signal in the mock lane in A for β-actin reflects the unexpected interaction of the probe with size markers. In other experiments, e.g., Fig. 2, the size marker was run in a lane distant from the mock lane.
This experiment was repeated and extended with mRNAs purified from the cytoplasmic fraction of infected cells and compare to the stability of those purified from mock-infected cells (Fig. 2). In mock- and in ΔUL41 mutant virus-infected cells, GAPDH and β-actin transcripts were stable in presence of actinomycin D (lanes 4, 5, 14, and 15). Conversely, both mRNAs quickly disappeared in the cytoplasmic fraction of cells infected with WT virus (lanes 9 and 10), even in absence of actinomycin D (lanes 6–8). It is noteworthy that the persistence of the mRNAs in the presence of actinomycin D in mock-infected cells is similar to that extracted from the cytoplasm of treated cells infected with UL41 mutant.
Fig. 2.
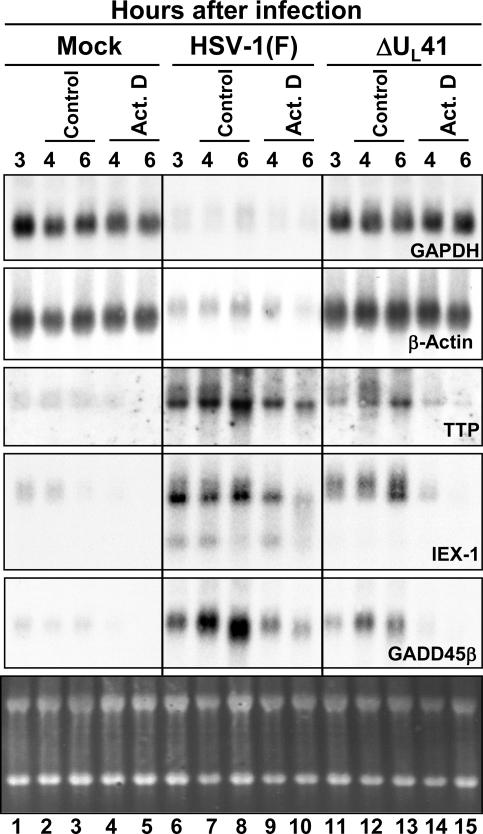
Cytoplasmic RNA accumulations in HeLa cells infected with WT and ΔUL41 mutant virus. HeLa cells were either mock-infected or exposed to 10 pfu of HSV-1(F) or ΔUL41 mutant virus per cell and incubated for 3 h. Transcription was then stopped by addition of 5 μg of actinomycin D (Act. D) per ml. In parallel, an identical series was incubated in medium containing 0.5% DMSO (control). The cells were harvested at the time intervals shown; cytoplasmic mRNA was extracted, and the amounts of GAPDH, β-actin, TTP, IEX-1, and GADD45β mRNAs were determined by Northern blotting. The ethidium bromide staining of cytoplasmic RNA samples is shown as a loading control.
To examine in greater detail the impact of HSV-1 infection on the accumulation of cellular mRNAs and to verify the evidence that the stability of the mRNAs was related solely to UL41 function and not to some other defect in the ΔUL41 mutant virus, we analyzed total mRNAs extracted from mock-infected cells and from cells infected with the ΔUL41R recombinant virus in which the deletion in the UL41 gene had been repaired. The procedures used in this experiment were identical to those described above for Fig. 1 A except that the cells were infected with the ΔUL41R recombinant. As in the case of the cells infected by the WT virus, Northern blot analyses indicated that GAPDH mRNA was totally degraded after infection with the repaired virus (Fig. 1B, lanes 2–8). Moreover, the pattern of accumulation of IEX-1 transcripts in ΔUL41R-infected cells was similar to that observed in WT virus-infected cells shown in Fig. 1 A (data not shown). These results are consistent with earlier results and indicate that the rapid disappearance of cellular mRNAs after infection is mediated by UL41.
The Rate of Turnover of mRNAs Induced After Infection with WT Virus Is Slower than in Mock-Infected or ΔUL41 Mutant Virus-Infected Cells. Earlier, we reported that in WT virus-infected cells several mRNAs up-regulated after infection appeared to linger, that the amounts of mRNA accumulating in WT virus-infected cells were higher than in ΔUL41 mutant virus-infected cells, and finally, that in the WT virus-infected cells in some instances there was an accumulation of truncated forms of the mRNAs that were absent from infected cells (6, 7).
The mRNAs comprising this group were those encoding IEX-1, IκBα, c-fos, etc. Other mRNAs (e.g., TTP and GADD45β) either did not form stable 5′-truncated products or did not linger in the cytoplasm of WT virus-infected cells. A question central to these studies was whether the mRNAs of representative members of each group differ with respect to stability. The mRNAs examined in these studies were the mRNAs encoding IEX-1, GADD45β, and TTP. As reported earlier, although IEX-1 protein could not be detected in infected cells past the first hour of infection, both TTP and GADD45β proteins were readily detected in infected cells (6, 7). The mRNAs analyzed by Northern blotting were the same as those described above and are shown in Fig. 1 A. The results indicate that IEX-1 and GADD45β mRNAs were up-regulated in both WT virus-infected (lanes 2–5) or in ΔUL41 mutant virus-infected (lanes 9–12) cells. Actinomycin D treatment revealed difference in the stability of IEX-1 and GADD45β RNAs in cells infected by the two viruses. These mRNAs persisted in actinomycin D-treated cells infected by WT virus (lanes 6–8), whereas they were rapidly degraded in cells infected by ΔUL41 mutant virus, as early as 1 h after actinomycin D exposure (lanes 13–15).
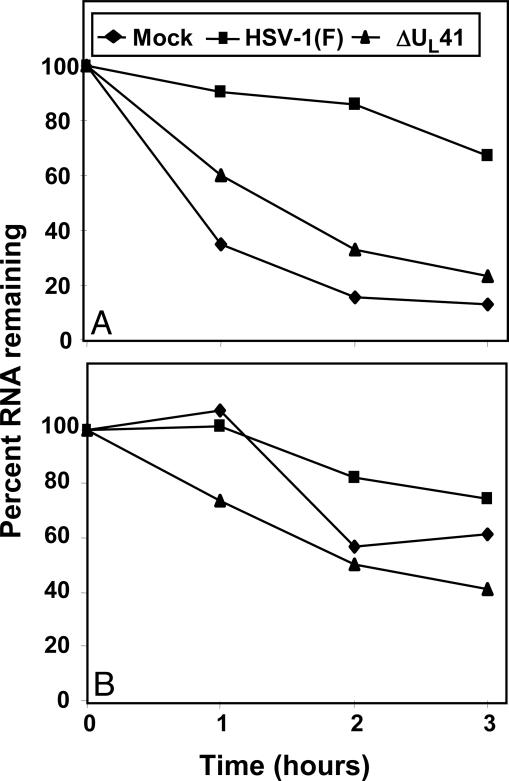
To quantify the half-life of up-regulated cellular transcripts, a real-time PCR analysis of total cellular RNA was done as described in Materials and Methods by using primers specific for IEX-1 and TTP mRNAs. HeLa cells were either mock-infected or exposed to 10 pfu of HSV-1(F) or ΔUL41 mutant virus for 1 h. After 3 h (time 0) of incubation, the cultures were replenished with medium containing actinomycin D (5 μg/ml) or DMSO (0.5%). Total RNAs extracted 1, 2, or 3 h after exposure to the drug were reverse transcribed by using random hexamers and normalized with respect to reverse transcribed 18S ribosomal RNA. The amounts of RNAs from cells harvested at time 0 after mock, HSV-1(F), and ΔUL41 infection were used as calibrators. The results are shown in Fig. 3 and can be summarized as follows: In mock-infected cells, IEX-1 mRNA was highly unstable, with a half-life <1 h. On the other hand, the amount of IEX-1 mRNA in cells infected by WT HSV-1(F) remained stable during the first 2 h of treatment, slightly decreasing after 3 h. In ΔUL41 mutant virus-infected cells, the half-life of IEX-1 mRNA was shorter and more similar to that observed in mock-infected cells. In fact, the amount of IEX-1 mRNA in ΔUL41-infected cells was already reduced 1 h after treatment to 60%, dropping to 30% after 3 h (Fig. 3A). The primers specific for TTP (Fig. 3B) indicated that the degradation of TTP mRNA in ΔUL41-infected cells appeared to proceed at slower rate compared to that of IEX-1 mRNA. These results showed that the half-life of the transcripts whose expressions are induced after infection is prolonged in HSV-1 infected cells and is mediated by UL41.
Fig. 3.
Evaluation of IEX-1 and TTP mRNA stability in total RNA. HeLa cells were either mock-infected or exposed to HSV-1(F) or ΔUL41 mutant viruses (10 pfu per cell) for 3 h. The cells were then incubated in medium containing actinomycin D (5 μg/ml). Total cellular RNA was harvested at the indicated time points. IEX-1 (A) and TTP (B) mRNA levels were measured by using real-time PCR and normalized with respect to the amounts of 18S RNA. The amount of total IEX-1 or TTP mRNA remaining in cells at different times are shown as a percentage of the RNA present at the time of addition of actinomycin D.
A second series of experiments was done to specifically examine the accumulation of mRNAs in the cytoplasm. The Northern blot hybridizations shown in Fig. 2 indicate that IEX-1, GADD45β, and TTP mRNAs were either not present or barely detectable in mock-infected cells (lanes 1–5). The mRNAs appeared to be relatively stable in untreated, WT virus-infected (lanes 6–8) or ΔUL41 mutant virus-infected (lanes 11–13) cells. As noted earlier, the amounts of mRNA detected by Northern blots were higher in WT virus-infected cells than in mutant virus-infected cells (compare lanes 6–8 to lanes 11–13). In WT virus-infected cells treated with actinomycin D, the disappearance of TTP, IEX-1, or GADD45β mRNAs was significantly slower than those of mRNAs in actinomycin D-treated ΔUL41 mutant virus-infected cells (compare lanes 9–10 to lanes 14–15). The decreased amounts of cytoplasmic mRNA detected in untreated, ΔUL41 mutant virus-infected cells may be due to a more rapid degradation of the accumulating mRNAs than in WT virus-infected cells.
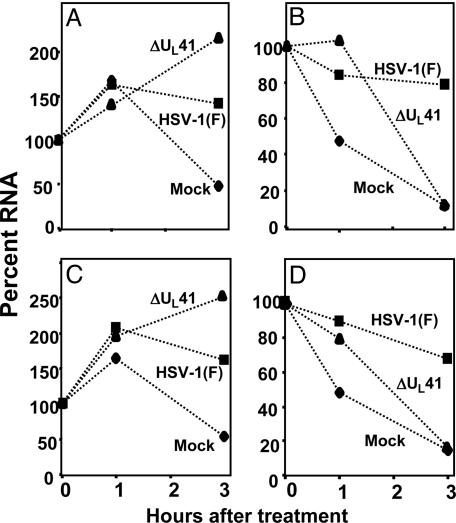
The results of the real-time PCR analyses on cytoplasmic mRNAs shown in Fig. 4 were done with primers for IEX-1 and TTP mRNAs. The overall patterns are similar to those shown in Fig. 3. In the absence of actinomycin D, both IEX-1 and TTP mRNAs accumulate in ΔUL41 mutant virus-infected cells but, the amounts of mRNA accumulating in the cytoplasm of these cells were lower than those detected in WT virus-infected cells. In the presence of actinomycin D, both mRNAs decreased in mock-infected and in ΔUL41 mutant virus-infected cells more precipitously than in WT virus-infected cells.
Fig. 4.
Evaluation of IEX-1 and TTP mRNA stability in cytoplasmic RNA. HeLa cells were either mock-infected or exposed to HSV-1(F) or ΔUL41 mutant viruses (10 pfu per cell) for 3 h. The cells were then mock-treated (A and C) or exposed to actinomycin D (5 μg/ml) (B and D). The cells were harvested and cytoplasmic RNA extracted at indicated time points after treatment. IEX-1 (A and B) and TTP (C and D) mRNA levels were measured by using real-time PCR and normalized with respect to 18S RNA levels. The amount of cytoplasmic IEX-1 or TTP mRNA remaining in cell at different times is given as a percentage of the RNA present at the time of addition of actinomycin D.
The fundamental conclusion from these studies is that steady-state levels of mRNAs induced after infection, as exemplified by TTP, GADD45β, and IEX-1 mRNAs, reflects both synthesis and degradation of the mRNAs, and that the rate of degradation of these mRNAs was faster in the ΔUL41 virus-infected cells than in WT virus-infected cells.
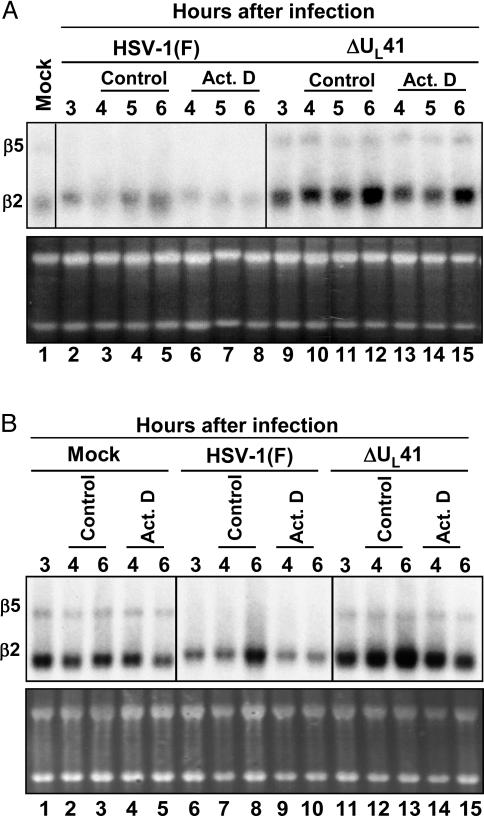
β2- and β5-Tubulin mRNAs Are Differentially Affected in HSV-1-Infected Cells. The results of the microarray analyses previously reported indicated that β2-tubulin (GenBank accession no. X79535) is up-regulated in infected cells at late times after infection (10). β2- and β5-tubulin show 84% sequence identity and differ primarily in the length of the 3′ UTR. We designed a probe template able to detect the two mRNAs and analyzed total and cytoplasmic RNA extracted from infected cells by Northern blot hybridization by using the RNAs described earlier (see Figs. 1 and 2). The pattern of β5-tubulin mRNA accumulation (Fig. 5) was similar to the one observed for the constitutively expressed mRNAs, GAPDH and β-actin (see Fig. 3). Specifically, β5-tubulin mRNA was rapidly degraded in HSV-1(F)-infected cells but accumulated in mock- or in ΔUL41 mutant virus-infected cells. Conversely, β2-tubulin transcript, even if not up-regulated at the time point tested, appeared to be more stable after WT virus infection. β2-tubulin mRNA persisted in the cytoplasm of HSV-1(F)-infected cells even in presence of actinomycin D (Fig. 5B, lanes 9 and 10). These results indicate that the stabilization of cellular mRNAs after HSV-1 infection is not limited to stress-inducible genes. In addition, β2-tubulin mRNA accumulated at higher levels in ΔUL41 mutant virus-infected cells as compared to the amounts detected in mock-infected cells. At this time it is not clear whether the rate of transcription of β2-tubulin is higher in ΔUL41 mutant infected cells or whether the steady-state levels in WT virus-infected cells reflect a low level of degradation of the mRNA.
Fig. 5.
Total and cytoplasmic β2- and β5-tubulin mRNA accumulations in HSV-1(F)- and ΔUL41-infected cells. HeLa cells were either mock-infected or infected with 10 pfu of HSV-1(F) or ΔUL41 mutant virus per cell and incubated for 3 h. Transcription was then stopped by addition of 5 μg of actinomycin D (Act. D) per ml. In parallel, an identical series was incubated in medium containing 0.5% DMSO (control). The cells were harvested, and total (A) or cytoplasmic (B) mRNAs were extracted at the time intervals shown. The amounts of β2- and β5-tubulin transcripts were determined by Northern blotting. The ethidium bromide staining of total and cytoplasmic RNA is shown as a loading control.
Discussion
The UL41 protein enters cells in the course of infection and it is active during the first few hours of infection, before the synthesis of late proteins. The consensus of the literature published before the initiation of our studies is that UL41 mediates indiscriminate degradation of mRNA, 5′ to 3′ either by acting directly or through its association with two cellular proteins, eIF4H and eIF4B (11–13). The key finding that precipitated this and earlier studies is that contrary to published data, UL41 protein does not indiscriminately mediate the degradation of mRNA. The earlier findings (6, 7, 14) indicated that in WT virus-infected cells, some mRNAs containing AREs in their 3′ UTRs were deadenylated and subjected to endonucleolytic cleavage and 3′ to 5′ degradation, in a process that resembles the degradation of AREs-containing mRNAs in uninfected cells (15). However, the major difference was that this process was extremely slow. Each individual degradation step was readily monitored and, moreover, the 5′-truncated mRNA fragments persist in WT virus-infected cells for many hours. These mRNAs, exemplified by IEX-1, IκBα, etc., did not appear to function effectively as mRNA because the protein product was not detectable past 1 h after infection. The 5′-truncated RNA fragments were not detectable in either mock-infected or ΔUL41 mutant-infected cells. We also reported that two other mRNAs, i.e., the AREs-containing TTP mRNA (16) and GADD45β mRNA, which lacks AREs, did not appear to be subjected to similar degradation inasmuch as GADD45β was neither degraded 3′ to 5′ nor deadenylated (7). Moreover, both TTP and GADD45β proteins were readily detected in WT virus-infected cells. In all of these studies, we examined the steady-state mRNA. To dissect the events occurring in ΔUL41 mutant-infected cells, we examined the relative rates of degradation of several of the mRNAs examined in detail in earlier publications. The salient feature of the results may be summarized as follows.
(i) We have examined in detail three constitutively expressed mRNAs in mock-, WT virus-, and ΔUL41 mutant virus-infected cells. These were GAPDH, β-actin, and β5-tubulin. These mRNAs are not induced after infection and, as reported for β-actin mRNA (2), all three mRNAs were rapidly degraded after infection in a manner dependent on UL41. By 3 h after infection, the levels of the mRNAs were significantly below the levels present at the time of infection. Although it is tempting to conclude that they exemplify the fate of constitutively expressed mRNAs only, this may not be the case. As illustrated in Fig. 5, β2-tubulin mRNA appears to be constitutively expressed and is not degraded in WT virus-infected cell.
(ii) IEX-1, c-fos, TTP, IκBα, and GADD45β, etc., mRNAs are induced after infection and accumulate preferentially in the cytoplasm of WT virus-infected cells and to a lesser extent in the cytoplasm of ΔUL41 mutant virus-infected cells. The significant observation reported here is that, in cells infected with the ΔUL41 mutant virus, the rate of degradation of representative mRNAs of this group is rapid and similar to that of mock-infected cells. This observation permits two conclusions: First, it explains the reason why, in contrast to β-actin or GAPDH mRNAs, the IEX-1 mRNA accumulates in smaller amounts in ΔUL41 mutant virus-infected cells than in WT virus-infected cells. The RNAs steady-state level in ΔUL41 mutant virus-infected cells reflects a faster rate of degradation of these mRNAs than in WT virus-infected cells. The second conclusion, based on these data, is that, in WT virus-infected cells, the rate of overall degradation of the mRNAs is decreased relative to that of mock-infected cells or cells infected with the ΔUL41 mutant virus. Consequently, the steady-state levels in WT virus-infected cells are higher than in ΔUL41 mutant-infected cells. Lastly, the results indicate that the fate of the mRNAs is not governed solely by the presence of AREs in the 3′ UTR because GADD45β, β2-tubulin, etc., are bereft of such sequences.
(iii) The question arises as to why some 5′ fragments of some mRNAs linger, whereas others do not, and why some mRNAs are translated, whereas others are not. The unambiguous involvement of the UL41 protein suggests a unifying model whereby UL41 protein interferes with the normal mRNA decay pathway. The first step in decay of mRNAs is removal of the poly(A) tail, which can be catalyzed by several different enzymes. After deadenylation, the body of the mRNA is open to attack from either the 5′ or 3′ ends. In yeast, a 5′- to 3′-decay pathway is initiated by removal of the cap structure by the decapping enzymes Dcp1 and Dcp2 (17), followed by 5′ to 3′ decay by the Xrn1 exonuclease (18). Decay from the 3′ end is catalyzed by a large complex of exonucleases known as the exosome (19). In mammalian cells, the available evidence implies that 3′ to 5′-decay by the exosome is much more active than the decapping 5′ to 3′ pathway (15). In HSV-1-infected cells, UL41 protein could sequester specific cellular nucleases involved in the exosome-dependent 3′ to 5′ degradation of mRNAs. The consequence of such a process would be a decrease in the rate of 3′ to 5′ degradation and selectivity of the degradation based on secondary structure and sequence of the mRNA. This model could explain the presence in the cytoplasm of WT virus-infected cells of cleavage products of IEX-1 mRNA, which is not translated, and their absence in the case of TTP mRNA, which is translated.
(iv) Finally, it seems clear that the shut-off of protein synthesis deduced from the rapid decrease in amino acid incorporation into cellular proteins reflects the degradation of mRNAs like those of β-actin and GAPDH as well as mRNAs up-regulated after infection but not translated (e.g., IEX-1, IκBα, etc.). A question central to the biology of HSV-1 infections is whether the differential fate of GADD45β or TTP mRNAs and the synthesis of these proteins reflect viral functions evolutionarily selected because they confer a benefit to the virus. The studies on the biology of HSV-1 suggest that, in colloquial terms, HSV-1 is a “control freak.” The cumulative evidence to date is that discriminatory functions expressed by HSV-1 benefit the virus.
Acknowledgments
We thank Dr. Beatrice Fineschi and the Biological Sciences Collegiate Division for making available the real-time PCR machine. These studies were aided by National Cancer Institute Grants CA78766, CA71933, CA83939, CA87661, and CA88860 of the U.S. Public Health Service.
Author contributions: A.E. and B.T. designed research, performed research, and analyzed data.
Abbreviations: HSV-1, herpes simplex virus 1; pfu, plaque-forming units; TTP, tristetraprolin; AREs, AU-rich elements.
References
- 1.Roizman, B. & Knipe, D. M. (2001) in Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), pp. 2399–2459.
- 2.Strom, T. & Frenkel, N. (1987) J. Virol. 61, 2198–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Read, G. S. & Frenkel, N. (1983) J. Virol. 46, 498–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwong, A. D. & Frenkel, N. (1987) Proc. Natl. Acad. Sci. USA 84, 1926–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smiley, R. J. (2004) J. Virol. 78, 1063–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taddeo, B., Esclatine, A., Zhang, W. & Roizman, B. (2003) J. Virol. 77, 6178–6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esclatine, A., Taddeo, B., Evans, L. & Roizman, B. (2004) Proc. Natl. Acad. Sci. USA 101, 3603–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ejercito, P. M., Kieff, E. D. & Roizman, B. (1968) J. Gen. Virol. 2, 357–364. [DOI] [PubMed] [Google Scholar]
- 9.Poon, A. P. & Roizman, B. (1997) Virology 229, 98–105. [DOI] [PubMed] [Google Scholar]
- 10.Taddeo, B., Esclatine, A. & Roizman, B. (2002) Proc. Natl. Acad. Sci. USA 99, 17031–17036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karr, B. M. & Read, G. S. (1999) Virology 264, 195–204. [DOI] [PubMed] [Google Scholar]
- 12.Feng, P., Everly, D. N., Jr., & Read, G. S. (2001) J. Virol. 75, 10272–10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doepker, R. C., Hsu W. L., Saffran, H. A. & Smiley, J. R. (2004) J. Virol. 78, 4684–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taddeo, B., Esclatine, A. & Roizman, B. (2004) Biochem. Soc. Trans. 32, 697–701. [DOI] [PubMed] [Google Scholar]
- 15.Mukherjee, D., Gao, M., O'Connor, J. P., Raijmakers, R., Pruijn, G., Lutz, C. S. & Wilusz, J. (2002) EMBO J. 21, 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brooks, S. A., Connolly, J. E. & Rigby, W. F. (2004) J. Immunol. 172, 7263–7271. [DOI] [PubMed] [Google Scholar]
- 17.Steiger, M., Carr-Schmid, A., Schwartz, D. C., Kiledjian, M. & Parker, R. (2003) RNA 9, 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larimer, F. W., Hsu, C. H., Maupin, M. K. & Stevens, A. (1992) Gene 120, 51–57. [DOI] [PubMed] [Google Scholar]
- 19.van Dijk, E., Le Hir, H. & Séraphin, B. (2003) Proc. Natl. Acad. Sci. USA 100, 12081–12086. [DOI] [PMC free article] [PubMed] [Google Scholar]