Abstract
Estrogens play a crucial role in the causation and development of sporadic human breast cancer (BC). Chromosomal instability (CIN) is a defining trait of early human ductal carcinoma in situ (DCIS) and is believed to precipitate breast oncogenesis. We reported earlier that 100% of female ACI (August/Copenhagen/Irish) rats treated with essentially physiological serum levels of 17β-estradiol lead to mammary gland tumors with histopathologic, cellular, molecular, and ploidy changes remarkably similar to those seen in human DCIS and invasive sporadic ductal BC. Aurora-A (Aur-A), a centrosome kinase, and centrosome amplification have been implicated in the origin of aneuploidy via CIN. After 4 mo of estradiol treatment, levels of Aur-A and centrosomal proteins, γ-tubulin and centrin, rose significantly in female ACI rat mammary glands and remained elevated in mammary tumors at 5–6 mo of estrogen treatment. Centrosome amplification was initially detected at 3 mo of treatment in focal dysplasias, before DCIS. At 5–6 mo, 90% of the mammary tumor centrosomes were amplified. Comparative genomic hybridization revealed nonrandom amplified chromosome regions in seven chromosomes with a frequency of 55–82% in 11 primary tumors each from individual rats. Thus, we report that estrogen is causally linked via estrogen receptor α to Aur-A overexpression, centrosome amplification, CIN, and aneuploidy leading to BC in susceptible mammary gland cells.
More than 90% of all human breast cancer (BC) cases are sporadic (1). Numerous epidemiological and animal studies show that both endogenous and exogenously ingested estrogens (Es) play a central, if not paramount, role in the causation and development of human sporadic BC (2–6). Recent epidemiological studies show only a minimal rise in BC risk in postmenopausal women taking E replacement therapy over varying periods of use (7–9). In premenopausal women, however, all of the well established risk factors clearly implicate Es in the causation of BC (2–6). In this latter group, 17β-estradiol (E2) concentrations, all in the low picogram range, within narrow limits of serum and breast tissue levels (10–13), are sufficient to increase sporadic BC risk. Therefore, it is essential to gain a better understanding of how Es, at these physiological concentrations, elicit their oncogenic effects in susceptible target tissues.
Chromosomal instability (CIN) and aneuploidy are defining traits of early human BC ductal carcinoma in situ (DCIS) and primary invasive ductal BCs. These distinguishing characteristics of human BC have been seen in 55–78% of the DCISs and in 85–92% of invasive ductal BCs (14–17). Aneuploidy has been a reliable biomarker for BC for many decades. However, it has not been realized until now that it provides an important clue to the causation of sporadic human BC and the involvement of Es in its etiology.
Overexpression of a centrosome kinase, Aurora-A (Aur-A), centrosome amplification, and CIN invariably occur together (18, 19). Centrosome amplification, found in human BC, may play a key role in the origin of CIN and aneuploidy (20–22). Errors in centrosome duplication/separation, frequently found (>90%) in DCIS and in primary invasive ductal BC, are characterized by the development of supernummery centrosomes resulting in the assembly of multipolar spindles during mitosis (21–23). As a result, the maintenance of the diploid genome is lost through CIN leading to the development of tumor cell heterogeneity in BC. Moreover, it has been suggested that Aur-A may have a role in controlling chromosomal segregation events because it specifically associates with interphase centrosomes, mitotic spindle poles and microtubules, and the spindle midbody (24, 25). Overexpression of Aur-A has been shown to effect neoplastic transformation of mammalian cells, both in vivo and in vitro (25, 26), and occurs with high frequency (>90%) in human DCISs and in primary invasive ductal BCs (27, 28). The studies presented herein link E exposure to the overexpression of Aur-A, γ-tubulin, and centrin, as well as to centrosome amplification, CIN, and aneuploidy, and ultimately to mammary gland tumor (MGT) development. Importantly, these events occur at or nearly physiological, albeit constant, serum E2 concentrations in susceptible murine breast cells.
Materials and Methods
Animals and Treatment. Intact, cycling female ACI (August/Copenhagen/Irish) rats, 6 weeks old and weighing 90–110 g (Harlan–Sprague–Dawley, Indianapolis), were housed in facilities certified by the AAALAC. The rats were acclimated for 1 week before treatment. They were maintained on a 12-h light/dark cycle, fed ad libitum Teklad Rodent Diet 8604 and tap water, and divided into three groups of 20 rats each. Group 1 received either no treatment or a 20-mg pellet of cholesterol. Groups 2 and 3 received either a single E2 pellet containing 2 or 3 mg of E2 plus 18 or 17 mg of cholesterol, respectively. The pellets (Hormone Pellet Press, Shawnee Mission, KS) were implanted in the shoulder region as reported in refs. 5 and 14. The rats were killed at 3, 4, 5, and 6 mo of treatment. Over this period, the serum E2 concentrations ranged from 55 to 85 pg/ml and from 110 to 140 pg/ml after a 2- or 3-mg dose of E2, respectively, as reported in ref. 5. At either E2 dose, 100% MGT incidences were obtained, albeit the MGTs were modestly larger and more numerous at the higher dose. Rats were killed by decapitation and immediately subjected to macroscopic examination for the presence of MGTs, and the number and site were recorded. The abdominal inguinal mammary glands (MGs) were quickly removed. Portions of the MGs and MGTs were immediately frozen in liquid nitrogen and stored at –80°C, whereas others were fixed in 10% buffered formalin, embedded in paraffin, sectioned at 5–6 μm, and stained with hematoxylin/eosin. The spleens of the untreated rats were rapidly placed in 5 ml of RPMI medium 1640 (BioWhittaker) with l-glutamine for immediate processing and cell culture.
Immunoprecipitation and Western Blot Analysis. All immunoblots were performed according to standard procedures by using individual MG and tumor cytosolic fractions from six to eight female ACI rats per group. The tissue samples were homogenized with a polytron in a lysate buffer containing 50 mM Tris·HCl (pH 7.4), 0.2 M NaCl, 2 mM EDTA, and a mixture of protease and phosphatase inhibitors, as reported in ref. 5. The total protein concentration was determined with bicinchoninic acid reagents (Pierce), and equal amounts of protein (100 μg) were resolved by SDS/PAGE and electrotransferred to nitrocellulose membranes. Membranes were probed with a γ-tubulin rabbit polyclonal antibody (H-183, Santa Cruz Biotechnology). Proteins were detected by using chemiluminescence (Amersham Pharmacia). Equal loading was confirmed by Coomassie blue staining and immunolabeling of β-actin (I-19, Santa Cruz Biotechnology) of the same membranes. For Aur-A immunoprecipitation, equal amounts of protein (1,000 μg) were precleared with the appropriate IgG corresponding to the host species of the primary antibody, together with agarose conjugate, and incubated at 4°C for 30 min. Following the standard Santa Cruz Biotechnology protocol, immune complexes were discarded, and the supernatant was incubated by mixing at 4°C overnight with 2 μg of Aur-A-1 antibody (L-18, Santa Cruz Biotechnology) and A/G-agarose beads. Proteins were transferred onto nitrocellulose membranes. For centrin immunoprecipitation, protein aliquots (1,000 μg) were incubated with 2 μg of MC1 anti-centrin (prepared in the laboratory of J.L.S.) and precipitated with A/G agarose as described above. After SDS/PAGE, the gels were treated as described by Errabolu et al. (29). The blots were visualized by chemiluminescence. Densitometry of Western blots and immunoprecipitation analyses were quantitated by using a Molecular Dynamics Personal Densitometer and imagequant software.
RT-PCR. Tissue total RNA was extracted by polytron homogenization, using 1 ml of TRIzol per 100 mg of tissue. After centrifugation, each sample was treated with 0.2 vol of chloroform, shaken, and centrifuged. The total RNA was precipitated with isopropanol and dissolved in diethyl pyrocarbonate-water. RNA concentration was determined by 260-nm absorbance. The exponential range of amplification was determined by varying the number of PCR cycles for each cDNA and a set of two primers, forward 5′-GGCGAATGCTTT GTCCTACT and reverse 5′-CCGTCACAAAGTCAGGGAAT. These primers represent a specific sequence of the catalytic domain of Aur-A. RT-PCR was conducted by using the Invitrogen standard protocol for Moloney murine leukemia virus reverse transcriptase and Taq recombinant DNA polymerase, according to the manufacturer's instructions. PCR products were separated on 1.5% agarose gels and visualized with ethidium bromide. The PCR product size was 350 bases. For actin, the forward primer was GGCATCCTGACCCTGAAGTA, the reverse primer was GCCATCTCTTGCTCGAAGTC, and the PCR product was 497 bases.
Centrosome Amplification: Number and Volume. Centrosome amplification was assessed by confocal microscopy of paraffin sections immunolabeled with a monoclonal antibody against the centrosomal protein γ-tubulin (Sigma clone gtu-88C) (30, 31). Determinations made were based on average values of centrosomes in four randomly selected fields of view. A minimum of 100 centrosomes were analyzed from untreated ACI rat MG epithelial tissue, ductal cells with and without dysplasia, cells confined to DCIS, and primary E2-induced tumors, derived from three to five individual rats. Although fields of E2-induced primary MGT cells largely exhibited uniform centrosome amplification, robust centrosome amplification in fields of lobular–alveolar hyperplasia was only seen when it coincided with groups of focal dysplastic cells. Centrosomes were scored as amplified if they were larger in size and/or number than seen in normal, untreated rat mammary epithelial tissue. Centrosome area (size) was measured in maximum intensity projections of seven consecutive 0.5-μm optical sections. Centrosomes were scored as amplified in size when their measured area was more than two times the average seen in untreated MGs. A cutoff of 2 times larger was established to exclude centrosomes equivalent in size to late G2 centrosomes of normal tissues. Additionally, centrosomes were scored as amplified in number when a cluster of more than two centrosomes was associated with a single nucleus.
Spleen Culture and Chromosome Preparation. Spleens from untreated, female ACI rats were cultured as reported in ref. 14. Cell culture suspensions (25 ml) were incubated at 37°C for 4–5 days, with 150 μl of Con A and 30 μl of 0.5% 2-mercaptoethanol. Before harvesting, the spleen cell suspensions were treated with colcemid.
DNA Isolation and Labeling for Comparative Genomic Hybridization (CGH) Analysis. Individual untreated MGs and MGTs were quickly frozen in liquid N, and the DNA was extracted by the LiCl protocol (32). A nick translation kit (Vysis) was used for direct DNA labeling for CGH, according to the manufacturer's recommendations. The probe preparation, hybridization, and posthybridization steps were carried out according to the University of Colorado Health Science Center, Cancer Center Cytogenetics Core FISH Protocol 12 “CGH with Directly Labeled Probes.” CGH analyses were performed in an average of 10 metaphase spreads per individual MGT as we have reported (33). Chromosomes were identified by using digitally inverted images of DAPI-banded metaphases and an ACI rat idiogram implemented in the image analysis software. For CGH detection of regional gains and losses, thresholds of 1.20 and 0.80 for over- and underrepresentation were used, respectively. Ratio profiles were generated with the cgh package of quips software (Vysis) and displayed along with idiograms of rat chromosomes. Fluorescence ratio values exceeding the thresholds were regarded as copy gains (fluorescence ratio >1.2) or losses (fluorescence ratio <0.8). As a precaution against region- or band-specific ratio fluctuations, CGH hybridizations were verified by exchanging fluorescent labels in the tumor and reference DNA, as described in refs. 34 and 35. For CGH analyses, a criterion of ≥30% frequency of occurrence within any given tumor was considered as a nonrandom/consistent amplified/deleted region.
Statistical Analysis. The significance of differences in protein/RNA expression and centrosome amplification between experimental groups was determined by using Student's t test. The data for the CGH analysis were analyzed by the exact binomial distribution test that determines the occurrence/nonoccurrence of an event.
Results
Aur-A Kinase mRNA and Protein Expression. Aur-A mRNA (Fig. 1A) and protein (Fig. 1B) expression were assessed in control MGs and primary MGTs from 4-mo E2-treated ACI rats. Employing RT-PCR, a 1.4- and a 1.5-fold increase in Aur-A mRNA was detected after 4 mo of E2 treatment and in primary MGTs, respectively, compared with age-matched untreated MGs (Fig. 1 A). Western blot analyses showed a 7.2-fold increase in Aur-A protein expression after 4 mo of E2 treatment, compared with age-matched control MGs, whereas primary MGTs exhibited a 7.4-fold rise (Fig. 1B).
Fig. 1.
Expression of Aur-A gene (A) and protein (B) in untreated MGs, E2-treated MGs, and E2-induced MGTs. Representative individual samples of age-matched, untreated control (MC1–2), 4.0-mo (ME41–2), and E2-induced MGTs (MGT1–2) were used. (A) Electrophoretic image of RT-PCR of Aur-A and β-actin mRNA, used as an internal control. RNA processing, primer details, and RT-PCR conditions are described in Materials and Methods. B Upper shows a Western blot of the relative expression of immunoprecipitated Aur-A in the same treatment groups. B Lower shows the Aur-A mRNA (▪) and protein (□) relative expression. The values represent the mean ± SE (n = 8).
Centrosome Amplification. Untreated cycling female ACI rat MG epithelial tissue exhibited normal position and complement of centrosomal protein γ-tubulin by immunofluorescence staining. The γ-tubulin labeling was confined to the pair of centrioles apical to the nucleus and proximate to the luminal membrane (Fig. 2A). All untreated ACI rat MG tissues showed comparable levels of γ-tubulin immunostaining, including epithelial, myoepithelial, and stromal cells. Similarly, unaffected MG tissue adjacent to E2-induced MGTs also exhibited normal centrosome staining distribution. However, E2-induced MGTs showed a markedly elevated number of centrosomes, which were larger and lacking the organized distribution seen in untreated control MG cells (Fig. 2 F and F′). Centrosome amplification was evident in dysplasia as early as 3moafterE2 treatment (Fig. 2 B, B′, C, and C′). After 4 and 5 mo of E2 treatment, a significant increase (P < 0.002) in centrosome amplification was detected in groups of cells residing in DCISs (Fig. 2 D, D′, E, and E′). However, other cells within the same DCISs exhibited a normal complement of centrosomes. The DCISs after 4 and 5 mo of E2 treatment were predominantly cribriform (ER+/PR+), whereas papillary, comedo, and solid types were less common. With respect to the γ-tubulin staining marker (Fig. 3), 30% of centrosomes were amplified in areas of dysplasias at 3 mo (P < 0.002), whereas 38% were amplified in DCIS after 4 mo (P < 0.002). In primary MGTs, ≈90% of the centrosomes observed were amplified (P < 0.001). Conversely, in ducts without atypia, only 7% of the centrosomes were amplified after 3 mo of E2 treatment. These results indicate that centrosome amplification is an early event that becomes more pronounced during progression to frank MGTs.
Fig. 2.
Centrosome amplification. Centrosomes and nuclei were observed by confocal microscopy of sections labeled with an antibody against γ-tubulin (red) and DNA dye Hoechst 33342 (blue) (A′–F′) in areas corresponding to hematoxylin/eosin-stained sections (A–F). (A and A′) Untreated rats. Centrosomes in ductal cells are apical to the nucleus and often appear as a pair of adjacent spots. The size of the centrosome spots, including those in the fibroblast at the bottom of A′, is uniform. (B and B′) dysplasia after 3.0 mo of E2 treatment. Centrosomes are often larger than those from untreated rats. (C and C′) Dysplasia after 4 mo of treatment. Centrosomes are often amplified in number and size. (D and D′) DCIS after 5 mo of treatment. Centrosome amplification in size and number is evident. (E and E′) DCIS after 6 mo of treatment. Many centrosomes are much larger and often more numerous than centrosomes in control tissues. (F and F′) E2-induced MGT. Centrosomes are consistently amplified in both size and number. (Black scale bar, 45 μm; white scale bar, 20 μm.)
Fig. 3.
The percentage of amplified centrosomes relative to untreated MG controls in ductal cells without atypia after 3 mo of E2 treatment is 7% compared with 30% in dysplastic cells (DYSPL) after 3 mo of treatment. Amplification increased from 30% to 38% in DCIS after 4 and 5 mo of E2 treatment, respectively, while 90% of MGT centrosomes were amplified.
Centrosome Protein Expression. A single γ-tubulin band was evident in untreated MGs (Fig. 4A). Two forms of γ-tubulin (γf: fast, 50 kDa; and γs: slow, 52 kDa) were observed in MGTs. After 4 mo of E2-treatment, the γf-tubulin was reduced and the γs-tubulin was increased (Fig. 4A). In E2-induced MGTs, a 2.4-fold rise was observed in total (γf + γs) γ-tubulin when compared with untreated MGs, whereas a rise of 1.4-fold was detected after 4 mo of E2 treatment. Centrin levels were increased 11.2-fold in the MGTs compared with untreated MGs (Fig. 4B). These results are consistent with the centrosome amplification detected in both E2-induced DCISs and primary MGTs reported here.
Fig. 4.
Immunoblot analysis of the relative expression of γ-tubulin (A) and centrin (B) in untreated MGs, E2-treated MGs, and E2-induced MGTs. Whole-cell lysates from HeLa cells (HC), used as a positive control; age-matched untreated control, MC; 4-mo treated, ME41–2; and E2-induced MGT1–2 were prepared. (A) For γ-tubulin, 100-μg protein fractions were examined by Western analysis. (B) For centrin, 1,000-μg protein fractions were immunoprecipitated as described in Material and Methods. Lower shows the relative expression of γs-tubulin + γf-tubulin (▪) and centrin (□). The values represent the mean ± SE (n = 6).
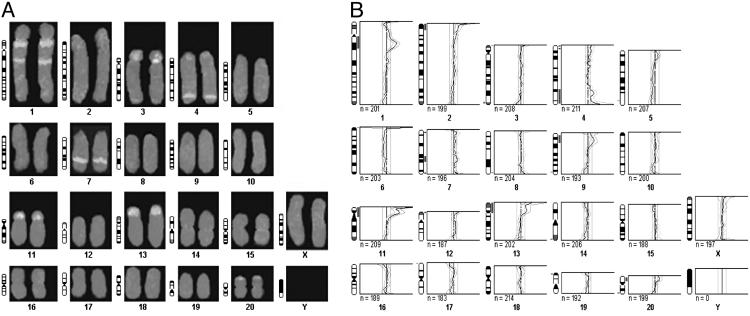
CIN: CGH Analysis. CGH analyses, employing MGT DNA from female ACI rats, showed consistent regional genomic alterations in 11 E2-induced MGTs derived from individual rats. Analysis of 10–12 CGH metaphase spreads from each E2-induced primary MGT revealed nonrandom amplified regions in chromosomes 1, 4, 7, 9, 11, 13, and 20. The consistently amplified regions seen in all MGTs examined were present in 54.5–81.9% of the metaphase spreads analyzed (Table 1). Amplification of chromosomal regions 1q21–22 and 7q33 include the loci for cyclin E1 (D.P., unpublished work) and c-myc (14), respectively (Fig. 5 A and B and Table 1). These results indicate that cyclin E1, in addition to c-myc, may also be amplified in E2-induced MGTs.
Table 1. Frequency of genomic alterations determined by CGH analysis in female ACI rat E2-induced mammary tumors.
| Chromosome | Genetic alteration | Region* | Locus* | Ratio† | Frequency, % | P value‡ |
|---|---|---|---|---|---|---|
| 1 | q11-q22 | 1q21-22 | Ccne1 | 7/11 | 63.6 | 0.017 |
| 4§ | q41-q44 | 4q32-44 | Ccnd2 | 7/11 | 63.6 | 0.005 |
| 7§ | q31-q33 | 7q33 | c-myc | 9/11 | 81.9 | 0.005 |
| 9 | q11-q13 | 6/11 | 54.5 | 0.056 | ||
| 11§ | p-q11 | 6/11 | 54.5 | 0.056 | ||
| 13§ | q | 7/11 | 63.6 | 0.017 | ||
| 20§ | p12 | 9/11 | 81.9 | 0.005 |
Eleven individual MTs were examined with an average of 10 metaphases per MT.
From Entrez Genome, Ratus norvegicus Map View (www.ncbi.nlm.nih.gov/mapview/maps.cgi).
Frequencies ≥30%.
Analyzed by binomial distribution.
Chromosomal alterations identified by karyotype analysis.
Fig. 5.
CGH of an ACI rat mammary gland tumor. (A) Representative CGH rat spleen karyotype employing female ACI rat E2-induced primary MGTs. DNA was detected in green (SpectrumGreen FITC, Vysis) and untreated MG DNA in red (SpectrumRed TRITC, Vysis). Note the regional gains on chromosomes 1, 3, 4, 7, 11, 13, and 20. (B) Ratio profiles of green-to-red fluorescence intensities after CGH from female ACI rat E2-induced MGTs. Profiles from 11 individual MGTs were normalized to an average green-to-red ratio of 1.0. The right and left lines depict the upper and lower threshold of 1.2 for overrepresentation (copy gain) and 0.80 for underrepresentation (copy loss), respectively.
Discussion
Centrosome amplification, CIN, and aneuploidy are striking features of human DCIS and BC (21, 22, 30, 14–17). Recently, we have shown that Fisher and SD female rats administered with synthetic chemical and environmental carcinogens yielded primarily diploid MGTs, whereas MGTs induced by E2 alone in ACI rats were highly aneuploid (14). The molecular alterations reported here, in E2-treated female ACI rat MGs and MGTs, which precede aneuploidy, are similar to those seen in early preinvasive human sporadic breast lesions and thus likely to be distinctively related to the causation of this disease. The prevention of E2-induced MGTs in female ACI rats by the concomitant administration of tamoxifen (5) strongly indicates that the MGTs induced are driven and mediated by E via ERα. Moreover, both histological changes and the induction of ERα and progesterone receptor isoforms were also prevented by the concomitant tamoxifen treatment (5).
The high frequencies of E2-mediated centrosome amplification observed in dysplasias, DCISs, and primary MGTs in female ACI rats reported herein link these centrosome defects to the CIN and aneuploidy in DCISs and MGTs shown previously by us (14). Furthermore, the centrosome amplification found in most but not all cells within a given DCIS suggests that the development of these MGTs is clonal. Interestingly, hamster tumors in the kidney and their early tumorous lesions induced by E treatment alone also exhibited high frequencies of centrosome amplification, CIN, and aneuploidy (33, 36, 37). These molecular changes appear to be common features of E-induced oncogenic processes. Although the precise sequence of these events leading to tumor aneuploidy is unresolved (26, 38, 39), it is evident that there is an early loss of normal centrosome homeostasis, shown here, in E2-mediated dysplasias, DCISs, and primary MGTs in female ACI rats. This process may involve the E-mediated c-myc overexpression and the down-stream deregulation of the cell cycle (40), indicated by the overexpression of cyclin E.cdk2 and eventual amplification of c-myc (14). This sequence is similar to that reported by us in the E2-induced tumors in the hamster kidney (37, 41). These data suggest there is an intimate causal relationship between the deregulation of cell cycle components and centrosome amplification, leading to CIN. Consistent with our findings, MYC protein overexpression elicits CIN and increased tumorigenicity in rat 1A cells (42). In these cells, ectopic expression of c-myc perturbs the coupling of DNA replication and mitosis (43). Moreover, rat 1A-Myc ER cells in the presence of E exhibited irreversible chromosomal aberrations, including numerical changes (44). Downstream, overexpression of cyclin E.cdk2, but not cyclin D1 or A, in immortalized rat embryo fibroblasts and human breast epithelial cells resulted in CIN (45).
Centrosome-associated kinases are key regulators of centrosome maturation, chromosome segregation, and cytokinesis (18, 46–48). Overexpression of Aur-A in NIH 3T3 cells has been shown to induce centrosome amplification, aneuploidy, and transformation (25). After ectopic overexpression of Aur-A in MCF-7 cells, similar changes were detected (26). These results clearly indicate that Aur-A when overexpressed behaves as an oncogene. Aur-A overexpression has been seen in 94% of 33 invasive ductal BC samples, irrespective of the histopathology type, when compared with normal ductal breast tissue (27). This finding is comparable to the frequency of Aur-A overexpression seen here in solely E2-induced primary ACI rat MGTs. The finding that in 4-mo E2-treated MGs, Aur-A expression, both mRNA and protein, rose to a level essentially equal to that of primary MGTs indicates that this increase is likely due to the coincident rise in DCISs found at this treatment interval. These data indicate that the high levels of Aur-A in DCISs may be a crucial event during early MGT development. Our finding is consistent with a recent report showing markedly high levels of Aur-A in human breast DCISs (49).
Although Aur-A overexpression and centrosome amplification were detected in N-nitrosourea-induced rat MGTs (50), the frequency of occurrence of these changes in synthetic chemical carcinogen-induced MGTs would be very low because the vast majority (>85%) of these MGT cells are diploid (14, 51, 52). It remains to be seen whether other centrosomal kinases (18, 27, 46, 48, 53, 54) might also be involved in effecting centrosome amplification in solely E2-induced murine breast oncogenesis.
The presence of a single γ-tubulin isoform in control tissue and two γ-tubulin isoforms in E2-treated MGs and MGTs may be due to differential posttranslational modification, as has been reported in non-mammalian species (55–57). These two γ-tubulins have been implicated in having distinct roles in nucleation, organization, and stabilization of microtubules based on their differential binding to centrosomes and to mitotic spindle poles (56). The slow γ-tubulin isoform seen in the present study in MGTs may explain the increased microtubule nucleating capacity of amplified centrosomes observed in human breast tumors (30).
The CIN and aneuploidy generated by E2-induced ACI rat MGTs yielded both nonrandom and random chromosome changes. The consistent regional chromosome alterations detected by CGH reported here largely coincided with the numerical chromosome alterations (i.e., gains in chromosomes 7, 11, 13, 19, and 20) detected by conventional karyotype analyses, described earlier by us, for primary ACI rat MGTs (14). Remarkably, the overexpression and amplification of the c-myc gene was found in two distinctive tumors that have in common E as the sole etiologic agent (14, 37). Similar alterations in the c-myc gene have been commonly found in human breast DCISs and invasive ductal BCs (58, 59). A sequence of cascading events is proposed for ACI rat breast oncogenesis, beginning with E interacting with its receptor, ERα, followed by c-myc/MYC protein overexpression, subsequent cyclin E.cdk2 and Aur-A overexpression, centrosome amplification, CIN, aneuploidy, and ultimately BC. Thus, E is intimately linked for the first time to these aforementioned molecular changes leading to tumor development. These findings may provide targets for the prevention and therapeutic intervention of human sporadic BC.
Acknowledgments
This work was supported by National Institutes of Health–National Cancer Institute Grants CA87591 (to S.A.L.), CA102849 (to J.J.L.), and CA72836 (to J.L.S.) and Department of Defense Breast Cancer Research Program Grant DAMD 17-01-1-0753 (to W.L.L.).
Author contributions: J.J.L. designed research; S.J.W., W.L.L., and D.P. performed research; J.L.S. contributed new reagents/analytic tools; J.J.L., S.J.W., W.L.L., J.L.S., and S.A.L. analyzed data; and J.J.L., W.L.L., J.L.S., and S.A.L. wrote the paper.
Abbreviations: Aur-A, Aurora-A; BC, breast cancer; CGH, comparative genomic hybridization; CIN, chromosomal instability; DCIS, ductal carcinoma in situ; E, estrogen; E2, 17β-estradiol; MG, mammary gland; MGT, MG tumor.
References
- 1.King, M.-C., Rowell, S. & Love, S. M. (1993) J. Am. Med. Assoc. 269, 1975–1980. [PubMed] [Google Scholar]
- 2.Clemons, M. & Gross, P. (2001) N. Engl. J. Med. 344, 276–285. [DOI] [PubMed] [Google Scholar]
- 3.Feigelson, H. S. & Henderson, B. E. (1996) Carcinogenesis 17, 2279–2284. [DOI] [PubMed] [Google Scholar]
- 4.Adami, H.-O., Persson, I., Ekbom, A., Wolk, A., Ponten J. & Trichopoulas, D. (1995) Mutat. Res. 333, 29–35. [DOI] [PubMed] [Google Scholar]
- 5.Li, S. A., Weroha, S. J. & Li, J. J. (2002) J. Endocrinol. 175, 297–305. [DOI] [PubMed] [Google Scholar]
- 6.Blankenstein, M. A., Broerse, J. J., deVries, J. B., vandenBerg, K. K., Knaan, S. & van der Molen, H. J. (1977) Eur. J. Cancer 13, 1437–1443. [DOI] [PubMed] [Google Scholar]
- 7.Daling, J. R., Malone, K. E., Doody, D. R., Voigt, L. F., Bernstein, L., Coates, R. J., Marchbanks, P. A., Norman, S. A., Weiss, L. K., Ursin, G., et al. (2002) Cancer 95, 2455–2464. [DOI] [PubMed] [Google Scholar]
- 8.Li, C. I., Weiss, N. S., Stanford, J. L. & Daling, J. R. (2000) Cancer 88, 2570–2577. [DOI] [PubMed] [Google Scholar]
- 9.Colditz, G. A., Stampfer, M. J., Willett, W. C., Hennekens, C. H., Rosner, B. & Speizer, F. E. (1990) J. Am. Med. Assoc. 264, 2648–2653. [PubMed] [Google Scholar]
- 10.Chetrite, G. S., Cortes-Prieto, J., Philippe, J. C., Wright, F. & Pasqualini J. R. (2000) J. Steroid Biochem. Mol. Biol. 72, 23–27. [DOI] [PubMed] [Google Scholar]
- 11.Vermeulen, A., Deslypere, J. P., Paridaens, R., LeClereq, G., Roy, F. & Heuson, J. C. (1986) Eur. J. Cancer 22, 515–525. [DOI] [PubMed] [Google Scholar]
- 12.Thijssen, J. H. H. & Blankenstein, M. A. (1989) Eur. J. Cancer Clin. Oncol. 25, 1933–1959. [DOI] [PubMed] [Google Scholar]
- 13.Thijssen, J. H. H., van Landeghem, A. A. J. & Poortman, J. (1986) Ann. N.Y. Acad. Sci. 484, 106–116. [DOI] [PubMed] [Google Scholar]
- 14.Li, J. J., Papa, D., Davis, M. F., Weroha, J. S., Aldaz, C. M., El-Bayoumy, K., Ballenger, J., Tawfik, O. & Li, S. A. (2002) Mol. Carcinog. 3, 56–65. [DOI] [PubMed] [Google Scholar]
- 15.Arnerlov, C., Emdin, S. O., Cajander, S., Bengtsson, N.-O., Tavelin, B. & Roos, G. (2001) Anal. Cell. Pathol. 23, 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makris, A., Allred, D. C., Powles, T. J., Dowsett, M., Fernando, I. N., Trott, P. A., Ashley, S. E., Ormerod, M. G., Titley, J. C. & Osborne, C. K. (1997) Breast Cancer Res. Treat. 44, 65–74. [DOI] [PubMed] [Google Scholar]
- 17.Leal, C. B., Schmitt, F. C., Bento, M. J., Maia, N. C. & Lopez, C. S. (1995) Cancer 75, 2123–2131. [DOI] [PubMed] [Google Scholar]
- 18.Dutertre, S., Descamps, S. & Prigent, C. (2002) Oncogene 21, 6175–6183. [DOI] [PubMed] [Google Scholar]
- 19.Kramer, A., Neben, K. & Ho, A. D. (2002) Leukemia 16, 767–775. [DOI] [PubMed] [Google Scholar]
- 20.Pihan, G. A., Wallace, J., Zhou, Y. & Doxsey, S. J. (2003) Cancer Res. 63, 1398–1404. [PubMed] [Google Scholar]
- 21.Lingle, W. L., Barrett, S. L., Negron, V. C., D'Assoro, A. B., Boeneman, K., Liu, W., Whitehead, C. M., Reynolds, C. & Salisbury, J. L. (2002) Proc. Natl. Acad. Sci. USA 99, 1978–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D'Assoro, A. B., Lingle, W. L. & Salisbury, J. L. (2002) Oncogene 21, 6146–6153. [DOI] [PubMed] [Google Scholar]
- 23.Salisbury, J. L. (2001) J. Mamm. Gland Biol. Neoplasia 6, 203–212. [DOI] [PubMed] [Google Scholar]
- 24.Katayama, H., Brinkley, W. R. & Sen, S. (2003) Cancer Met. Rev. 22, 451–464. [DOI] [PubMed] [Google Scholar]
- 25.Brinkley, B. R., Goepfert, T. M. (1998) Cell Motil. Cytoskeleton 41, 281–288. [DOI] [PubMed] [Google Scholar]
- 26.Zhou, H., Kuang, J., Xhow, L., Kuo, W. L., Gray J. W., Suhin, A., Brinkley, B. R. & Sen, S. (1998) Nat. Genet. 20, 189–193. [DOI] [PubMed] [Google Scholar]
- 27.Miyoshi, Y., Iwao, K., Egawa, C. & Noguchi, S. (2001) Int. J. Cancer 92, 370–373. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka, T., Kimura, M., Matsunaga, K., Fukada, D., Mori, H. & Okano, Y. (1999) Cancer Res. 59, 2041–2044. [PubMed] [Google Scholar]
- 29.Errabolu, R., Sanders, M. A. & Salisbury, J. L. (1994) J. Cell Sci. 107, 9–16. [DOI] [PubMed] [Google Scholar]
- 30.Lingle, W. L., Lutz, W. H., Ingle, J. N., Maihle, N. J. & Salisbury, J. L. (1998) Proc. Natl. Acad. Sci. USA 96, 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carroll, P. E., Okuda, M., Horn, H. F., Biddinger, P., Stambrook, P. J., Gleich, L. L., Li, Y. Q., Tarapore, P. & Fukasawa, K. (1999) Oncogene 18, 1935–1944. [DOI] [PubMed] [Google Scholar]
- 32.Gemmell, N. J. & Akyiama, S. (1996) Trends Genet. 12, 338–339. [DOI] [PubMed] [Google Scholar]
- 33.Papa, D., Li, S. A. & Li, J. J. (2003) Mol. Carcinog. 38, 97–105. [DOI] [PubMed] [Google Scholar]
- 34.Kallioniemi, A., Kallioniemi, O.-P. & Piper, J. (1994) Genes Chromosomes Cancer 10, 231–243. [DOI] [PubMed] [Google Scholar]
- 35.Mohapatra, G., Kim, D. H. & Feuerstein, B. G. (1995) Genes Chromosomes Cancer 13, 86–93. [DOI] [PubMed] [Google Scholar]
- 36.Li, J. J., Weroha, S. J., Davis, M. F., Hou, X., Tawfik, O. & Li, S. A. (2001) Endocrinology 142, 4006–4014. [DOI] [PubMed] [Google Scholar]
- 37.Li, J. J., Hou, X., Banerjee, S. K., Liao, D. J., Maggouta, F., Norris, J. S. & Li, S. A. (1999) Cancer Res. 59, 2340–2346. [PubMed] [Google Scholar]
- 38.Salisbury, J. L., Whitehead, C. M., Lingle, W. L. & Barrett, S. L. (1999) Biol. Cell 91, 451–460. [PubMed] [Google Scholar]
- 39.Li, R., Sonik, A., Stindl, R., Rasnick, D. & Duesberg, P. (2000) Proc. Natl. Acad. Sci. USA 97, 3236–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weroha, J. S. (2004) Proc. Am. Assoc. Cancer Res. 45, 12. [Google Scholar]
- 41.Liao, D. J., Hou, X., Bai, S., Li, S. A. & Li, J. J. (2000) Carcinogenesis 21, 2167–2173. [DOI] [PubMed] [Google Scholar]
- 42.Felsher, D.W. & Bishop, J. M. (1999) Proc. Natl. Acad. Sci. USA 96, 3940–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, Q. & Dang, C. V. (1999) Mol. Cell. Biol. 19, 5339–5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mai, S., Fluri, M., Siwarski, D. & Hupp, I. (1996) Chromosome Res. 4, 365–371. [DOI] [PubMed] [Google Scholar]
- 45.Spruck, C. H., Won, K.-A. & Reed, S. I. (1999) Nature 401, 297–300. [DOI] [PubMed] [Google Scholar]
- 46.Nigg, E. A. (2001) Nat. Rev. Mol. Cell Biol. 2, 21–32. [DOI] [PubMed] [Google Scholar]
- 47.Warner, S. L., Bearss, D. J., Han, H. & Von Hoff, D. D. (2003) Mol. Cancer Ther. 2, 589–595. [PubMed] [Google Scholar]
- 48.Carmena, M. & Earnshaw, W. C. (2003) Nat. Rev. Mol. Cell Biol. 4, 842–854. [DOI] [PubMed] [Google Scholar]
- 49.Hoque, A., Carter, J., Xia, W., Hung, M. C., Sahin, A. A., Sen, S. & Lippman, S. M. (2003) Cancer Epidemiol. Biomarkers Prev. 12, 1518–1522. [PubMed] [Google Scholar]
- 50.Goepfert, T. M., Adigum, Y. E., Zhong, L., Gay, J., Medina, D. & Brinkley, W. R. (2002) Cancer Res. 62, 4115–4122. [PubMed] [Google Scholar]
- 51.Aldaz, C. M., Gollahon, L. S. & Chen, A. (1992) Prog. Clin. Biol. Res. 376, 137–153. [PubMed] [Google Scholar]
- 52.Haag, J. D., Hsu, L. C., Newton, M. A. & Gould, M. N. (1996) Mol. Carcinog. 17, 134–143. [DOI] [PubMed] [Google Scholar]
- 53.Smith, M. R., Wilson, M. L., Hamanaka, R., Chase, D., Kung, H., Longo D. L. & Ferris, D. K. (1997) Biochem. Biophys. Res. Commun. 234, 397–405. [DOI] [PubMed] [Google Scholar]
- 54.Fry, A. M. (2002) Oncogene 21, 6184–6194. [DOI] [PubMed] [Google Scholar]
- 55.Oakley, C. E. & Oakley, B. R. (1989) Nature 338, 662–664. [DOI] [PubMed] [Google Scholar]
- 56.Lajoie-Mazenc, I., Detraves, C., Rotaru, V., Gares, M., Tollon, Y., Jean, C., Julian, M., Wright, M. & Raynaud-Messina, B. (1996) J. Cell Sci. 109, 2483–2492. [DOI] [PubMed] [Google Scholar]
- 57.Starita, L. M., Machida, Y., Sankaran, S., Elias, J. E., Griffin, K., Schlegel, B. P., Gygi, S. P. & Parvin, J. D. (2004) Mol. Cell. Biol. 24, 8457–8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hynes, N. E. & Lane, H. A. (2001) J. Mamm. Gland Biol. Neoplasia 6, 141–150. [DOI] [PubMed] [Google Scholar]
- 59.Deming, S. L., Nass, S. J., Dickson, R. B. & Trock, B. J. (2000) Br. J. Cancer 83, 1688–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]