Abstract
In eukaryotes, the initiation of DNA replication requires the interaction between origin sequences and the origin recognition complex (ORC), which is highly conserved. In this report, atomic force microscopy (AFM) was used to examine the binding of Schizosaccharomyces pombe (sp) ORC and the spOrc4 protein with the sp autonomously replicating sequence 1 (ars1). AFM imaging revealed that spORC binding to ars1 occurred solely through spOrc4p and depended on the N-terminal AT-hook domains present in spOrc4p. At high molar ratios of spORC (or spOrc4p alone) to DNA (6:1), all of the input ars1 was bound in a one protein complex to one plasmid manner. Restriction digestion and AFM analysis of protein–DNA fragments revealed the presence of two binding sites in ars1. One site mapped to a region centered at nucleotide 838 of ars1 previously detected by DNase I protection that was reported to be essential for the autonomously replicating sequence activity of ars1. The second site mapped to a previously uncharacterized region centered at nucleotide 1148. AFM showed that the length of the DNA fragment complexed with either spORC or spOrc4p was shortened by ≈140 bp, suggesting the wrapping of two turns of the DNA around the spOrc4p alone as well as the spOrc4p in spORC. We also show that treatment of the spORC (spOrc4p)–ars1 complex with topoisomerase I induced a negative shift in the topoisomer distribution. These findings suggest that the binding of spORC to origin DNA alters the structure of the DNA. Thus, in the case of spORC, due to its unusual spOrc4p, at least two factors are likely to influence ars1 activation. These include the selective binding of the complex to A- and T-rich regions and the alteration of the DNA structure due to its wrapping around spOrc4p.
Keywords: atomic force microscopy, topoisomers, replication
Chromosomal replication in eukaryotes requires the origin recognition complex (ORC), a heteromeric six-subunit complex that acts as the initiator for the assembly of other key initiator factors such as Cdc18 (Cdc6), Cdt1, and the MCM2–7 complex (the probable replicative helicase) (1). The ORC was isolated initially from Saccharomyces cerevisiae (Sc) based on its origin binding activity (2). Replication origins of Sc are relatively short AT-rich sequences (≈150 bp) that include an essential autonomously replicating sequence (ARS) with an 11-bp consensus sequence (ACS) (3–5) that acts as the key binding site for ScORC in a reaction requiring ATP (2). Additional sequences in Sc ARSs, referred to as B domains, enhance the efficiency of origin activity (3, 6).
The structure and initiator role of ORC is conserved in all eukaryotes, and six-subunit complexes have been purified from species including Schizosaccharomyces pombe (sp), Drosophila melanogaster (Dm), Xenopus laevis, and humans (7–11). In vitro and in vivo studies indicate that although ORC is essential for DNA replication in these species (1), the sequences recognized by these ORC complexes differ. In metazoans, initiation of replication occurs at multiple sites extended over broad replication zones, suggesting that sequences required for initiation may be less specific than in budding yeast (12, 13). Detailed studies with a well characterized DmORC have not revealed any DNA sequence binding specificity other than a propensity to bind to AT-rich regions and an increased association with negatively supercoiled DNA (14). Like the ScORC, ATP is required for these interactions.
Specific replication origins have been detected in sp, but they differ from those of Sc. They range between 0.5 and 1 kbp in length, lack a consensus sequence analogous to the ARS A element that is essential for ScORC binding, and contain redundant AT-rich regions (15–17). The spORC possesses a unique Orc4p that contains multiple AT-hook domains that bind specifically to the minor groove of AT-rich regions (18). Previous studies showed that spORC binding to different origin sequences is mediated solely through the N-terminal AT-hook domains of Orc4p and that these interactions were not dependent on ATP (7, 18–20). A stable five-subunit spORC lacking Orc4p has been isolated as well as a six-subunit spORC containing an Orc4p derivative devoid of the N-terminal AT-hook domains (7, 19, 20). Such spORC derivatives do not bind to sp origin DNAs. Furthermore, the spOrc4p alone binds to DNA as efficiently as spORC. The importance of the N-terminal AT hook domain was further substantiated by the finding that this region of spOrc4p is essential for viability in vivo (8).
In this report, the interaction of spOrc4p and the six-subunit spORC with the AT-rich spars1 DNA was examined by using atomic force microscopy (AFM). Similar to previous reports, AFM studies indicated that the AT-hook domain of spOrc4p is essential for DNA binding. AFM measurements also revealed that the length of restriction fragments bound to spOrc4p or spORC were shorter than identical free fragments. The decrease in length of the DNA fragment bound by spOrc4p alone or by the six-subunit ORC was identical, suggesting that DNA is wrapped solely about the spOrc4p subunit in both complexes. In keeping with the wrapping of DNA around spOrc4p and spORC, their interaction with circular DNA containing ars1 lowered the average linking number of the plasmid topoisomer distribution after treatment with topoisomerase I (topo I). No evidence was obtained that this change involved the generation of single-stranded DNA.
Materials and Methods
Proteins and DNAs. SpORC, spORC containing ΔN-Orc4p (spORC-ΔN-Orc4p), spOrc4p, and ΔN-spOrc4p were isolated from baculovirus-infected Sf9 cells as described in ref. 7. The spOrc4p derivative in which the N-terminal region was truncated (ΔN-spOrc4p) contained amino acids 516–972 of spOrc4p. Homogenous preparations of vaccinia topo I were a gift from Stewart Shuman (Sloan–Kettering Institute, New York). The DNA substrates used, pUC19 (2.7 kbp) and pUR19N (5.7 kbp), containing pUC19, the sp ura4 gene (1.8 kbp), and the complete sp ars1 sequence (1.2 kbp), were as described in ref. 7. Additional plasmids used included Δars1 pUR19N (4.5 kbp; pUR19N devoid of the ars1 sequence) and pUR19N (devoid of nucleotides 766–865 or nucleotides 974-1201 of ars1). The spars1 sequence, corresponding to nucleotides 4360–3157 in GenBank entry Z67961, has been renumbered as nucleotides 1–1204.
AFM Analysis of Protein Binding to DNA. Reaction mixtures (1–3 μl), containing binding buffer (20 mM Hepes-NaOH, pH 7.5/20 mM KCl/10 mM magnesium acetate), 250 nM pUR or other specified plasmids, protein preparations as indicated, and 1 mM ATP (where indicated), were incubated at 25°C for 10 min. Aliquots were diluted 5-fold with 0.25× binding buffer (“imaging buffer”) and immediately analyzed by AFM. This procedure was used to score for the binding of proteins to DNA. The sites in ars1-containing protein and the length of DNA ars1 fragments complexed with protein (or free of protein) were analyzed as follows. After binding, pUR19N was restricted with both PstI (80 units) and EcoRV (80 units; New England Biolabs) in NEB4 buffer for 1 h at 37°C. Samples (3 μl) were then subjected to agarose gel electrophoresis to confirm that restriction was complete. The rest of the sample was gel-filtered through a Superose 6 column (Pharmacia) equilibrated with NEB4 at 4°C to enrich fragments bound with spORC or spOrc4p and separate them from restriction enzymes. Restriction fragments in eluted fractions were detected by UV absorption. The collected fractions containing DNA fragments, stored on ice, were subjected to AFM analysis after 2–10× dilution in imaging buffer. The time elapsed between restriction, filtration, and microscopy was usually ≈2 h. In general, spORC was found attached to DNA fragments 2–4 h after gel filtration, whereas after 12 h only free DNA and free spORC were visible. Restriction fragments with bound spOrc4p were less stable; complexes were detected after storage on ice for ≈1 h after gel filtration. At later times, only free DNA and free spOrc4p were observed.
Imaging was carried out with 2 μl of diluted samples deposited on freshly cleaved mica surface. After 2 min, the mica was washed twice with double-distilled H2O, dried under a stream of nitrogen, and mounted in a Nanoscope IIIa atomic force microscope (Digital Instruments/Veeco). Imaging was carried out in tapping mode in air with TESP probes (Veeco Probes). Resonant frequency of the cantilevers was tuned to 270–330 kHz, with an amplitude of 30–60 mV and a setpoint between 1.2 and 1.6 V. Areas ranging from 0.36 (600 × 600 nm) to 4 μm2 were scanned in height mode with rates of 2.4–3.0 Hz, and trace and retrace images were collected. Standard flattening and plainfit were applied to images by using nanoscope iiia software. Bearing analysis and 3D plots from the nanoscope iiia software were used to dissect the topography of DNA–protein complexes.
The length of restriction fragments was measured after correcting the tip broadening effect by using the public domain image sxm v.1.67–3p software (S. Barrett, Surface Science Research Center, University of Liverpool, Liverpool, U.K.), a version of the public domain image analysis software nih image (developed at the U.S. National Institutes of Health and available at http://rsb.info.nih.gov/nih-image). DNA molecules were hand-traced on the computer screen, and the program computed the lengths of the trace in nm. The “tails” of DNA were traced from the middle of the protein “bead” to the shorter end of the DNA strand. Tracings were repeated at least twice for each fragment, and average results were recorded. Lengths expressed in nm were recalculated to bp by using the 0.34-nmper-base-pair coefficient applicable to B-form DNA.
Imaging of spORC and spOrc4 proteins was carried out in “wet mode”: the samples deposited on mica were not washed and dried. Instead, they were overlaid with 30 μl of imaging buffer, so both the sample and the probe were always submerged. Oxide-sharpened silicon nitride tips with cantilevers of nominal spring constant 0.32 N/m (Veeco Probes) mounted in a “wet chamber” were used for the wet method.
Results
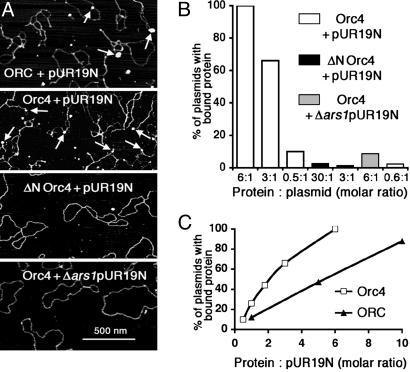
Selective Interaction of spOrc4p and spORC with DNA Containing ars1. We and others have shown that spORC binds to ars1 and other spARS sequences in an ATP-independent (7, 8, 20, 21) but relatively sequence-specific manner (7, 20, 21). The N-terminal AT-hook motifs of spOrc4p are essential for this interaction. We investigated this interaction by using AFM to visualize complexes formed between DNA and spOrc4p and spORC. In these experiments, two plasmids were used, pUC19 containing the 1.2-kbp ars1 and spura4 sequences (pUR19N) and Δars1 pUR19N. These DNAs were incubated with spORC or spOrc4p at various molar ratios of protein to DNA and their interactions monitored by AFM (Fig. 1). Incubation of spOrc4p with pUR19N at a molar ratio of protein:DNA of three resulted in the detection of spOrc4p bound to ≈65% of the plasmid molecules (Fig. 1 A and B). When the input ratio of spOrc4p to pUR19N was lowered to 0.5, plasmids containing bound protein decreased to 10% (Fig. 1B), whereas at a ratio of six, all pUR19N molecules contained protein. The binding of spOrc4p to DNA was linear up to a molar ratio of protein:pUR19N of two (Fig. 1C) at which ≈50% of the DNA molecules contained one molecule of protein. In contrast to these findings, spOrc4p bound to Δars1pUR19N was barely detectable (≈10%) at a molar ratio of six. When ΔN-spOrc4p was used, virtually no interactions were detected with either pUR19N or Δars1pUR19N even at high protein:DNA ratios (Fig. 1 A and B). In all experiments, AFM imaging indicated that ars1-containing plasmids or restriction fragments (see below) bound a single ORC or Orc4p particle.
Fig. 1.
Interaction of spOrc4p, ΔN-spOrc4p, and spORC with pUR19N and Δars1pUR19N. (A) AFM visualization of protein–DNA complexes. Reaction mixtures were prepared and imaged by AFM as described in Materials and Methods. The molar ratio of protein to DNA was always 3:1. Arrows show protein particles (spORC, spOrc4p) bound to plasmids. The images are presented in height scale, with white corresponding to 5 nm. (B) Quantitation of the binding of spOrc4 and ΔN-spOrc4 to pUR19N or Δars1pUR19N. Reactions, as described in Materials and Methods at protein to plasmid ratios indicated, were subjected to AFM analysis. The total number of plasmids analyzed in each case was as follows: spOrc4+pUR19N at molar ratios of 6:1, 3:1, and 0.5:1 were 10, 76, and 309, respectively; ΔN-spOrc4+pUR19N at molar ratios of 30:1 and 3:1 were 78 each; spOrc4+Δars1pUR19N at molar rates of 6:1 and 0.6:1 were 162 and 130, respectively. The percentage of DNA bound with protein is indicated. (C) Comparison of the binding of spORC and spOrc4p to pUR19N. Reactions were carried out as in A. The total number of plasmids analyzed by AFM (starting from the lowest molar ratio) was as follows: spOrc4p, 309, 46, 18, 76, and 10; in experiments with spORC, the number of plasmids analyzed was 42, 17, and 16.
Identical experiments carried out with the six-subunit spORC yielded results similar to those with spOrc4p alone, except that the efficiency of binding to pUR19N was reduced ≈60% (Fig. 1C). Because spORC contained almost equimolar levels of each subunit (7), the reasons for this discrepancy are unclear. Binding reactions carried out in the presence of ATP did not alter this discrepancy (data not shown). As previously observed, spORC containing the ΔN-spOrc4p subunit did not bind to either pUR19N or pUC (7). Thus, visualization of protein–DNA complexes by AFM corroborated previous studies that spORC and spOrc4p bind selectively to ars1 solely through the AT-hook motifs of spOrc4p (7, 20).
Determination of Sites in ars1 That Bind spORC and spOrc4. The above results indicate that spOrc4p and ORC bind selectively to the ars1 region in pUR19N. To determine the site(s) in ars1 to which spORC and spOrc4p bind, pUR19N–protein complexes were formed first and then digested with two restriction enzymes, PstI and EcoRV, that cleaved DNA at sites bordering the ars1 region. Complete restriction reproducibly yielded fragments of the expected sizes in the presence or absence of spORC and spOrc4p. In subsequent experiments, after incubation of spORC or spOrc4p with pUR19N (ATP additions made no difference in the results), reaction mixtures were incubated with PstI and EcoRV for 1 h at 37°C and digests were subjected to Superose 6 gel filtration at 4°C to isolate protein-bound DNA fragments for AFM. DNA molecules were hand-traced on the computer screen, and their lengths were computed in nm. The accuracy of measurements was tested by comparing theoretical and experimental lengths of free DNA fragments. pUR19N was expected to be 1,922 nm long, and the measurements resulted in the length of 1,920 ± 20 nm (mean ± SD; n = 12). The ars1 fragment formed from pUR19N by ClaI should be 409 nm long, and we observed a length of 408 ± 18 nm (n = 41). We concluded that our procedure determined the length of DNA fragments with high accuracy.
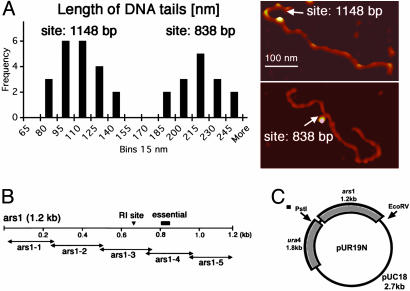
In Fig. 2A, lengths of short-tailed DNA fragments bound to spORC or spOrc4p are presented as a frequency histogram. On the right, examples of AFM images of two restriction fragments complexed with spOrc4p at different sites are presented. Based on the observed frequencies, two discrete peaks were detected, presumably reflecting the presence of two distinct binding sites in ars1. The average length of DNA from the middle of the imaged protein–DNA complex to the shorter-tailed fragment end was 102 ± 17 nm (n = 23), whereas the average length of the longer tailed fragment was 207 ± 19 nm (n = 17). Identical results were obtained with spORC. Furthermore, the length of the restriction fragment bound to spORC or spOrc4p was identical.
Fig. 2.
Determination of the binding sites of spORC and spOrc4p in ars1. Reaction mixtures, scaled-up to 200 μl, were prepared as described in Materials and Methods with a protein:DNA molar ratio of 5:1. Reactions were restricted with PstI plus EcoRV, gel-filtered, and AFM-imaged as described. (A Left) frequency histogram for the measurements of the distance between DNA-bound spORC or spOrc4p and ends of the restriction fragment (“tail”). The distance from the protein complex to the ends of the shorter tail was measured as described in Materials and Methods. The statistical significance of the existence of two groups, as indicated, was tested with cluster analysis (centroid method). Measurements of both groups followed a normal distribution (Shapiro–Wilk test). (A Right) Examples of AFM images of two restriction fragments with spOrc4 bound at distinct sites. The shorter tail in top image was 115 nm long, indicating the binding site at 1,148 bp (left cluster on the histogram). The shorter tail in the bottom image was 211 nm long, indicating the binding site at ≈838 bp (right histogram cluster). The total lengths of restriction fragments shown were 620 nm (Upper) and 635 nm (Lower). The images are presented in height scale, with white corresponding to 10 nm. (B) Schematic diagram of various segments of ars1 (1.2 kbp) and position of regions reported as essential for ARS activity and the site of initiation of replication (RI). The 1.2-kbp ars1 region was subdivided into specific fragments that are described in ref. 7. The binding sites at nucleotides 838 and 1148 are located within ars1–4 and ars1–5 fragments, respectively. (C) Positions of the PstI and EcoRV restriction sites in pUR19N. Map of pUR19N, which contains, in addition to pUC19 (2.7 kbp), spura4 (1.8 kbp) and spars1 (1.2 kbp). Restriction with PstI (site located in ura4) and EcoRV (site located in pUC19) yields a 2-kbp fragment with the complete ars1 region.
The lengths of the DNA, measured in nm, were converted to bp with the relationship that 0.34 nm of B-form DNA corresponds to 1 bp. Thus, the measured tail lengths of 102 ± 17 nm and 207 ± 19 nm correspond to 300 ± 51 bp and 610 ± 56 bp, respectively. Because the size of the ars1-bound restriction fragment generated by PstI and EcoRV consists of ars1 (1,204 bp) flanked on the left by 553 bp and on the right by 244 bp (a fragment of 2,001 bp in total), the shorter tail must extend to the right from ars1 to place the spORC binding sites in ars1 (Fig. 2C). Based on these considerations, the binding sites in ars1 could be centered at 1,148 bp [(2,001–553)–300 bp = 1,148 bp] and the longer tail could extend to the right or to the left of ars1. In the first case, the site bound would be centered at 838 bp [(2,001 – 553) – 610 bp = 838 bp]. If the longer tail extended to the left, the position bound would be centered at 57 bp (610–553 bp = 57 bp), or the very beginning of ars1.
To verify that the protein binding regions were centered about 838 and 1,148 bp (located within fragments previously called ars1–4 and ars1–5, respectively, as shown in Fig. 2B), plasmids were constructed in which either the region between nucleotides 766 and 865 or the region between nucleotides 974 and 1201 of ars1 was deleted. These mutated plasmids were analyzed for their ability to bind spOrc4 and spORC by AFM. All plasmids, including those containing the deleted regions, bound a single spORC or spOrc4p particle as did the control pUR19N. As expected, deletion of the 838- or 1,148-bp regions resulted in restriction fragments that when visualized by AFM were devoid of protein “beads” at the deleted sites, whereas the remaining site was liganded (data not shown). These findings support the assignment of the spOrc4/ORC protein–DNA complexes to the sites indicated above.
The binding site centered in the 838-bp region (in ars1–4) was identified previously as a region required for ARS activity (16) as well as a site protected by spORC and spOrc4p in DNase I footprinting experiments (7, 20). However, the binding site centered at 1,148 bp was not identified previously as a region critical for ARS activity or an ORC binding site. Previous in vitro DNA binding studies with spOrc4p and spORC were carried out in the presence of a DNA competitor [poly(dA-dC), ≈3 kbp], whereas the experiments described in Fig. 2 A lacked competitor. Thus, it is possible that the site located at 1,148 bp may represent a region competed by the presence of a DNA competitor. Technical reasons prevented us from carrying out such competitor experiments by using AFM because the presence of a large excess of poly(dA-dC) competitor swamped out the visualization of the pUR19N restriction fragment.
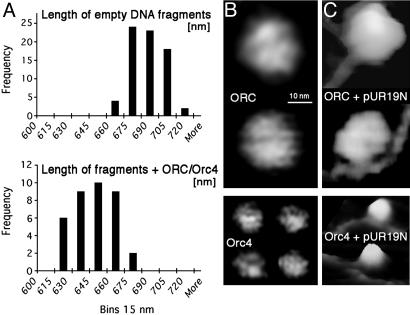
Interaction of Orc4p/ORC with ars1 Results in the Wrapping of DNA Around spOrc4p. The lengths of the PstI–EcoRV DNA fragment either free or complexed with spOrc4p or spORC are summarized in Fig. 3A as a frequency histogram. The theoretical length of the restriction fragment containing ars1 is 2,001 bp (Fig. 2C), equivalent to 680 nm. AFM measurements indicated that the average length of the free DNA restriction fragment was 682 ± 14 nm (n = 75; Fig. 3A), in agreement with the theoretical value.
Fig. 3.
DNA fragments are shortened upon binding ORC/orc4p. (A) Measurement of the length of the DNA restriction fragments free and bound by spORC and spOrc4p. Reactions were carried out as described above and contained protein:DNA at a molar ratio of 5:1 where applicable. A frequency histogram of the lengths of DNA fragments free and bound to spORC and spOrc4p is presented. The lengths of fragments bound to spORC or to spOrc4p were identical, and the average lengths of fragments containing either protein are presented. Quantitation of the data, including the number of DNA molecules analyzed, is presented in the text. (B) AFM images of spORC (Upper) and spOrc4p (Lower). The observed diameter of the spORC complex was 25–30 nm, which after a correction for “tip broadening effects” decreased to 12–16 nm. The diameter of spOrc4p after similar corrections was 5–7 nm. The images are presented in height scale, with white corresponding to 20 nm (Upper) or 7nm(Lower). (C) AFM images of spORC and spOrc4p bound to ars1. The scale in C is the same as in B.
The measured lengths of bound DNA shown in Fig. 3A represent cumulative observations made with both spOrc4p and spORC. The average length of the DNA fragment complexed with spORC was 637 ± 16 nm (n = 18), while the average length of DNA fragment with spOrc4p was 634 ± 17 nm (n = 22). These findings indicate that there was no difference in the length of the DNA fragment complexed with spORC or spOrc4p. Based on these measurements, the difference in length between the mean values of free and protein-complexed DNA was 682 nm – 636 nm = 47 nm, corresponding to 137 bp. This difference is statistically significant with P > 0.001 (t test). We suggest that the shortening of the length of the DNA fragment reflects the wrapping of DNA around the bound spORC/Orc4p.
The diameters of spOrc4p and spORC were determined by AFM imaging (Fig. 3B). SpORC appears rounded and somewhat pentagonal with a visible shallow groove (up to 0.5 nm deep). The diameter of the complex was 12–16 nm after correction for “AFM tip broadening”; this range of values and the appearance of the complex were identical in all AFM experiments. The diameter of spOrc4p was measured to be 5–7 nm after the correction, a value observed reproducibly in all AFM experiments. The size difference between Orc4p and ORC was observed even when they were bound to DNA (see Figs. 1 A and 3C), although the structure of Orc4 bound to DNA appeared to be higher (more white) than that for free Orc4, presumably because of the wrapped DNA (Fig. 3C).
The length of DNA wrapped around the spOrc4p appears to be more than one full turn. One turn around a spherical protein would be expected to have a maximal length of 2 × π × (approximated radius of spOrc4p + radius of DNA) = 6.28 × (3 nm + 1 nm) = 25 nm, equivalent to 74 bp. The experimentally determined shortening of DNA corresponded to 137 bp, suggesting that two turns of the DNA are wrapped around spOrc4p. Similar calculations of the length of DNA wrapped about spORC (radius 7 nm) corresponded to 148 bp, indicating that only one turn of the DNA may be wrapped completely around the spORC. Although this possibility cannot be excluded, we suggest that the observed shortening of the spORC–DNA complex is due to the wrapping of DNA around the Orc4 subunit in the complex. This notion is based on the findings that the shortening of the DNA fragments complexed with spORC and spOrc4p are identical and that spORC interactions with DNA occur solely through spOrc4p.
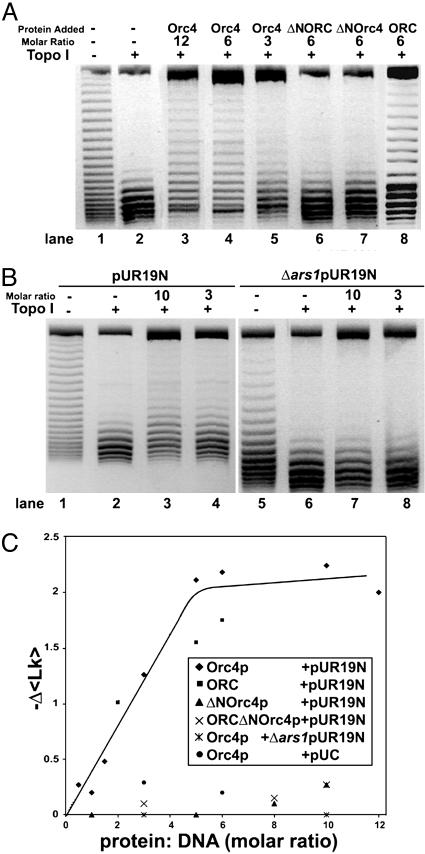
Binding of spOrc4p and spORC Induces Topological Changes in pUR19N. We used a biochemical approach to examine whether the binding of spOrc4p and spORC to DNA altered the structure of DNA. For this purpose, changes in the linking number of pUR19N complexed with spOrc4p or spORC were examined after exposure to eukaryotic topo I. After deproteinization, topoisomers were resolved by agarose gel electrophoresis in the presence of chloroquine. As shown in Fig. 4A, treatment of pURN19 with topo I substantially removed the negative supercoils present in the DNA (compare lanes 1 and 2). When DNA was complexed first with Orc4p and then exposed to topo I, the topoisomer distribution was shifted upward and the extent of the shift depended on the molar ratio of spOrc4p:DNA. It was detectable at a molar ratio of ≈1 (see Fig. 4C), clearly evident at a ratio of 3 (Fig. 4A, lane 5) and maximal at an spOrc4p:DNA ratio of 6 (compare lanes 3 and 4). Because slower-migrating topoisomers possess a lower linking number than faster-migrating species, the binding of spOrc4p reduced the average linking number of the topoisomer population. In the absence of topo I, this effect was not observed, indicating that Orc4p alone did not alter the distribution of topoisomers (data not shown). The same alterations in topoisomer distribution observed in Fig. 4A were evident over a wide concentration of topo I (0.1–2 pmol of vaccinia topo I) as well as with HeLa topo I in place of the vaccinia enzyme (data not shown). When ΔN-spOrc4p or spORC-ΔN-Orc4p was used in place of spOrc4p, the topoisomer distribution was hardly affected even at molar ratios of protein:pUR19N of six (Fig. 4A, lanes 6 and 7, respectively). The molar ratio of spOrc4p:pURN19 (6:1) required to observe the maximal change in topoisomer distribution was in keeping with the AFM binding studies shown in Fig. 1, where it was noted that at a molar ratio of spOrc4p:pUR of 3, ≈65% of the input DNA contained bound protein, whereas at a ratio of 6:1, 100% of the input plasmid was liganded. Similar results were obtained with spORC, although the changes in linking number observed with the six-subunit complex were somewhat lower than those observed with spOrc4p (Fig. 4 A, lane 8, and C).
Fig. 4.
SpORC/Orc4p binding to pUR19N results in topological changes in DNA. (A) Influence of spOrc4p, ΔN-spOrc4p, and spORC-ΔN-Orc4p on the topology of pUR19N. Reaction mixtures (20 μl) containing 20 mM Hepes-NaOH (pH 7.5), 10 mM magnesium acetate, 40 μg/ml BSA, 20 mM KCl, 80 mM NaCl, 200 fmol of pUR19N, and spOrc4p (lanes 3–5), spORC-ΔN-Orc4p (lane 6), ΔN-spOrc4p (lanes 7), or spORC (lane 8), at the molar ratios indicated, were incubated for 30 min at 30°C. Vaccinia topo I (0.75 pmol) was added to reactions in lanes 2–8, and after 30 min at 30°C, 2.5 μl of a stop solution (0.18 M EDTA/3.6% SDS/1.2 mg/ml tRNA/12 μg of proteinase K) was added. After 30 min at 30°C, the solution was adjusted to 0.25 M sodium acetate (pH 5.2) and 2.5 vol of ethanol added and the precipitated DNA collected. Pellets suspended in 10 μl of TE and loading buffer (containing no SDS) were then subjected to vertical gel electrophoresis through 1.5% agarose in TAE plus 1.5 μg of chloroquine per ml with a TAE running buffer containing 1.5 μg of chloroquine per ml at 5 V/cm for 15 h. Inverse images of gels stained with ethidium bromide are presented because of their sharper contrast. (B) SpOrc4p selectively induces topological changes in DNA containing ars1. Reactions were as described in A with 200 fmol of pUR19N in lanes 1–4 and 200 fmol of Δars1 pUR19N in lanes 5–8, at the molar ratios of spOrc4p:DNA indicated. (C) Summary of the Δ〈Lk〉 observed at different molar ratios of various spORC and spOrc4p derivatives to pUR19N, Δars1 pUR19N, and pUC. Reactions were as described in A with 200 fmol of each DNA at molar ratios of protein:DNA indicated. Δ〈Lk〉 was calculated as described in the text.
In keeping with the selective interaction of spOrc4p with the ars1 sequence in pUR19N (Fig. 1), when pUR19N devoid of ars1 (Δars1 pUR19N) was used in place of pUR19N (Fig. 4B), no shift in topoisomer distribution was detected even in the presence of an spOrc4p:Δars1 pUR19N ratio of 10. Virtually no shift in topoisomers was observed when the parent plasmid pUC was used in lieu of pUR19N (data not shown; Fig. 4C).
Calculations of linking number changes (ΔLk) were carried out by using the image gauge science lab 2003 4.1 program (Fuji). Each topoisomer band was assigned an integral number, representing the linking number of that particular topoisomer. The fraction of the total DNA in each band was then multiplied by this integral value, and the products were summed to yield the relative average linking number (Δ〈Lk〉) of the DNA in that sample. The difference in the Δ〈Lk〉 between individual experiments could then be calculated and compared. Results from a number of experiments in which the Δ〈Lk〉 was determined at varying molar ratios of spOrc4p, ΔN-spOrc4p, spORC and spORC-ΔN-Orc4p to pUR19N, Δars1 pUR19N, and pUC are summarized in Fig. 4C. A maximal Δ〈Lk〉 of –2.25 was observed at a molar ratio of spOrc4p:pUR19N of ≈6, a value slightly higher than that observed with spORC (–1.75). Changes in the linking number observed with spOrc4p + pUC or ΔN-spOrc4p + pUR19N were considerably lower (maximum –0.20). In all experiments, the Δ〈Lk〉 observed with spORC or spOrc4p were unaffected by the presence of ATP (data not shown).
Because both DNA wrapping of proteins and DNA unwinding can contribute to changes in ΔLk (22), we investigated whether the binding of spORC or spOrc4p to pUR19N yielded single-stranded regions. Production of single-stranded regions was monitored by KMnO4 oxidation and by P1 nuclease digestion. In both assays, formation of single-stranded DNA was not detected (data not shown).
Discussion
In this study, in vitro interactions of spORC and spOrc4p with ars1 were examined by AFM. In keeping with previous findings, interactions were detected with pUR19N, the plasmid containing the 1.2-kbp ars1, but not with Δars1pUR19N, a plasmid lacking this sp origin sequence. It should be noted that both plasmids contain spura4, which, like ars1, is nearly 70% AT-rich. Interactions with pUR19N were not observed with ΔN-spOrc4p or spORC-ΔN-Orc4p. Thus, spORC binding to the spars1 origin, measured by AFM, depends on the N terminus of the spOrc4p subunit that contains multiple AT-hook motifs. Identical observations have been made by using nitrocellulose filter binding, mobility shift, and DNase I protection assays (7, 8, 20, 21).
DNase I footprinting and competition experiments identified a number of sites in sp origins that bind spORC or spOrc4p. In some cases, these sites corresponded to those important for ARS activity. All binding sites identified contained clusters of A or T residues on one strand and were devoid of GC-rich sequences (7, 20, 21). AFM imaging of spORC or spOrc4p bound to ars1 also revealed a number of interactive regions. The sites detected represented the binding of a single protein molecule to one DNA molecule, and none of the DNA molecules examined contained more than a single protein. Approximately 40% of the liganded plasmid molecules contained a protein particle centered in the ars1–4 region (838 bp), and ≈60% of the liganded molecules were centered in the ars1–5 region (1,148 bp). Deletion of each of these sites in ars1 abrogated spORC (Orc4p) binding to the region deleted without affecting the binding to the region present and served to verify the location of the proteins in ars1. Previous studies demonstrated that a site in ars1–4, which contains a region critical for ARS activity in vivo, was protected by spORC in DNase I footprinting experiments (7, 20). The AFM-imaged binding site located in the ars1–5 region, however, was not observed in previous experiments. In some experiments, a third site in ars1, centered in the 549-bp region, was also detected. However, the experimental conditions used that resulted in binding to this site (located between the ars1–2 and ars1–3 regions) differed from those described in Fig. 2. These included binding of proteins to DNA after restriction and experiments carried out in the presence of a 100-fold molar excess of poly(dA-dC) 100 bp in length. The efficiency of spORC binding to DNA after restriction was markedly lower than its binding to DNA before restriction. We have no explanation for the variations in the binding sites. However, it appears that the interaction of spORC or spOrc4p with a particular site in ars1 prevents protein binding to other sites within the same DNA molecule.
Analysis of the length of the restriction fragment containing spORC or spOrc4p complexed with ars1 revealed that the DNA was 137 bp shorter than the restriction fragment without protein. All protein-bound DNA molecules, irrespective of the position of the protein on DNA, exhibited identical shortening and contained a single spORC or spOrc4p molecule because the dimensions of the proteins on DNA or free were similar to those observed in Fig. 3B. These findings are best explained by assuming that the DNA wrapped about spORC is due to its selective wrapping around the spOrc4p subunit of the complex. This idea suggests that the N-terminal region of spOrc4p, even when complexed with the other spOrc subunits, may be exposed. This notion is in keeping with the finding that the five-subunit complex, devoid of spOrc4p, is converted to the six-subunit complex upon addition of spOrc4p (19, 20).
We also demonstrated that the interaction of spORC or spOrc4p with pUR19N, followed by topo I relaxation, resulted in a decrease in the linking number relative to the unliganded plasmid. Increasing levels of either spOrc4p or spORC resulted in a change that peaked at a ΔLk equivalent to approximately –2. A 〈ΔLk〉 of –2 is in accord with the wrapping of two turns of the DNA helix around spOrc4p. In contrast, the maximal 〈ΔLk〉 found with pUR19N incubated with either ΔN-spOrc4p or spORC-ΔN-Orc4p was –0.29. These relatively smaller changes may represent weak interactions undetected by other techniques. We also examined whether interactions of spORC and spOrc4p with pUR19N resulted in the unwinding of the duplex. Unwinding was monitored with the P1 nuclease, which would cleave single-stranded DNA if formed and by KMnO4 oxidation, which would oxidize T and/or C residues present in single-stranded DNA. Both assays failed to detect DNA unwinding.
The major finding reported here suggests that ≈140 bp of DNA is wrapped about the spOrc4p subunit of the spORC complex. DNA wrapping of origin binding proteins has been observed with λ0 (22), DnaA (23), and DmORC (14). DmORC was shown to bind to negatively supercoiled DNA with a much higher affinity than to linear or nicked duplex DNA (14). Binding measurements carried out by AFM also indicated that spORC has a higher binding affinity to supercoiled pUR19N than linear pUR19N. Nitrocellulose filter binding competition assays indicated that supercoiled pUR19N reduced the binding of spORC to labeled ars1 DNA 5-fold more efficiently than linear pUR19N (data not shown). Thus, at least two factors may contribute to the binding of spORC to origins, the superhelical structure of the DNA, and the AT-hook domain of spOrc4p that bind to multiple A and T-rich regions.
It has been suggested that multiple spORC binding sites within an origin are required for origin firing in sp (21, 25). A number of spARSs that have been examined in detail have revealed that the progressive shortening of their length gradually decreased their replication efficiency rather than an all or none response (16, 17, 26). Extensive studies of four spARS elements, ars2004, ars3002, ars1, and ars3001, indicated that regions required for ARS activity contain clusters of T or A residues. Footprinting analysis of spORC binding to ars1 and ars3002 (both in vitro and in vivo) suggests that A- or T-rich clusters are important (summarized in ref. 24). In the case of ars2004, spORC binding was mapped to two clustered A or T regions essential for its ARS activity (25). These regions spanned ≈0.7 kbp, suggesting that multiple spORC binding sites may be collectively required for efficient activation of this origin. If DNA wrapping, as observed with ars1, is a general property of spORC–origin complexes, spORC binding to two sites would require a minimal length of ≈300 bp, excluding spacer regions.
Although spORC contains weak ATPase activity and three of its subunits, spOrc1, -4, and -5, contain core AAA+ domains, its interaction with DNA is unaffected by ATP, whereas all other ORCs require ATP for this step (1). Possibly, the strong spOrc4p-mediated DNA interaction prevents detection of weaker ATP-dependent interactions between other spOrc subunits and DNA. Other origin binding proteins (DnaA, SV40 T antigen, etc.) play multiple roles in the activation of replication origins leading to localized DNA unwinding and the assembly of other replication proteins at origins (27, 28). The chief role of ORC in replication is the generation of the prereplication complex that includes the assembly of Cdc18 (Cdc6), Cdt1, and the MCM complex (29–31). This complex leads to the stable association of the MCM complex (the replicative helicase) with chromatin, which probably requires a DNA unwinding reaction. Although the wrapping of DNA around spORC changes the DNA structure, it does not result in duplex unwinding. The role played by the other prereplication proteins that leads to DNA unwinding, however, remains to be elucidated.
Acknowledgments
We thank Dr. Frank Dean for helpful discussions regarding topology experiments. This work was supported by National Institutes of Health Grants GM 069819 (to M.G.) and GM 38559 (to J.H.).
Author contributions: M.G., P.A.O., Y.J., J.-K.L., V.B., and J.H. designed research, performed research, contributed new reagents/analytic tools, analyzed data, and wrote the paper.
Abbreviations: ARS, autonomously replicating sequence; ORC, origin recognition complex; Sc, Saccharomyces cerevisiae; sp, Schizosaccharomyces pombe; topo I, topoisomerase I.
References
- 1.Bell, S. P. & Dutta, A. (2002) Annu. Rev. Biochem. 71, 333–374. [DOI] [PubMed] [Google Scholar]
- 2.Bell, S. P. & Stillman, B. (1992) Nature 357, 128–134. [DOI] [PubMed] [Google Scholar]
- 3.Marahrens, Y. & Stillman, B. (1992) Science 255, 817–823. [DOI] [PubMed] [Google Scholar]
- 4.Marahrens, Y. & Stillman, B. (1994) EMBO J. 13, 3395–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Theis, J. F. & Newlon, C. S. (1994) Mol. Cell. Biol. 14, 7652–7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newlon, C. S. (1996) in DNA Replication in Eukaryotic Cells, ed. DePamphillis, M. L. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 873–914.
- 7.Lee, J.-K., Moon, K.-Y., Jiang, Y. & Hurwitz, J. (2001) Proc. Natl. Acad. Sci. USA 24, 13589–13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chuang, R. Y., Chrétien, L., Dai, J. & Kelly, T. J. (2002) J. Biol. Chem. 277, 16920–16927. [DOI] [PubMed] [Google Scholar]
- 9.Gossen, M., Pak, D. T., Hansen, S. K., Acarya, J. K. & Botchan, M. R. (1995) Science 270, 1674–1677. [DOI] [PubMed] [Google Scholar]
- 10.Rowles, A., Chong, J. P., Brown, L., Howell, M., Evand, G. I. & Blow, J. J. (1996) Cell 87, 287–296. [DOI] [PubMed] [Google Scholar]
- 11.Vashee, S., Simancek, P., Challber, M. D. & Kelly, T. J. (2001) J. Biol. Chem. 276, 26666–26678. [DOI] [PubMed] [Google Scholar]
- 12.Blow, J. J., Gilespe, P. J., Francis, D. & Jackson, D. A. (2001) J. Cell Biol. 152, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyrien, O., Marheineke, K. & Goldar, A. (2003) BioEssays 25, 116–125. [DOI] [PubMed] [Google Scholar]
- 14.Remus, D., Beall, E. L. & Botchan, M. R. (2004) EMBO J. 23, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubey, D. D., Kim, S. M., Todorov, I. T. & Huberman, J. A. (1996) Curr. Biol. 6, 467–473. [DOI] [PubMed] [Google Scholar]
- 16.Clyne, R. K. & Kelly, T. J. (1995) EMBO J. 14, 6348–6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okuno, Y., Satoh, H., Sekeguchi, M. & Masukata, H. (1999) Mol. Cell. Biol. 19, 6699–6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuang, R. Y. & Kelly, T. J. (1999) Proc. Natl. Acad. Sci. USA 96, 2656–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moon, K.-Y., Kong, D., Lee, J.-K., Raychaudhuri, S. & Hurwitz, J. (1999) Proc. Natl. Acad. Sci. USA 96, 12367–12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong, D. & DePamphillis, M. L. (2001) Mol. Cell. Biol. 21, 8095–8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong, D. & DePamphillis, M. L. (2002) EMBO J. 21, 5667–5676. [Google Scholar]
- 22.Dodson, M., Roberts, J., McMacken, R. & Echols, H. (1985) Proc. Natl. Acad. Sci. USA 82, 4678–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bramhill, D. & Kornberg, A. (1988) Cell 52, 743–755. [DOI] [PubMed] [Google Scholar]
- 24.Segurado, M., deLuis, A. & Antequera, F. (2003) EMBO Rep. 4, 1048–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi, T., Ohara, E., Nishitani, H. & Masukata, H. (2003) EMBO J. 22, 904–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubey, D. D., Zhu, J., Carlson, D. L., Sharma, K. & Huberman, J. A. (1994) EMBO J. 13, 3638–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kornberg, A. & Baker, T. A. (1992) DNA Replication (Freeman, New York).
- 28.Waga, S. & Stilllman, B. (1998) Annu. Res. Biochem. 67, 721–751. [DOI] [PubMed] [Google Scholar]
- 29.Rowles, A., Tada, S. & Blow, J. J. (1999) J. Cell Sci. 112, 2011–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huan, X. H. & Newport, J. (1998) J. Cell Biol. 140, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seki, T. & Diffley, J. F. (2000) Proc. Natl. Acad. Sci. USA 97, 14115–14120. [DOI] [PMC free article] [PubMed] [Google Scholar]