Abstract
TBX21 encodes for the transcription factor T-bet (T-box expressed in T cells), which influences naïve T lymphocyte development and has been implicated in asthma pathogenesis. Specifically, the T-bet knockout mouse spontaneously develops airway hyperresponsiveness and other changes consistent with asthma. Because airway responsiveness is moderated by the use of inhaled corticosteroids in asthma, it is conceivable that genetic variation in TBX21 may alter asthma phenotypes in a treatment-specific fashion. Here we demonstrate that the nonsynonymous variation in TBX21 coding for replacement of histidine 33 with glutamine is associated with significant improvement in the PC20 (a measure of airway responsiveness) of asthmatic children in a large clinical trial spanning 4 years. We note that this increase occurs only in the children randomized to inhaled corticosteroids and that it dramatically enhances the overall improvement in PC20 associated with inhaled corticosteroid usage. The average PC20 at trial end for subjects on inhaled corticosteroids possessing a variant allele was in the normal range for nonasthmatics. In cellular models, we show that the TBX21 variant increases T helper 1 and decreases T helper 2 cytokine expression comparably with wild type. TBX21 may thus be an important determinant pharmacogenetic response to the therapy of asthma with inhaled corticosteroids.
Keywords: PC20, pharmacogenetics, T-bet, interaction
Corticosteroids mediate a variety of immunological actions and are commonly used in the treatment of a diverse number of diseases. Inhaled corticosteroids are the most effective and commonly used therapy in the management of asthma (1, 2) but may be associated with serious adverse reactions (3). In evaluating asthma therapy response, measures of lung function, such as forced expiratory volume at 1 second (FEV1), and of airway responsiveness, as measured by the provocative concentration of methacholine causing a 20% decrement in FEV1 (PC20) are commonly used. However, there is large interindividual variation in the FEV1 and PC20 responses to inhaled corticosteroids (4, 5). Thus, identifying those patients most likely to benefit from this treatment would be valuable. Because the intraindividual response to inhaled corticosteroid treatment in patients with asthma is highly repeatable (6) and because both FEV1 and PC20 are heritable traits (7, 8), a genetic basis for the heterogeneity of this therapeutic response is plausible.
The gene TBX21 (GenBank accession no. NM_013351) encodes for transcription factor T-bet (T-box expressed in T cells), which is responsible for the induction of T helper (Th)1 cells and the repression of Th2 cells from naïve T lymphocytes (9). T-bet has been implicated in the pathogenesis of asthma (10, 11). Because the T-bet knockout mouse develops spontaneous airway hyperresponsiveness (10), a phenotype that is modulated by corticosteroids, we assessed the relationship of TBX21 with PC20 outcomes in asthma. Only one common (estimated heterozygosity ≥ 5%) nonsynonymous TBX21 SNP has been described to date, rs2240017, which codes for a replacement of histidine 33 with glutamine (H33Q). We evaluated the role of this SNP in the therapeutic response to inhaled corticosteroids in children enrolled in the Childhood Asthma Management Program (CAMP). The CAMP study design allowed us to investigate this pharmacogenetic association both within those children randomized to inhaled corticosteroids and between the corticosteroid group and those not randomized to corticosteroids. Here, we demonstrate a significant interaction between H33Q and inhaled corticosteroid use resulting in improvement of airway responsiveness to nonasthmatic levels. We also provide a context for the cellular function of H33Q and a rationale for the use of interaction terms in pharmacogenetic analyses.
Materials and Methods
Study Population. The CAMP study is a multicenter, randomized, double-blinded clinical trial testing the safety and efficacy of inhaled budesonide versus nedocromil versus placebo over a mean of 4.3 years. Trial design and methodology have been published (12, 13). CAMP enrolled 1,041 children ages 5–12 years with mild to moderate asthma. DNA was collected from families representing >700 probands. Entry criteria included asthma symptoms and/or medication use for ≥6 months in the previous year and airway responsiveness with PC20 ≤ 12.5 mg/ml.
Spirometry and methacholine testing were performed at least 4 hours after the use of a short-acting bronchodilator and 24 hours after the last use of a long-acting bronchodilator. Spirometry was performed on a Collins Stead–Wells dry-seal Survey III spirometer interfaced to a Dell desktop personal computer and met or exceeded the American Thoracic Society standards.
Airway responsiveness was determined by methacholine testing with the Wright nebulizer technique after the method of Cockcroft et al. (14) modified for methacholine administration and by Juniper et al. (15). Methacholine sensitivity was expressed in terms of PC20 with the following equation: PC20 = antilog[loge C1 + (loge C2 – loge C1)(20 – R1)/(R2 – R1)], where C1 is the second to last concentration tested, C2 is the last concentration tested, R1 is the percentage of decrease in FEV1 from diluent after C1, and R2 is the percentage of decrease in FEV1 from diluent after C2.
Follow-up visits with spirometry occurred at 2 and 4 months and every 4 months thereafter. Methacholine studies were performed during the run-in period, at 8 months, then yearly. Our primary focus was on the CAMP children randomized to the corticosteroid group, evaluated at their 1- and 4-year follow-up visits for change in prebronchodilator FEV1 and at their 4-year (44 month) visit for difference in PC20 from baseline. These time points were chosen because the 1-year visit represented the time that maximal steroid effects were noted in the primary clinical trial (13) and the 4-year visit represented the last visit with nearly complete follow-up. The CAMP Genetics Ancillary Study was approved by each study center's internal review board, and informed consent/assent was obtained from all participants and their parents.
Genotyping. Genotyping was performed by using the SEQUENOM MassARRAY MALDI-TOF mass spectrometry platform (Sequenom, San Diego). Primers were designed by using a semiautomated program (spectrodesigner, Sequenom). We used the very short extension method (16), whereby sequencing products are extended by only one base for three of the four nucleotides and by several additional bases for the fourth nucleotide (representing one of the alleles), permitting clearly delineated mass separation of the two allelic variants at a given locus.
To assess for possible population stratification, we also genotyped a random panel of 49 SNPs across the genome (17) obtained through the SNP Consortium database. SNPs were chosen to exclude the gene proper (promoter, genomic UTR, exons, and introns) of known genes and be widely distributed throughout the genome. Genotyping quality control consisted of at least 90% of successful calls, regenotyping of ≈10% of genotypes, and documentation of Hardy–Weinberg equilibrium for each SNP.
Functional Analysis of H33Q. To investigate possible functional consequences of the H32Q T-bet polymorphism, which is the mouse homologue of human H33Q, we used a QuickChange site-directed mutagenesis kit (Stratagene) to generate a T-bet cDNA encoding a protein for which the histidine residue at position 32 is replaced with glutamine. Wild-type (wt) T-bet or the H32Q version of T-bet H32Q were cotransfected into EL-4 cells with an IFN-γ promoter reporter gene and a β-gal reporter gene. Relative luciferase activity was assayed, standardized with β-gal activity, and shown as fold induction. We also tested the ability of the H32Q version of T-bet to control cytokine production in primary T cells by taking advantage of our T-bet-deficient mouse strain (9). The wt and polymorphic versions of T-bet cDNAs were cloned into a bicistronic retroviral vector (RV), and the wt (RV-T-bet) and H32Q T-bet (RV-H32Q) GFP retroviruses were transduced between 24–36 hours into CD4+ Th cells isolated from T-bet–/– lymph node. Cells were stimulated with anti-CD3 and anti-CD28 and cultured for an additional 5 days under nonpolarizing conditions. On day 6, cells were sorted for GFP expression and restimulated with anti-CD3; 24 h later, expression of wt and H32Q T-bet was measured by Western blot analysis and cytokine production was assessed by ELISA, according to the manufacturer's protocol (Pharmingen).
Cellular Effect of Corticosteroids on T-bet Expression. Human CD4 T cells were isolated from peripheral blood by using human CD4 microbeads (Miltenyi Biotec). We stimulated these CD4 cells with plate-bound anti-CD3 and anti-CD28 under nonpolarizing and Th1 polarizing conditions in the presence or absence of 10–7 M dexamethasone. Relative gene transcript levels for human T-bet were measured by real-time RT-PCR. As a control, we measured transcripts encoding IRF-4, a known steroid-regulated gene. Because of the rarity of the H33Q minor allele, no human CD4 cells with the 33Q variant were available for examination.
Statistical Methodology. The percentage of change in FEV1 was defined as (FEV1 at the end of a period – FEV1 at randomization)/(FEV1 at randomization × 100). Values of PC20 were log-transformed before analysis. Change in PC20 was defined as PC20 at 4 years – PC20 at the beginning of the trial. Geometric mean values were calculated by using the exponentiated mean of the log-transformed PC20 at a given time.
Linear regression models were used to assess the association between H33Q genotypic status and the outcome of interest. Univariate and multivariable analyses adjusting for age, sex, and baseline FEV1 (as a percent of predicted) were performed. Height was incorporated into the multivariable models involving FEV1 outcomes. To test for interaction between steroid treatment and H33Q genotypic status, we included interaction terms in the models. Student's t tests were used to evaluate the comparability between the nedocromil and placebo groups before their combination as a singular “nonsteroid” group. Given the small number of individuals with the genotype of interest, our primary analysis was repeated by using the nonparametric exact Mann–Whitney U test. Additionally, to account for the censored nature of individuals exceeding the maximal threshold for methacholine dosing, we used nonparametric survival methods as incorporated into Kaplan–Meier curves and log-rank test statistics to evaluate differences in the PC20 over time. Such procedures have been demonstrated to be adequate for the analysis of airway responsiveness in population-based studies (18).
Analysis of the stratification of SNPs proceeded in two steps. First, for each individual SNP, an allelic χ2 test statistic was obtained in standard fashion by using 2 × 2 contingency tables that allocated the totals of the wt and variant alleles for the highest and lowest quartiles of FEV1 and log PC20 response to each of four cells. An overall summary χ2 test statistic was then obtained by summing the individual χ2 test statistics and setting the degrees of freedom to the number of tests. All analyses were performed in sas v.8 (SAS Institute, Cary, NC).
Results
Population Characteristics and Response to Corticosteroids. The characteristics of the CAMP children included in our analyses are shown in Table 1. Of all the CAMP children, 701 were successfully genotyped at H33Q and had data for our outcomes of interest; ≈30% were randomized to the corticosteroid group. Baseline age, gender, ethnic distributions, FEV1, and PC20 were similar between the steroid and nonsteroid groups (Table 1). Over time, utilization of corticosteroids was associated with significant improvements in FEV1 and PC20. There were no significant differences between children randomized to nedocromil or placebo (P = 0.62 and 0.39 for 1-year FEV1 and 4-year PC20 change, respectively). Therefore, these children were combined into one larger, nonsteroid group.
Table 1. Population characteristics.
| Steroid | Nonsteroid | |
|---|---|---|
| Total no. of subjects | 195 | 506 |
| Age | 9.0 ± 2.1 | 8.9 ± 2.1 |
| Sex, n (%) | ||
| Male | 128 (65.6) | 312 (61.7) |
| Female | 67 (34.4) | 194 (38.3) |
| Race, n (%) | ||
| Caucasian* | 139 (71.3) | 377 (74.5) |
| African American | 19 (9.7) | 52 (10.3) |
| Hispanic | 18 (9.2) | 38 (7.5) |
| Other | 19 (9.7) | 39 (7.7) |
| Baseline FEV1, % predicted | 93.5 ± 14.7 | 93.4 ± 14.2 |
| Geometric mean baseline PC20, mg/ml | 1.08 ± 3.25 | 1.09 ± 3.24 |
| Treatment response† | ||
| Change in FEV1, % | 5.13 ± 11.4 | -0.18 ± 11.4 |
| Difference in log PC20 | 1.10 ± 1.4 | 0.64 ± 1.3 |
Values are mean ± SD where applicable.
Because of concerns over possible population stratification, only genotypic information from Caucasians was analyzed.
P < 0.001 for differences between steroid and nonsteroid groups for FEV1 at 1 year and PC20 at 4 years.
Analyses were confined to those children whose parents identified their ethnicity as Caucasians, because of concerns about possible population stratification and the small sample sizes of the other ethnic groups. In the Caucasian children, no evidence of overt population stratification was found (P > 0.05 for each of our three outcomes).
Association of TBX21 with Improved PC20 in the Corticosteroid Group. The minor (G) allele frequency for the TBX21 SNP, which codes for glutamine, was 4.5% in the CAMP Caucasians (n = 24); no homozygotes were expected or observed. The SNP was in Hardy–Weinberg equilibrium.
For children on inhaled corticosteroids, the change in log-transformed PC20 over 4 years was relatively normally distributed (Fig. 1), suggesting that factors other than treatment type contributed to this response. H33Q was present in five of these children and was significantly associated with this change in both univariate (β = 2.97, P = 0.0002) and multivariate (β = 3.03, P = 0.0001) models. The estimated mean increase in log-transformed PC20 on inhaled steroids over 4 years was ≈3.5-fold more in those children with glutamine variants compared with those homozygous for wt histidine 33 TBX21 (Fig. 2). H33Q was not associated with either 1-year or 4-year change in FEV1 (data not shown).
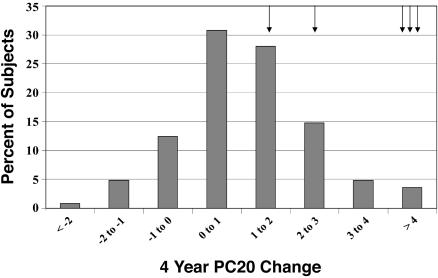
Fig. 1.
Distribution of PC20 response over time. The 4-year change in airway responsiveness in CAMP, as measured by log-transformed PC20, to inhaled corticosteroid therapy varies by individual and is approximately normally distributed. This finding suggests that factors other than treatment, including genetics, may have contributed to the treatment response. Arrows indicate the individuals containing the H33Q variant allele.
Fig. 2.
The 4-year change in log-transformed PC20 stratified by treatment group assignment and H33Q genotype. Steroid usage by itself is associated with an improvement in the PC20 over time (P < 0.001), whereas the rs2240017 genotype is not (P = 0.90). However, individuals on corticosteroids who also possessed a minor allele encoding for glutamine (33Q) demonstrated dramatic improvement in their PC20 over and above that associated with corticosteroids alone (interaction, P = 0.0002). Values shown are mean ± SE.
To explore the robustness of these findings, our primary analysis of the children on inhaled corticosteroids was repeated by using the Mann–Whitney U test. In this nonparametric comparison, H33Q remained significantly associated with the 4-year log PC20 change (exact P = 0.003).
Interaction of TBX21 with Corticosteroids. We performed stratified and interaction analyses to evaluate whether the observed associations between H33Q with improvement in PC20 were due to TBX21 influence on the natural history of asthma or whether a true pharmacogenetic effect involving the enhancement of a therapeutic response was present. H33Q was not associated with the change in PC20 over time in the children randomized to the nonsteroid group (P = 0.90) but was, as noted above, strongly associated with PC20 change in those randomized to steroids (P = 0.0001) (Fig. 2).
Given these treatment-specific differences, we incorporated a formal interaction term into the multivariate regression model. After adjustment for age, gender, and baseline level of lung function, the estimated mean (± SE) improvement for a CAMP subject on inhaled corticosteroids with the H33Q variant was 3.85 ± 0.90, whereas it was 1.22 ± 0.75 for a wt subject on inhaled corticosteroids and 0.66 ± 0.75 for a wt subject not taking inhaled corticosteroids (interaction P = 0.0002). Overall, for subjects randomized to inhaled corticosteroids, individuals homozygous for histidine demonstrated gradual improvement over time, whereas 33Q carriers demonstrated marked improvement over 1 year, with a multiplicative effect over time (Fig. 3). After 4 years, the geometric mean PC20 value for 33Q carriers who were randomized to the corticosteroid group was at least 27.7 mg/ml. Physiologically, any value >25 mg/ml corresponds to a lack of significant airways responsiveness, placing these subjects into the typical range of individuals without asthma. Individual values at 4 years for 33Q carriers on steroids ranged from 11.3–37.5 (the value ascribed to the upper limits of testing).
Fig. 3.
Geometric mean value of PC20 at various time points over the 4-year follow-up, stratified by treatment group assignment and H33Q genotype. The mean PC20 of individuals on corticosteroids who also possessed a copy of the 33Q variant improved significantly by the end of the first year of the study and continued to improve substantially with time and corticosteroid usage. The mean PC20 value at 4 years is within the range normally ascribed to individuals without the diagnosis of asthma.
To account for the group of individuals exceeding the maximal threshold for methacholine dosing (18) and the small number of individuals with the variant genotype, we verified the findings in our between-group analyses by using the log rank test to evaluate differences in the maximal PC20 achieved during the trial (P = 0.01). Kaplan–Meier curves illustrating these differences are shown in Fig. 4. Each of the 33Q carriers on inhaled corticosteroids normalized his PC20 at some point during the 4-year follow-up. Similar results were noted in the analysis of final PC20 (P = 0.05).
Fig. 4.
Maximal PC20 dose achieved, stratified by treatment group assignment and H33Q genotype. Despite similar PC20 values at baseline, each of the individuals on corticosteroids who also possessed a copy of the 33Q variant fully normalized his PC20, exceeding the upper limits of methacholine testing at least once during the 4-year follow-up. In the other groups, few individuals ever normalized PC20.
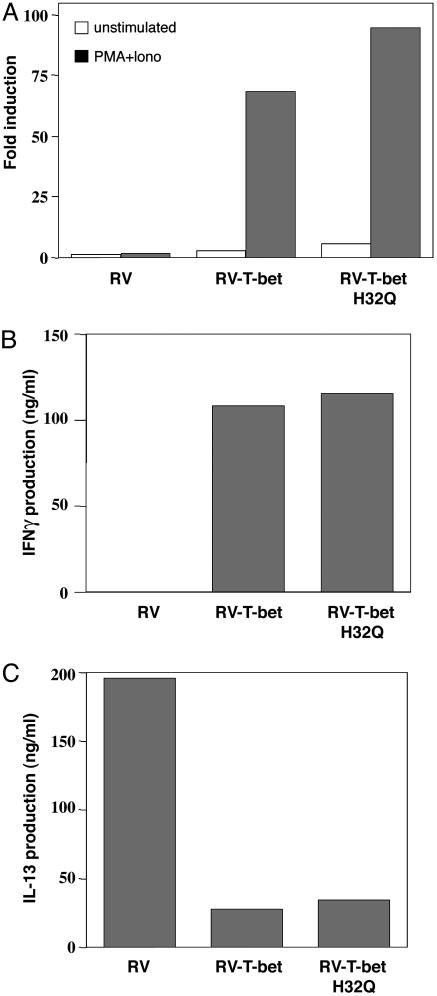
Functional Characteristics of H32Q. The wt and H32Q (the mouse homologue to human H33Q) versions of T-bet are potent transactivators of the IFN-γ gene as assessed by expression of the luciferase reporter gene (Fig. 5A), with no significant differences observed between the two. We next tested the ability of the H32Q version of T-bet to control cytokine production in primary T cells (9, 19). T-bet–/– Th cells transduced with control GFP retrovirus produced very low amounts of IFN-γ but large amounts of the Th2 cytokines IL-4, IL-5, and IL-13. RV-T-bet (wt) transduced T-bet–/– Thp cells, as expected, produced large amounts of IFN-γ and exhibited a significant repression of Th2 cytokine production. The H32Q version of T-bet was equally effective in performing these functions (Fig. 5 B and C).
Fig. 5.
Functional characteristics of H32Q, the mouse analog of H33Q. (A) The H32Q polymorphism is equally effective in the transactivation of the IFN-γ gene regulatory region. T-bet or the H32Q version of T-bet were cotransfected into IL-4 cells with an IFN-γ promoter reporter gene and a β-gal reporter gene. Relative luciferase activity was assayed, standardized with β-gal activity, and shown as fold induction. (B and C) H32Q was as effective at modifying cytokine gene expression in primary T cells as wt T-bet. CD4+ T cells were transduced with GFP-RV, RV-T-bet, and RV-T-bet H32Q. Cells were expanded for 5 days, sorted for GFP, and restimulated with anti-CD3 for 24 hours. ELISA was performed to measure IFN-γ and IL-13 production. The results are from three independent experiments. TB, wt T-bet.
Inhibition of T-bet by Corticosteroids. The correlation between T-bet polymorphisms and PC20 response to steroid therapy raised the possibility that steroid treatment might control the expression of T-bet. As noted by others (20), after stimulating human CD4 cells in the presence or absence of dexamethasone, we found that glucocorticoids inhibited the induction of T-bet (Fig. 6).
Fig. 6.
Dexamethasone represses the expression of T-bet in human Th cells. Human CD4 T cells isolated from peripheral blood by using microbeads (Miltenyi Biotec) were stimulated with plate-bound anti-CD3 and anti-CD28 under nonpolarizing (NS) and Th1-skewed conditions in the absence or presence of 10–7 M dexamethasone (DEX). Relative gene transcripts for human T-bet were measured by real time RT-PCR. In both groups, glucocorticoids inhibited the induction T-bet.
Discussion
Our results demonstrate that a nonsynonymous SNP in the TBX21 gene substantially enhances the effects of inhaled corticosteroid usage on airway responsiveness in asthmatic children. The glutamine 33 carriers randomized to corticosteroids demonstrated substantial improvement in their PC20 over time, with an average value at the end of the study in the normal range for nonasthmatics. These findings are consistent with both known association of corticosteroid usage with improvement in PC20 (21, 22) and the known biology of TBX21 (10). Moreover, the pharmacogenetic association reported represents a true gene-by-environment interaction, in that the change noted in subjects possessing both the genetic variant of interest and treatment of interest was considerably greater than that represented by genetic variation, treatment effect, or the combination of these effects alone (Figs. 2 and 3).
TBX21 is a member of the T-box gene family, a phylogenetically ancient series of transcription factors characterized by a highly conserved DNA-binding domain (9, 23, 24). T-bet was first described by Szabo et al. (9), as a transcription factor responsible for the commitment of naïve Th cells into the Th1 lineage. Specifically, T-bet induces the production of IFN-γ and represses IL-4 and IL-5 (9, 10). Altering the early life balance from a Th1 to a Th2 lymphocyte predominance may enhance a child's risk for atopic diseases, including asthma: the so-called hygiene hypothesis (25, 26). Given that T-bet can enhance the development of Th1 cells and is expressed in the lung (9, 27), a role for TBX21 in the pathogenesis of asthma has been postulated and investigated (10, 11). Airways of human asthmatic subjects demonstrate decreased numbers of CD4+ T cells expressing TBX21 when compared with controls (10). TBX21 gene deletions in mice result in airway hyperresponsiveness (10). The airways of TBX21 knockout mice also demonstrate enhanced eosinophilia and remodeling (10). Because the PC20 is a measure of airway hyperresponsiveness that is related to the degree of pulmonary eosinophilia and airway remodeling over time (28), our pharmacogenetic association of TBX21 with the PC20 over time is biologically both relevant and plausible. Consistent with this association, our cellular models indicate that H33Q potently activates IFN-γ expression, increases Th1, and decreases Th2 cytokine production.
In CAMP, worsening airway hyperresponsiveness, as manifested by decrements in PC20, was associated with decrements in pulmonary function, increased eosinophilia, and increased asthma symptoms, duration, and severity (29). Use of inhaled corticosteroids improves airway inflammation, hyperresponsiveness, obstruction, and clinical symptoms in asthma (21, 22, 30). Improvement in PC20 in the CAMP corticosteroid arm has been documented (13) and is reflected in our results (Table 1 and Fig. 2). However, we report a significant enhancement to the improvement in each of the subjects on corticosteroids with the H33Q variant, when compared with inhaled steroids or wt genotype alone (Figs. 2 and 3). Although the exact biological mechanism for such strong genetic effect modification remains to be elucidated, after administration of oral corticosteroids, bronchial biopsy specimens of asthmatic patients demonstrated a reduction of cellular mRNA expression of IL-4 and IL-5 and an increase in cellular mRNA expression of IFN-γ (31). As demonstrated (Fig. 5), the transduced TBX21 H33Q parallels the cytokine changes resulting from corticosteroid usage precisely. Although we might have expected that steroids would increase T-bet expression concordant with their ability to ameliorate asthma, our data indicate that examination of gene expression in vitro does not necessarily approximate an in vivo situation. Additionally, cells with the H33Q variant may respond differently to the administration of exogenous corticosteroids.
A potential limitation to our study is sample size. Although our sample size is reasonable for testing relatively common alleles, the minor allele frequency was only 4.5% for H33Q, and we did not observe any minor allele homozygotes. Interestingly, heterozygous TBX21 knockout mice demonstrate the same phenotypic features as homozygous knockout mice (10), including the spontaneous development of airway hyperresponsiveness., which suggests that even slight changes in gene expression, such as with H33Q heterozygotes, may contribute to altered phenotypic expression and is consistent with our observed association. Moreover, we feel that the strength and robustness of the association within the steroid group, the demonstration of a true gene by environment interaction, and the biologic plausibility all strongly support the validity of our findings.
In conclusion, our findings of a TBX21 genotype-specific improvement in airway responsiveness within asthmatics on inhaled corticosteroids provide insights into asthma therapy and may allow the identification of at least a small group of individuals with asthma most likely to benefit from this form of therapy. Replication studies are warranted and should likely focus on the interrogation of PC20 in a larger cohort of asthmatics using inhaled corticosteroids over a relatively shorter period of time. These results may also be pertinent to the myriad of diseases treated with corticosteroids and the increasing number of diseases in which T-bet is believed to play a role. Finally, our demonstration of a true gene by environment interaction provides for an alternative statistical approach toward the elucidation of what comprises a true pharmacogenetic effect. Formal interaction terms should be considered when the primary outcome of interest is readily measured and may vary over time either as a result of therapeutic intervention or as part of the natural history of the underlying disease.
Acknowledgments
We thank all the families for their enthusiastic participation in the CAMP Genetics Ancillary Study, which was supported by National Heart, Lung, and Blood Institute Grant N01-HR-16049, and the CAMP investigators and research team, who were supported by the National Heart, Lung, and Blood Institute, for collection of CAMP Genetic Ancillary Study data. This work was supported by the Pharmacogenetics of Asthma Treatment project from the National Heart, Lung, and Blood Institute under the auspices of National Institutes of Health Grant U01 HL65899; National Institute of Allergy and Infectious Diseases Grant AI31541 (to L.H.G.); and National Heart, Lung, and Blood Institute Grants N01 HR16044, HR16045, HR16046, HR16047, HR16048, HR16049, HR16050, HR16051, and HR16052. E.S.H. is a recipient of an Arthritis Foundation Postdoctoral Fellowship.
Author contributions: K.G.T., E.S.H., B.A.R., S.L.P., J.M.D., L.H.G., and S.T.W. designed research; K.G.T., E.S.H., B.A.R., S.L.P., L.H.G., and S.T.W. performed research; K.G.T., E.S.H., S.L.L., B.G.R., L.H.G., and S.T.W. analyzed data; K.G.T., E.S.H., B.A.R., E.S.S., J.M.D., L.H.G., and S.T.W. wrote the paper; and B.G.R. contributed new reagents/analytic tools.
Abbreviations: FEV1, forced expiratory volume at 1 second; PC20, provocative concentration of methacholine causing a 20% decrement in the FEV1; T-bet, T-box expressed in T cells; CAMP, Childhood Asthma Management Program; RV, retroviral vector; Th, T helper; wt, wild type.
References
- 1.Barnes, P. J. (1996) Am. J. Respir. Crit. Care Med. 154, S21–S27. [DOI] [PubMed] [Google Scholar]
- 2.Bousquet, J. (2000) Clin. Exp. Allergy 30, Suppl. 1, 2–5. [DOI] [PubMed] [Google Scholar]
- 3.Lemanske, R. F., Jr., & Allen, D. B. (1997) Am. J. Respir. Crit. Care Med. 156, 685–687. [DOI] [PubMed] [Google Scholar]
- 4.Malmstrom, K., Rodriguez-Gomez, G., Guerra, J., Villaran, C., Pineiro, A., Wei, L. X., Seidenberg, B. C. & Reiss, T. F. (1999) Ann. Intern. Med. 130, 487–495. [DOI] [PubMed] [Google Scholar]
- 5.Szefler, S. J., Martin, R. J., King, T. S., Boushey, H. A., Cherniack, R. M., Chinchilli, V. M., Craig, T. J., Dolovich, M., Drazen, J. M., Fagan, J. K., et al. (2002) J. Allergy Clin. Immunol. 109, 410–418. [DOI] [PubMed] [Google Scholar]
- 6.Drazen, J. M., Silverman, E. K. & Lee, T. H. (2000) Br. Med. Bull. 56, 1054–1070. [DOI] [PubMed] [Google Scholar]
- 7.Wilk, J. B., Djousse, L., Arnett, D. K., Rich, S. S., Province, M. A., Hunt, S. C., Crapo, R. O., Higgins, M. & Myers, R. H. (2000) Genet. Epidemiol. 19, 81–94. [DOI] [PubMed] [Google Scholar]
- 8.Palmer, L. J., Burton, P. R., James, A. L., Musk, A. W. & Cookson, W. O. (2000) Eur. J. Hum. Genet. 8, 853–860. [DOI] [PubMed] [Google Scholar]
- 9.Szabo, S. J., Kim, S. T., Costa, G. L., Zhang, X., Fathman, C. G. & Glimcher, L. H. (2000) Cell 100, 655–669. [DOI] [PubMed] [Google Scholar]
- 10.Finotto, S., Neurath, M. F., Glickman, J. N., Qin, S., Lehr, H. A., Green, F. H., Ackerman, K., Haley, K., Galle, P. R., Szabo, S. J., et al. (2002) Science 295, 336–338. [DOI] [PubMed] [Google Scholar]
- 11.Robinson, D. S. & Lloyd, C. M. (2002) Curr. Biol. 12, R322–R324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The Childhood Asthma Management Program (1999) Controlled Clin. Trials 20, 91–120. [PubMed] [Google Scholar]
- 13.The Childhood Asthma Management Program Research Group (2000) N. Engl. J. Med. 343, 1054–1063. [DOI] [PubMed] [Google Scholar]
- 14.Cockcroft, D. W., Killian, D. N., Mellon, J. J. & Hargreave, F. E. (1977) Clin. Allergy 7, 235–243. [DOI] [PubMed] [Google Scholar]
- 15.Juniper, E. F., Frith, P. A., Dunnett, C., Cockcroft, W. & Hargreave, F. E. (1978) Thorax 33, 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun, X., Ding, H., Hung, K. & Guo, B. (2000) Nucleic Acids Res. 28, E68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pritchard, J. K. & Rosenberg, N. A. (1999) Am. J. Hum. Genet. 65, 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munoz, A. & Sunyer, J. (1996) Am. J. Respir. Crit. Care Med. 154, S234–S239. [DOI] [PubMed] [Google Scholar]
- 19.Mullen, A. C., Hutchins, A. S., High, F. A., Lee, H. W., Sykes, K. J., Chodosh, L. A. & Reiner, S. L. (2002) Nat. Immunol. 3, 652–658. [DOI] [PubMed] [Google Scholar]
- 20.Refojo, D., Liberman, A. C., Giacomini, D., Carbia Nagashima, A., Graciarena, M., Echenique, C., Paez Pereda, M., Stalla, G., Holsboer, F. & Arzt, E. (2003) Ann. N.Y. Acad. Sci. 992, 196–204. [DOI] [PubMed] [Google Scholar]
- 21.Olivieri, D., Chetta, A., Del Donno, M., Bertorelli, G., Casalini, A., Pesci, A., Testi, R. & Foresi, A. (1997) Am. J. Respir. Crit. Care Med. 155, 1864–1871. [DOI] [PubMed] [Google Scholar]
- 22.Chung, K. F. & O'Byrne, P. (2003) in Asthma, ed. Fabbri, L. M. (Eur. Respir. Soc. J., Sheffield, U.K.), Vol. 8, pp. 458. [Google Scholar]
- 23.Horton, A. C. & Gibson-Brown, J. J. (2002) J. Exp. Zool. 294, 112–121. [DOI] [PubMed] [Google Scholar]
- 24.Hatton, R. D. & Weaver, C. T. (2003) Science 302, 993–994. [DOI] [PubMed] [Google Scholar]
- 25.Strachan, D. P. (1989) Br. Med. J. 299, 1259–1260.2513902 [Google Scholar]
- 26.Romagnani, S. (1992) Int. Arch. Allergy Immunol. 98, 279–285. [DOI] [PubMed] [Google Scholar]
- 27.Shier, P., Hofstra, C. L., Ma, X. J., Wu, Y., Ngo, K. & Fung-Leung, W. P. (2000) Immunogenetics 51, 771–778. [DOI] [PubMed] [Google Scholar]
- 28.Sont, J. K., Willems, L. N., Bel, E. H., van Krieken, J. H., Vandenbroucke, J. P. & Sterk, P. J. (1999) Am. J. Respir. Crit. Care Med. 159, 1043–1051. [DOI] [PubMed] [Google Scholar]
- 29.Weiss, S. T., Van Natta, M. L. & Zeiger, R. S. (2000) Am. J. Respir. Crit. Care Med. 162, 50–56. [DOI] [PubMed] [Google Scholar]
- 30.Barnes, P. J. (1999) in Anti-inflammatory Drugs in Asthma, ed. Church, M. K. (Birkhauser, Basal), pp. 33–85.
- 31.Bentley, A. M., Hamid, Q., Robinson, D. S., Schotman, E., Meng, Q., Assoufi, B., Kay, A. B. & Durham, S. R. (1996) Am. J. Respir. Crit. Care Med. 153, 551–556. [DOI] [PubMed] [Google Scholar]