ABSTRACT
Axonal degeneration is a key pathological feature of several neurological disorders. Emerging evidence has suggested a pathological connection between axonal degeneration and autophagy, a lysosomal degradation pathway. We recently reported that GSK3B-mediated phosphorylation of MCL1 regulates axonal autophagy to promote axonal degeneration. GSK3B–MCL1 pathway affects ATP production locally in degenerating axons and the exposure of phosphatidylserine (PS), an “eat-me” signal for phagocytes, on degenerating axons, resulting in the failed engulfment of axonal debris in vivo. Here we showed that the PS exposure is accomplished by phospholipid scramblase activity. This finding provides a novel mechanism that local ATP production through autophagy promotes PS exposure on degenerating axons. In addition, it opens new perspectives for the understanding of axonal autophagy to regulate Wallerian degeneration.
KEYWORDS: autophagy, phospholipid scramblase, ubiquitin-proteasome system, Wallerian degeneration
Axonal degeneration is recognized as a key pathological feature of many neurological disorders, including Alzheimer disease and Parkinson disease.1,2 A typical form of pathological axonal degeneration is Wallerian degeneration, which has been observed in segments distal to the site of injury. Emerging evidence has suggested pathological connections between Wallerian degeneration and autophagy, a lysosomal degradation pathway. Previous reports have revealed increased expression of autophagy markers and formation of autophagosomes in degenerating axons.3,4 However, the pathophysiological significance and regulation of axonal autophagy remain elusive.
We previously reported that glycogen synthase kinase 3B (GSK3B) is one of the critical mediators to regulate Wallerian degeneration.5,6 Using Wallerian degeneration models in vitro and in vivo, we recently demonstrated that myeloid cell leukemia 1 (MCL1), a BCL2 family protein, negatively regulates axonal autophagy by binding to BECLIN1, a key regulator of autophagy, and also that GSK3B-mediated phosphorylation of MCL1 serves as an initiating signal to induce axonal autophagy.7 Interestingly, the perturbation of axonal autophagy by inhibition of the GSK3B–MCL1 pathway affected the exposure of phosphatidylserine (PS), which serves as the “eat-me” signal for phagocytes, on degenerating axons, and resulted in the failed engulfment of axonal debris. These results raised the next question on the connecting mechanism between the local ATP production by autophagy and PS exposure in degenerating axons.
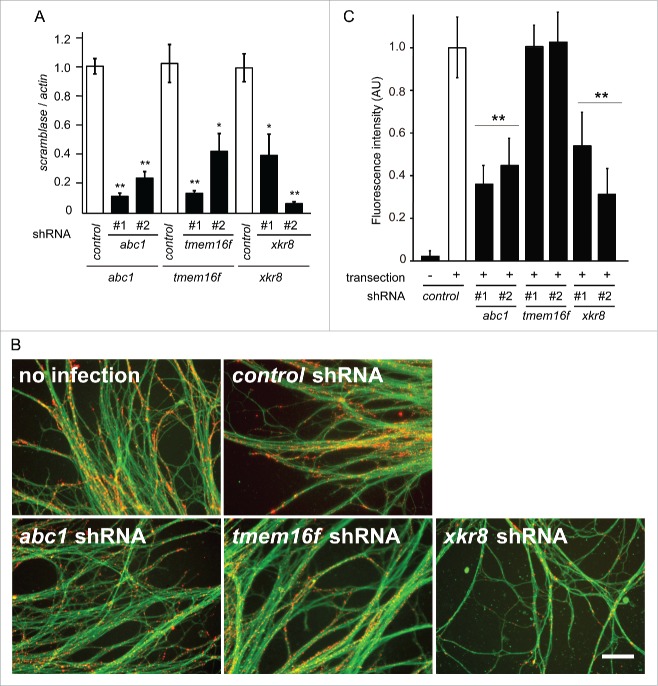
Dying cellular structures, including axonal debris, are rapidly cleared by phagocytes. In this process, phagocytes recognize “eat-me” signals, including PS on degenerate cells.8 Autophagy has been implicated in PS exposure on apoptotic cells.9,10 Using in vitro Wallerian degeneration experiments, we recently reported a significant increase in PS exposure on axons 3 h post-transection,7 and reduction of autophagic activity by shRNAs for the autophagy genes including atg5 or atg7 prevented PS exposure, implicating autophagy in PS exposure.7 PS exposure is known to be accomplished by the activity of phospholipid scramblases, including ATP-binding-cassette transporter 1 (ABC1), Transmembrane protein 16F (TMEM16F), and XK-related protein 8 (XKR8).11-13 To demonstrate the involvement of phospholipid scramblase in PS exposure during axonal degeneration, we examined PS exposure under the RNAi-mediated downregulation of a phospholipid scramblase in axons after transection in vitro. We found reduced PS exposure on axons expressing shRNAs against abc1 and xkr8, but not tmem16f (Fig. 1B–C). These results suggest that ABC1 and XKR8 are the phospholipid scramblases responsible for PS exposure on degenerating axons.
Figure 1.
Scramblase activity is responsible for PS exposure on degenerating axons. (A) The shRNA-mediated downregulation of abc1, tmem16f, or xkr8 expression was performed in cultured DRG neurons using lentivirus vectors. The downregulated expression of abc1, tmem16f, or xkr8 by 2 independent shRNAs used in this study was confirmed by quantitative RT-PCR. The expression levels of each molecule normalized to β-actin are shown relative to that of the control (mean ± SEM, n = 5). (B, C) PS exposure on the transected axons was monitored using the specific probe Alexa594-labeled Annexin V (red). DTAF (green) was used to stain axons. Representative photomicrographs of the fluorescent signal in transected axons expressing the indicated molecules are shown in B. Scale bar = 25 μm. Quantified levels of fluorescent intensities for Annexin V normalized to DTAF relative to the level in the control (labeled as “no infection“) are shown in (C)(mean ± SEM, n = 5). Note that PS exposure is reduced on axons expressing shRNAs for abc1 or xkr8, but not tmem16f. Significant differences from the control (*, P < 0.05; **, P < 0.01) were determined by a one-way ANOVA with Tukey's post-hoc test.
The precise role of autophagy during axonal degeneration has been debated.13,14-16 We recently showed that upregulation of axonal autophagy is promotes degradation of axonal structures.7 There, we demonstrated that axonal autophagy contributes to the local production of ATP, which is required to promote axonal degeneration, and also to the PS exposure on degenerating axons to recruit phagocytes to the injury site. Thus, our results provide a novel significance of axonal autophagy to regulate Wallerian degeneration. Combined with the additional evidence we showed here, our data further support the model that axonal autophagy contributes to the local ATP production and subsequent PS exposure on degenerating axons through activation of the specific phospholipid scramblases.
Materials and methods
Animals
All animals were maintained in accordance with the guidelines of the National Center for Neurology and Psychiatry. The technical protocols for animal experiments in this study were approved by a review committee for Animal Resources in the National Center for Neurology and Psychiatry.
Dorsal root ganglia (DRG) explant culture and in vitro Wallerian degeneration
Murine DRG explants dissected from embryonic day (E) 13 C56BL/6J mice were cultured on poly-L-lysine (PLL) and laminin-coated 24-well plates in Neuro-medium (Miltenyi Biotec) containing 10% FBS and 25 ng/ml nerve growth factor (NGF) (Harlan Bioproducts). After 24 h, the culture medium was changed to Neuro-medium supplemented with 2% Neuro-Brew-21 (Miltenyi Biotec), 25 ng/ml NGF, and 1 mM Glutamine in addition to a mixture of 1 mM 5′-fluoro-2′-deoxyuridine and 1 mM uridine to remove non-neuronal cells. The in vitro Wallerian degeneration of axons was introduced by removing cell bodies at 10–14 d in vitro using a pipette tip.5,6
Viral vectors and infection
In experiments using lentiviral vectors, regarding specific short hairpin RNAs (shRNAs) against the phospholipid scramblase genes abc1, tmem16f, and xkr8, each target nucleotide sequence was designed using BLOCK-iT RNAi Designer (Thermo Fischer) and cloned into the pLKO.1 puromycin-resistant lentiviral vector. The target nucleotide sequences for each shRNA were as follows: abc1 #1, 5′-GCAGCACAATTTGTCCCTTCC-3′; abc1 #2, 5′-GGGACAGAATTGCCATCATTT-3′; tmem16f #1, 5′-GCTCCTCATACTCCTCTACCA-3′; tmem16f #2, 5′-GCTCAAGAGTTCACAGTATTT-3′; xkr8 #1, 5′-GCTACCTGTATAGGTGTTTGC-3′; and xkr8 #2, 5′-GCCATGTGATGCCCAGTAAGT-3′. Non-target control shRNAs in the pLKO.1 vector (Cat No. SHC002) was purchased from Sigma-Aldrich. Lentiviral packaging was performed using HEK293FT cells, as described previously.17
Quantitative reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted from cultured DRG neurons using the RNeasy MiniKit (Qiagen). Real-time quantitative RT-PCR was performed as described previously3,4 using the Applied Biosystems Prism model 7300 sequence detection instrument and a standard SYBR green detection protocol. The sequences of the PCR primers using the SYBR green method are as follows: glyceraldehyde-3-phosphate dehydrogenase (gapdh) forward, 5′-CCCCCAATGTATCCGTTGTG-3′; gapdh reverse, 5′-TAGCCCAGGATGCCCTTTAGT-3′; abc1 forward, 5′- CTTCTTGGAAAACGATGGGGG-3′; and abc1 reverse, 5′- TGTGGTGTTGGCAAATATGTCTT-3′; tmem16f forward, 5′-AGGAGCGCATCCCATTTACC-3′; tmem16f reverse, 5′-CGATCACAGAGGCGATGATGA-3′;xkr8forward,5′-GGCCGTTGTCCAGTACGTG-3′; and xkr8 reverse, 5′-GCAAACACCTATACAGGTAGCC-3′. The samples were run in duplicate. The expression level for each sample of interest was normalized to that of gapdh.
Evaluation of PS exposure
PS exposure on transected axons of cultured DRG neurons was evaluated using Alexa594-labeled Annexin V (A13203, Thermo Fisher Scientific). Axons were co-stained with 5-(4,6-dichlorotriazinyl) aminofluorescein (DTAF) (D-16, Life Technologies) for visualization. Three non-overlapping images per explant were randomly collected. More than 5 explants were examined for each experimental condition. To evaluate PS exposure on transected axons, the fluorescent intensities of each image were measured using ImageJ software (National Institutes of Health). Quantified levels of fluorescent intensities for PS normalized to DTAF were averaged and compared with the control.
Statistical analysis
Quantification data were shown as the mean ± SEM. Differences between groups were examined for significance using one-way ANOVA with Tukey's post hoc test.
Abbreviations
- ANOVA
analysis of variance
- CRMP2
collapsin response mediator protein 2
- DRG
dorsal root ganglion
- DTAF
5-(4,6-dichlorotriazinyl) aminofluorescein
- GSK3B
glycogen synthase kinase 3B
- MCL1
myeloid cell leukemia 1
- PS
phosphatidylserine
- SEM
standard error of mean
- UPS
ubiquitin proteasome system
- WT
wild-type
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported in part by a Grant-in-Aid for Scientific Research on Innovative Areas (“Brain Environment”) from the Ministry of Education, Culture, Sports, Science and Technology (S.W.); a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (S.W.); Intramural Research Grant for Neurological and Psychiatric Disorders of NCNP (T.A.); grants from Takeda Science Foundation (S.W., and T.A.), and Pfizer academic contributions (T.A.).
References
- [1].Conforti L, Gilley J, Coleman MP. Wallerian degeneration: an emerging axon death pathway linking injury and disease. Nat Rev Neurosci 2014; 15:394-409; PMID:24840802; http://dx.doi.org/ 10.1038/nrn3680 [DOI] [PubMed] [Google Scholar]
- [2].Wang JT, Medress ZA, Barres BA. Axon degeneration: molecular mechanisms of a self-destruction pathway. J Cell Biol 2012; 196:7-18; PMID:22232700; http://dx.doi.org/ 10.1083/jcb.201108111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wong YC, Holzbaur EL. Autophagosome dynamics in neurodegeneration at a glance. J Cell Sci 2015; 128:1259-67; PMID:25829512; http://dx.doi.org/ 10.1242/jcs.161216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yang Y, Coleman M, Zhang L, Zheng X, Yue Z. Autophagy in axonal and dendritic degeneration. Trends Neurosci 2013; 36:418-28; PMID:23639383; http://dx.doi.org/ 10.1016/j.tins.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wakatsuki S, Furuno A, Ohshima M, Araki T. Oxidative stress-dependent phosphorylation activates ZNRF1 to induce neuronal/axonal degeneration. J Cell Biol 2015; 211:881-96; PMID:26572622; http://dx.doi.org/ 10.1083/jcb.201506102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wakatsuki S, Saitoh F, Araki T. ZNRF1 promotes Wallerian degeneration by degrading AKT to induce GSK3B-dependent CRMP2 phosphorylation. Nat Cell Biol 2011; 13:1415-23; PMID:22057101; http://dx.doi.org/ 10.1038/ncb2373 [DOI] [PubMed] [Google Scholar]
- [7].Wakatsuki S, Tokunaga S, Shibata M, Araki T. GSK3B-mediated phosphorylation of MCL1 regulates axonal autophagy to promote Wallerian degeneration. J Cell Biol 2017; 216:477-93; PMID:28053206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ravichandran KS. Beginnings of a good apoptotic meal: the find-me and eat-me signaling pathways. Immunity 2011; 35:445-55; PMID:22035837; http://dx.doi.org/ 10.1016/j.immuni.2011.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mellen MA, de la Rosa EJ, Boya P. The autophagic machinery is necessary for removal of cell corpses from the developing retinal neuroepithelium. Cell Death Differ 2008; 15:1279-90; PMID:18369370; http://dx.doi.org/ 10.1038/cdd.2008.40 [DOI] [PubMed] [Google Scholar]
- [10].Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell 2007; 128:931-46; PMID:17350577; http://dx.doi.org/ 10.1016/j.cell.2006.12.044 [DOI] [PubMed] [Google Scholar]
- [11].Tarling EJ, de Aguiar Vallim TQ, Edwards PA. Role of ABC transporters in lipid transport and human disease. Trends Endocrinol Metab 2013; 24:342-50; PMID:23415156; http://dx.doi.org/ 10.1016/j.tem.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nagata S, Suzuki J, Segawa K, Fujii T. Exposure of phosphatidylserine on the cell surface. Cell Death Differ 2016; 23:952-61; PMID:26891692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].negawa K, Suzuki J, Nagata S. Constitutive exposure of phosphatidylserine on viable cells. Proc Natl Acad Sc USA 2011; 108:19246-51; http://dx.doi.org/ 10.1073/pnas.1114799108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Maday S, Holzbaur EL. Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev Cell 2014; 30:71-85; PMID:25026034;http://dx.doi.org/ 10.1016/j.devcel.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med 2013; 19:983-97; PMID:23921753; http://dx.doi.org/ 10.1038/nm.3232 [DOI] [PubMed] [Google Scholar]
- [16].Menzies FM, Fleming A, Rubinsztein DC. Compromised autophagy and neurodegenerative diseases. Nat Rev Neurosci 2015; 16:345-57; PMID:25991442; http://dx.doi.org/ 10.1038/nrn3961 [DOI] [PubMed] [Google Scholar]
- [17].Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science 2004; 305:1010-3; PMID:15310905; http://dx.doi.org/ 10.1126/science.1098014 [DOI] [PubMed] [Google Scholar]