Abstract
Quorum sensing (QS) is a cell density-dependent signaling system used by bacteria to coordinate gene expression within a population. QS systems in Gram negative bacteria consist of transcription factors of the LuxR family and their acyl homoserine lactone (AHL) ligands. We describe here a method for examining QS signaling systems in mammalian cells that uses engineered LuxR-type proteins from the opportunistic pathogen, Pseudomonas aeruginosa, which can function as AHL-dependent transcription factors. The engineered proteins respond to their cognate ligands and display sequence specific DNA binding properties. This system has several potential biotechnological and biological applications. It may be used to characterize any LuxR-type protein, screen animal and plant cell extracts or exudates for compounds that mimic or interfere with AHL signaling or to screen different cell types for AHL inactivating activities.
Keywords: Pseudomonas aeruginosa autoinducer, LuxR protein
Introduction
Intercellular signaling is a vital process for coordinating cell functions in all multicellular organisms. The discovery of the phenomenon of quorum sensing (QS) revealed that bacteria, despite being unicellular organisms, can also communicate via hormone-like signaling molecules. QS is a cell-density dependent signaling mechanism used by many species of bacteria to coordinate gene expression within a population (1). Numerous Gram-negative species of bacteria synthesize N-acyl-homoserine lactones, also called autoinducers, that function as the signal molecules of QS. The first QS system was discovered through the study of bioluminescence regulation in the Gram-negative marine bacterium Vibrio fischeri (2). In this system, an inducer protein, LuxI, catalyzes the synthesis of the autoinducer molecule that binds to the receptor protein, LuxR. When bound to its autoinducer molecule, LuxR dimerizes and binds to a 20 base pair inverted repeat, the Lux box, located in the promoter region of target genes, and activates transcription of QS-regulated genes. Several LuxR homologues have been identified in many other species of bacteria and share similarity in their C-terminal DNA binding domains (3, 4).
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen that utilizes QS to cause infection (5, 6), reviewed in (7). In P. aeruginosa there are at least two complete QS systems named the Las and Rhl sytems (7). In these systems, the LasI and RhlI proteins synthesize N-3-oxododecanoyl-homoserine lactone (3O-C12-HSL) and N-butyryl-homoserine lactone (C4-HSL), respectively. 3O-C12-HSL and C4-HSL bind to their respective receptors, LasR and RhlR, which are members of the LuxR family of helix-turn-helix transcription factors (4). When bound to autoinducer, LasR and RhlR can bind to specific DNA sequences, termed las boxes, that are located in the promoter region of target genes. Many of the genes controlled by LasR and RhlR encode virulence factors and proteins involved in pathogenesis (7). Several investigators have demonstrated that 3O-C12-HSL not only controls gene expression in bacterial cells but may also modulate host immune responses both in vitro and in vivo, apparently by interacting with specific mammalian receptor proteins (8-11). Thus, elucidating the mechanisms by which bacterial autoinducers manipulate gene expression in mammalian cells may yield new therapeutic approaches for abrogating bacterial infections.
As a first step towards identifying mammalian autoinducer receptor proteins, we recently tested whether the autoinducers synthesized by P. aeruginosa could enter mammalian cells. To do this we designed a eukaryotic intracellular autoinducer sensor system (12). Our strategy was to modify the LuxR-type proteins LasR and RhlR so that they could function as autoinducer-activated transcription factors in mammalian cells. We demonstrated that the function of LasR-based chimeric proteins was dependent on the presence of its cognate autoinducer in the culture medium. These data therefore confirmed that the P. aeruginosa autoinducer that exhibits the most potent effects on mammalian gene expression can indeed diffuse across the cell membrane and suggest that mammalian autoinducer receptors may reside in the cytoplasm and/or nucleus of mammalian cells. However, this sensor system has several additional potential applications including the screening of mammalian and plant cells for production of autoinducer mimics or antagonists, and the identification of autoinducer-degrading activities in cell extracts. We describe here the design and characterization of sensor systems based on three members of the LuxR family from P. aeruginosa, LasR, RhlR and the recently identified QscR (13). We anticipate that this system will be of significant utility to others wishing to characterize QS systems and their constituent parts in other bacterial species, particularly those that are difficult to maintain in the laboratory.
Materials and Methods
Construction of expression plasmids of LuxR homologues and reporter plasmids
The coding regions of LasR, RhlR and QscR were amplified by PCR either from the mP63.9 plasmid (LasR (14)) or P. aeruginosa genomic DNA. The primers were designed based on the sequences of these genes reported in the complete sequence of the PAO1 strain of P. aeruginosa (accession number NC_002516) (see Table 1). In general, PCR primers were designed to introduce a BglII site at the 5’ end of the amplified region and a HindIII site at the 3’ end (Table 1). In certain cases primers included the sequence CCCAAGAAGAAGCGCAAGGTG, which encodes the minimal nuclear localization signal (NLS, but designated as N throughout this manuscript) from the simian virus 40 (SV40) T antigen (PKKKRKV) (15). The amplified regions were sequenced in their entirety to preclude inclusion of unintended mutations and transferred as BglII-HindIII fragments into the pMEX-FLAG expression (LasR and RhlR) plasmid that attaches an N-terminal FLAG epitope tag to expressed proteins. QscR-derived sequences were inserted into the pcDNA 3.1 Myc His B plasmid (Invitrogen Corporation, Carlbad, CA) in frame with a C-terminal Myc epitope tag. The VP16 transcriptional activation domain (TAD, but designated as T throughout this manuscript, encoding amino acids 429 to 456) has been described previously (16). Multimerized versions of the VP16 T were generated by directional cloning of individual T modules as BglII-BamHI fragments. T3 or T3N modules were transferred into pMEX-FLAG- or pcDNA 3.1-based plasmids as NcoI-BamHI fragments.
Table 1.
PCR primers used in this study
| Primers used to isolate coding sequences from P. aeruginosagenomic DNA or plasmids | ||
| LasR | GGCAGATCTCATGGCCTTGGTTGACGGT | GGCAAGCTTTCAGAGAGTAATAAGACC |
| RhlR | CGGTGCTGGCATAACAGATA | CTGCGCTTCAGATGAGACC |
| QscR | GGCAGATCTCGCCATGCATGATGAGAGAGAG | GCCAAGCTTGGGTTGAGCAGGCCGATGGC |
| Primers used to generate RhlR-based constructs | ||
| RhlRNLSBgl 1 | GGCAGATCTCCCCAAGAAGAAGCGCAAGGTGATGAGGAATGACGGAGGC | |
| RhlRDIII 241 | GGCAAGCTTTCAGATGAGACCCAGCGC | |
| RhlR Bgl 125 | GGCAGATCTCGCCATGCGCGCGCCGAACAATTTG | |
| RhlR Bgl 159 | GGCAGATCTCGCCATGCTGCGTTGCATGATCGAG | |
| RhlR Bgl 182 | GGCAGATCTCGCCATGCCGGTCTGCCTGAGC | |
| Primers used to generate QscR-based plasmids | ||
| PhzR Bgl 173 | GGCAGATCTCGCCATGCTGCTCTGGATCACCAGC | |
| The coding sequences of LasR, RhlR and QscR were isolated by PCR from either plasmid (LasR) or P. aeruginosa genomic DNA (RhlR and QscR). The primer used for isolating LasR and QscR coding regions contained BglII and HindIII restriction sites (underlined) but the RhlR primers did not. In all constructs BglII sites were introduced at the 5’ end and HindIII sites at the 3’ end. The primers used to generate RhlR and QscR-based constructs are named according to the restriction site introduced and the position at which this site was introduced. Thus the RhlR NLSBgl 1 primer introduced a BglII site prior to amino acid one of RhlR. This primer also contained the coding sequence for the T antigen NLS. | ||
Reporter plasmids were constructed by inserting two copies of either the LasBOX 1 or LasBOX 2 sequences (Table 2) upstream of the minimal promoter from the rat albumin gene and the firefly luciferase gene by a strategy described previously (16). The LasBOX sequences were designed according to either a previously reported LasR consensus recognition sequence (LasBOX 1 (17)) or a variant sequence located in the RhlA gene (LasBOX 2 (18)).
Table 2.
Sequences of LasR responsive elements in luciferase reporter plasmids
| Sequence Name | Sequence | Gene1 | Accession No. | # matches2 |
| Consensus | NNCTNNNNNNNNNNNNAGNN | |||
| lasBOX 1 | ACCTACCAGATCTGGCAGGT | |||
| RhlI | AE004768 | 19/20 | ||
| VsmI | U15644 | 19/20 | ||
| PA0122 | AE04450 | 18/20 | ||
| PA2069 | AE004633 | 17/20 | ||
| RmlB | AE004929 | 15/20 | ||
| lasBOX 2 | TCCTGTGAAATCTGGCAGTT | 14/20 | ||
| RhlA | L28170 | 20/20 | ||
|
1This is the gene located immediately downstream of the las box-like sequence in the P. aeruginosa genome. In each case listed, the las box-like sequence is located immediately 3’ to a predicted σ54 recognition site. 2This refers to the number of matches of the specific sequence found adjacent to each gene and the idealized lasBOX 1 sequence except for the number listed for RhlA which refers to its match to the lasBOX 2 sequence (i.e. it is identical to this sequence). | ||||
Cell culture and transfections (Protocol I)
The monkey kidney COS-1 cell line was cultured in Dulbecco’s modified Eagle’s medium (BioWhittaker, Walkersville, MD) supplemented with 10% Cosmic Calf Serum (HyClone, Logan, UT). The cells were transfected with 1 μg (6-well dishes) or 0.6 μg (24-well dishes) of plasmid DNA at approximately 30 to 40% confluency for immunocytochemical analysis and luciferase assays, respectively. Transfections were performed with the Effectene transfection reagent under conditions recommended by the manufacturer (Qiagen, Valencia, CA). Luciferase assays were performed with the Luciferase Assay Kit (Promega, Madison, WI). Transfections were performed in triplicate at least three times, and the results of representative experiments are presented.
Autoinducers (Protocol II)
C4-HSL and C12-HSL were obtained from Sigma (St. Louis, MO). C12-HSL has previously been shown to be an efficient ligand for LasR (19, 20) and functioned similarly to 3O-C12-HSL in all of our assays, except that higher concentrations of C12-HSL were required. 3O-C12-HSL was synthesized following a previously described procedure (19). In the synthesis, a 5-acyl Meldrum’s acid derivative was obtained at first by performing an acylation reaction in the presence of DCCI/DMAP as the catalyst, which was then followed by an amide formation method. 1H and 13C spectra were consistent with published data. Autoinducers were prepared for addition to culture as described previously (20). Briefly, autoinducer was first dissolved in ethyl acetate supplemented with 0.001% glacial acetic acid for stable storage at -20°C. The amount of dissolved autoinducer required to give the final concentrations indicated in each figure was aliquoted into a 1.5 ml microcentrifuge tube and placed under a stream of nitrogen gas to evaporate the ethyl acetate. Following evaporation, the autoinducers were dissolved in the culture medium described above immediately prior to use. Autoinducers were added to culture medium at least 4 hours after transfection, and control cells were cultured in the presence of fresh culture medium added to evaporated vehicle alone.
Immunofluorescence analysis (Protocol III)
Cells were plated on cover slips in six-well dishes and transfected as described above. The coverslips were collected 48 h after transfection, and fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) for 30 min. The cells were washed twice with PBS and then incubated with a 1:250 dilution of an anti-Myc monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 to 3 h. After washing in PBS/0.1% Tween 20, the cells were incubated with fluorescently labeled secondary antiserum (AlexaFluor 488 donkey anti-mouse immunoglobulin G [Molecular Probes, Eugene, OR]) and 0.5μg/ml 4’,6’-diamidino-2-phenylindone (DAPI) for 1 h, followed by three washes in PBS/0.1% Tween 20. The cells were observed by epifluorescence microscopy with an Axiovert 135 TV microscope (Zeiss, Göttingen, Germany). Anti-Myc immunoreactivity and DAPI staining were observed with standard fluorescein and DAPI filters (Zeiss). Images were captured with a Kodak DC290 zoom camera and analyzed with MetaMorph 6.0 (Universal Imaging Corporation, Downingtown, PA).
Results and Discussion
Construction of LuxR-based transcriptional activator proteins
The protein components of AHL-based QS systems in Gram negative bacteria are transcription factors that belong to the LuxR subfamily of NarL-type helix-turn-helix proteins (21, 22). The structure of the founding member of this family, LuxR from Vibrio fischeri (2), is shown in Fig. 1A. A domain search using the SMART algorithm revealed the presence of two predicted domains within LuxR, an N-terminal autoinducer binding domain (AIBD) and a C-terminal HTH-type DNA binding domain. The HTH domain is responsible for sequence specific DNA binding and, according to structural studies on the TraR protein from Agrobacterium tumefaciens, is also likely to include residues required for dimer-/multimerization (23). The AIBD is responsible for recognition and binding of a specific ligand (3O-C6-HSL in the case of LuxR (2)) and may contain additional residues that contribute to dimer-/multimerization (23). To date, greater than 60 members of the LuxR subfamily have been identified in a variety of bacterial species (based on a search of the non-redundant GenPept database, SCW unpublished observations); however, specific DNA recognition sequences and cognate AHL ligands are only known for a subset of these proteins.
Fig. 1. LuxR-type proteins and constructs used in this study.
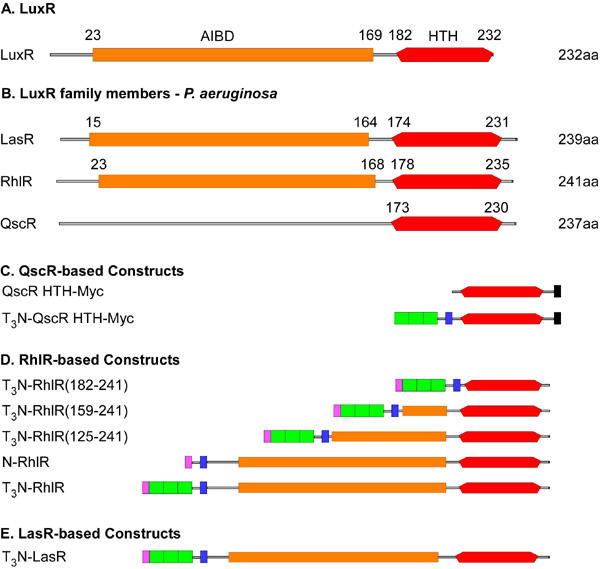
A. A schematic representation of the domain structure of the founding member of the LuxR family, the LuxR protein from Vibrio fischeri. The coordinates of the autoinducer binding domain (AIBD, red) and helix turn helix DNA binding domain (HTH, orange) were determined using the Simple Modular Architecture Research Tool (SMART) algorithm (Letunic et al., 2004). B. The domain structure of three LuxR-type proteins from P. aeruginosa is shown with domain coordinates determined as described above. Note that the SMART algorithm did not detect a consensus AIBD sequence in QscR. C. QscR-based chimeric proteins. The structures of chimeric proteins that contain the HTH domain from QscR are shown. Both proteins contain a C-terminal Myc epitope tag (black) and the T3N-QscR HTH-Myc protein contains three copies of the VP16 TAD (green) and the NLS (blue) from the SV40 T antigen (the T3N module) fused to the N-terminus of the HTH domain. D. RhlR-based constructs. The coordinates of the segments of RhlR included in the first three constructs are listed in their name. Each protein contains the T3N module except for N-RhlR, which only contain the N domain. Each protein also includes an N-teminal FLAG epitope tag (purple). E. LasR-based constructs. The structure of the single LasR-based protein used in these studies is shown.
A search of the P. aeruginosa genome revealed the presence of three LuxR-type proteins, LasR, RhlR and QscR (also known as PhzR) (4, 13). As mentioned above, the cognate ligands for LasR and RhlR are 3O-C12-HSL and C4-HSL, respectively, which are specifically bound by the respective AIBDs of each protein (Fig. 1B). A ligand for QscR has not yet been identified and the SMART algorithm failed to recognize the consensus sequence for an AIBD in QscR (Fig. 1B). While it is certainly possible to examine the mechanisms that control the intrinsic activities of these proteins in bacteria, we have developed a method for studying their mode of action in eukaryotic cells, where confounding effects of interacting bacterial proteins should be absent. We describe below the strategies we have developed to examine the function of LuxR-type proteins in mammalian cell lines, using the three proteins from P. aeruginosa as examples. We propose that these approaches will provide reliable assays for examining many aspects of QS systems.
We first developed strategies for expressing bacterial LuxR-type proteins that could function as transcription factors in mammalian cells, and the structures of the resultant chimeric proteins described in this study are shown schematically in Fig. 1C-E. This strategy is similar to one employed to convert the TraR protein into a transcriptional activator capable of functioning in eukaryotic cells (24). There are two functions required of eukaryotic transcription factors that would normally be absent in their bacterial counterparts. As bacteria lack nuclei, bacterial proteins do not contain nuclear localizations signals (NLS), except in rare, presumably serendipitous, cases. Therefore, we have incorporated sequences encoding the NLS from the SV40 T antigen protein into the expression plasmids. Second, eukaryotic transcription factors usually contain transcriptional activation domains (TAD) that facilitate communication between the DNA-bound transcription factor and the general transcriptional machinery located at the proximal promoter. Eukaryotic TADs are generally defined based on the presence of a preponderance of a specific amino acid or class of amino acids. In these constructs, we have included three copies of the potent acidic TAD from the Herpes Simplex Virus VP16 protein. We previously demonstrated that including multiple copies of a TAD in these chimeric proteins results in high level, synergistic activation when compared to proteins with a single TAD (12).
Nuclear localization of QscR-based chimeric proteins
Transcription factors are generally modular proteins and thus we predicted that chimeric proteins containing just the HTH domains of LuxR-type proteins should function as constitutive activators of transcription, assuming they contain the requisite eukaryotic peptide domains. To test this proposal, we first examined the effect of including a NLS on the subcellular localization of proteins containing the HTH domain of QscR. Two constructs were generated that contained amino acids 173-237 of QscR fused to a C-terminal Myc epitope to facilitate immunological detection. One construct also contained the SV40 NLS and three copies of the VP16 TAD (T3 module). We previously determined that the T3 module does not affect subcellular location of chimeric proteins (EKP, unpublished observations). The subcellular location of each protein was monitored by immunofluorescence after expression in COS-1 cells. The QscR HTH-Myc protein was detected almost entirely in vesicular structures within the cytoplasm of transfected cells and was excluded from the nucleus (detected by staining with DAPI, Fig. 2). This pattern is similar to that seen with both RhlR- and LasR-based proteins (12). In contrast, the T3N-QscR HTH-Myc protein was efficiently localized to the nucleus as indicated by the overlap between staining in the immunofluorescence and DAPI images (Fig. 2). Thus the presence of a NLS permits efficient localization of chimeric proteins to the nucleus.
Fig. 2. Visualization of the subcellular location of QscR-based chimeric proteins expressed in COS-1 cells.
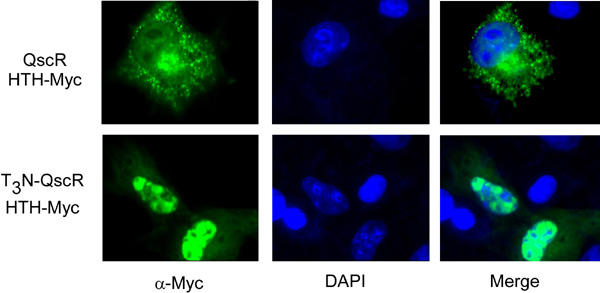
COS-1 cells expressing either of the indicated QscR-based proteins were fixed and QscR proteins were detected using an antiserum directed against the Myc epitope as described in materials and methods. The nuclei of the cells were stained with, 6’-diamidino-2-phenylindone (DAPI, blue image in central panel). The left and center images were merged to detect overlap between the signals.
Transcriptional activity of RhlR-based chimeric proteins
At present, the DNA sequence recognized by QscR has not been identified, thus we have not yet examined the transcriptional activity of QscR-based chimeric proteins. However, binding sites for both LasR and RhlR are known and thus we performed transcription assays with these proteins. The LuxR transcriptional activator protein activates transcription of target genes by binding to a 20 bp region of dyad symmetry, called the lux box, which is located in the promoter region of LuxR-regulated genes (17). Consensus lux box-like sequences have subsequently been identified within the promoters of other genes regulated by LuxR homologues (25). Lux box-like elements have been identified in the promoter regions of several LasR- and RhlR-regulated genes in P. aeruginosa (17). These “las boxes” were palindromic in nature and contained a loose consensus sequence of NNCT-(N)12-AGNN (Table 2) (17). We constructed two luciferase-based reporter plasmids containing two variations of the las box consensus sequence (Table 2). The first contained two copies of lasBox 1 ((lasBOX 1)2-Luc), which is a perfect palindrome that matches 19/20 bases of a sequence found in the promoter of the RhlI gene, a major transcriptional target of LasR (26). Similar sequences that contain exact matches of at least 15 adjacent bases within this idealized las box are present in the promoter of four other genes in P. aeruginosa (Table 2). The second reporter plasmid ((lasBOX 2)2-Luc) contained two copies of a las box-like sequence located in the promoter of the RhlA gene (18). This sequence matches the loose consensus but differs from the lasBox 1 sequence at several internal bases.
The transcriptional activities of three proteins that contain successive greater segments of the C-terminus of RhlR fused to the T3N module were tested on both reporter plasmids in COS-1 cells (Fig. 3). A fusion protein containing just the HTH domain of RhlR (T3N-RhlR(182-241)) increased activation of the (lasBOX 1)2-Luc reporter approximately 10 fold when compared to cells transfected with the reporter plasmid alone.
Fig. 3. Sequence-specific transcriptional activation by RhlR-based proteins.
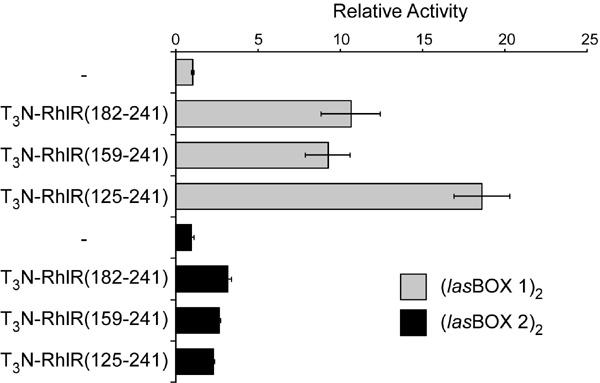
Mammalian expression vectors encoding the indicated RhlR-based chimeric proteins were co-transfected into COS-1 cells with either the (lasBOX 1)2-Luc or (lasBOX 2)2-Luc reporter plasmids and luciferase values were measured. The activity detected in cells transfected with the (lasBOX 1)2-Luc plasmid alone was set to 1.0 and the activity measured for the other transfections were calculated relative to this value. All data points were performed in triplicate and data representative of three repetitions are shown.
A fusion protein containing an additional 23 additional amino acids (T3N-RhlR(159-241)) exhibited similar activity to the protein containing the HTH domain alone; however, inclusion of an additional 34 amino acids (T3N-RhlR(125-241)) resulted in an approximately two-fold increase in luciferase activity. Each of these three proteins displayed significantly lower activity on the (lasBOX 2)2-Luc reporter plasmid and the enhanced activity of T3N-RhlR(125-241) was not observed on this reporter. These data corroborate our earlier studies on LasR (12) indicating that the HTH domains of LuxR-type proteins contain sequences necessary and sufficient for DNA binding and dimer-/multimerization (23). Earlier structural studies on the TraR protein suggested that additional sequences within the N-terminal AIBD may also contribute to dimer-/multimer formation and the increased activity of the T3N-RhlR(125-241) protein may result from the inclusion of analogous sequences from RhlR. Finally, the difference in activities of the RhlR proteins on the two reporters suggests that this assay may be useful in defining the contribution of DNA sequences within the Las BOX and specific domains within RhlR to promoter recognition and activation. Additional studies are currently underway to extend these observations.
Autoinducer-dependent activation of RhlR chimeric proteins
To test whether autoinducer-dependent gene regulation could be reconstituted in mammalian cells, we next tested whether chimeric proteins containing the full length RhlR coding sequence were sensitive to addition of its cognate autoinducer, C4-HSL. In this experiment, we also examined the contribution of the T3 module to transcriptional activation by chimeric proteins. COS-1 cells were cotransfected with the (lasBOX 1)2-Luc reporter plasmid and an expression vector encoding either N-RhlR or T3N-RhlR (see Fig. 1). The transfected cells were cultured either in the absence or presence of 250 μM C4-HSL for 40 hours and luciferase values were measured. Expression of the N-RhlR protein resulted in a small increase in luciferase activity compared to control and this activity was unchanged by the addition of C4-HSL (Fig. 4). In contrast, expression of T3N-RhlR resulted in significantly higher activity in the absence of C4-HSL and addition of C4-HSL yielded an approximately three-fold increase in activity. Although most of the unliganded T3N-RhlR is found in the cytoplasm (12), its relatively high activity in the absence of C4-HSL was not entirely unexpected as RhlR has been reported to form homodimers and bind DNA in the absence of autoinducer (27, 28). Therefore, these data demonstrate that RhlR functions as a ligand-regulated transcription factor in this heterologous system.
Fig. 4. Autoinducer-dependent activation of RhlR-based chimeric proteins.
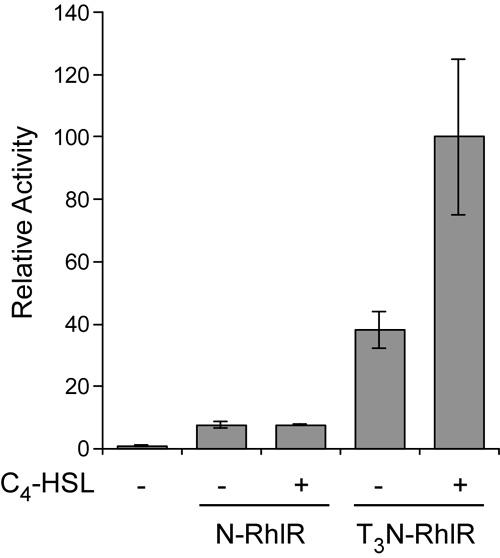
COS-1 cells were cotransfected with expression vectors encoding the indicated RhlR-based chimeric proteins and the (lasBOX 1)2-Luc reporter plasmid in the absence or presence of C4-HSL for 40 hours. Luciferase values were measured and expressed relative to the activity of the reporter plasmid alone.
Autoinducer-specific activation of LasR-based fusion proteins
The activation of different members of the LuxR family by acyl homoserine lactones that differ in acyl chain length and modification is the underlying basis of the specificity of QS responses. Previous studies in bacteria indicated that the presence of a carbonyl group at position 3 on the acyl side chain of 3O-C12-HSL is critical for efficient activation of LasR (20). To test whether this specificity is recapitulated in eukaryotic cells, we examined the responsiveness of the T3N-LasR protein to synthetic autoinducers that differed at position 3 on the acyl chain. COS-1 cells were cotransfected with the (lasBOX 1)1-Luc reporter plasmid and the T3N-LasR expression plasmid and cultured in the absence or presence of either 3O-C12-HSL or C12-HSL. Each AHL was used at concentrations previously determined to be maximally effective. In addition, the cells were exposed to each autoinducer for different length of time to assess the most effective exposure times. Although the T3N-LasR protein exhibited low basal activity, exposure to both synthetic autoinducers results in significant enhancement of luciferase activity, with maximal enhancement (>10 fold) observed after 8 hours of exposure (Fig. 5). Exposure for longer time periods results in a drop in autoinducer-dependent enhancement with only minor effects observed after 48 hours. Recently, it was shown that airway epithelial cells produce AHL-inactivating enzymes (29) and the time-dependent decrease in AHL-responsiveness observed here may be due to similar inactivating mechanisms. Alternatively, exposure to 3O-C12-HSL has been associated with apoptosis in macrophages and neutrophils (30) and thus the time-dependent decrease in activity may be due to progressive cell death. These data indicate that the mammalian expression system recapitulates autoinducer specificity of LasR responsiveness to synthetic AHLs and extend and corroborate our previous studies using 3O-C12-HSL purified from bacterial cell cultures (12).
Fig. 5. Temporal and autoinducer-specific activation of LasR-based chimeric proteins.
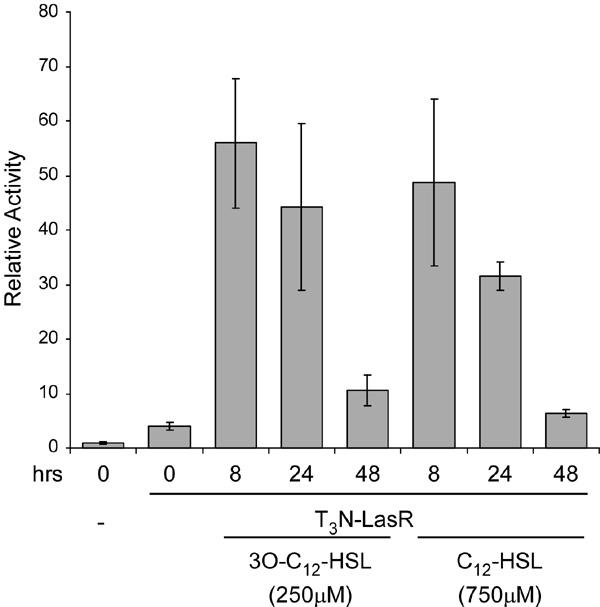
COS-1 cells were cotransfected with a mammalian expression plasmid encoding T3N-LasR and the (lasBOX 1)2-Luc reporter plasmid. The cells were incubated in the absence or presence of either 250μM 3O-C12-HSL or 750μM C12-HSL for three different time periods and luciferase activities were measured. The data are presented relative to the activity of the reporter plasmid alone. The highest activity of T3N-LasR was observed after 8 hours incubation with either 3O-C12-HSL or C12-HSL.
In conclusion, we describe a mammalian cell culture-based system for examining the mechanisms of action of components of bacterial QS systems. Specifically, we demonstrate that the attachment of specific domains required for nuclear function of eukaryotic proteins permits bacterial LuxR-type proteins to function as ligand-activated transcription factors in mammalian cells. Furthermore, these chimeric proteins display autoinducer specificities predicted from studies in bacteria and sequence specific transcriptional activity. We propose that this system can be adapted to examine several aspects of QS systems. First, as demonstrated by our studies on three members of the LuxR family from P. aeruginosa, this strategy can be adapted to identify functional domains within any LuxR-type protein, to test the responsiveness of LuxR proteins to autoinducers of different structures, and to identify DNA sequences recognized by each protein. By expressing different members of the LuxR family together, it should be possible to determine whether some family members can heterodimerize and whether heterodimers possess unique functions when compared to each homodimer. Furthermore, this system can be used to screen for compounds that function as AHL mimics or that interfere with QS systems, such as those identified in many plant species (31, 32). Finally, this system may also be useful for screening different cell lines for endogenous cell products that block QS, such as the lactonase activity identified in airway epithelial cells (29).
Acknowledgments
The expertise of Vijian Dhevan with fluorescence imaging is greatly appreciated. This work was supported in part by TTUHSC SOM Seed Grant and South Plains Foundation Grant to K.P.R and by an NIH grant (CA 99995-1) to G.L. This research was supported in part by a Howard Hughes Medical Institute grant through the Undergraduate Biological Sciences Education Program to Texas Tech University. The authors have no conflicts of interest to declare related to this publication.
Abbreviations
- QS
quorum sensing
- AHL
acylhomoserine lactone
- 3O-C12-HSL
N-3-oxodocanoyl-homoserine lactone
- C4-HSL
N-butyryl homoserine lactone
- 3O-C6-HSL
N-3-oxoheptanoyl homoserine lactone
- T3N
protein module containing three copies of the VP16 transcriptional activation domain (TAD) and the nuclear localization signal (NLS) from SV40 T antigen
- DAPI
4’6’-diamidino-2-phenylindone
- SMART
Simple Modular Architecture Research Tool
- AIBD
autoinducer binding domain
- HTH
helix-turn-helix DNA binding domain
Appendix
Protocols
Protocol I: Transfection and luciferase assays
Seed 40,000 COS-1 cells into each well of a 24-well tissue culture plate and incubate overnight.
At ~40% confluency, transfect each well of cells with 0.4 μg of effector plasmid DNA and 0.2 μg of reporter plasmid DNA, running each experimental condition in triplicate.
Transfect the cells according to the recommendation of the manufacturer of the transfection reagents.
4 h following transfection, change the media on the cells.
Incubate the transfected cells for 48 h and then lyse the cells with the lysis buffer and determine the luciferase activity using a commercially available kit (e.g. Luciferase Assay Kit (Promega, Madison, WI)).
Protocol II: Preparation of autoinducer
Dissolve the autoinducer in ethyl acetate supplemented with 0.001% glacial acetic acid to a final concentration of 20mM. This can be stored stably at -20°C.
To stimulate cells in culture with autoinducer, place the amount of dissolved autoinducer that is calculated to give the final concentration in the cell culture in a microcentrifuge tube.
Evaporate off the ethyl acetate solution by placing under a stream of nitrogen gas. Keep the dried autoinducer on ice.
To dissolve the autoinducer into the culture medium, draw the volume of pre-warmed culture medium appropriate to the culture conditions into a conical tube (e.g. for a 6-well dish, the volume will be 1.5 ml per well).
Using a pipet, transfer 500 μl of the medium from the conical tube to the microcentrifuge tube containing the dried autoinducer.
Pipette up and down to dissolve the autoinducer, taking care to remove any autoinducer adhering to the sides of the microcentrifuge tube, and return the medium to the conical tube.
Repeat steps 4-6 until all the autoinducer is removed from the microcentrifuge tube. Some autoinducer molecules, particularly those with longer acyl chains, are relatively insoluble and care must be taken to ensure that all of the autoinducer has been removed.
Vortex the conical tube for 15-60 sec to ensure complete dissolution of the autoinducer.
Aspirate the spent medium from the cell culture and add the medium containing the dissolved autoinducer. The length of time required for autoinducer effects to be observed must be determined empirically.
Protocol III: Immunofluorescence analysis
Seed 150,000 COS-1 cells onto 22 mm x 22 mm glass cover slips placed at the bottom of each well of a 6-well tissue culture plate and incubate overnight.
At ~30% confluency, transfect the cells with 1 μg of plasmid DNA according to standard transfection procedure of the laboratory.
After two days of incubation, remove the glass coverslips and place into a coverslip staining rack.
Fix the cells onto the coverslips by incubating the cells in PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.3)/3.7% formaldehyde at room temperature (it is best to use 100 ml of solution in a 250 ml beaker and to place the boat containing the coverslips into the beaker for all the steps of this protocol, except for probing with antibody).
All of the following steps can be carried out at room temperature. The volume of buffers should be sufficient to completely submerge the coverslips.
Incubate the coverslips in PBS/1% Triton-X-100 for 10 min.
Wash the coverslips 2 times in PBS/0.1% Tween 20 (PBS-T) for 10 min.
Carefully place the coverslips onto parafilm on a flat surface. Dilute the primary antibody solution according to the manufacturer’s specifications in PBS-T and pipet the solution directly onto the coverslips (150-200μl/slide).
Incubate for 1-3 hrs.
Return the coverslips into the boat and wash 3 times with PBS-T for 10 min.
Place the coverslips onto parafilm on a flat surface and pipette secondary antibody and 0.5 μg/ml 4’,6’-diamidino-2-phenylindone (DAPI, Molecular Probes) in PBS-T. Incubate at room temperature for 1 h.
Return the coverslips to the boat and wash 3 times in PBS-T for 10 min.
Wash the coverslips in ddH2O for 5 min.
Mount the coverslips onto microscope slides using 20 μl mounting solution (e.g. Mowiol (Polyvinyl alcohol, Calbiochem, San Diego, CA)).
Visualize the cells using epifluorescence microscopy in order to determine the expression and subcellular localization of the protein of interest.
References
- Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: Acyl-Homoserine Lactone Quorum Sensing. Annual Review of Genetics. 2001;35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- Fuqua C, Winans SC, Greenberg EP. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- Slock J, VanRiet D, Kolibachuk D, Greenberg EP. Critical regions of the Vibrio fischeri luxR protein defined by mutational analysis. J Bacteriol. 1990;172:3974–3979. doi: 10.1128/jb.172.7.3974-3979.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens AM, Greenberg EP. Transcriptional Activation by LuxR. In: Cell-Cell signaling in bacteria. Edited by Dunny GM, Winans SC. Washington, D.C.: American Society for Microbiology, 1999: 231-242.
- Rumbaugh KP, Griswold JA, Hamood AN. Contribution of the regulatory gene lasR to the pathogenesis of Pseudomonas aeruginosa infection of burned mice. J Burn Care Rehabil. 1999;20:42–49. doi: 10.1097/00004630-199901001-00008. [DOI] [PubMed] [Google Scholar]
- Rumbaugh KP, Griswold JA, Hamood AN. The role of quorum sensing in the in vivo virulence of Pseudomonas aeruginosa. Microbes Infect. 2000;2:1721–1731. doi: 10.1016/s1286-4579(00)01327-7. [DOI] [PubMed] [Google Scholar]
- Smith RS, Iglewski BH. P. aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol. 2003;6:56–60. doi: 10.1016/S1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- Telford G, Wheeler D, Williams P, Tomkins PT, Appleby P, Sewell H, Stewart GS, Bycroft BW, Pritchard DI. The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-L-homoserine lactone has immunomodulatory activity. Infect Immun. 1998;66:36–42. doi: 10.1128/iai.66.1.36-42.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RS, Fedyk ER, Springer TA, Mukaida N, Iglewski BH, Phipps RP. IL-8 production in human lung fibroblasts and epithelial cells activated by the Pseudomonas autoinducer N-3-oxododecanoyl homoserine lactone is transcriptionally regulated by NF-kappa B and activator protein-2. J Immunol. 2001;167:366–374. doi: 10.4049/jimmunol.167.1.366. [DOI] [PubMed] [Google Scholar]
- Smith RS, Harris SG, Phipps RP, Iglewski BH. The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)homoserine lactone contributes to virulence and induces inflammation in vivo. J Bacteriol. 2002;184:1132–1139. doi: 10.1128/jb.184.4.1132-1139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RS, Kelly R, Iglewski BH, Phipps RP. The Pseudomonas autoinducer N-(3-oxododecanoyl) homoserine lactone induces cyclooxygenase-2 and prostaglandin E2 production in human lung fibroblasts: implications for inflammation. J Immunol. 2002;169:2636–2642. doi: 10.4049/jimmunol.169.5.2636. [DOI] [PubMed] [Google Scholar]
- Williams SC, Patterson EK, Carty NL, Griswold JA, Hamood AN, Rumbaugh KP. Pseudomonas aeruginosa autoinducer enters and functions in mammalian cells. J Bacteriol. 2004;186:2281–2287. doi: 10.1128/JB.186.8.2281-2287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugani SA, Whiteley M, Lee KM, D'Argenio D, Manoil C, Greenberg EP. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. PNAS. 2001;98:2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambello MJ, Iglewski BH. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Williams SC, Baer M, Dillner AJ, Johnson PF. CRP2 (C/EBPb) contains a bipartite regulatory domain that controls transcriptional activation, DNA-binding activity and cell specificity. EMBO J. 1995;14:3170–3183. doi: 10.1002/j.1460-2075.1995.tb07319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley M, Greenberg EP. Promoter specificity elements in Pseudomonas aeruginosa quorum-sensing-controlled genes. J Bacteriol. 2001;183:5529–5534. doi: 10.1128/JB.183.19.5529-5534.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahim R, Ochsner UA, Olvera C, Graninger M, Messner P, Lam JS, Soberon-Chavez G. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol Microbiol. 2001;40:708–718. doi: 10.1046/j.1365-2958.2001.02420.x. [DOI] [PubMed] [Google Scholar]
- Chhabra SR, Harty C, Hooi DS, Daykin M, Williams P, Telford G, Pritchard DI, Bycroft BW. Synthetic analogues of the bacterial signal (quorum sensing) molecule N-(3-oxododecanoyl)-L-homoserine lactone as immune modulators. J Med Chem. 2003;46:97–104. doi: 10.1021/jm020909n. [DOI] [PubMed] [Google Scholar]
- Passador L, Tucker KD, Guertin KR, Journet MP, Kende AS, Iglewski BH. Functional analysis of the Pseudomonas aeruginosa autoinducer PAI. J Bacteriol. 1996;178:5995–6000. doi: 10.1128/jb.178.20.5995-6000.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch JA, Silhavy TJ. Two component signal transduction. Washington, DC: American Society of Microbiology; 1995.
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Vannini A, Volpari C, Gargioli C, Muraglia E, Cortese R, De Francesco R, Neddermann P, Marco SD. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 2002;21:4393–4401. doi: 10.1093/emboj/cdf459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neddermann P, Gargioli C, Muraglia E, Sambucini S, Bonelli F, De Francesco R, Cortese R. A novel, inducible, eukaryotic gene expression system based on the quorum-sensing transcription factor TraR. EMBO Rep. 2003;4:159–165. doi: 10.1038/sj.embor.embor734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua WC, Winans SC, Greenberg EP. Quo rum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner UA, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventre I, Ledgham F, Prima V, Lazdunski A, Foglino M, Sturgis JN. Dimerization of the quorum sensing regulator RhlR: development of a method using EGFP fluorescence anisotropy. Mol Microbiol. 2003;48:187–198. doi: 10.1046/j.1365-2958.2003.03422.x. [DOI] [PubMed] [Google Scholar]
- Medina G, Juarez K, Diaz R, Soberon-Chavez G. Transcriptional regulation of Pseudomonas aeruginosa rhlR, encoding a quorum-sensing regulatory protein. Microbiology. 2003;149:3073–3081. doi: 10.1099/mic.0.26282-0. [DOI] [PubMed] [Google Scholar]
- Chun CK, Ozer EA, Welsh MJ, Zabner J, Greenberg EP. Inactivation of a Pseudomonas aeruginosa quorum-sensing signal by human airway epithelia. Proc Natl Acad Sci USA. 2004;101:3587–3590. doi: 10.1073/pnas.0308750101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateda K, Ishii Y, Horikawa M, Matsumoto T, Miyairi S, Pechere JC, Standiford TJ, Ishiguro M, Yamaguchi K. The Pseudomonas aeruginosa autoinducer N-3-oxododecanoyl homoserine lactone accelerates apoptosis in macrophages and neutrophils. Infect Immun. 2003;71:5785–5793. doi: 10.1128/IAI.71.10.5785-5793.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U, Mulders S, Gao M, Teplitski M, Caetano-Anolles G, Rolfe BG, Bauer WD. Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc Natl Acad Sci USA. 2003;100:1444–1449. doi: 10.1073/pnas.262672599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplitski M, Robinson JB, Bauer WD. Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol Plant Microbe Interact. 2000;13:637–648. doi: 10.1094/MPMI.2000.13.6.637. [DOI] [PubMed] [Google Scholar]


