Abstract
Chlamydomonadalean green algae are no stranger to linear mitochondrial genomes, particularly members of the Reinhardtinia clade. At least nine different Reinhardtinia species are known to have linear mitochondrial DNAs (mtDNAs), including the model species Chlamydomonas reinhardtii. Thus, it is no surprise that some have suggested that the most recent common ancestor of the Reinhardtinia clade had a linear mtDNA. But the recent uncovering of circular-mapping mtDNAs in a range of Reinhardtinia algae, such as Volvox carteri and Tetrabaena socialis, has shed doubt on this hypothesis. Here, we explore mtDNA sequence and structure within the colonial Reinhardtinia algae Yamagishiella unicocca and Eudorina sp. NIES-3984, which occupy phylogenetically intermediate positions between species with opposing mtDNA mapping structures. Sequencing and gel electrophoresis data indicate that Y. unicocca has a linear monomeric mitochondrial genome with long (3 kb) palindromic telomeres. Conversely, the mtDNA of Eudorina sp., despite having an identical gene order to that of Y. unicocca, assembled as a circular-mapping molecule. Restriction digests of Eudorina sp. mtDNA supported its circular map, but also revealed a linear monomeric form with a matching architecture and gene order to the Y. unicocca mtDNA. Based on these data, we suggest that there have been at least three separate shifts in mtDNA conformation in the Reinhardtinia, and that the common ancestor of this clade had a linear monomeric mitochondrial genome with palindromic telomeres.
Keywords: Eudorina, linear mitochondrial DNA, telomere, Volvox, Yamagishiella
Introduction
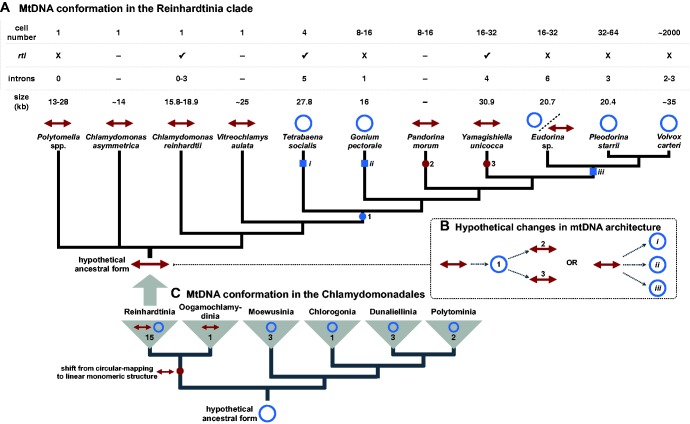
One of the first linear mitochondrial genomes to be studied in detail was that of the model unicellular green alga Chlamydomonas reinhardtii (Boer et al. 1985; Gray and Boer 1988; Vahrenholz et al. 1993), which belongs to the Reinhardtinia clade of the Chlamydomonadales (Chlorophyceae, Chlorophyta) (Nakada et al. 2008) (fig. 1A). At the time that it was first characterized, the linear conformation and defined telomeres of the C. reinhardtii mitochondrial DNA (mtDNA) differed drastically from the circular maps typical of animal (Boore 1999) and yeast mtDNAs (Foury et al. 1998).
Fig. 1.—
Variation in mitochondrial genome structure across Reinhardtinia-clade (A) and chlamydomonadalean (C) algae. Blue circles and red arrowed lines represent circular-mapping and linear-mapping mtDNA conformations, respectively. Organismal and mitochondrial genome features are shown in the table above the tree in (A); rtl: reverse-transcriptase-like gene; dash (–): data not available. Squares and circles on the branches of the trees denote hypothetical shifts from linear to circular-mapping conformations (blue) and from circular-mapping to linear structures (red). Two hypothetical scenarios to explain the evolution of mtDNA conformation in Reinhardtinia algae (B); for others see supplementary figure S2, Supplementary Material online. Branching orders based on published phylogenetic analyses, including Nozaki et al. (2006), Nakada et al. (2008), Herron et al. (2009), and Lemieux et al. (2015), as well as the phylogenetic analysis shown in supplementary figure S3, Supplementary Material online. Note, some Polytomella and Oogamochlamydinia species can have linear-fragmented mtDNAs (Borza et al. 2009; Smith, Hua, et al. 2013).
Decades of further organelle genomic research, however, have proven that linear mtDNAs are not nearly as uncommon or exceptional as once thought, and can be found in a wide diversity of groups, including animals, fungi, land plants, and various protists (Nosek et al. 1998; Smith and Keeling 2013). Moreover, in addition to C. reinhardtii, other Reinhardtinia algae are now known to have linear mitochondrial genomes (fig. 1A), such as Chlamydomonas asymmetrica (Laflamme and Lee 2003), the colonial volvocine Pandorina morum (Moore and Coleman 1989), and all known members of the nonphotosynthetic genus Polytomella (Smith, Hua, et al. 2013). Some Polytomella species even have linear fragmented mitochondrial genomes (Smith, Hua, et al. 2013), as does Lobochlamys culleus (Borza et al. 2009), which belongs to the Oogamochlamydinia, a sister clade to the Reinhardtinia (Nakada et al. 2008) (fig. 1C).
But linear mitochondrial genomes are by no means ubiquitous throughout the Reinhardtinia or its sister clades (fig. 1A and fig. 1C). In fact, many close relatives of C. reinhardtii have circular-mapping mtDNAs, including Tetrabaena socialis (Featherston et al. 2016), Gonium pectorale (Hamaji et al. 2013), Pleodorina starrii (Smith, Hamaji, et al. 2013), and Volvox carteri (Smith and Lee 2010). Altogether, the Reinhardtinia—and the Chlamydomonadales as a whole—is a hodgepodge of mtDNA architectures, which has led to speculation about the origins and evolution of linear mitochondrial chromosomes within this group (Borza et al. 2009; Featherston et al. 2016). For example, how many changes in mtDNA conformation have occurred within the Reinhardtinia? And did the ancestor of the clade have a linear- or circular-mapping genome?
Here, we address these questions by exploring the mtDNA sequence and structure of the colonial (16–32 cells) Reinhardtinia algae Yamagishiella unicocca NIES-3982 and Eudorina sp. NIES-3984. Both algae come from the volvocine line (Herron et al. 2009) and occupy phylogenetically intermediate positions between species with opposing mtDNA structures, namely Pandorina morum and Pleodorina starrii, which have linear- and circular-mapping mtDNAs, respectively (fig. 1A). Together, the Y. unicocca and Eudorina sp. mtDNA architectures provide a surprisingly dynamic picture of mitochondrial genomic architecture within the Reinhardtinia, one involving multiple shifts from linear- to circular-mapping forms, and potentially the reverse process.
Materials and Methods
Yamagishiella unicocca strain NIES-3982 (mating type plus) and Eudorina sp. strain NIES-3984 (female) were grown in 300 ml SVM medium (Kirk and Kirk 1983) at 25 °C on a 14:10 h light–dark cycle (150–180 mmol photons/m2/s). Total DNA from each species was isolated following the protocol of Miller et al. (1993) and sequenced using PacBio (RS II system; SMRTbell 10 and 20 kb library preparations) and Illumina (HiSeq 2000 and 2500 systems; paired-end TruSeq library prep kit) technologies. The Eudorina sp. PacBio subreads were mapped to the C. reinhardtii and G. pectorale mitochondrial genomes (GenBank accessions NC_001638 and NC_020437) using BLASR (Pacific Biosciences, Menlo Park, CA) and then assembled de novo with HGAP3 (Pacific Biosciences). The Illumina data were then mapped against the PacBio assembly using BWA-MEM Release 0.7.7 (Li and Durbin 2010), including error correction with the samtools/bcftools/vcfutils.pl program v0.1.19 (http://samtools.sourceforge.net/), ultimately giving a complete Eudorina sp. mitochondrial genome sequence. Similarly, the Y. unicocca PacBio subreads were mapped against the Eudorina sp. mtDNA using BLAST v2.2.26 (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and then assembled and corrected as described above, yielding a linear-mapping Y. unicocca mitochondrial genome sequence. Any unmapped regions were PCR-amplified from genomic DNA and sequenced using an ABI 3730xl DNA Analyzer (Applied Biosystems, Waltham, MA).
Restriction digests of genomic DNA were electrophoresed in agarose gels and blotted onto Hybond N+ membrane (GE Healthcare, Buckinghamshire, UK) using a 785 vacuum blotter (Bio-Rad, Hercules, CA). Probes used in this study were labeled and hybridized to samples with the PCR DIG Probe Synthesis Kit and DIG Luminescent Detection Kit (Roche, Penzberg, Germany) following manufacturer’s instructions. Label-associated chemiluminescence was detected by ChemiDoc XR (Bio-Rad). The acquired images were processed with a median filter (radius: 2 pixels) in FIJI (Schindelin et al. 2012) to remove random noise from long exposures (2 h).
Yamagishiella unicocca Has a Linear Monomeric mtDNA with Palindromic Telomeres
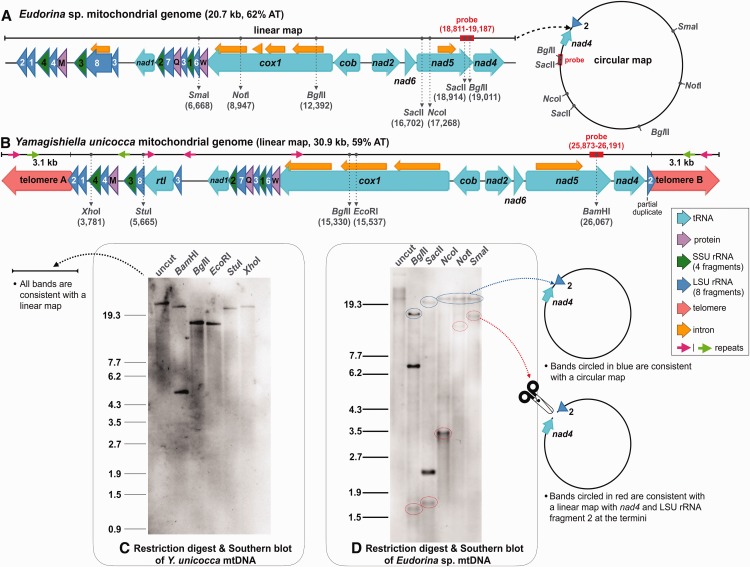
We used a combination of single molecule real time (SMRT) sequencing, Illumina sequencing, restriction digest, gel electrophoresis, and Southern blotting to investigate the mitochondrial genomic architectures of Y. unicocca and Eudorina sp. (fig. 2A–D). The Y. unicocca mitochondrial genome (GenBank accession KY442293) assembled as a 30.9 kb, AT-rich (59%) linear molecule with 3 kb telomeres, which together form a palindromic repeat (fig. 2B). Linear mtDNAs, because of their often elaborate terminal structures (e.g., single-stranded closed loops), can be difficult to sequence (Vahrenholz et al. 1993; Smith and Lee 2008), but the recent advent of SMRT (i.e., PacBio) sequencing (Eid et al. 2009) has made them much easier to decode. This is because PacBio technology can yield long reads (10-15 kb), long enough, in some cases, to capture an entire mitochondrial genome in a few or even a single read, which was the case for Y. unicocca. In fact, the entire “A” and “B” telomeric sequences of the Y. unicocca mtDNA (fig. 2B), which are identical, were clearly distinguishable from the PacBio data. There were also no traces of circular or concatemeric (i.e., tandemly repeated) mtDNAs in either the PacBio or Illumina reads, nor could such forms be amplified via PCR, further supporting the linear monomeric architecture of this genome.
Fig. 2.—
Mitochondrial genome maps and mtDNA restriction digest analyses for Eudorina sp. (A and D) and Yamagishiella unicocca (B and C). Gel electrophoresis was performed with OneSTEP Marker 6 (λ/Sty I digest; Nippon Gene, Tokyo, Japan). See Materials and Methods for details on gel electrophoresis and blotting procedures.
Restriction digests of the Y. unicocca mtDNA were consistent with the linear map derived from the sequencing data (fig. 2C). The genome was independently digested with five different single-cutter restriction enzymes, the products were run on an agarose gel, and then blotted with a probe built from the mitochondrial nad5 gene (fig. 2B). All the bands highlighted in the Southern blot supported the linear map, including the uncut Y. unicocca mtDNA, which comigrated with linear markers at its expected size based on the genome assembly (fig. 2C).
When ignoring the reverse transcriptase-like gene rtl (discussed below), the mtDNA gene content of Y. unicocca is identical to that of other available volvocine algae (figs. 1A and 2B), including the presence of fragmented and scrambled large-subunit (LSU) and small-subunit (SSU) rRNA-coding regions (Boer and Gray 1988a). The second coding fragment of the LSU rRNA (rnl2) is located at the “A” terminus of the genome and 94 nt of this 114 nt region are integrated into telomere A. Consequently, a partial duplicate of rnl2 is found at the “B” terminus (telomere B) (fig. 2B). It is well documented that gene conversion between opposing telomeres can cause telomere expansion and the duplication of subtelomeric loci in linear mitochondrial genomes (Smith and Keeling 2013), and this is likely how rnl2 was duplicated and incorporated into the telomeres.
Two sections of the telomeres share near-perfect sequence identity with certain noncoding regions within the genome. For example, a 368 nt segment of intergenic mtDNA between trnM and rns3 (see green arrows on fig. 2B) is 99.7% identical to telomeric DNA, and a 266 nt segment of the third intron of cox1 is 100% identical to another segment of telomeric DNA (see pink arrows on fig. 2B). The latter repeat is also present in two other intergenic locations. Together, these data suggest that there is a lot of intra and intergenomic recombination and gene conversion occurring within the Y. unicocca mitochondrion.
The mtDNA of Eudorina sp. Can Be Found in Both Circular- and Linear-Mapping Forms
Unlike for Y. unicocca, the mtDNA of Eudorina sp. assembled as a circular-mapping molecule (20.7 kb, 62% AT, GenBank accession KY442294) (fig. 2A). Moreover, no mitochondrial telomeric-like sequences, or long palindromic repeats of any kind, were detected in the assembly or raw sequencing reads. However, a modest number (12) of short (60–140 nt) palindromes were found in the intergenic and intronic mtDNA. Despite their contrasting mapping structures, the mitochondrial gene order and content of Eudorina sp. and Y. unicocca are identical (rtl notwithstanding), including the shift in transcriptional polarity at the cob-nad2 junction (fig. 2A and B).
The Eudorina sp. mitochondrial intergenic sequence between nad4 and rnl2, which corresponds to the linearization point in Y. unicocca (fig. 2A and B), is 219 nt, AT-rich (76%), and contains no obvious repeats. These 219 nt also show no detectable similarity to the telomeres or any other regions of the Y. unicocca mtDNA. PacBio reads spanned the entire nad4-rnl2 intersection in Eudorina sp. and we could successfully PCR across this junction, indicating that it is not an artifact from the assembly.
A circular genetic map does not necessarily reflect a circular genomic architecture (Bendich 2004, 2010). For instance, the assembly of a genome that exits primarily as a linear head-to-tail concatemer, like the Plasmodium falciparum mtDNA (Hikosaka et al. 2013), typically yields a circular map, as does a restriction digest of such a structure. Nevertheless, genomes that exist predominantly in a linear monomeric form, such as the C. reinhardtii mtDNA (Laflamme and Lee 2003), do not generally give circular maps when assembled or digested.
Restriction digests of Eudorina sp. mtDNA indicated that the genome occurs in circular-mapping and linear monomeric forms. Eudorina mtDNA was independently digested with five different restriction enzymes, including three single and two double cutters, and the resulting bands were visualized through agarose gel electrophoresis followed by a Southern blot with a nad5-derived probe (fig. 2D). Each enzyme gave a “mixed” banding pattern, indicative of a heterogeneous population of mtDNAs comprising a circular-mapping and a linear monomeric form (fig. 2A and D). What’s more, all the bands corresponding to the linear monomeric conformation supported an architecture in which nad4 and rnl2 were located at the extreme ends. In other words, the restriction digest data implied that the Eudorina sp. mtDNA can exist in a genome-sized linear form with a genomic arrangement identical to that predicted for Y. unicocca. There was a significant decrease in mapping coverage of the PacBio and Illumina reads to the Eudorina sp. mtDNA at the putative ends of the genome, further supporting the existence of a linear monomeric form with nad4 and rnl2 positioned at the telomeres (supplementary fig. S1, Supplementary Material online). Read coverage for nearly all types of linear DNAs, including mtDNAs and plastid DNAs, typically drops towards the ends of the chromosome (Voigt et al. 2008; Janouškovec et al. 2013)
Gel electrophoresis and blotting of uncut Eudorina sp. mtDNA gave a sharp band that comigrated with linear markers at the expected genome size (20.7 kb) as well as a short smear and a dark band migrating at a size much larger than the estimated genome length (fig. 2D). Although far from conclusive, these data support the hypothesis that the Eudorina sp. mitochondrial genome occurs in complex multigenomic, circular-mapping structures as well as unit-sized linear forms. Similar findings have come from analyses of organelle DNA in plants and yeast (Bendich 2004; Gerhold et al. 2010). The mode of replication of the Eudorina sp. mitochondrial genome as well as its telomeric structure and maintenance strategy will need to be explored in future studies.
Setting Straight the mtDNA Conformation of Reinhardtinia Algae
The data presented here for Y. unicocca and Eudorina sp. when combined with available mtDNA sequence information from closely related species paint a dynamic picture of mitochondrial genomic architecture in the Reinhardtinia (fig. 1A). There are now ten Reinhardtinia-clade species known to have linear mitochondrial genomes (not including Eudorina sp.), six of which have had their mtDNAs and mitochondrial telomeres sequenced. There are also five known members of the Reinhardtinia with circular-mapping genomes (including Eudorina sp.), all of which belong to the volvocine line (fig 1A). It is important to stress, however, that in all these examples of circular-mapping mtDNA, multigenomic concatenated structures have not been ruled out, and in some cases gel electrophoresis data are completely lacking, which is a recurring theme throughout organelle genomic research (Sanitá Lima et al. 2016). Moreover, for Eudorina sp. there is evidence of linear monomeric mtDNA in addition to a circular-mapping form. But unlike the linear mtDNAs from other Reinhardtinia algae, that identified in Eudorina sp. does not appear to have palindromic telomeres.
Thus, we can confidently say that there are at least two distinctive types of mitochondrial genomic architectures in the Reinhardtinia clade: those that assemble as unit-sized linear chromosomes with palindromic telomeres and those that assemble as circular-mapping molecules, which may or may not exist as multigenomic concatemers. Based on the phylogenetic relationships of Reinhardtinia algae for which mtDNA sequence and/or structure have been explored (fig. 1A), we hypothesize that throughout the evolution of this clade there has been no fewer than three separate transitions between these two architectures (fig. 1B;supplementary fig. S2, Supplementary Material online), and that the ancestor of the clade likely had a linear monomeric mitochondrial genome with telomeres—a feature that might have existed before the split of the Reinhardtinia and Oogamochlamydinia (fig. 1C).
Another varying feature of Reinhardtinia mtDNAs is the presence/absence of a freestanding (i.e., independent of an intron) reverse-transcriptase-like gene (rtl), located downstream of the rnl3 coding region. This gene was first identified in the C. reinhardtii mtDNA (Boer and Gray 1988b), and more recently in the mtDNAs of Chlamydomonas incerta (Popescu and Lee 2007) and T. socialis (Featherston et al. 2016). We found an rtl gene in the mtDNA Y. unicocca, but not in that of Eudorina sp. (fig. 1A). The Y. unicocca rtl is 1,365 nt long and shows 42.5% and 36.2% amino acid sequence identity with the rtl genes from T. socialis and C. reinhardtii, respectively. The function and evolutionary origin of rtl are unknown, but there is reason to believe that it originated from a group II intron (Nedelcu and Lee 1998). Although no group II introns have yet been found in the Reinhardtinia mtDNAs that harbour rtl, both the V. carteri and Pleodorina starrii mtDNAs contain a group II intron that encodes a putative reverse transcriptase. The mitochondrial intron content can vary within Reinhardtinia species (fig. 1A). For instance, some strains of C. reinhardtii have introns, whereas others do not (Smith and Lee 2009). Thus, even though group II introns have not been identified in rtl-containing species, it is possible that some members of the population do contain such introns and that rtl has an intron-related function.
Vahrenholz et al. (1993) proposed that rtl is involved in the maintenance of the mitochondrial telomeres in C. reinhardtii, but its presence in a circular-mapping genome (T. socialis) and absence from various telomere-containing mtDNAs (e.g., Polytomella spp.), is indicative of an alternative function. Reverse-transcriptase genes are sometimes found in linear mitochondrial plasmids (Galligan et al. 2011), which also typically contain palindromic telomeres (Smith and Keeling, 2013). One theory for the origin of a linear mitochondrial genomic architecture is the recombination of a circular mtDNA with a linear plasmid (Schardl et al. 1984; Sakurai et al. 2000; Nosek et al. 1998). No mitochondrial plasmids have been found in Reinhardtinia algae, but it is intriguing to think that a linear plasmid might be responsible for the linear structure, palindromic telomeres, and rtl gene found in the mtDNAs of various species in this clade. Finally, on a broader note, the data presented here underscore that one should not assume a circular organelle genome conformation based on DNA assembly data alone. As has been emphasized before (Bendich 2010; Sanitá Lima et al. 2016), many mtDNAs likely have more complicated and multifarious architectures than genome assembly data would suggest.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada (to D.R.S.), a Grants-in-Aid for Scientific Research on Innovative Areas “Genome Science” (grant number 221S0002; to A.T. and A.F.), and Scientific Research (A) (grant number 16H02518; to H.N.) and Research Activity Startup grants (grant number 16H06734 to T.H.) from MEXT/JSPS KAKENHI.
Literature Cited
- Bendich AJ. 2010. The end of the circle for yeast mitochondrial DNA. Mol Cell. 39:831–832. [DOI] [PubMed] [Google Scholar]
- Bendich AJ. 2004. Circular chloroplast chromosomes: the grand illusion. Plant Cell 16:1661–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer PH, Bonen L, Lee RW, Gray MW. 1985. Genes for respiratory chain proteins and ribosomal RNAs are present on a 16-kilobase-pair DNA species from Chlamydomonas reinhardtii mitochondria. Proc Natl Acad Sci U S A. 82:3340–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer PH, Gray MW. 1988a. Scrambled ribosomal RNA gene pieces in Chlamydomonas reinhardtii mitochondrial DNA. Cell 55:399–411. [DOI] [PubMed] [Google Scholar]
- Boer PH, Gray MW. 1988b. Genes encoding a subunit of respiratory NADH dehydrogenase (ND1) and a reverse transcriptase-like protein (RTL) are linked to ribosomal RNA gene pieces in Chlamydomonas reinhardtii mitochondrial DNA. EMBO J. 7:3501–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borza T, Redmond EK, Laflamme M, Lee RW. 2009. Mitochondrial DNA in the Oogamochlamys clade (Chlorophyceae): high GC content and unique genome architecture for green algae. J Phycol. 45:1323–1334. [DOI] [PubMed] [Google Scholar]
- Eid J, et al. 2009. Real-time DNA sequencing from single polymerase molecules. Science 323:133–138. [DOI] [PubMed] [Google Scholar]
- Featherston J, Arakaki Y, Nozaki H, Durand PM, Smith DR. 2016. Inflated organelle genomes and a circular-mapping mtDNA probably existed at the origin of coloniality in volvocine green algae. Eur J Phycol. 51(4):369–377. [Google Scholar]
- Foury F, Roganti T, Lecrenier N, Purnelle B. 1998. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 440:325–331. [DOI] [PubMed] [Google Scholar]
- Galligan JT, Marchetti SE, Kennell JC. 2011. Reverse transcription of the pFOXC mitochondrial retroplasmids of Fusarium oxysporum is protein primed. Mob DNA 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhold JM, Aun A, Sedman T, Jõers P, Sedman J. 2010. Strand invasion structures in the inverted repeat of Candida albicans mitochondrial DNA reveal a role for homologous recombination in replication. Mol Cell. 39:851–861. [DOI] [PubMed] [Google Scholar]
- Gray MW, Boer PH. 1988. Organization and expression of algal (Chlamydomonas reinhardtii) mitochondrial DNA. Philos Trans R Soc Lond B Biol Sci. 319:135–147. [DOI] [PubMed] [Google Scholar]
- Hamaji T, et al. 2013. Mitochondrial and plastid genomes of the colonial green alga Gonium pectorale give insights into the origins of organelle DNA architecture within the Volvocales. PLoS One 8:e57177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron MD, Hackett JD, Aylward FO, Michod RE. 2009. Triassic origin and early radiation of multicellular volvocine algae. Proc Natl Acad Sci U S A. 106:3254–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikosaka K, Kita K, Tanabe K. 2013. Diversity of mitochondrial genome structure in the phylum Apicomplexa. Mol Biochem Parasitol. 188:26–33. [DOI] [PubMed] [Google Scholar]
- Janouškovec J, et al. 2013. Split photosystem protein, linear-mapping topology, and growth of structural complexity in the plastid genome of Chromera velia. Mol Biol Evol. 30:2447–2462. [DOI] [PubMed] [Google Scholar]
- Kirk DL, Kirk MM. 1983. Protein synthetic patterns during the asexual life cycle of Volvox carteri. Dev Biol. 96:493–506. [DOI] [PubMed] [Google Scholar]
- Laflamme M, Lee RW. 2003. Mitochondrial genome conformation among CW-group chlorophycean algae. J Phycol. 39:213–220. [Google Scholar]
- Lemieux C, Vincent AT, Labarre A, Otis C, Turmel M. 2015. Chloroplast phylogenomic analysis of chlorophyte green algae identifies a novel lineage sister to the Sphaeropleales (Chlorophyceae). BMC Evol Biol. 15:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LJ, Coleman AW. 1989. The linear 20 kb mitochondrial genome of Pandorina morum (Volvocaceae, Chlorophyta). Plant Mol Biol. 13:459–465. [DOI] [PubMed] [Google Scholar]
- Miller SM, Schmitt R, Kirk DL. 1993. Jordan, an active Volvox transposable element similar to higher plant transposons. Plant Cell 5:1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada T, Misawa K, Nozaki H. 2008. Molecular systematics of Volvocales (Chlorophyceae, Chlorophyta) based on exhaustive 18S rRNA phylogenetic analyses. Mol Phylogenet Evol. 48:281–291. [DOI] [PubMed] [Google Scholar]
- Nedelcu AM, Lee RW. 1998. A degenerate group II intron in the intronless mitochondrial genome of Chlamydomonas reinhardtii: evolutionary implications. Mol Biol Evol. 15:918–922. [DOI] [PubMed] [Google Scholar]
- Nozaki H, Ott FD, Coleman AW. 2006. Morphology, molecular phylogeny and taxonomy of two new species of Pleodorina (Volvoceae, Chlorophyceae). J Phycol. 42:1072–1080. [Google Scholar]
- Nosek J, Fukuhara H, Suyama Y, Kováč L. 1998. Linear mitochondrial genomes: 30 years down the line. Trends Genet. 14:184–188. [DOI] [PubMed] [Google Scholar]
- Sakurai R, Sasaki N, Takano H, Abe T, Kawano S. 2000. In vivo conformation of mitochondrial DNA revealed by pulsed-field gel electrophoresis in the true slime mold, Physarum polycephalum. DNA Res. 7:83–91. [DOI] [PubMed] [Google Scholar]
- Sanitá Lima M, Woods LC, Cartwright MW, Smith DR. 2016. The (in)complete organelle genome: exploring the use and nonuse of available technologies for characterizing mitochondrial and plastid chromosomes. Mol Ecol Res. 16:1279–1286. [DOI] [PubMed] [Google Scholar]
- Schardl CL, Lonsdale DM, Pring DR, Rose KR. 1984. Linearization of maize mitochondrial chromosomes by recombination with linear episomes. Nature 310:292–296. [Google Scholar]
- Schindelin J, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Hamaji T, et al. 2013. Organelle genome complexity scales positively with organism size in volvocine green algae. Mol Biol Evol. 30:793–797. [DOI] [PubMed] [Google Scholar]
- Smith DR, Hua J, Archibald JM, Lee RW. 2013. Palindromic genes in the linear mitochondrial genome of the nonphotosynthetic green alga Polytomella magna. Genome Biol Evol. 5:1661–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Keeling PJ. 2013. Gene conversion shapes linear mitochondrial genome architecture. Genome Biol Evol. 5:905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Lee RW. 2008. Mitochondrial genome of the colorless green alga Polytomella capuana: a linear molecule with an unprecedented GC content. Mol Biol Evol. 25:487–496. [DOI] [PubMed] [Google Scholar]
- Smith DR, Lee RW. 2009. Nucleotide diversity of the Chlamydomonas reinhardtii plastid genome: addressing the mutational-hazard hypothesis. BMC Evol Biol. 9:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Lee RW. 2010. Low nucleotide diversity for the expanded organelle and nuclear genomes of Volvox carteri supports the mutational-hazard hypothesis. Mol Biol Evol. 27:2244–2256. [DOI] [PubMed] [Google Scholar]
- Popescu CE, Lee RW. 2007. Mitochondrial genome sequence evolution in Chlamydomonas. Genetics 175:819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahrenholz C, Riemen G, Pratje E, Dujon B, Michaelis G. 1993. Mitochondrial DNA of Chlamydomonas reinhardtii: the structure of the ends of the linear 15.8-kb genome suggests mechanisms for DNA replication. Curr Genet. 24:241–247. [DOI] [PubMed] [Google Scholar]
- Voigt O, Erpenbeck D, Wörheide G. 2008. A fragmented metazoan organellar genome: the two mitochondrial chromosomes of Hydra magnipapillata. BMC Genomics 9:350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.