Abstract
Drug-induced toxicity is a major public health concern that leads to patient morbidity and mortality. To address this problem, the Food and Drug Administration is working on the PredicTox initiative, a pilot research program on tyrosine kinase inhibitors, to build mechanistic and predictive models for drug-induced toxicity. This program involves integrating data acquired during preclinical studies and clinical trials within pharmaceutical company development programs that they have agreed to put in the public domain and in publicly available biological, pharmacological, and chemical databases. The integration process is accommodated by biomedical ontologies, a set of standardized vocabularies that define terms and logical relationships between them in each vocabulary. We describe a few programs that have used ontologies to address biomedical questions. The PredicTox effort is leveraging the experience gathered from these early initiatives to develop an infrastructure that allows evaluation of the hypothesis that having a mechanistic understanding underlying adverse drug reactions will improve the capacity to understand drug-induced clinical adverse drug reactions.
Keywords: Biomedical ontologies, data integration, PredicTox, adverse drug reaction
Introduction
Adverse drug reactions (ADRs) are unintended and undesired medical outcomes associated with administration of drugs. Adverse drug reactions are a major challenge to drug development and can lead to withdrawal of marketed drugs or failure of drugs during development.1,2 Therefore, identification and prediction of ADRs is a major focus of the pharmaceutical industry and regulatory agencies, such as the Food and Drug Administration (FDA).
During drug development, large amounts of data are acquired during the discovery, preclinical, and clinical phases. A small fraction of these data are published when a drug is approved for marketing. However, substantial amounts of data remain unpublished within the relevant pharmaceutical company. In addition to this, data from genome-wide studies and high-throughput studies acquired by academic institutions remain in isolated silos in different biomedical and chemical databases, such as Gene Expression Omnibus (GEO), NCBI Database of Genotypes and Phenotypes (dbGaP), The Pharmacogenomics Knowledge Base (PharmGKB), Connectivity Map, and European Bioinformatics Institute (EBI) Array Express Archive.3–5 Effectively integrating and analyzing these data using current systems pharmacology approaches may allow understanding of ADR mechanisms and improve ADR evaluation and prediction.6,7
Current Framework for Capturing Medical Diagnosis and ADRs
One of the challenges in building a systems pharmacology model for ADRs is in capturing medical information related to drug administration and adverse events. Most of the medical information exists as natural language text, which means that it is not easily accessible to computational analyses.8 In addition, different databases annotate clinical phenotypes using different vocabularies. For example, FDA and the European Medical Agency use Medical Dictionary for Regulatory Activities (MedDRA)9 to capture adverse events. The MedDRA is a standardized medical terminology established by the International Conference on Harmonization of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH) and is used by regulatory agencies, pharmaceutical companies, and health care systems. Another standardized medical vocabulary is Systematized Nomenclature of Medicine—Clinical Terminology (SNOMED-CT)10 that is used in direct patient care and is implemented in electronic medical records (EMRs). The SNOMED-CT is maintained by the International Health Terminology Standards Development Organization (IHTSDO) and is structured into 19 hierarchies that are further subdivided to describe a patient’s health. Another classification system that is used for billing purposes in the United States is International Statistical Classification of Diseases and Related Health Problems (ICD). The ICD is a medical classification system for diseases, laboratory findings, and causes of injury and diseases. It is owned and published by World Health Organization. All of these dictionaries and classification systems are capturing clinical phenotypes. However, to be able to use the information in these medical records for mechanistic modeling, these data need to be expanded from a simple vocabulary and be translated to a knowledge framework that will provide information about mechanism and causes of adverse events.4 Therefore, the terminologies used in these vocabularies have to be standardized to a common vocabulary for the cross-communication between content coded in different formats and standards.
Ontologies to Integrate Biomedical Data
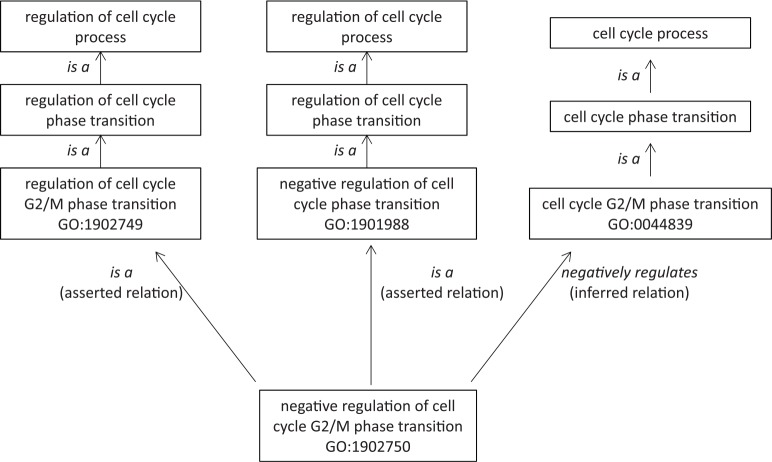
Ontologies are being effectively used to systematically classify biomedical terms from model organisms.11 The earliest of these ontologies is the Gene Ontology (GO) which was initiated in 1998 to annotate genes and gene products from Drosophila melanogaster (fruit fly), Mus musculus (mouse), and Saccharomyces cerevisiae (baker’s yeast).11 An ontology is defined by Kohler et al12 as “a computational representation of a domain of knowledge based upon a controlled, standardized vocabulary for describing entities and the semantic relationship between them.” Ontology allows one to define terms and define the relationship between terms after rigorous discussions among the biomedical researchers and bioinformaticians on how to capture this relationship such that a computer program using this ontology produces reliable results based on the agreed-upon term definitions and semantic relations among the terms. Therefore, an ontology is built around a specific subject matter and is characterized by classes that are well defined. Hierarchical organization of classes in an ontology is based on both direct asserted parent-child relationships and indirect inferred logical relationships that go from a general concept (broader/upper level class) to a specific concept (narrower/lower level class). For example, in GO, certain terms are asserted as a parent-child relationship via an “is a” relation. At the same time, certain terms are classified via the inferred relations (ie, negatively regulates, positively regulates) in the ontology. An example of this is shown in Figure 1. The term negative regulation of cell cycle G2/M phase transition is a direct asserted child of 2 parents: “regulation of cell cycle G2/M phase transition” and “negative regulation of cell cycle phase transition.” At the same time, the logical relation “negatively regulates” property puts this class as an inferred subclass of “cell cycle G2/M phase transition.” The multiple classifications without having to explicitly reinstate the same class at various places of the hierarchy is one of the useful features of ontology that allows clear definition and properties of one unique class to be expressed multidimensionally. Complex information linked to a biological concept can be annotated with this multidimensional ontology definition and semantic logics for further computation.
Figure 1.
Asserted versus inferred relations. In Gene Ontology (GO), the term negative regulation of cell cycle G2/M phase transition (GO:1902750) is asserted as a child of 2 parents: “regulation of cell cycle G2/M phase transition” (GO:1902749) and “negative regulation of cell cycle phase transition” (GO:1901988). At the same time, “negative regulation of cell cycle G2/M phase transition” (GO:1902750) is inferred as a subclass of “cell cycle G2/M phase transition” via the logical relation “negatively regulates.”
The biomedical community has built ontologies to capture different spheres of biomedical knowledge. This process has produced several ontologies to describe subjects such as genes and gene products (GO),11 cell types (Cell Ontology, CL),13,14 protein modifications (Protein Ontology),15 anatomical description (Uber anatomy ontology [UBERON]),16 human phenotypes (Human Phenotype Ontology [HPO]),17 and diseases (Disease Ontology [DO]).18 Each of these ontologies is updated and refined continuously. With the development of these domain-specific ontologies, it is essential to be able to reuse terms that have already been defined by the experts of each domain. Therefore, one of the challenges in ontology development is maintaining the interoperability between them such that terms defined in one ontology can be reused by another ontology without breaking the relational rules established for the term in the original ontology and in the new ontology. This process allows terms to be linked to other terms and allows coverage of a wider area of biology than would have been possible with any one of these ontologies alone. This produces ontologies that are interoperable such that terms defined in an ontology are logically consistent and compatible with other ontologies.4 For example, interoperability between the Cell Ontology (CL) and UBERON allows a computer program to infer that the term cardiac muscle cell (described in CL) is part of “heart” (described in UBERON) (Figure 2).
Figure 2.
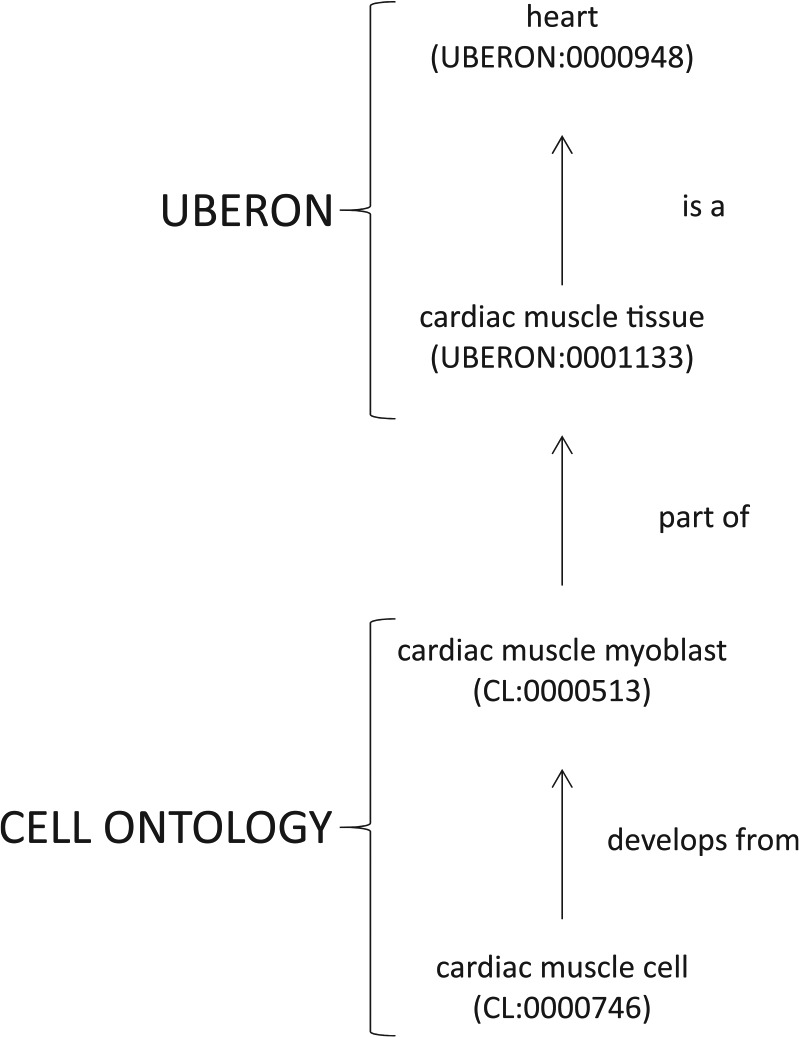
Interoperability between ontologies allows relations to be inferred across ontologies. Interoperability between the Cell Ontology and UBERON allows a computer reasoner to infer that “cardiac muscle cell” (CL:0000746) is located in the “heart” (UBERON:0000948).
Implementation of Interoperability Between Ontologies
To achieve interoperability between ontologies, ontology developers historically created the Open Biomedical Ontologies (OBO) file format to ensure that ontologies that they are developing are open, orthogonal, and follow a structured, controlled vocabulary. To allow for a more complex annotation that requires higher computational expressivity, the OBO developers later adopted the Web Ontology Language (OWL) format, whereas the name OBO remains the reference to the community.19 The OBO developers initiated the OBO Foundry that has developed a set of principles for ontology developers to follow.20 One of the key principles is to use a common upper-level ontology. This ontology, called Basic Formal Ontology, facilitates the organization of biomedical terms using a standardized categorization process that helps integrate data from different biomedical domains.21 The OBO Relation Ontology (RO) allows ontology developers to use a consistent format to describe relational logic between terms.22 These ontologies share a common semantic linking mechanism with each other using the relational properties from RO, by either the direct reuse of RO’s relations or the creation of a new relation with common attributes inherited from RO’s relations.
Modeling Data That Represent “Sometimes Associated” Relationship
One challenge in using ontologies to discover the mechanism of drug-induced toxicity lies in defining the relationship between adverse event and drug. When a patient experiences adverse events related to a specific drug administration, not all patients will experience an adverse event. In addition, patients might experience the adverse events at different levels of severity. This “sometimes associated” phenotype is a challenge for ontologies to handle, as all classes and relations stored and reasoned in an ontology have an implication, by ontology rules, that every class-relation statement must hold true at all times. Ontologists have handled this problem using OBAN (Open Biomedical AssociatioN),23 dealing with the association at the individual instance level of a class, rather than at the class level itself. For example, in rare diseases, disease-phenotype relationships have to be captured in a way such that not every instance of the disease is associated with all the observed phenotypes of the disease.23 The relationship between disease and its phenotypes has been separated by introducing an intermediate relationship called an OBAN association that associates the source of evidence for the phenotype. An OBAN association is “true for a given disease and a phenotype” where one has a subject role (disease) and one has an object role (phenotype) because the OBAN model qualifies the association between disease and phenotype with evidence for that association.23
Therefore, in this drug safety scenario, the association of drug-adverse event is handled based on a case-by-case basis of evidence. The OBAN is a data representation model that describes the relationship between a subject and an object by stating that the subject is associated with an object. This individual association is supported by specific evidence. For example, OBAN model allows the statement that “[sunitinib] is associated with [higher incidence of hypertension] via the supporting evidence of [PubMed_ID 24930624]” to be computationally represented.
Modeling Data to Capture Temporal Relationships
In the case of adverse events, capturing the temporal relationship between the medical intervention, such as drug administration or vaccination, and the adverse event has to be accounted for in a comprehensive analysis of the drug-drug response (improved condition, or adverse event). This information is important because adverse events can occur due to the drug. However, the same clinical phenotype can also occur due to factors other than the drug being administered to the patient. There are many confounding factors that can contribute to the adverse event, such as the patient’s previous medical history or concomitant medications taken by the patient. These data exist in unstructured narratives in a patient’s case report form or medical record. Therefore, establishing a causal association between the drug and adverse event is not an easy task. Being able to capture data on when a drug was administered, when an adverse event started and ended, the patient’s medical history including time of diagnosis of other diseases, concomitant medications that the patient was on, and other temporal information at the patient level may reveal patterns in the data that can help build the causal link between the drug administration and the adverse event.
The phenotype of an adverse event also evolves over time. To be able to capture this change, we have to be able to define the time period between the drug administration and when the adverse event was first reported. If we can compare this information between different patients, we should be able to detect a pattern in the adverse events experienced by patients on a specific drug. This would give strength to building a hypothesis of causal association between the drug and adverse event. For example, in the case of heart disease associated with administration of a drug, a patient might manifest symptoms that evolve over time beginning with shortness of breath which changes over time to pedal edema, ascites (fluid in the abdomen), and eventually ventricular arrhythmia. All of the patients might not show the same adverse event at any one point in time. However, seeing this pattern of adverse events in different patients who are on the drug can provide a signal of a causal link between the drug and adverse event. In exploring the mechanism of this sequence of adverse events, it is expected that the evolution of the cellular signaling networks involved in the process will progress in parallel with the clinical phenotypes. An ontology that is being used to model adverse event needs to capture all of the data described above. Therefore, ontology relations have to be developed to effectively capture these temporal relationships.
Ontologists have been working on mechanisms to model temporal information. The Ontology of Adverse Events (OAE) was developed to represent adverse event data in an ontological format. In OAE, an “adverse event” is defined as “a pathological bodily process that occurs after a medical intervention.”24 According to this definition, the adverse event occurs temporally at time T1 after a medical intervention at time T0. This definition does not assume a causal association between the administered drug and adverse event, but rather a temporal drug-adverse event observation. To make the causal association, OAE defines the term causal adverse event to represent a subtype of adverse event for which clear causal relationship has been established between the adverse event and the drug exposure as confirmed by the clinical record. In this way, OAE has made it possible to capture a hypothesis of the adverse event process.
The unstructured clinical narratives in case report forms contain temporal information on the patient’s disease, age, medication dose, and other features. To be able to capture this information in an ontological format, the Clinical Narrative Temporal Relation Ontology (CNTRO) was developed which eventually evolved into the Time Event Ontology (TEO) to model temporal information for all domains and not just the biomedical domain.25,26 This ontology can model timing events, time instants, intervals, durations, and temporal relations. The TEO is currently being used in combination with OAE and the Vaccine Ontology (VO) to represent unstructured clinical narratives from adverse event reports to make the data accessible to computer programs for further querying and analysis.25 These ontologies can be applied to discover time trends in adverse event reports, be integrated with statistical tools, and eventually be used to build a causal link between drug administration and adverse events.25
Development of OAE to Capture Adverse Event in an Ontological Format
Ontologies are useful in data integration. They facilitate the integration of data annotated using different vocabularies.27 In addition, the ontological relations between terms in one ontology and the links between ontologies allow more information to be extracted about a term than would have been otherwise possible. For identifying the mechanism of adverse events, the adverse event reports in FDA Adverse Event Reporting System (FAERS) and EMRs are linked to chemical and biomedical databases on drugs. This integration is being aided by the development of the OAE. The OAE defines adverse events with cross-references to MedDRA, SNOMED-CT, and ICD terms. The OAE was built using OBO foundry principles and is therefore interoperable with other ontologies that follow OBO foundry principles. If these ontologies have sufficient coverage of the biomedical and chemical space, it is expected that we can use computers to infer the relationship between terms in one ontology to terms in another ontology. This method of integration will be useful in discovering the mechanism of ADRs.
The OAE has been extended to include cardiovascular adverse events that are associated with a class of drugs that target tyrosine kinases (TKs) called tyrosine kinase inhibitors (TKIs) or monoclonal antibodies that target TKs (mAbs).28 The cardiovascular adverse events associated with 5 TKIs/mAbs, namely, dasatinib, imatinib, lapatinib, cetuximab, and trastuzumab, were extracted from reports in FAERS. This analysis produced 1053 cardiovascular MedDRA terms, of which 884 were unique terms that were not in OAE. These terms were curated with the help of clinicians to classify them accurately within OAE and they were all cross-referenced to MedDRA terms. In addition, they were linked to the HPO, UBERON, and GO to facilitate discovery of biological processes involved in the adverse event. The development of OAE to include cardiovascular adverse events in an ontological format will help in learning the molecular mechanism of adverse events associated with TKIs/mAbs and will help the PredicTox project which is described in the next section.
Examples of Studies Working on Integrating Biological Knowledge
The preceding discussion on OAE demonstrates that the direction of ontology development depends on the projects that will use them. Therefore, ontologies are in a continuous development process. To highlight this process, we have described several ongoing initiatives using ontologies to integrate biomedical data to discover the genetic causes of diseases and mechanism of adverse event, and predict drug-induced toxicity. Examples of these initiatives—eTOX, MONARCH Initiative, Open Targets, and PredicTox—are described in the following sections.
eTOX
The eTOX project (http://www.etoxproject.eu/) was developed to build a data warehouse for drug safety data to predict nonclinical toxicity associated with small-molecule drugs. This project is part of the European Union–funded innovative medicines initiative. eTOX is a public-private partnership involving 11 academic institutions, 6 small- and medium-sized enterprises, and 13 pharmaceutical companies. These groups contributed data that are believed to have important safety information that could be used to predict drug-induced toxicity. These data were converted into a machine-readable format to be reused and extracted for information for developing predictive algorithms. This activity involved a major curation effort that required using a standardized terminology. Biomedical ontologies with their term reusability were used for this task as many existing ontologies are in place, and where needed, development of new ontologies that were interoperable with the current ones were implemented. The curation itself involved confirming that the verbatim terms were converted to a standard terminology such as SEND (Standard for Exchange of Nonclinical Data) and a CDISC Standard Data Tabulation Model (SDTM) standard for presenting nonclinical data.29 The eTOX consortium developed a curation tool called OntoBrowser that maps terms from their database to ontologies.29 In addition to OntoBrowser, the consortium developed a number of tools that they have made publicly available. These include LIBRARY that provides articles, journal, and links on drug toxicity; eTOXLab that can be used to predict biological properties of small molecules; LiMTox, which is a text mining software, that focuses on association between drugs and drug toxicity; and others.29 The eTOX consortium has been able to bring together different groups and develop secure data-sharing agreements to build a data warehouse and analytical framework to address questions around drug toxicity. The eTOX project is at its completion date (2016).
The Monarch Initiative
The Monarch Initiative (https://monarchinitiative.org/) was developed to integrate data acquired from different species to discover new genotype-phenotype relationships for both model organisms and human disease.27,30 The goal was to integrate phenotypic data that are captured in textual descriptions from different model organisms. A computer algorithm could then be used to identify the genetic associations of human diseases based on similarity in the phenotypic descriptions. The Monarch Initiative integrates information, including data on genes, genotypes, gene variants, model systems, pathways, orthologs, phenotypes, and publications. The data from different model systems are linked using ontologies that semantically map the data across different levels of biological organization and across different species. For example, genes are mapped to NCBI gene identifiers, diseases to DO,18 and phenotypes to unified phenotype ontology (Uberpheno).16 These integrated data are then available to run queries to answer a specific biological question. When investigators query a specific phenotype, it will retrieve all the data related to that query from the system.
The Monarch Initiative therefore relies heavily on ontologies to semantically link the data. It has also developed tools called OWLSim to run semantic similarity searches that use ontologies and computational reasoners to analyze similarities between phenotypes. One of the advantages of the methodology developed by the Monarch Initiative is that because it uses information from different model organisms, the opportunities to more robustly characterize specific diseases are available than would have been available through analysis of human data alone.
Open Targets
Open Targets (http://www.opentargets.org/) is a public-private partnership involving GlaxoSmithKline, Biogen, European Bioinformatics Institute (EMBL-EBI), and The Wellcome Trust Sanger Institute to create a platform that investigators can use to prioritize potential therapeutic targets for pharmacological intervention. Evidence for the role of the molecular target in disease is obtained by integrating data from genome-wide studies such as RNAi screens, genome-wide association studies, microarray studies, and genetic data on rare diseases and cancer. These data are used to generate an evidence score that is used to prioritize the target.
Biomedical ontologies were used to normalize the data annotation at Open Targets. The target-disease associations were generated in the ontology-assisted pipeline. The target such as a gene was linked to the disease associated with the gene with terms obtained from Experimental Factor Ontology (EFO). The target-disease association was made using association terms from the OBAN ontology model (Open Biomedical AssociatioN).23 The type of evidence for the association (eg, computationally predicted) was made using terms from the Evidence Code Ontology (ECO).31 Each disease-target association was linked to its corresponding supporting evidence, the datasource for the evidence, and an ECO evidence code. Each piece of evidence was provided a score based on the statistical significance of the evidence. An overall association score was calculated based on the evidence, datasource, and data type. This overall score can be used by investigators to decide whether their favorite target can be developed for therapeutic intervention for the disease that they are studying.
PredicTox
PredicTox is a public-private partnership that was spearheaded by FDA and the Reagan-Udall Foundation for the FDA to build a systems pharmacology model to predict the occurrence of drug-induced toxicity. For its pilot project, PredicTox is focusing on understanding the mechanism of heart failure cardiac adverse events that are associated with TKIs. The project team is working closely with specific pharmaceutical companies to acquire data generated during the development of small-molecule inhibitors and monoclonal antibodies that target TKs. These data acquired during the discovery, preclinical pharm/tox, and clinical phases of development that contributors agree to share publicly will be integrated with genomic, proteomic, and metabolomic data in a knowledge environment (KE). The KE will be placed at a site external to FDA such that contributors and investigators will have access to the data. The data will be shared using a data-sharing model that is agreed upon by the data partners and PredicTox team members.32 The KE will also include data analysis tools for investigators interested in addressing specific scientific questions related to the toxicity of TKIs.
To aid with the data integration, the PredicTox team is also developing an application ontology that reuses terms from domain-specific ontologies so that the clinical and nonclinical data can be linked to information in different pharmacological, biological, and chemical databases. A major challenge for the PredicTox project was in integrating adverse event terms from the FAERS that is annotated using MedDRA. This was addressed in the development of OAE in which the adverse event terms are captured using MedDRA or are cross-referenced to MedDRA.28 Therefore, with the development of OAE, adverse event terms can be semantically linked to other ontologies.
Conclusions
As can be seen with the above examples, many projects are using ontologies to integrate data that are annotated using different vocabularies. This integration effort requires continuous development of the domain-specific ontologies to complete coverage of the biomedical and relevant chemical space. There is also further research required to advance ontology development. For example, as described above, in the past, ontology was not able to capture the “sometimes associated” relationship. With the development of the OBAN model in which an intermediate relationship was introduced that provided sources of evidence for the assertion of relationship between 2 terms, the problem in capturing “sometimes associated” relationships has been addressed. However, further development is required to be able to capture temporal relationships. Ontologies may therefore provide one solution to better integrate data for analysis and problem solving related to ADRs.
Footnotes
Peer review:Seven peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1395 words, excluding any confidential comments to the academic editor.
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of conflicting interests:The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: The views expressed are those of the authors and do not necessarily represent the position of, nor imply endorsement from, the US Food and Drug Administration or the US government.
Author Contributions: SZ wrote the first draft of the manuscript. SZ, SS, and DRA contributed to the writing of the manuscript. SZ, SS, and DRA made critical revisions and approved the final version. All authors reviewed and approved the final manuscript.
References
- 1. Wilke RA, Lin DW, Roden DM, et al. Identifying genetic risk factors for serious adverse drug reactions: current progress and challenges. Nat Rev Drug Discov. 2007;6:904–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hennessy S, Strom BL. PDUFA reauthorization—drug safety’s golden moment of opportunity? N Engl J Med. 2007;356:1703–1704. [DOI] [PubMed] [Google Scholar]
- 3. Abernethy DR, Bai JP, Burkhart K, Xie HG, Zhichkin P. Integration of diverse data sources for prediction of adverse drug events. Clin Pharmacol Ther. 2011;90:645–646. [DOI] [PubMed] [Google Scholar]
- 4. Zhichkin PE, Athey BD, Avigan MI, Abernethy DR. Needs for an expanded ontology-based classification of adverse drug reactions and related mechanisms. Clin Pharmacol Ther. 2012;91:963–965. [DOI] [PubMed] [Google Scholar]
- 5. Garcia-Serna R, Vidal D, Remez N, Mestres J. Large-scale predictive drug safety: from structural alerts to biological mechanisms. Chem Res Toxicol. 2015;28:1875–1887. [DOI] [PubMed] [Google Scholar]
- 6. Berger SI, Iyengar R. Role of systems pharmacology in understanding drug adverse events. Wiley Interdiscip Rev Syst Biol Med. 2011;3:129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boland MR, Jacunski A, Lorberbaum T, Romano JD, Moskovitch R, Tatonetti NP. Systems biology approaches for identifying adverse drug reactions and elucidating their underlying biological mechanisms. Wiley Interdiscip Rev Syst Biol Med. 2016;8:104–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Robinson PN, Mungall CJ, Haendel M. Capturing phenotypes for precision medicine. Cold Spring Harb Mol Case Stud. 2015;1:a000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown EG, Wood L, Wood S. The medical dictionary for regulatory activities (MedDRA). Drug Saf. 1999;20:109–117. [DOI] [PubMed] [Google Scholar]
- 10. Rothwell DJ, Cote RA, Cordeau JP, Boisvert MA. Developing a standard data structure for medical language—the SNOMED proposal. Proc Annu Symp Comput Appl Med Care. 1993;695–699. Available at: https://www.ncbi.nlm.nih.gov/pmc/issues/161398/ [PMC free article] [PubMed]
- 11. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kohler S, Schulz MH, Krawitz P, et al. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am J Hum Genet. 2009;85:457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Diehl AD, Meehan TF, Bradford YM, et al. The Cell Ontology 2016: enhanced content, modularization, and ontology interoperability. J Biomed Semantics. 2016;7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meehan TF, Masci AM, Abdulla A, et al. Logical development of the cell ontology. BMC Bioinformatics. 2011;12:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Natale DA, Arighi CN, Blake JA, et al. Protein Ontology: a controlled structured network of protein entities. Nucleic Acids Res. 2014;42:D415–D421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mungall CJ, Torniai C, Gkoutos GV, Lewis SE, Haendel MA. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 2012;13:R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kohler S, Doelken SC, Mungall CJ, et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–D974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schriml LM, Arze C, Nadendla S, et al. Disease Ontology: a backbone for disease semantic integration. Nucleic Acids Res. 2012;40:D940–D946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moreira DA, Musen MA. OBO to OWL: a protege OWL tab to read/save OBO ontologies. Bioinformatics. 2007;23:1868–1870. [DOI] [PubMed] [Google Scholar]
- 20. Smith B, Ashburner M, Rosse C, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25:1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Simon J, Dos Santos M, Fielding J, Smith B. Formal ontology for natural language processing and the integration of biomedical databases. Int J Med Inform. 2006;75:224–231. [DOI] [PubMed] [Google Scholar]
- 22. Smith B, Ceusters W, Klagges B, et al. Relations in biomedical ontologies. Genome Biol. 2005;6:R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sarntivijai S, Vasant D, Jupp S, et al. Linking rare and common disease: mapping clinical disease-phenotypes to ontologies in therapeutic target validation. J Biomed Semantics. 2016;7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. He Y, Sarntivijai S, Lin Y, et al. OAE: the Ontology of adverse events. J Biomed Semantics. 2014;5:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tao C, He Y, Yang H, Poland GA, Chute CG. Ontology-based time information representation of vaccine adverse events in VAERS for temporal analysis. J Biomed Semantics. 2012;3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tao C, Wei WQ, Solbrig HR, Savova G, Chute CG. CNTRO: a semantic web ontology for temporal relation inferencing in clinical narratives. AMIA Annu Symp Proc. 2010;2010:787–791. [PMC free article] [PubMed] [Google Scholar]
- 27. Washington NL, Haendel MA, Mungall CJ, Ashburner M, Westerfield M, Lewis SE. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 2009;7:e1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sarntivijai S, Zhang S, Jagannathan DG, et al. Linking MedDRA®-coded clinical phenotypes to biological mechanisms by the Ontology of adverse events: a pilot study on tyrosine kinase inhibitors. Drug Saf. 2016;39:697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cases M, Briggs K, Steger-Hartmann T, et al. The eTOX data-sharing project to advance in silico drug-induced toxicity prediction. Int J Mol Sci. 2014;15:21136–21154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mungall CJ, Washington NL, Nguyen-Xuan J, et al. Use of model organism and disease databases to support matchmaking for human disease gene discovery. Hum Mutat. 2015;36:979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chibucos MC, Mungall CJ, Balakrishnan R, et al. Standardized description of scientific evidence using the Evidence Ontology (ECO). Database (Oxford). 2014;2014:bau075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Altshuler JS, Balogh E, Barker AD, et al. Opening up to precompetitive collaboration. Sci Transl Med. 2010;2:52cm26. [DOI] [PubMed] [Google Scholar]