Abstract
1[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole (CDDO-Im), a synthetic derivative of oleanolic acid that exhibits antioxidant and anti-inflammatory activity in several animal and in vitro models, has been shown to be beneficial if given after injury. Although induction of heme oxygenase 1 appears to be a major effector of cytoprotection, the mechanism by which the overall effect is mediated is largely unknown. This study evaluated temporal gene expression profiles to better characterize the early transcriptional events and their relationship to the dynamics of the cytoprotective response in human umbilical vein endothelial cells (HUVEC) to CDDO-Im. Time-course gene expression profiling was performed on HUVEC treated with CDDO-Im for 0.5, 1, 3, 6, and 24 hours. More than 10 000 genes were statistically altered in their expression in at least 1 time point across the time course. Large alterations in immediate-early gene expression were readily detectable within 0.5 hour after administration of CDDO-Im.
Keywords: Microarray, time-course gene expression, CDDO-IM, cytoprotection
Introduction
The synthetic oleanane triterpenoid 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO) and its derivatives 1[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole (CDDO-Im) and methyl 2-cyano-3,12-dioxooleana-1,9(11)dien-28-oate (CDDO-Me) have been shown to protect against oxidative stress in various cell and animal models1–5 and exhibit anti-inflammatory properties.6 In addition, these 2 compounds have been shown in some studies to provide a chemopreventive effect against certain tumors, largely by reducing the viability of these cells.7–10 Comparing these 2 structurally different forms of CDDO, we previously determined that there is a similar degree of cytoprotection between the 2 compounds; however, significant differences in gene expression were seen between the 2 following a 6-hour treatment on human umbilical vein endothelial cells (HUVEC).11 The derivative CDDO-Im was shown to induce more gene expression changes than CDDO-Me and resulted in significantly higher expression levels of key cytoprotective genes, including heme oxygenase 1 (HMOX1).11
Both CDDO-Im and CDDO-Me have been shown to elicit protective effects largely through the activation of the transcription factor nuclear factor erythroid 2–related factor 2 (NRF2).12–14 Our previous work with CDDO-Im and CDDO-Me supported published work that demonstrated the central role of NRF2 in homeostasis11,15 and its role in HMOX1 induction; in addition, it identified potentially new genes that provide additional cytoprotection. A recent study evaluated the kinetics of HMOX1 induction by 4 different transcription factors (HSF-1, AP-1, NRF2, and NF-κB) and showed that they differ in their mechanism of action and kinetics of inducing HMOX1.16 That study highlighted the need for a better understanding of protective mechanisms of action that accurately takes into account the temporal nature of gene response over time; through the use of mathematical modeling, the study revealed multiple pathways for HMOX1 induction. The study presented here expands on that approach by studying transcriptional kinetics of a relevant system that not only allows for modeling with experimentally derived data but also extends the study of mechanism of action by using further analysis to identify upstream initiators of observed gene expression.
The CDDO-Im cytoprotection of HUVEC was investigated by studying gene expression over a time course (0.5, 1, 3, 6, and 24 hours). The purpose of this study was to determine whether alterations in gene expression induced by CDDO-Im over time, particularly at early time points, could uncover the proximal mechanism responsible for the cytoprotective effect.11,15 In addition, analysis with Expression2Kinases (X2K) was used to identify transcription factors, kinases, and complexes that drive changes in global gene expression.17 Expression2Kinases integrates chromatin immunoprecipitation (ChIP)-seq/chip and position weight matrix data, protein-protein interactions, and kinase-substrate phosphorylation reactions to better identify regulatory mechanisms upstream of genome-wide differences in gene expression.
Observing and measuring temporal changes provide for a better understanding of dynamic responses and provide insight into the actual regulatory mechanisms responsible for cytoprotection; critical processes required for downstream initiation of cytoprotective genes will be investigated by focusing on early time points. This study also sought to provide methods that allow for the discovery of mechanisms of action involved in cytoprotection from CDDO-Im in an approach that can be applied to other drug model systems.
Materials and Methods
Chemicals and reagents
The derivative CDDO-Im (96% purity) was purchased from Toronto Research Chemicals Inc. (Toronto, ON, Canada). Dimethyl sulfoxide (DMSO) was from Sigma-Aldrich (Saint Louis, MO, USA).
Cell culture
Stock cultures of gender-mixed HUVEC (Lifeline Cell Technology, Walkersville, MD, USA) pooled from 10 different donors were cultivated on T75 flasks (Corning Inc., Corning, NY, USA) in MCDB 131 medium at 37°C in a humidified atmosphere of 92% nitrogen, 3% oxygen, and 5% CO2 with medium changes every 2 days until confluence is reached.18,19 MCDB 131 medium, trypsin/EDTA, and antibiotic/antimycotic were obtained from Life Technologies (Carlsbad, CA, USA). Endothelial supplements were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Prior to an experiment, HUVEC were subcultivated with trypsin/EDTA onto Costar 96-well or 12-well multiplates (Corning Inc.) at 5000 cells/cm2, grown to confluence, and kept for 72 hours to produce a quiescent cell layer. Only the second through fifth population doublings of cells were used as described in a study by Wang et al.20 The derivative CDDO-Im was diluted (1000-fold) after dissolving in DMSO (0.1%) with medium to a final concentration of 200 nM. Human umbilical vein endothelial cells were grown to confluence at which time they were pretreated with CDDO-Im for their respective incubation times (0.5, 1, 3, 6, and 24 hours). Gene expression profiling of CDDO-Im–treated HUVEC was compared with vehicle control (0.1% DMSO).
Total RNA isolation
RNA from cultured HUVEC grown on multiplates was extracted using TRI reagent according to the manufacturer’s instructions (Molecular Research Center, Cincinnati, OH, USA) as previously described.15 Using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA), RNA yield was quantified and its quality assessed by electrophoresis on 1% agarose gels containing 1:10 000 SYBR Gold in the loading buffer (Invitrogen, Carlsbad, CA, USA). The resulting gels were scanned with a Kodak Image Station 2000R Digital Imaging System and the resulting digitized image scanned with NIH Image to determine the density in each band. The ratio of 28S to 18S was 2 to 1, indicating high-quality RNA.
Quantitative real-time polymerase chain reaction
Complementary DNA (cDNA) was prepared from 1 µg of total RNA from each sample using the High-Capacity cDNA Archive Kit (Life Technologies, Grand Island, NY, USA). Real-time polymerase chain reaction (RT-PCR) was performed on a Roche LightCycler 480 thermal cycler (Roche Diagnostics, Indianapolis, IN, USA) with Roche LightCycler TaqMan Master Kit (Roche Diagnostics) for confirmation of microarray results. The 18S primer/probe was used as an endogenous control for each sample and measured simultaneously with each labeled sample for the purpose of normalization; relative quantification was determined by the comparative CT method.21 Primer/probes of interest (JUNB, FOS, DUSP1, and EGR3) and 18S primer sets were purchased from TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA, USA). Dimethyl sulfoxide–treated (vehicle) HUVEC at time 0 were used as the comparator for all CDDO-Im–treated samples.
Polyacrylamide gel electrophoresis and Western blotting
Protein was isolated from the HUVEC after incubation with CDDO-Im (200 nM) for each of the time points represented in the microarray experiment (0.5, 1, 3, 6, and 24 hours), by addition of 50 µL of lysis buffer (Life Technologies) containing 10 mM Tris (carboxyethyl) phosphine hydrochloride (Sigma-Aldrich). About 15 µL, containing approximately 5 µg of protein, from each treatment was run on E-PAGE 96-well 6% gels (Life Technologies) and then transferred to a nitrocellulose membrane (Life Technologies). After blocking in blocking buffer (LI-COR Biosciences, Lincoln, NE USA), the blots were then incubated with rabbit HMOX1, HSP105, DUSP1, DYRK3, or HSP70 primary antibodies (1:5000; Assay Designs Inc., Ann Arbor, MI, USA) for 1 hour. In addition, each blot was dual-labeled with β-actin as a loading control that was used for normalizing antibody intensity of each protein of interest. The blots were washed 3 times with 0.1% Tween 20 in phosphate-buffered saline and incubated with donkey anti-rabbit secondary antibody (LI-COR Biosciences) for 30 minutes before 3 more washes and then allowed to dry for visualization. Visualization was performed using the Odyssey Imaging System (LI-COR Biosciences) that allowed for dual labeling of 2 different proteins and was quantified by direct measurement of background-subtracted infrared florescent signals.
Gene expression analysis
RNAs (500 ng) from 4 replicates for each time point (0.5, 1, 3, 6, and 24 hours) were labeled following Agilent’s Low RNA Input Linear Amplification Kit (Agilent, Santa Clara, CA, USA). All sample labeling, hybridization, washing, and scanning steps were conducted according to the manufacturer’s specifications. For each group, 200 ng of complementary RNA from each labeling reaction was hybridized to the Agilent Whole Human Genome Oligo Microarray (Agilent). The Whole Human Genome Oligo Microarray is in an 8× 60K slide format and microarray interrogates all known genes. After hybridization, the slides were washed and then scanned with the Agilent G2505C Microarray Scanner System (Agilent). The fluorescence intensities on scanned images were extracted and preprocessed by Agilent Feature Extraction Software. These data have been submitted to the Gene Expression Omnibus under accession no. GSE71622.
Statistical analysis
Microarray data analysis and statistical analysis were performed with BRB-ArrayTools.22 Genes were considered statistically significant with P < .001 and false discovery rate value of <5%. Filtered data analyzed by BRB-ArrayTools were submitted to GeneMANIA for network analysis. This program represents the complexity of biological regulatory systems as networks whose topology provides a manageable technique for analyzing the principal components of a complex system.23–25
Principal components (genes or proteins) are nodes, and interactions are depicted as edges.24 Similar patterns of expression were observed between certain individual time points after BRB-ArrayTools analysis and were consolidated into early (0.5 and 1 hour), intermediate (3 and 6 hours), and late (24 hours) defined groups for more consistent representation of the global expression response over time. Western blot data are presented as the mean ± SD. A 2-class comparison was performed to identify genes that were differentially expressed using a random-variance t test (http://linus.nci.nih.gov/BRB-ArrayTools.html).22
X2K analysis
The immediate-early time-course gene expression data (30 minutes and 1 hour) were submitted to X2K to identify upstream, proximal regulators likely responsible for later observed patterns of genome-wide gene expression.17 The most likely transcription factors that regulate the differences in gene expression are inferred, then known protein-protein interactions are connected to the identified transcription factors using additional proteins for building transcriptional regulatory subnetworks centered on these factors, and finally, kinase-substrate protein phosphorylation reactions are used to identify and rank candidate protein kinases that most likely regulate the formation of the identified transcriptional complexes.
Results
Time-course gene expression profiling of CDDO-Im reveals different phases of response
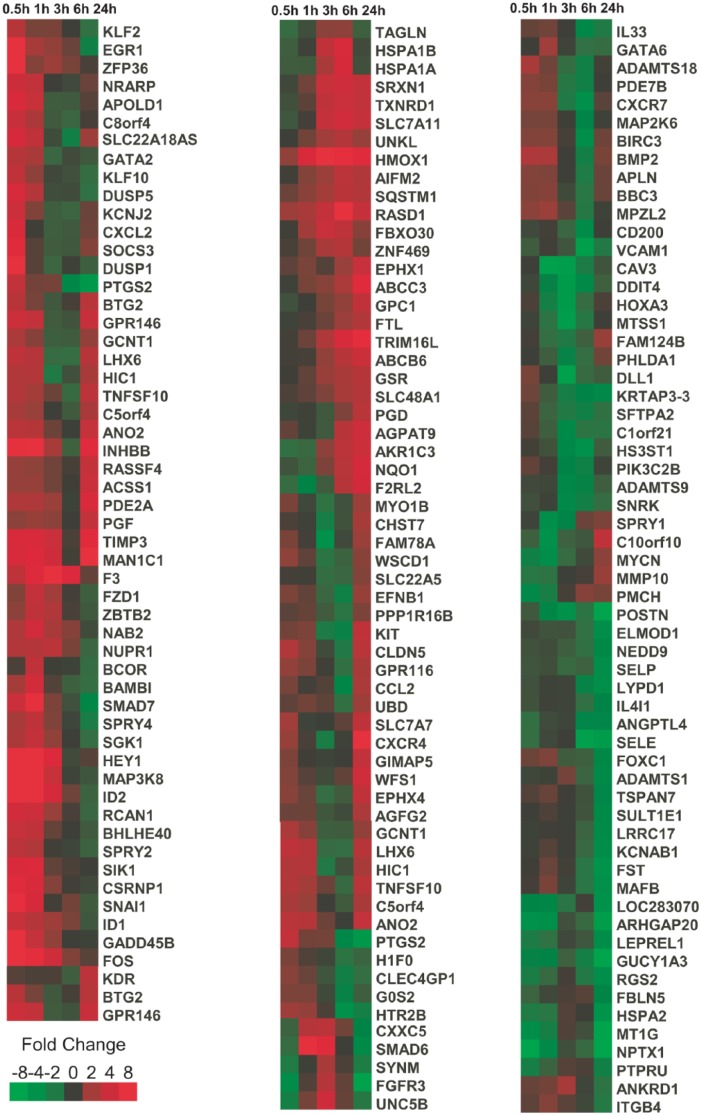
Gene expression profiling of CDDO-Im treated for 0.5, 1, 3, 6, and 24 hours was performed using Agilent Whole Human Genome Microarrays. A heat map of genes statistically altered in their expression upregulated or downregulated more than 8-fold across the time following clustering is shown in (Figure 1). The derivative CDDO-Im induced genes as quickly as 0.5 hour, including genes involved in early growth response (EGR1, EGR2, and EGR3) as well as genes such as FOS (regulators of cell proliferation, differentiation, and transformation in the early phases [0.5-1 hour] of gene expression response induced by CDDO-Im (Table 1)). These genes were rapidly upregulated by 0.5 hour and in several cases returned to near basal levels by 3 hours (Figure 1).
Figure 1.
Hierarchical agglomerative clustering of genes exhibiting more than an 8-fold statistical alteration in expression (false discovery rate < 10%) based on Pearson correlation coefficient.
Table 1.
Top 10 most highly expressed genes in the “early” phase (0.5-1 hour) of induction resulting from treatment with a 200-nM dose of CDDO-Im in HUVEC.
| Symbol | Name | 0.5 h | 1 h | 3 h | 6 h | 24 h |
|---|---|---|---|---|---|---|
| EGR3 | Early growth response 3 | 80.6 | 42.4 | 1.6 | 0.3 | 0.3 |
| NR4A1 | Nuclear receptor subfamily 4, group A, member 1 | 63.9 | 22.9 | 0.6 | 0.4 | 0.4 |
| FOS | FBJ murine osteosarcoma viral oncogene homolog | 45.3 | 7.0 | 4.2 | 1.5 | 1.2 |
| EGR2 | Early growth response 2 | 26.1 | 10.8 | 0.8 | 0.9 | 0.2 |
| FOSB | FBJ murine osteosarcoma viral oncogene homolog B | 23.5 | 24.3 | 3.7 | 1.0 | 2.8 |
| EGR4 | Early growth response 4 | 15.6 | 16.9 | 3.1 | 1.0 | 1.0 |
| EGR1 | Early growth response 1 | 8.5 | 2.1 | 1.8 | 1.3 | 0.4 |
| DUSP1 | Dual specificity phosphatase 1 | 8.4 | 1.2 | 0.6 | 0.9 | 0.7 |
| KLF4 | Kruppel-like factor 4 (gut) | 8.1 | 8.4 | 1.8 | 1.0 | 0.6 |
| JUNB | Jun B proto-oncogene | 7.8 | 4.6 | 1.9 | 1.2 | 1.3 |
Abbreviations: CDDO-Im, 1[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole; HUVEC, human umbilical vein endothelial cells.
Values at each time point represent fold change compared with control (dimethyl sulfoxide vehicle–treated HUVEC at time 0) after normalization to housekeeping genes.
Genes identified in the intermediate phase (3- and 6-hour expression levels) of the global gene expression analysis included known protective-related genes such as HMOX1, NQO1, HSPA1A, HSPA1B TAGLN, SRXN1, PTPRU, and UNC5B. Table 2 lists the top 10 most highly expressed genes in this intermediate phase (3-6 hours). Several of these genes, as illustrated in Table 3, are directly related to the NRF2 or Antioxidant Response Element (ARE) pathways and are known to be downstream protective genes induced through signaling and phosphorylating events.
Table 2.
Top 10 most highly expressed genes in the “intermediate” phase (3-6 hours) of induction resulting from treatment with a 200-nM dose of CDDO-Im in HUVEC.
| Symbol | Name | 0.5 h | 1 h | 3 h | 6 h | 24 h |
|---|---|---|---|---|---|---|
| NOG | Noggin | 12.2 | 31.5 | 24.6 | 0.7 | 0.7 |
| F3 | Coagulation factor III (thromboplastin, tissue factor) | 2.9 | 5.7 | 9.2 | 5.6 | 1.3 |
| DACT1 | Dapper, antagonist of β-catenin, homolog 1 (Xenopus laevis) | 2.3 | 8.5 | 8.2 | 1.4 | 0.9 |
| HMOX1 | Heme oxygenase (decycling) 1 | 1.7 | 5.2 | 8.0 | 7.7 | 6.2 |
| ATOH8 | Atonal homolog 8 (Drosophila) | 1.9 | 7.9 | 7.2 | 1.4 | 1.4 |
| CLDN23 | Claudin 23 | 6.1 | 13.0 | 5.6 | 1.4 | 2.2 |
| HEY1 | Hairy/enhancer of split related to YRPW motif 1 | 43.6 | 55.7 | 5.4 | 0.9 | 1.2 |
| ALPK2 | Alpha-kinase 2 | 1.8 | 1.0 | 5.4 | 1.0 | 2.1 |
| FZD7 | Frizzled family receptor 7 | 6.8 | 20.8 | 5.3 | 4.1 | 0.9 |
| RASL1B | RAS-like, family 11, member B | 8.4 | 16.9 | 5.1 | 0.9 | 0.4 |
Abbreviations: CDDO-Im, 1[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole; HUVEC, human umbilical vein endothelial cells.
Values at each time point represent fold change compared with control (dimethyl sulfoxide vehicle–treated HUVEC at time 0) after normalization to housekeeping genes.
Table 3.
Altered nuclear factor erythroid 2–related factor 2 (NRF2)-mediated gene set.
| Symbol | Name | 0.5 h | 1 h | 3 h | 6 h | 24 h |
|---|---|---|---|---|---|---|
| HMOX1 | Heme oxygenase (decycling) 1 | 1.7 | 5.2 | 8.0 | 7.7 | 6.2 |
| NQO1 | NAD(P)H dehydrogenase, quinone 1 | 0.8 | 0.7 | 1.5 | 2.6 | 4.5 |
| JUNB | Jun B proto-oncogene | 7.8 | 4.6 | 1.9 | 1.2 | 1.3 |
| FOS | FBJ murine osteosarcoma viral oncogene homolog | 45.3 | 7.0 | 4.2 | 1.5 | 1.2 |
| DNAJA4 | DnaJ (Hsp40) homolog, subfamily A, member 4 | 0.9 | 1.2 | 1.8 | 1.9 | 0.9 |
| DNAJB11 | DnaJ (Hsp40) homolog, subfamily B, member 11 | 0.7 | 0.8 | 1.1 | 1.0 | 0.8 |
| GCLC | Glutamate-cysteine ligase, catalytic subunit | 1.1 | 0.8 | 0.7 | 1.4 | 1.3 |
| GCLM | Glutamate-cysteine ligase, modifier subunit | 1.0 | 0.8 | 1.1 | 1.4 | 1.3 |
This table shows gene expression values in response to 1[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole treatment throughout the time course that are known to be mediated through the NRF2-mediated oxidative stress response pathway. Values at each time point represent fold change compared with control (dimethyl sulfoxide vehicle–treated human umbilical vein endothelial cells at time 0) after normalization to housekeeping genes.
Expression of genes at 24 hours served as a reference for activity of early initiated gene expression in response to CDDO-Im treatment and included TIMP3, CXCR4, and TFEC, among others, which largely resembled the control (DMSO-treated) gene expression profile (Table 4).
Table 4.
Top 10 most highly expressed genes in the “late” phase (24 hours) of induction resulting from treatment with a 200-nM dose of CDDO-Im in HUVEC.
| Symbol | Name | 0.5 h | 1 h | 3 h | 6 h | 24 h |
|---|---|---|---|---|---|---|
| GPIHBP1 | Glycosylphosphatidylinositol anchored high-density lipoprotein–binding protein 1 | 1.6 | 2.1 | 1.8 | 1.5 | 12.6 |
| CXCR4 | Chemokine (C-X-C motif) receptor 4 | 1.8 | 1.1 | 0.5 | 0.9 | 12.0 |
| MAN1C1 | Mannosidase, alpha, class 1C, member 1 | 4.9 | 5.4 | 4.0 | 1.1 | 9.8 |
| TFEC | Transcription factor EC | 1.7 | 0.8 | 1.7 | 3.4 | 7.1 |
| FLJ34503 | Uncharacterized FLJ34503 | 3.5 | 2.6 | 1.7 | 1.0 | 6.7 |
| AFAP1L2 | Actin filament–associated protein 1-like 2 | 5.1 | 4.9 | 2.6 | 1.6 | 6.6 |
| TIMP3 | TIMP metallopeptidase inhibitor 3 | 4.8 | 5.0 | 3.7 | 1.1 | 6.2 |
| SEMA6A | Sema domain, transmembrane domain (TM), and cytoplasmic domain, semaphorin 6A | 5.3 | 1.6 | 0.7 | 0.7 | 5.9 |
| INHBB | Inhibin, beta B | 7.7 | 9.0 | 2.0 | 0.7 | 5.9 |
| APCDD1 | Adenomatous polyposis coli downregulated 1 | 1.6 | 1.9 | 0.8 | 0.6 | 5.4 |
Abbreviations: CDDO-Im, 1[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole; HUVEC, human umbilical vein endothelial cells.
Values at each time point represent fold change compared with control (dimethyl sulfoxide vehicle–treated HUVEC at time 0) after normalization to housekeeping genes.
Profiling reveals peak times for maximal protection provided by NRF2-mediated genes
Examining the results over time showed the time point at which maximal expression is achieved. Early genes such as JUNB and FOS exhibit rapid increases in gene expression (7.81 and 45.26, respectively) at the 0.5-hour time point (Table 1). Other NRF2-mediated genes, such as HMOX1 and NQO1, obtain peak expression later in the time course (7.96 at 3 hours for HMOX1 and 4.52 at 24 hours for NQO1) (Table 2).
RT-PCR and Western blotting confirm microarray gene expression profiles
To validate the microarray results, quantitative RT-PCR was performed on several immediate-early key genes at 0.5, 1, and 3 hours. The results confirmed that genes such as JUNB, FOS, DUSP1, and EGR3 were rapidly induced by 0.5 hour and back to gene expression levels comparable with controls (DMSO) by 3 hours (Figure 2). To investigate translation of protein products over the time course, Western blotting was performed on several of the significantly expressed genes. Figure 3 shows protein expression for HSP105, DYRK3, DUSP1, HSP70, and HMOX1 at 1, 3, and 6 hours. HMOX1 showed 2-, 3-, and 12-fold increases in protein expression compared with DMSO at 1, 3, and 6 hours, respectively (P < .05) for all time points. HSP105 showed 1.2-, 1.4-, and 4.7-fold increases compared with DMSO (P < .05). DUSP1 exhibited protein expression levels that were all close to 1-fold increase compared with DMSO whose protein expression levels were consistent with messenger RNA (mRNA) levels from the microarray with the exception of 0.5 hour. DYRK3 had small but significant increases in mRNA levels in the gene expression profiles; this finding was confirmed by protein expression levels (1.0-, 1.3-, 1.8-fold) compared with DMSO (P < .05). HSPA1A showed similar trend such that the protein expression levels exhibited small increases over the time course (1.2, 1.4, and 1.6; P < .05). The mRNA expression levels obtained from the microarray analysis for HSPA1A at the same time points (1, 3, and 6 hours) were 0.74-, 2.1-, and 4.8-fold increased, respectively.
Figure 2.
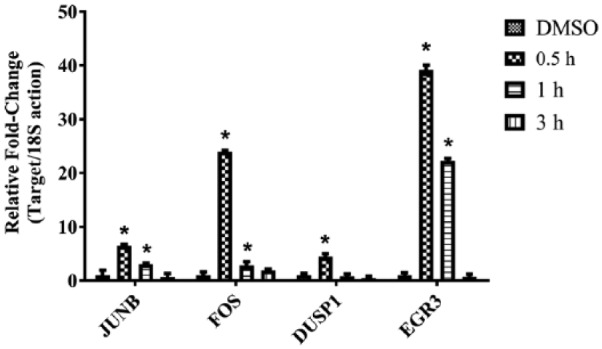
Immediate-early gene expression levels induced by 1[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole (CDDO-Im) treatment in human umbilical vein endothelial cells measured with quantitative real-time polymerase chain reaction. JUNB, FOS, DUSP1, and EGR3 expression levels were significantly increased by 30 minutes after CDDO-Im treatment compared with dimethyl sulfoxide (DMSO) control (n = 4; *P < .001). Gene expression levels for this set of genes were back to levels comparable with control (DMSO) by 3 hours.
Figure 3.
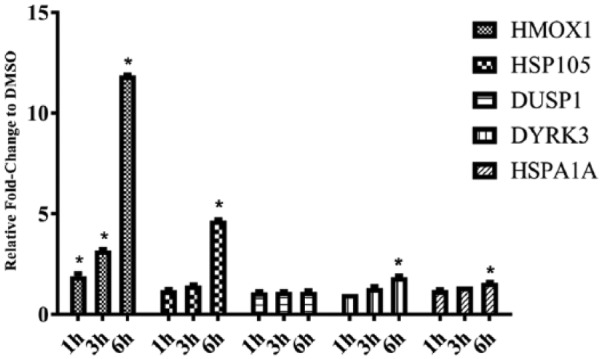
Protein induction by 1[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole (CDDO-Im) in human umbilical vein endothelial cells by relative Western blot analysis for time points 1, 3, and 6 hours. HMOX1, HSP105, DUSP1, DYRK3, and HSP70 were all compared with dimethyl sulfoxide (DMSO) control (0.1% final concentration). Values are presented as mean values with standard deviations (n = 4; *P < .05).
Correlation of functional and biological connectivity using network analysis
To focus on early initiators of the gene expression profile, network analysis was performed on the 0.5-hour samples following dosing with 200 nM CDDO-Im. The network graphically represents nodes (genes) and edges (the biological relationship between genes). This network included genes such as NR4A2, FOS, and JUNB; several EGR variations; and identified DUSP1 as a key immediate-early gene. This class of genes includes functions such as early growth stimulation, cell differentiation and growth, and signal transduction (Figure 4). Figure 5 demonstrates a network that highlights intermediate gene expression 3 hours after treatment with CDDO-Im.
Figure 4.
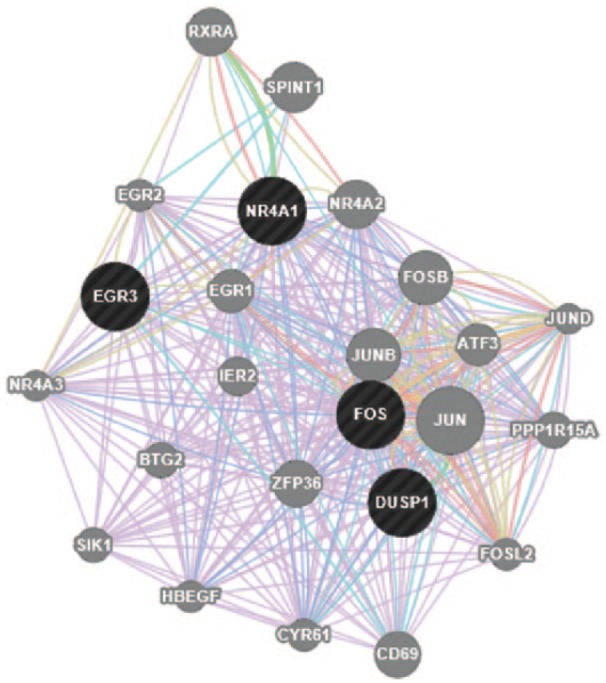
Network of genes constructed by GeneMANIA software on the basis of the functional and biological connectivity of genes. This network highlights the genes induced by 1[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole after treatment for 0.5 hour and represents the immediate-early gene response. The network is graphically represented as nodes (genes) and edges (the biological relationship between genes). Black nodes represent significantly expressed genes present in the input data submitted to GeneMANIA. Gray nodes represent genes returned by GeneMANIA. The size of each node is proportional to the degree of connectivity within the network, whereas the edge width is proportional to the confidence of the connection.
Figure 5.
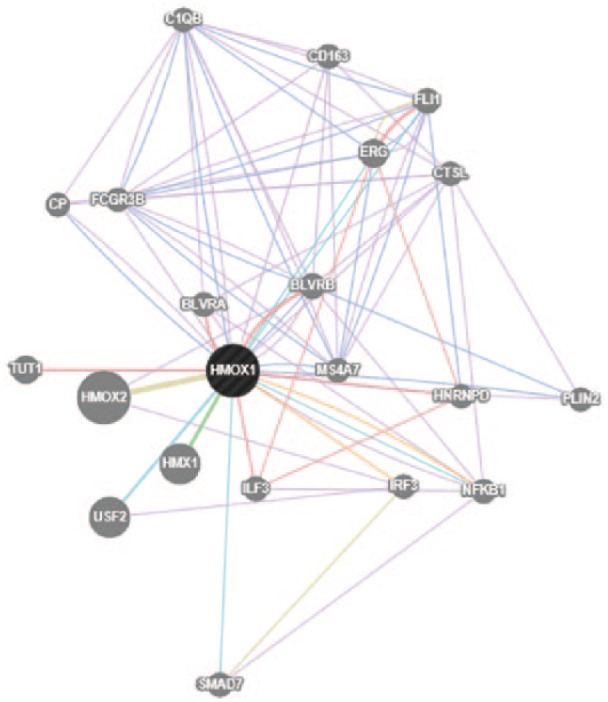
Network of genes constructed by GeneMANIA software on the basis of the functional and biological connectivity of genes. This network highlights the genes induced by 1[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole (CDDO-Im) after treatment for 3 hours and represents intermediate gene expression response to CDDO-Im. The network is graphically represented as nodes (genes) and edges (the biological relationship between genes). Black nodes represent significantly expressed genes present in the input data submitted to GeneMANIA. Gray nodes represent genes returned by GeneMANIA. The size of each node is proportional to the degree of connectivity within the network, whereas the edge width is proportional to the confidence of the connection.
X2K reveals key protein kinase events including possible role of mitogen-activated protein kinase kinase 1
To identify early regulators of cytoprotection provided by CDDO-Im, the time-course gene expression study was submitted to X2K for analysis of transcription factors, intermediate proteins, and protein kinases that regulate downstream induction of cytoprotective genes. Once the expression data were submitted for analysis, a list of predicted transcription factors was generated.
Using the top predicted transcription factors as seeds for building protein complexes, a subnetwork of intermediate proteins were identified; these proteins connect through known protein-protein associations to the transcription factors and to the known protein kinases involved that phosphorylate the protein complexes. Among these predicted kinases, dual specificity mitogen-activated protein kinase kinase 1 (MAP2K1) was identified as playing a compelling early role in the regulation of subsequent gene expression (Figure 6).
Figure 6.
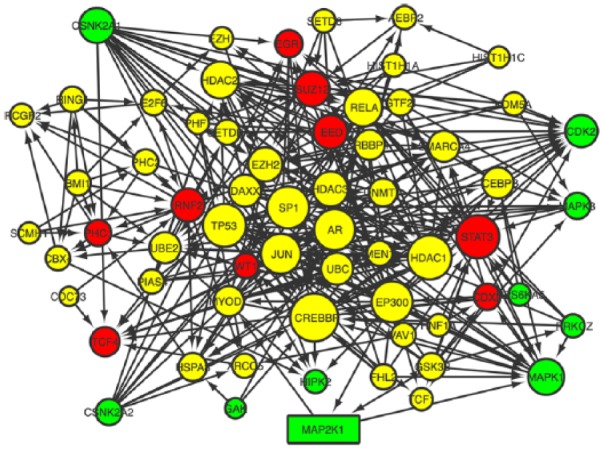
Expression2Kinases network constructed on the basis of the transcription factors, protein-protein interactions, and protein kinases acting within 30 minutes of treatment with 1[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole . Transcription factor nodes (red), protein-protein interaction nodes (yellow), and kinase (green). Network generated with Cytoscape 3.1.1.
Discussion
Time-course gene expression through transcriptomic and proteomic data analysis has increasingly become a plausible approach to study cellular responses to stimuli for drug discovery, development, and drug therapeutics.26,27 The technology is well suited for the dynamic nature of gene expression over time and allows for detailed examination of underlying mechanisms. This study highlights a multileveled use of bioinformatics tools that incorporate human time-course gene expression data in a systems pharmacology approach to illustrate a method for detailing mechanisms for observed genome-wide gene expression patterns.
Time-course gene expression analysis of HUVEC treated with 200 nM CDDO-Im revealed distinct phases of gene expression as a function of time; the dynamic nature of the most significantly expressed genes is illustrated in the clustering analysis and heat map presented in this work. Of particular interest is the early response (0.5 hour), which was key for elucidating mechanisms that drive downstream gene expression responses and regulate critical cellular signaling events. To the best of our knowledge, this work is the first of its kind to examine CDDO-Im in HUVEC from a systems pharmacology approach using time-course gene expression and applying further novel analysis to truly elucidate initiators of upstream gene expression.
Analysis of this gene expression data set with integration of data obtained from protein-protein interactions and kinase-substrate phosphorylation reactions showed that CDDO-Im initiates rapid responses as early as 0.5 hour and causes early key phosphorylation events that lead to activation of several initiators of downstream expression. Consistent with our previous work, as well as others, several genes involved in the NRF2 pathway were reconfirmed in this study.11,13,15 Many of these genes have previously been identified to play crucial roles in downstream cytoprotection, but the underlying mechanism initiating this response has not been fully elucidated.28,29 This analysis identified a key role provided by MAP2K1, also commonly known as MEK1. Mitogen-activated protein kinase kinase 1 lies upstream of mitogen-activated protein kinases (MAPKs) and regulates this class of kinases through a wide variety of extracellular and intracellular signals.30 Mitogen-activated protein kinases, also known as extracellular signal–regulated kinases (ERK), are well known to act as initiation points for further signaling events.31 Mitogen-activated protein kinase kinase 1 has been shown to obtain activation through events such as the binding of extracellular ligands to cell-surface receptors which in turn activates RAS and RAF1.32 RAF1, through phosphorylation of threonine and tyrosine residues of MAP2K1, leads to further activation and transduction of signaling pathways such as MAPK/ERK. The MAPK/ERK pathway has widely been reported to mediate biological functions, such as survival, cell growth, and metabolism, largely through downstream gene transcription.33–35
The identification of the DUSP1 gene through gene expression analysis (GeneMANIA) of the 0.5-hour time point revealed a crucial link in the understanding of the mechanism related to CDDO-Im. DUSP1, also known as mitogen-activated protein kinase phosphatase-1 (MKP-1), is a key phosphatase that has recently been shown to be rapidly induced in response to several anti-inflammatory drugs including glucocorticoids.36 DUSP1 preferentially dephosphorylates both threonine and tyrosine residues on MAPK, thereby modulating inflammation.36,37 Consistent with other studies, here we show that activation of DUSP1 is mediated by upstream activation of MAP2K1.38,39 Several studies have shown the dependence on DUSP1 induction, including 2 anti-inflammatory cytokine drugs (interleukin 10 and transforming growth factor β), corticosteroids, and rapamycin.40–42 The data in this study identify the potential role of DUSP1 and the required upstream mediator MAP2K1 and further suggest that CDDO-Im may initiate its cytoprotective effects through similar signaling precursors as other anti-inflammatory compounds.
The model as presented in this study addresses several limitations that often exist in much of current gene expression research to date. First, researchers often focus on a single or final time point alone and fail to take full advantage of the temporal nature of genes over time.43,44 Understanding the transcriptional kinetics provided by time-course studies allows for more accurate profiling of not just a single gene but sets of related genes that potentially offer greater significance when studied from a series of time points. Second, using a multileveled approach such as the one presented here offers better understanding into the underlying mechanisms responsible for observed responses.45 The integration of programs such as X2K takes expression data one step further and allows for identification of regulatory mechanisms upstream of gene expression. Although this study highlights a novel approach for identification of upstream initiators of observed cytoprotection from CDDO-Im treatment, it is not without its own limitations. As shown in Table 2 and other data in this study, defining the “intermediate” response of any particular gene over time is challenging due to the temporal nature of gene expression and limited time point data in a particular time-course study. The use of bioinformatics programs such as those used in this study is largely based on inferred interactions of experimentally derived data. As such, the associated networks and connectivity of the data presented are only as good as the databases and literature that are mined. Future studies exploring the interactions identified in this systems approach are needed to confirm the actual roles of key initiators such as MAP2K1 and DUSP1.
Acknowledgments
This study was conducted under a protocol reviewed and approved by the US Army Medical Research and Materiel Command Institutional Review Board and in accordance with the approved protocol.
Footnotes
Peer review:Six peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1383 words, excluding any confidential comments to the academic editor.
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the US Army Medical Research and Materiel Command.
Declaration of conflicting interests:The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: The opinions or assertions contained herein are the private views of the author and are not to be construed as official or as reflecting the views of the Department of the Army or the Department of Defense.
Author Contributions: JAB designed the study; collected, analyzed, and interpreted data; performed literature review; and wrote the manuscript. XW collected and analyzed data and performed a critical review of the manuscript. PDB and SAS designed the study, interpreted data, performed literature review, and contributed to the writing of the manuscript.
References
- 1. Eskiocak U., Kim S. B., Roig A. I., Kitten E., Batten K., Cornelius C., Zou Y. S., Wright W. E., Shay J. W. (2010) CDDO-Me protects against space radiation-induced transformation of human colon epithelial cells. Radiat Res 174, 27–36. [DOI] [PubMed] [Google Scholar]
- 2. Kulkarni A. A., Thatcher T. H., Hsiao H. M., Olsen K. C., Kottmann R. M., Morrissette J., Wright T. W., Phipps R. P., Sime P. J. (2013) The triterpenoid CDDO-Me inhibits bleomycin-induced lung inflammation and fibrosis. PLoS One 8, e63798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sussan T. E., Rangasamy T., Blake D. J., Malhotra D., El-Haddad H., Bedja D., Yates M. S., Kombairaju P., Yamamoto M., Liby K. T., Sporn M. B., Gabrielson K. L., Champion H. C., Tuder R. M., Kensler T. W., Biswal S. (2009) Targeting Nrf2 with the triterpenoid CDDO-imidazolide attenuates cigarette smoke-induced emphysema and cardiac dysfunction in mice 106, 250–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang F., Wang S., Zhang M., Weng Z., Li P., Gan Y., Zhang L., Cao G., Gao Y., Leak R. K., Sporn M. B., Chen J. (2012) Pharmacological induction of heme oxygenase-1 by a triterpenoid protects neurons against ischemic injury. Stroke 43, 1390–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reisman S. A., Buckley D. B., Tanaka Y., Klaassen C. D. (2009) CDDO-Im protects from acetaminophen hepatotoxicity through induction of Nrf2-dependent genes. Toxicol Appl Pharmacol 236, 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Suh N., Wang Y., Honda T., Gribble G. W., Dmitrovsky E., Hickey W. F., Maue R. A., Place A. E., Porter D. M., Spinella M. J., Williams C. R., Wu G., Dannenberg A. J., Flanders K. C., Letterio J. J., Mangelsdorf D. J., Nathan C. F., Nguyen L., Porter W. W., Ren R. F., Roberts A. B., Roche N. S., Subbaramaiah K., Sporn M. B. (1999) A novel synthetic oleanane triterpenoid, 2-cyano-3,12-dioxoolean-1,9-dien-28-oic acid, with potent differentiating, antiproliferative, and anti-inflammatory activity. Cancer Res 59, 336–341. [PubMed] [Google Scholar]
- 7. Kim E. H., Deng C., Sporn M. B., Royce D. B., Risingsong R., Williams C. R., Liby K. T. (2012) CDDO-methyl ester delays breast cancer development in BRCA1-mutated mice. Cancer Prev Res (Phila) 5, 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim E. H., Deng C. X., Sporn M. B., Liby K. T. (2011) CDDO-imidazolide induces DNA damage, G2/M arrest and apoptosis in BRCA1-mutated breast cancer cells. Cancer Prev Res (Phila) 4, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deeb D., Gao X., Liu Y., Varma N. R., Arbab A. S., Gautam S. C. (2013) Inhibition of telomerase activity by oleanane triterpenoid CDDO-Me in pancreatic cancer cells is ROS-dependent. Molecules 18, 3250–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deeb D., Gao X., Liu Y., Kim S. H., Pindolia K. R., Arbab A. S., Gautam S. C. (2012) Inhibition of cell proliferation and induction of apoptosis by oleanane triterpenoid (CDDO-Me) in pancreatic cancer cells is associated with the suppression of hTERT gene expression and its telomerase activity. Biochem Biophys Res Commun 422, 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bynum J. A., Rastogi A., Stavchansky S. A., Bowman P. D. (2015) Cytoprotection of human endothelial cells from oxidant stress with CDDO derivatives: network analysis of genes responsible for cytoprotection. Pharmacology 95, 181–192. [DOI] [PubMed] [Google Scholar]
- 12. Deeb D., Gao X., Arbab A. S., Barton K., Dulchavsky S. A., Gautam S. C. (2010) CDDO-Me: A Novel Synthetic Triterpenoid for the Treatment of Pancreatic Cancer. Cancers 2, 1779–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu M., Reddy N. M., Higbee E. M., Potteti H. R., Noel S., Racusen L., Kensler T. W., Sporn M. B., Reddy S. P., Rabb H. (2014) The Nrf2 triterpenoid activator, CDDO-imidazolide, protects kidneys from ischemia-reperfusion injury in mice. Kidney Int 85, 134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reddy N. M., Suryanaraya V., Yates M. S., Kleeberger S. R., Hassoun P. M., Yamamoto M., Liby K. T., Sporn M. B., Kensler T. W., Reddy S. P. (2009) The triterpenoid CDDO-imidazolide confers potent protection against hyperoxic acute lung injury in mice. Am J Respir Crit Care Med 180, 867–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang X., Bynum J. A., Stavchansky S., Bowman P. D. (2014) Cytoprotection of human endothelial cells against oxidative stress by 1-[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole (CDDO-Im): application of systems biology to understand the mechanism of action. Eur J Pharmacol 734, 122–131. [DOI] [PubMed] [Google Scholar]
- 16. Pronk T. E., van der Veen J. W., Vandebriel R. J., van Loveren H., de Vink E. P., Pennings J. L. (2014) Comparison of the molecular topologies of stress-activated transcription factors HSF1, AP-1, NRF2, and NF-kappaB in their induction kinetics of HMOX1. Biosystems 124C, 75–85. [DOI] [PubMed] [Google Scholar]
- 17. Chen E. Y., Xu H., Gordonov S., Lim M. P., Perkins M. H., Ma’ayan A. (2012) Expression2Kinases: mRNA profiling linked to multiple upstream regulatory layers. Bioinformatics 28, 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen Q., Fischer A., Reagan J. D., Yan L. J., Ames B. N. (1995) Oxidative DNA damage and senescence of human diploid fibroblast cells. Proc Natl Acad Sci U S A 92, 4337–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Packer L., Fuehr K. (1977) Low oxygen concentration extends the lifespan of cultured human diploid cells. Nature 267, 423–425. [DOI] [PubMed] [Google Scholar]
- 20. Wang X., Stavchansky S., Zhao B., Bynum J. A., Kerwin S. M., Bowman P. D. (2008) Cytoprotection of human endothelial cells from menadione cytotoxicity by caffeic acid phenethyl ester: the role of heme oxygenase-1. Eur J Pharmacol 591, 28–35. [DOI] [PubMed] [Google Scholar]
- 21. Fleige S., Walf V., Huch S., Prgomet C., Sehm J., Pfaffl M. W. (2006) Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnol Lett 28, 1601–1613. [DOI] [PubMed] [Google Scholar]
- 22. Simon R., Lam A., Li M. C., Ngan M., Menenzes S., Zhao Y. (2007) Analysis of gene expression data using BRB-ArrayTools. Cancer informatics 3, 11–17. [PMC free article] [PubMed] [Google Scholar]
- 23. Montojo J., Zuberi K., Rodriguez H., Kazi F., Wright G., Donaldson S. L., Morris Q., Bader G. D. (2010) GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics 26, 2927–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mostafavi S., Ray D., Warde-Farley D., Grouios C., Morris Q. (2008) GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome biology 9 Suppl 1, S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zuberi K., Franz M., Rodriguez H., Montojo J., Lopes C. T., Bader G. D., Morris Q. (2013) GeneMANIA prediction server 2013 update. Nucleic Acids Res 41, W115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bai J. P., Alekseyenko A. V., Statnikov A., Wang I. M., Wong P. H. (2013) Strategic applications of gene expression: from drug discovery/development to bedside. The AAPS journal 15, 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bates S. (2011) The role of gene expression profiling in drug discovery. Current opinion in pharmacology 11, 549–556. [DOI] [PubMed] [Google Scholar]
- 28. Thimmulappa R. K., Fuchs R. J., Malhotra D., Scollick C., Traore K., Bream J. H., Trush M. A., Liby K. T., Sporn M. B., Kensler T. W., Biswal S. (2007) Preclinical evaluation of targeting the Nrf2 pathway by triterpenoids (CDDO-Im and CDDO-Me) for protection from LPS-induced inflammatory response and reactive oxygen species in human peripheral blood mononuclear cells and neutrophils. Antioxid Redox Signal 9, 1963–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Turpaev K. T. (2013) Keap1-Nrf2 signaling pathway: mechanisms of regulation and role in protection of cells against toxicity caused by xenobiotics and electrophiles. Biochemistry (Mosc) 78, 111–126. [DOI] [PubMed] [Google Scholar]
- 30. Dave S., Nanduri R., Dkhar H. K., Bhagyaraj E., Rao A., Gupta P. (2014) Nuclear MEK1 sequesters PPARgamma and bisects MEK1/ERK signaling: a non-canonical pathway of retinoic acid inhibition of adipocyte differentiation. PLoS One 9, e100862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Greulich H., Erikson R. L. (1998) An analysis of Mek1 signaling in cell proliferation and transformation. J Biol Chem 273, 13280–13288. [DOI] [PubMed] [Google Scholar]
- 32. Pearson G., Bumeister R., Henry D. O., Cobb M. H., White M. A. (2000) Uncoupling Raf1 from MEK1/2 impairs only a subset of cellular responses to Raf activation. J Biol Chem 275, 37303–37306. [DOI] [PubMed] [Google Scholar]
- 33. Kidambi S., Yarmush J., Berdichevsky Y., Kamath S., Fong W., Schianodicola J. (2010) Propofol induces MAPK/ERK cascade dependant expression of cFos and Egr-1 in rat hippocampal slices. BMC research notes 3, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang L., Xu T., Wang S., Yu L., Liu D., Zhan R., Yu S. Y. (2012) Curcumin produces antidepressant effects via activating MAPK/ERK-dependent brain-derived neurotrophic factor expression in the amygdala of mice. Behavioural brain research 235, 67–72. [DOI] [PubMed] [Google Scholar]
- 35. Besnard A., Laroche S., Caboche J. (2014) Comparative dynamics of MAPK/ERK signalling components and immediate early genes in the hippocampus and amygdala following contextual fear conditioning and retrieval. Brain structure & function 219, 415–430. [DOI] [PubMed] [Google Scholar]
- 36. Fischer A., Gluth M., Weege F., Pape U. F., Wiedenmann B., Baumgart D. C., Theuring F. (2014) Glucocorticoids regulate barrier function and claudin expression in intestinal epithelial cells via MKP-1. American journal of physiology. Gastrointestinal and liver physiology 306, G218–228. [DOI] [PubMed] [Google Scholar]
- 37. Chi H., Barry S. P., Roth R. J., Wu J. J., Jones E. A., Bennett A. M., Flavell R. A. (2006) Dynamic regulation of pro- and anti-inflammatory cytokines by MAPK phosphatase 1 (MKP-1) in innate immune responses. Proc Natl Acad Sci U S A 103, 2274–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Casteel M., Nielsen C., Kothlow S., Dietrich R., Martlbauer E. (2010) Impact of DUSP1 on the apoptotic potential of deoxynivalenol in the epithelial cell line HepG2. Toxicol Lett 199, 43–50. [DOI] [PubMed] [Google Scholar]
- 39. Hammer M., Mages J., Dietrich H., Servatius A., Howells N., Cato A. C., Lang R. (2006) Dual specificity phosphatase 1 (DUSP1) regulates a subset of LPS-induced genes and protects mice from lethal endotoxin shock. The Journal of experimental medicine 203, 15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hammer M., Mages J., Dietrich H., Schmitz F., Striebel F., Murray P. J., Wagner H., Lang R. (2005) Control of dual-specificity phosphatase-1 expression in activated macrophages by IL-10. Eur J Immunol 35, 2991–3001. [DOI] [PubMed] [Google Scholar]
- 41. Rastogi R., Jiang Z., Ahmad N., Rosati R., Liu Y., Beuret L., Monks R., Charron J., Birnbaum M. J., Samavati L. (2013) Rapamycin induces mitogen-activated protein (MAP) kinase phosphatase-1 (MKP-1) expression through activation of protein kinase B and mitogen-activated protein kinase kinase pathways. J Biol Chem 288, 33966–33977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xiao Y. Q., Malcolm K., Worthen G. S., Gardai S., Schiemann W. P., Fadok V. A., Bratton D. L., Henson P. M. (2002) Cross-talk between ERK and p38 MAPK mediates selective suppression of pro-inflammatory cytokines by transforming growth factor-beta. J Biol Chem 277, 14884–14893. [DOI] [PubMed] [Google Scholar]
- 43. Rowell J., Koitabashi N., Kass D. A., Barth A. S. (2014) Dynamic Gene Expression Patterns in Animal Models of Early and Late Heart Failure Reveal Biphasic-Bidirectional Transcriptional Activation of Signaling Pathways. Physiological genomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Iyer V. R., Eisen M. B., Ross D. T., Schuler G., Moore T., Lee J. C., Trent J. M., Staudt L. M., Hudson J., Jr., Boguski M. S., Lashkari D., Shalon D., Botstein D., Brown P. O. (1999) The transcriptional program in the response of human fibroblasts to serum. Science 283, 83–87. [DOI] [PubMed] [Google Scholar]
- 45. Huang S. (1999) Gene expression profiling, genetic networks, and cellular states: an integrating concept for tumorigenesis and drug discovery. Journal of molecular medicine 77, 469–480. [DOI] [PubMed] [Google Scholar]