Abstract
Background
MicroRNAs (miRNAs) are a class of small non-coding RNAs that are strongly involved in various types of carcinogenesis, including hepatocellular carcinoma (HCC). This study aimed to clarify whether miR-4417 promotes HCC growth by targeting TRIM35 and regulating PKM2 phosphorylation.
Material/Methods
Online software, including TargetScan and miRanda, was used to predict the potential target of miR-4417. Real-Time PCR (qRT-PCR) and Western blot assays were performed to detect the expression levels of mRNA and protein, respectively. Cell proliferation was measured by MTT assay and apoptosis in A549 cells was examined by flow cytometry.
Results
Bioinformatics reveal that TRIM35 mRNA contains 1 conserved target site of miR-4417. High level of miR-4417 and low levels of TRIM35 mRNA and protein were observed in HCC cells compared with a normal liver cell line. Biological function analysis showed that miR-4417 inhibitor inhibits cell proliferation and promotes apoptosis in HCC cells. Furthermore, we verified that TRIM35 is a functional target of miR-4417 by use of luciferase reporter assay, and TRIM35 overexpressing showed an elevation of proliferation and a reduction of apoptosis in HCC cells. We subsequently investigated whether miR-4417 and TRIM35 regulate HCC cell proliferation and apoptosis through PKM2 Y105 phosphorylation, and the results supported our speculation that miR-4417 targets TRIM35 and regulates the Y105 phosphorylation of PKM2 to promote hepatocarcinogenesis.
Conclusions
Our findings indicate that miR-4417 may function as an oncogene in HCC and is a potential alternative therapeutic target for this deadly disease.
MeSH Keywords: Carcinoma, Hepatocellular; Lim Kinases; MicroRNAs; Pyruvate Kinase
Background
Hepatocellular carcinoma (HCC) ranks as the third leading cause of cancer-related deaths worldwide. There are more than 250,000 new HCC cases and approximate 500,000 to 600,000 HCC deaths each year [1]. Chronic hepatitis B and C and alcoholic liver disease are the predominant risk factors for the initiation of HCC [2]. Despite advances in functional genomics providing a deeper understanding of viral-associated hepatocarcinogenesis, the molecular pathogenesis of HCC still remains elusive and 5-year survival rates have not changed much during the past several years. Therefore, a better understanding of the underlying molecular mechanisms of HCC development and growth is urgently needed to develop novel diagnostic biomarkers and effective therapeutic strategies for patients with HCC [3].
MicroRNAs (miRNAs) are small non-coding RNAs of about 19–25 nt in length that predominantly bind to 3′untranslated regions (3′UTR) of target genes, leading to either translational repression or mRNA degradation. miRNAs regulate about 30% of all human protein-coding genes and the levels of some miRNAs have arisen as biomarkers for various pathological situations [4]. Emerging evidence indicates that miRNAs may play important roles in diverse physiological and pathological processes like development, proliferation, apoptosis, fat metabolism, signal transduction, drug resistance, and oncogenesis [5]. Data from array analyses have clearly showed that the altered miRNA expression profile occurs frequently in cancer samples, including HCC [6,7]. Previous studies have revealed a tumor-promoting function for some highly expressed miRNAs by negatively regulating tumor suppressor genes, while low-expressed miRNAs could serve as tumor suppressors by suppressing oncogenes.
Dysregulated expression of miRNA is a frequent event in human hepatocarcinogenesis. miRNA is a major class of regulator genes in HCC development, and understanding the relevant regulatory mechanisms of miRNAs might provide novel targets for treatment of this deadly disease. For example, miR-26a exhibited decreased expression in HCC tissues, which is associated with HCC recurrence and metastasis, and it was also reported to inhibit in vitro cell proliferation, migration, and invasion, and restrain in vivo tumor growth and metastasis in nude mouse models with human HCC, indicating the potential applications of miR-26a as a novel prognostic marker and therapeutic target [8]. Serum miR-1 may act as a new independent parameter of overall survival in patients with HCC, thereby improving the predictive value of the classical HCC staging system [9]. Furthermore, miR-21 enhances cell proliferation, invasion, and metastasis in HCC cells and xenograft tumor growth in mouse models, providing a candidate target for cancer gene therapy [10]. miR-4417 is a newly discovered miRNA that was reported to be upregulated in HCC tissues compared with adjacent normal tissues [11]. It was also reported that the top 5 putative targets of miR-4417 were SOS1, FAF2, PAXIP1, C1orf95, and WDR48 [11]. However, the direct functional targets and biological roles of miR-4417 in HCC cells remain unclear.
In the present study we measured miR-4417 expression in HCC cell lines by quantitative Real-Time PCR (qRT-PCR), and the role and mechanism of miR-4417 in regulation of proliferation and apoptosis in HCC cells were investigated.
Material and Methods
Cell culture
Three HCC cell lines (Huh7, SMMC-7721, and SK-Hep1) and a normal liver cell line (HL-7702) were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Cells were cultured in DMEM medium (Invitrogen, Gaithersburg, MD) at 37°C with 5% CO2 and 95% humidity. The DMEM medium was supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin, and 100 mg/ml streptomycin.
Cell treatment
SMMC-7721 cells (3×106 cells/ml) were seeded into 24-well plates and incubated overnight, then transfected with miR-4417 inhibitors (miR-inhibitor) pcDNA-TRIM35, si-TRIM35, pCMV-HA-PKM2 Y105F, and their respective controls (GenePharma, Shanghai, China) using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer’s instructions. Mutations Y105F were introduced into PKM2 using the QuikChange-XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). The cells were used for further experiments 48 h post-transfection.
Real-time quantitative reverse transcription-PCR (qRT-PCR)
Total RNA was extracted from the SMMC-7721 cell line (6×106 cells/ml, total 1 ml) by using Trizol (Invitrogen) according to the manufacturer’s instructions, and was inversely transcribed to cDNA using PrimeScript RT-polymerase (Takara, Dalian, China). The expression level of miR-4417 was determined by a TaqMan MicroRNA assay kit (Applied Biosystems), and U6 snRNA was used as an internal control. The mRNA levels of TRIM35 and PKM2 were detected by using the SYBR PrimeScript RT-PCR kit (Takara), and GAPDH was used as an internal control.
Western blot analysis
Total proteins were isolated from cell lysates using RIPA buffer containing protease inhibitors and phosphatase inhibitors (Thermo scientific, Rockford, IL). The protein extractions were then resolved on SDS-PAGE and transferred onto nitrocellular membranes (Millipore, Billerica, MA). After blocking in 5% skim milk, the membrane was incubated with primary antibodies overnight at 4°C. After washing, the horseradish peroxidase (HRP)-conjugated secondary antibodies were added and incubated for 1 h at room temperature. The blots were visualized using an enhanced chemiluminescence detection kit (Thermo scientific, Rockford, IL). Primary antibodies against TRIM35 and phosphorylated PKM2 (p-PKM2(Tyr105)) (Cell Signaling Technology, Danvers, MA) were used in Western blot analysis.
MTT assay
Cell growth was measured using the MTT method. Briefly, cell suspensions with 4×103 cells/well were seeded in 96-well plates and incubated at 37°C for 48 h. At each time point (0, 24, 48, and 72 h), 20 μl MTT (5 mg/ml, Sigma-Aldrich, St. Louis, MO) was added in each well followed by incubation for 4 h at 37°C, then the culture medium was removed and 150 μl of DMSO was added to each well to stop the reaction. The absorbance of each well was measured at 490 nm by using a microplate reader (Molecular Devices, Sunnyvale, CA).
BrdU incorporation assay
Cellular proliferation of SMMC-7721 cells was measured by bromodeoxyuridine (BrdU) incorporation assay. Briefly, after the cells underwent various treatments for 48 h, cell proliferation was determined using the BrdU Cell Proliferation ELISA kit (Roche Molecular Biochemicals, Mannheim, Germany) according to the manufacturer’s instructions. The results are shown as BrdU incorporation (%) compared to that in cells at 0 h.
Apoptosis assay
The apoptosis assay was performed by fluorescence-activated cell sorter (FACS) using Annexin V-FITC/PI apoptosis detection kits (BD Pharmingen, San Diego, CA). After washing, cells (2×106 cells/ml) were stained using Annexin V-FITC and propidium iodide (PI) for 15 min in the dark. Cell apoptosis was then analyzed with a flow cytometer (FACSCalibur, Becton Dickinson, San Jose, CA).
Luciferase reporter assay
The fragments including the 3′UTR-WT or 3′UTR-MUT regions of TRIM35 were inserted into pMIR-Report Luciferase vector (Ambion, Austin, TX) with a Renilla and firefly luciferase reporter gene. SMMC-7721 cells (3×106 cells/ml) were placed in a 96-well plate and incubated for 24 h. Then, cells were transfected with miR-4417 mimics or miR-control and pMIR-Report Luciferase vector containing 3′UTR-WT or 3′UTR-MUT regions of TRIM35 using Lipofectamine 2000 (Invitrogen). Forty-eight hours later, cells were lysed and luciferase activity was detected using the Dual Luciferase Reporter System (Promega, Madison, WI).
Statistical analysis
SPSS 15.0 software (SPSS, Chicago, IL) was used for general statistical analyses. The experiments were repeated at least 3 times, and the results are expressed as the mean ± standard deviation (SD). Student’s t-tests and one-way analysis of variance were used to analyze the data. A P-value <0.05 was considered statistically significant.
Results
miR-4417 was increased and TRIM35 was decreased in HCC cell lines
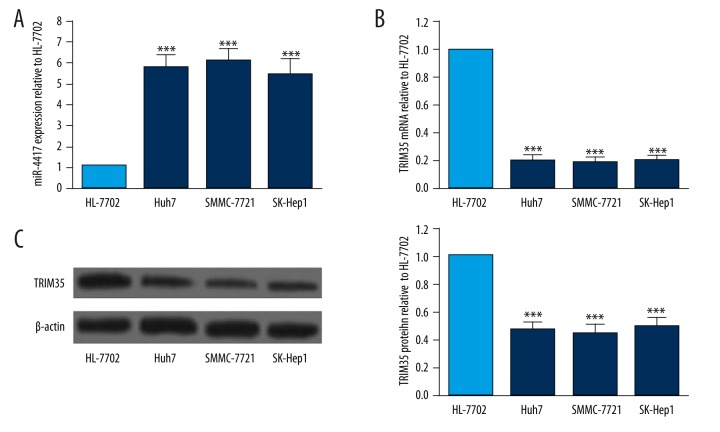
miR-4417 expression in HCC cell lines and a normal liver cell line was measured by qRT-PCR. We found that the level of miR-4417 significantly increased in HCC cell lines compared to the normal liver cell line HL-7702 (Figure 1A). In addition, the expression levels of TRIM35 mRNA and protein in HCC cell lines and HL-7702 were determined by qRT-PCR and Western blot, respectively. mRNA and protein levels of TRIM35 were both significantly decreased in HCC cell lines compared with those in the normal liver cell line (Figure 1B, 1C). The above results indicated that miR-4417 was upregulated and TRIM35 was downregulated in HCC cell lines. Since SMMC-7721 cells expressed the highest relative miR-4417 expression and the lowest TRIM35 level, this cell line was selected for the transfection of miR-4417 inhibitor.
Figure 1.
Levels of miR-4417 and TRIM35 in HCC cell lines. (A) Levels of miR-4417 were measured by qRT-PCR and normalized to U6 snRNA expression in 3 HCC cell lines and 1 normal liver cell line. miR-4417 expression was upregulated in HCC cell lines Huh7, SMMC-7721, and SK-Hep1 as compared with normal liver cell line HL-7702. (B, C) TRIM35 expression was determined by qRT-PCR and Western blot, and normalized to levels of GAPDH and β-actin, respectively. The expression levels of TRIM35 mRNA and protein were downregulated in HCC cell lines compared to normal liver cell line HL-7702. ** P<0.01; *** P<0.001.
Effects of miR-4417 and TRIM35 on proliferation and apoptosis of SMMC-7721 cells
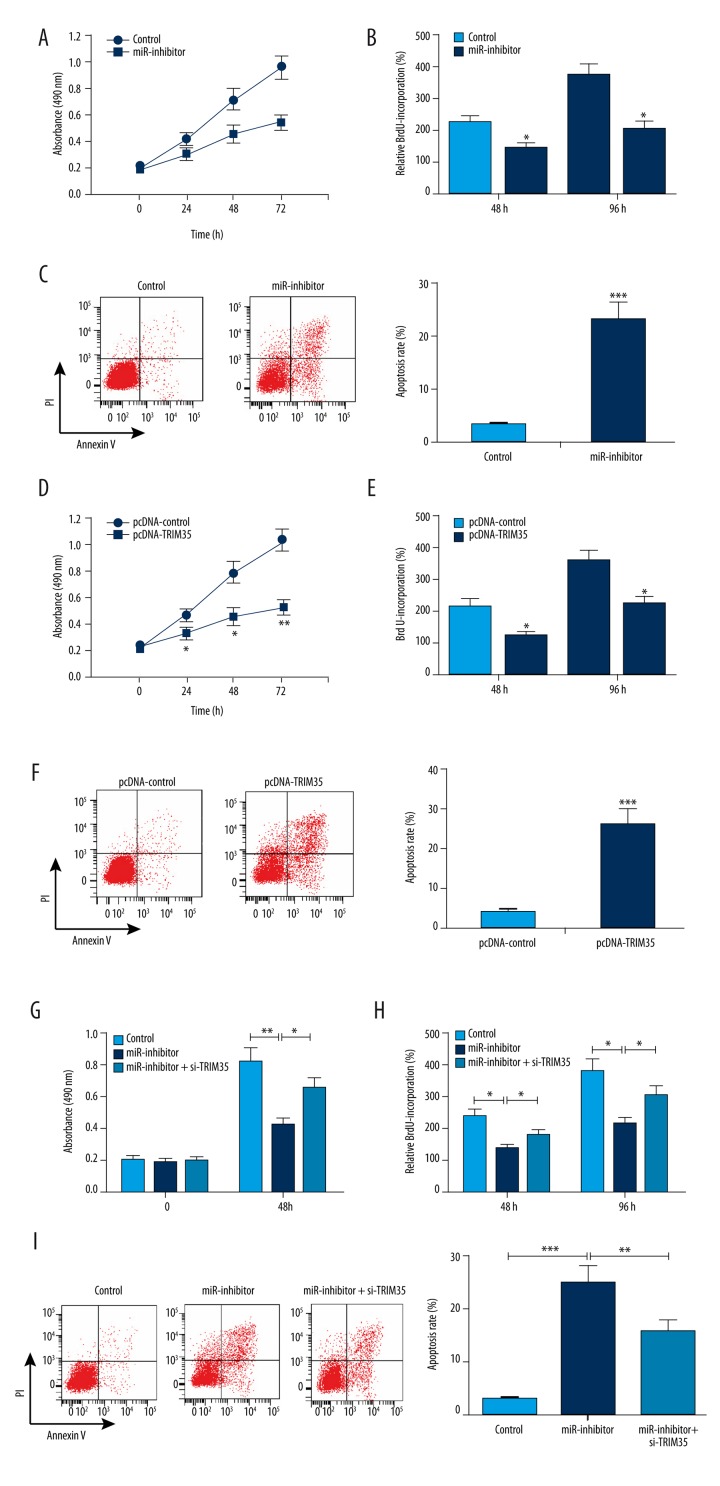
To investigate the effect of miR-4417 on HCC cell proliferation and apoptosis, we used miR-4417 inhibitor to inhibit miR-4417 expression in vitro, and then performed MTT, BrdU incorporation, and flow cytometry assays. MTT assay showed that cell growth was significantly impaired in SMMC-7721 cells transfected with miR-4417 inhibitor (Figure 2A). BrdU incorporation assay demonstrated that miR-4417 inhibitor inhibited cell proliferation compared with inhibitor control at 48 h and 72 h (Figure 2B). Flow cytometry showed that miR-4417 inhibitor promoted cell apoptosis compared with inhibitor control (Figure 2C). Furthermore, the effect of TRIM35 on proliferation and apoptosis of SMMC-7721 cells was determined by exogenous expression of TRIM35. As measured by MTT, BrdU incorporation, and flow cytometry assays, the upregulation of TRIM35 resulted in significant inhibitions in cell growth and proliferation, and a marked increase in apoptosis in SMMC-7721 cells (Figure 2D–2F). Additionally, miR-4417 inhibitor suppressed cell growth and promoted apoptosis in SMMC-7721 cells; however, si-TRIM35 significantly reversed these effects (Figure 2G–2I). Thus, downregulation of miR-4417 and upregulation TRIM35 suppressed proliferation and induced apoptosis in the SMMC-7721 cell line, and TRIM35 silencing could reverse the effects of decreased miR-4417 on SMMC-7721 cells.
Figure 2.
Effects of miR-4417 and TRIM35 on proliferation and apoptosis of SMMC-7721 cells. (A, B) Cell growth and proliferation were measured by MTT and BrdU assays, respectively. Cell growth and proliferation were suppressed by miR-4417 inhibitor. (C) Cell apoptosis was measured by flow cytometry. miR-4417 inhibitor significantly promoted cell apoptosis compared with inhibitor control. (D–F) Upregulation of TRIM35 resulted in significant inhibitions in cell growth and proliferation, and a marked increase in apoptosis in SMMC-7721 cells. (G–F) miR-4417 inhibitor suppressed cell growth and proliferation, and promoted apoptosis in SMMC-7721 cells; however, si-TRIM35 significantly reversed these effects. * P<0.05; ** P<0.01; *** P<0.001.
TRIM35 was a direct target of miR-4417
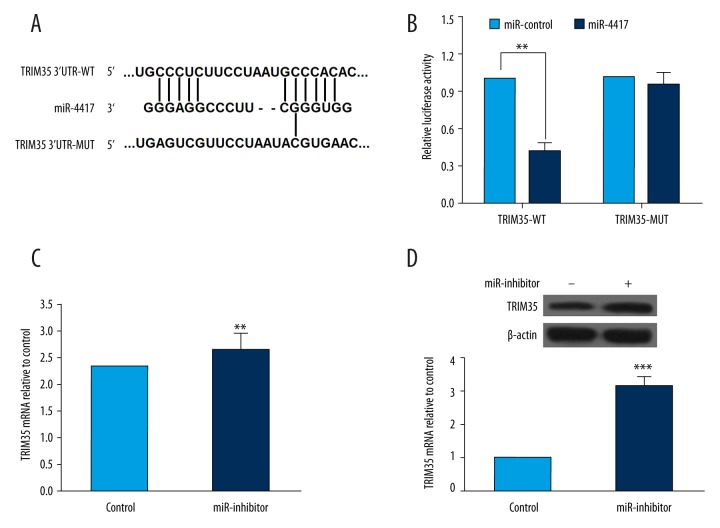
As described above, TRIM35 expression was associated with miR-4417, and miR-4417 might play a role in deregulation of TRIM35. Using online software, including TargetScan (www.targetscan.org) and miRanda (www.microrna.org), we found that tripartite motif-containing protein 35 (TRIM35) is a potential target of miR-4417. TRIM35 mRNA contains 1 conserved target site of miR-4417 (Figure 3A). Luciferase reporter assay was then performed to determine whether the 3′UTRs of TRIM35 is the binding site of miR-4417. The results from dual luciferase reporter assay showed that co-transfection with miR-4417 mimics significantly decreased the luciferase activity of the reporter containing wild-type 3′UTR of TRIM35 but not the mutant reporter gene, which indicated that TRIM35 is indeed a direct target of miR-4417 (Figure 3B). To further understand the regulatory effect of miR-4417 on TRIM35 expression, we detected the mRNA and protein levels of TRIM35 in SMMC-7721 cells transfected with miR-4417 inhibitor or inhibitor control. As illustrated in Figure 3C and 3D, compared with the controls, downregulation of miR-4417 led to increased levels of TRIM35 mRNA and protein. These data indicated that the expression of TRIM35 at both mRNA and protein levels was regulated by miR-4417 in vitro. miR-4417 regulates proliferation and apoptosis of HCC cells by directly targeting TRIM35.
Figure 3.
TRIM35 was a direct target of miR-4417 in SMMC-7721 cells. (A) Predicted miR-4417 target sequence in the 3′UTR of TRIM35 by online software, including TargetScan and miRanda. (B) Luciferase reporter assay showed the relative luciferase activities of TRIM35-WT and TRIM35-MUT co-transfected with miR-4417 mimics (miR-4417) or corresponding control (miR-control). (C, D) After SMMC-7721 cells were transfected with miR-4417 inhibitor, TRIM35 mRNA and protein levels were measured by qRT-PCR and Western blot, respectively. miR-4417 downregulation in SMMC-7721 cells was associated with increased TRIM35 mRNA and protein levels. ** P<0.01; *** P<0.001.
miR-4417 and TRIM35 regulated HCC cells through PKM2 Y105 phosphorylation
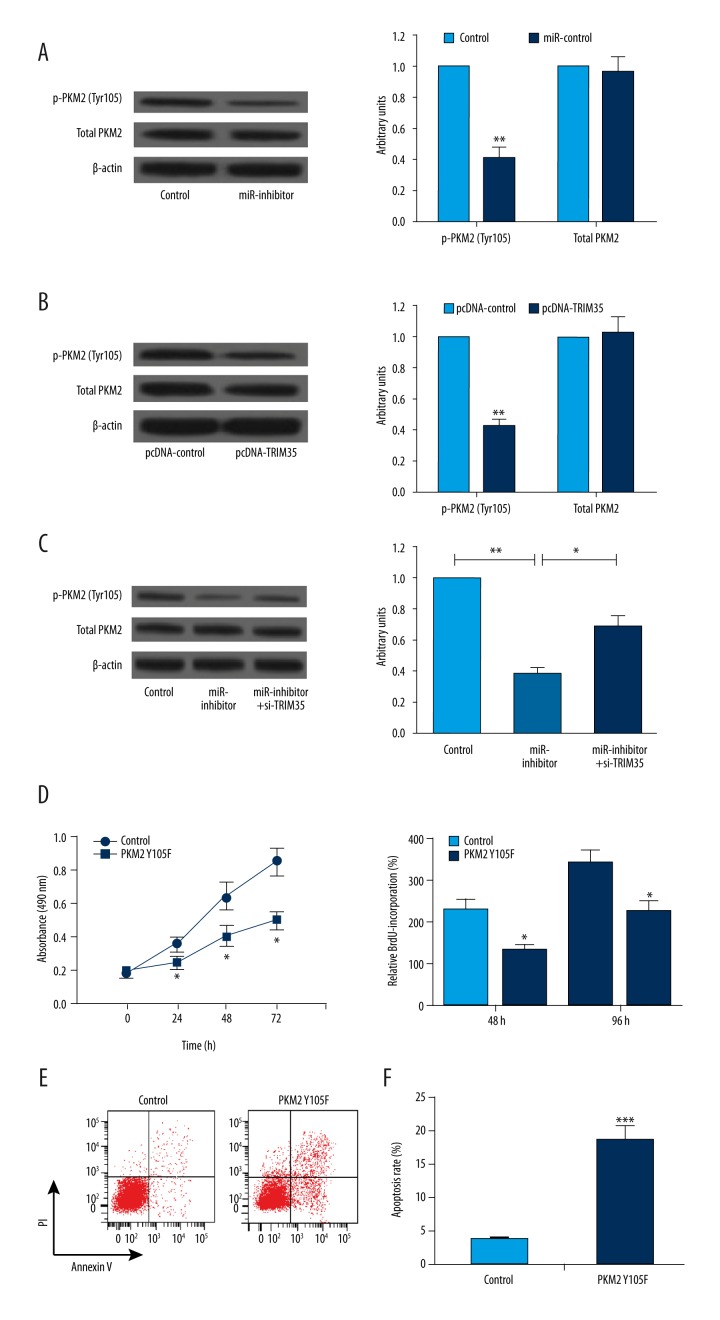
It was reported that TRIM35 inhibits tumorigenicity of HCC cells by regulating phosphorylation of PKM2 tyrosine residue 105 (Y105) in HCC [12]. To investigate whether miR-4417-induced upregulation of TRIM35 resulted in a reduction of PKM2 Y105 phosphorylation, we transfected miR-4417 inhibitor, pcDNA-TRIM35, si-TRIM35, and their respective controls into SMMC-7721 cells, respectively, and detected the Y105 phosphorylation of PKM2 expression using Western blot analysis. As shown in Figure 4A and 4B, decreased expression of miR-4417 and increased expression of TRIM35 significantly blocked PKM2 Y105 phosphorylation, but neither of them had an effect on total PKM2 levels. In addition, the inhibitory effect of miR-4417 inhibitor on PKM2 Y105 phosphorylation was overturned by TRIM35 deletion (Figure 4C). We next transfected SMMC-7721 cells with pCMV-HA vector containing mutant PKM2 Y105F, and then performed MTT, BrdU incorporation, and flow cytometry assays to determine the effect of PKM2 Y105F on SMMC-7721 cell proliferation and apoptosis. The results showed that PKM2 Y105F effectively inhibited cell proliferation and promoted apoptosis compared with the control (Figure 3D–3F). Taken together, our results suggest that miR-4417 inhibits cell proliferation and promotes apoptosis in HCC cells by targeting TRIM35 and blocking PKM2 Y105 phosphorylation.
Figure 4.
miR-4417 and TRIM35 regulated SMMC-7721 cell proliferation and apoptosis by regulating PKM2 phosphorylation. (A, B) miR-4417 inhibitor and TRIM35 overexpression significantly blocked PKM2 Y105 phosphorylation but not the total PKM2 levels compared with their corresponding controls. (C) The inhibitory effect of miR-4417 inhibitor on PKM2 Y105 phosphorylation was overturned by si-TRIM35. (D–F) SMMC-7721 cells transfected with PKM2 Y105F showed lower viable cell numbers and proliferation rate, and a higher apoptosis rate compared with that with PKM2 Y105 phosphorylation. * P<0.05; ** P<0.01; *** P<0.001.
Discussion
Hepatocarcinogenesis is a multistep process that is associated with the accumulation of molecular alterations from background liver disease. In recent years, miRNAs have gained increasing attention in cancer biology and the involvement of miRNA deregulation in human carcinogenesis has been potentially recognized. Aberrant expression of miRNAs results in altered expression of their target mRNAs and participates in development of several human cancers, including HCC. miRNAs can function as novel tumor suppressors or oncogenes in cancer cells according to their targets. Recently, high-throughput array analyses have clearly demonstrated that miR-4417 is upregulated in many human cancers, such as breast cancer, prostate cancer, and gastrointestinal cancer [13–16]. Keon Uk Park et al. reported that miR-4417 was upregulated in HCC tissues compared with non-cancerous liver tissue samples by using a microarray-based genome-wide miRNA analysis [11]. Here, we further examined the expression of miR-4417 in HCC cell lines, and confirmed that endogenous miR-4417 in HCC cell lines is indeed upregulated compared with the normal liver cell line HL-7702. Biological function analysis by MTT assay and flow cytometry showed that downregulation of miR-4417 significantly inhibits cell proliferation and promotes apoptosis in SMMC-7721 cell line. In addition, we investigated the mRNA targets and the molecular mechanisms of miR-4417 in SMMC-7721 cells, and bioinformatics revealed 3′UTR of TRIM35 contains 1 conserved target site of miR-4417.
TRIM35 (also named MAIR and Hls5) is a member of the tripartite motif-containing family of single-protein E3 ligases and has apoptosis-inducing activity [17]. Furthermore, TRIM35 has been identified as a tumor suppressor that can inhibit cell growth, clonogenicity, and tumorigenicity in various malignancies, including HCC [18]. In hemopoietic cells, enforced expression of TRIM35 was observed to inhibit cell growth, clonogenicity, and tumorigenicity, indicating the antitumor ability of TRIM35 [19]. In the present study, TRIM35 mRNA and protein were observed to be decreased in HCC cell lines compared with those in HL-7702 cells, and upregulation of TRIM35 by transfecting pcDNA-TRIM35 in SMMC-7721 cells significantly inhibited proliferation and promoted apoptosis. Moreover, the luciferase reporter assay confirmed that TRIM35 mRNA is a direct target of miR-4417, which verified that miR-4417 promotes cell proliferation and inhibits apoptosis in HCC cell, at least partly, by targeting TRIM35.
In patients with HCC, it has been revealed that TRIM35 expression is low and PKM2 expression is high in HCC tumor samples compared with matched adjacent non-tumor liver tissues, and PKM2/TRIM35 expression may functions as an independent and significant risk factor for recurrence and survival [20]. Chen et al. reported that TRIM35 can physically associate with PKM2 and decrease the Y105 phosphorylation, thereby inhibiting tumor growth in HCC [12]. Activation of PKM2 can suppress tumor growth, and the most active form of PKM2 is as a tetramer [21]. When PKM2 was phosphorylated on tyrosine 105, the tetramer assembly required for PKM2 activity was disrupted, which facilitated glucose metabolism in cancer [22]. Generally, PKM2 presents as an inactive monomer, and the forms of active dimer and active tetramer are reduced. The dimeric form redirects glucose-derived carbons towards biosynthesis through aerobic glycolysis, and the PKM2 tetramer enhances the flux of glycolysis and high capacity of ATP production by oxidative phosphorylation [23]. Therefore, the balance between the PKM2 dimers and tetramers is vital for tumorigenesis. Based on these findings, we hypothesize that miR-4417 might target TRIM35 and regulate the PKM2 phosphorylation to promote HCC growth. Our findings show that decreased expression of miR-4417 and increased expression of TRIM35 markedly restrain the Y105 phosphorylation of PKM2. In addition, the administration of PKM2 Y105F effectively suppresses cell proliferation and promotes apoptosis in SMMC-7721 cells, which are completely consistent with the experimental results of a previous study [21]. These results confirmed our speculation that miR-4417 targets TRIM35 and regulates PKM2 Y105 phosphorylation to promote HCC growth.
Numerous cancer cells present increased glucose uptake and lactate production, regardless of oxygen availability, which is known as aerobic glycolysis or the Warburg effect, facilitating tumor cell growth [24,25]. Thus, the glucose-metabolizing enzymes, including pyruvate kinase, are often elevated in cancer cells [26]. The pyruvate kinase M1 (PKM1) and M2 (PKM2) isoforms arise from alternate splicing of PKM (formerly PKM2) pre-mRNA, leading to inclusion of either exon 9 (PKM1) or exon 10 (PKM2), respectively. PKM2 is upregulated in multiple cancer types, including lung, gastric, cervical, colorectal cancers, and HCC [20,27–29]. A previous study showed that replacing PKM2 with PKM1 in human lung cancer cells fails to support the Warburg effect as well as tumor formation, suggesting the essential role of PKM2 in regulating anabolic metabolism of cancers [30].
Conclusions
The present study demonstrates that miR-4417 is upregulated and TRIM35 was downregulated in HCC cell lines, and suppression of miR-4417 inhibits proliferation and induces apoptosis of HCC cells. We also confirmed that miR-4417 regulates HCC cell proliferation and apoptosis by targeting TRIM35. Moreover, our findings provide further evidence that miR-4417 promotes HCC cell proliferation and inhibits apoptosis by targeting TRIM35 and enhances Y105 phosphorylation of PKM2, thereby providing a metabolic advantage to tumor cells. Our results suggest that inhibition of miR-4417 may be a promising therapeutic strategy for treatment of human HCC.
Footnotes
Conflicts of interest
The authors declare that no conflicts of interest exist.
Source of support: Departmental sources
References
- 1.Venook AP, Papandreou C, Furuse J, de Guevara LL. The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist. 2010;15:5–13. doi: 10.1634/theoncologist.2010-S4-05. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 3.Colecchia A, Schiumerini R, Cucchetti A, et al. Prognostic factors for hepatocellular carcinoma recurrence. World J Gastroenterol. 2014;20:5935–50. doi: 10.3748/wjg.v20.i20.5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jopling CL, Yi M, Lancaster AM, et al. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–81. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 5.Chen S, Chen X, Xiu YL, et al. MicroRNA-490-3P targets CDK1 and inhibits ovarian epithelial carcinoma tumorigenesis and progression. Cancer Lett. 2015;362:122–30. doi: 10.1016/j.canlet.2015.03.029. [DOI] [PubMed] [Google Scholar]
- 6.Murakami Y, Tanahashi T, Okada R, et al. Comparison of hepatocellular carcinoma miRNA expression profiling as evaluated by next generation sequencing and microarray. PloS One. 2014;9:e106314. doi: 10.1371/journal.pone.0106314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wojcicka A, Swierniak M, Kornasiewicz O, et al. Next generation sequencing reveals microRNA isoforms in liver cirrhosis and hepatocellular carcinoma. Int J Biochem Cell Biol. 2014;53:208–17. doi: 10.1016/j.biocel.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 8.Yang X, Liang L, Zhang XF, et al. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology. 2013;58:158–70. doi: 10.1002/hep.26305. [DOI] [PubMed] [Google Scholar]
- 9.Köberle V, Kronenberger B, Pleli T, et al. Serum microRNA-1 and microRNA-122 are prognostic markers in patients with hepatocellular carcinoma. Eur J Cancer. 2013;49:3442–49. doi: 10.1016/j.ejca.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Bao L, Yan Y, Xu C, et al. MicroRNA-21 suppresses PTEN and hSulf-1 expression and promotes hepatocellular carcinoma progression through AKT/ERK pathways. Cancer Lett. 2013;337:226–36. doi: 10.1016/j.canlet.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Park KU, Seo YS, Lee YH, et al. Altered microRNA expression profile in hepatitis B virus-related hepatocellular carcinoma. Gene. 2015;573:278–84. doi: 10.1016/j.gene.2015.07.053. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z, Wang Z, Guo W, et al. TRIM35 Interacts with pyruvate kinase isoform M2 to suppress the Warburg effect and tumorigenicity in hepatocellular carcinoma. Oncogene. 2015;34:3946–56. doi: 10.1038/onc.2014.325. [DOI] [PubMed] [Google Scholar]
- 13.Estal RM, Suela SP, de Juan Jiménez I, et al. MicroRNA signatures in hereditary breast cancer. Breast Cancer Res Treat. 2013;142:19–30. doi: 10.1007/s10549-013-2723-7. [DOI] [PubMed] [Google Scholar]
- 14.Fussek S, Rönnau C, Span P, et al. 246 The castration-resistant prostate cancer-associated miRNAs, miR-3687 and miR-4417, are involved in tumour cell hypoxia response and tumour cell migration. Eur Urol Suppl. 2016;15:e246. [Google Scholar]
- 15.Berillo OA, Issabekova AS, Régnier M, Ivashchenko AT. Characteristics of binding sites of intergenic, intronic and exonic miRNAs with mRNAs of oncogenes coding intronic miRNAs. Afr J Biotechnol. 2013;12(11) online. [Google Scholar]
- 16.Murria R, Palanca S, de Juan I, et al. Immunohistochemical, genetic and epigenetic profiles of hereditary and triple negative breast cancers. Relevance in personalized medicine. Am J Cancer Res. 2015;5:2330–43. [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura F, Suzu S, Nakamura Y, et al. Cloning and characterization of a novel RING-B-box-coiled-coil protein with apoptotic function. J Biol Chem. 2003;278:25046–54. doi: 10.1074/jbc.M303438200. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Yan S, Yang B, et al. TRIM35 negatively regulates TLR7-and TLR9-mediated type I interferon production by targeting IRF7. FEBS Lett. 2015;589:1322–30. doi: 10.1016/j.febslet.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 19.Lalonde J-P, Lim R, Ingley E, et al. HLS5, a novel RBCC (ring finger, B box, coiled-coil) family member isolated from a hemopoietic lineage switch, is a candidate tumor suppressor. J Biol Chem. 2004;279:8181–89. doi: 10.1074/jbc.M306751200. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Lu X, Wang Z, et al. Co-expression of PKM2 and TRIM35 predicts survival and recurrence in hepatocellular carcinoma. Oncotarget. 2015;6:2538–48. doi: 10.18632/oncotarget.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anastasiou D, Yu Y, Israelsen WJ, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8:839–47. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hitosugi T, Kang S, Vander Heiden MG, et al. Tyrosine phosphorylation inhibits KM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong N, Ojo D, Yan J, Tang D. PKM2 contributes to cancer metabolism. Cancer Lett. 2015;356:184–91. doi: 10.1016/j.canlet.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 24.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 25.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–37. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 26.Yang W, Zheng Y, Xia Y, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell biol. 2012;14:1295–304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng XC, Gong FM, Zhao YW, et al. Comparative proteomic approach identifies PKM2 and cofilin-1 as potential diagnostic, prognostic and therapeutic targets for pulmonary adenocarcinoma. PLoS One. 2011;6:e27309. doi: 10.1371/journal.pone.0027309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar Y, Tapuria N, Kirmani N, Davidson BR. Tumour M2-pyruvate kinase: A gastrointestinal cancer marker. Eur J Gastroenterol Hepatol. 2007;19:265–76. doi: 10.1097/MEG.0b013e3280102f78. [DOI] [PubMed] [Google Scholar]
- 29.Haug U, Hundt S, Brenner H. Sensitivity and specificity of faecal tumour M2 pyruvate kinase for detection of colorectal adenomas in a large screening study. Br J Cancer. 2008;99:133–35. doi: 10.1038/sj.bjc.6604427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christofk HR, Vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–33. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]