Abstract
Thaumarchaeota have been detected in several industrial and municipal wastewater treatment plants (WWTPs), despite the fact that ammonia-oxidizing archaea (AOA) are thought to be adapted to low ammonia environments. However, the activity, physiology and metabolism of WWTP-associated AOA remain poorly understood. We report the cultivation and complete genome sequence of Candidatus Nitrosocosmicus exaquare, a novel AOA representative from a municipal WWTP in Guelph, Ontario (Canada). In enrichment culture, Ca. N. exaquare oxidizes ammonia to nitrite stoichiometrically, is mesophilic, and tolerates at least 15 mm of ammonium chloride or sodium nitrite. Microautoradiography (MAR) for enrichment cultures demonstrates that Ca. N. exaquare assimilates bicarbonate in association with ammonia oxidation. However, despite using inorganic carbon, the ammonia-oxidizing activity of Ca. N. exaquare is greatly stimulated in enrichment culture by the addition of organic compounds, especially malate and succinate. Ca. N. exaquare cells are coccoid with a diameter of ~1–2 μm. Phylogenetically, Ca. N. exaquare belongs to the Nitrososphaera sister cluster within the Group I.1b Thaumarchaeota, a lineage which includes most other reported AOA sequences from municipal and industrial WWTPs. The 2.99 Mbp genome of Ca. N. exaquare encodes pathways for ammonia oxidation, bicarbonate fixation, and urea transport and breakdown. In addition, this genome encodes several key genes for dealing with oxidative stress, including peroxidase and catalase. Incubations of WWTP biofilm demonstrate partial inhibition of ammonia-oxidizing activity by 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl 3-oxide (PTIO), suggesting that Ca. N. exaquare-like AOA may contribute to nitrification in situ. However, CARD-FISH-MAR showed no incorporation of bicarbonate by detected Thaumarchaeaota, suggesting that detected AOA may incorporate non-bicarbonate carbon sources or rely on an alternative and yet unknown metabolism.
Introduction
Nitrification is an important process for municipal and industrial wastewater treatment plants (WWTPs) because it prevents the negative impacts of releasing ammonia to receiving waters, including toxicity to fish, eutrophication and increased oxygen demand. Ammonia-oxidizing bacteria (AOB) were traditionally believed to mediate ammonia oxidation in soils, aquatic habitats and engineered environments. However, several studies have implicated recently discovered ammonia-oxidizing archaea (AOA) of the phylum Thaumarchaeota as the dominant ammonia oxidizers in many environments, including the open ocean (Wuchter et al., 2006), soils (Leininger et al., 2006; Stopnisek et al., 2010; Yao et al., 2011; Zhang et al., 2012) and engineered environments such as aquaculture operations and aquarium biofilters (Sauder et al., 2011; Brown et al., 2013; Bagchi et al., 2014). However, there is evidence that AOB are numerically dominant in some environments, and that AOB may mediate ammonia oxidation in several soils, despite a numerical dominance of AOA (Di et al., 2009; Banning et al., 2015; Sterngren et al., 2015).
The role of AOA detected in WWTPs remains unclear. Compared with many natural environments, WWTPs contain relatively high levels of ammonia, which should favor AOB over AOA (Martens-Habbena et al., 2009; Schleper, 2010). Indeed, many studies have reported a numerical dominance of AOB in municipal and industrial WWTPs (Wells et al., 2009; Mussmann et al., 2011; Gao et al., 2013). Nevertheless, AOA have been detected in several WWTPs (Park et al., 2006; Zhang et al., 2009; Gao et al., 2013, 2014), and in some cases outnumber AOB (Kayee et al., 2011; Bai et al., 2012). Although the abundance and diversity of AOA and AOB have been assessed in several WWTPs, no previous studies have analyzed the relative contributions of these groups to nitrification in any municipal WWTPs. Intact polar lipids originating from thaumarchaeol have been identified in WWTP biofilms (Sauder et al., 2012), indicating that the detected Thaumarchaeota were viable in the system, though not necessarily oxidizing ammonia. Mussman et al. (2011) were unable to demonstrate bicarbonate assimilation by Thaumarchaeota in nitrifying sludge and called into question a strictly chemolithoautotrophic lifestyle of thaumarchaeotes in the examined industrial WWTP.
Although all cultured members of the Thaumarchaeota oxidize ammonia and fix inorganic carbon autotrophically, their metabolism in the environment may be more complex. AOA genomes encode transporters for a variety of organic compounds (for example, Hallam et al., 2006; Walker et al., 2010; Blainey et al., 2011; Spang et al., 2012) and archaea in the ocean incorporate amino acids (Ouverney and Fuhrman, 2000). In addition, supplementation of AOA cultures with pyruvate and alpha-ketoglutarate stimulates growth (Tourna et al., 2011; Qin et al., 2014), although this arises in several cultures from non-enzymatic detoxification of hydrogen peroxide (Kim et al., 2016).
Although several Group I.1a Thaumarchaeota representatives have been reported in laboratory cultures (for example, Könneke et al., 2005; Jung et al., 2011; Mosier et al., 2012; Lebedeva et al., 2013; Li et al., 2016), comparatively few Group I.1b AOA have been cultivated. Group I.1b representatives include Nitrososphaera viennensis, Nitrososphaera evergladensis, Nitrosocosmicus franklandus and Nitrosocosmicus oleophilus, which originate from soils or sediments (Tourna et al., 2011; Zhalnina et al., 2014; Lehtovirta-Morley et al., 2016; Jung et al., 2016), and Nitrososphaera gargensis, which originates from a hotspring (Hatzenpichler et al., 2008; Palatinszky et al., 2015). A phylogenetic analysis of archaeal amoA gene sequences demonstrated five major AOA lineages, represented by the genera Nitrosopumilus, Nitrosocaldus, Nitrosotalea, Nitrososphaera, and a fifth clade that forms a sister cluster to the Nitrososphaera (Pester et al., 2012). This clade contains most amoA sequences obtained from WWTPs (for example, Mussmann et al., 2011; Limpiyakorn et al., 2011; Sauder et al., 2012), manure composting facilities (Yamamoto et al., 2010), landfill sites (Im et al., 2011) and human skin (Probst et al., 2013). Only one AOA enrichment culture exists from a wastewater treatment system, belonging to the Group I.1a Thaumarchaeota (Li et al., 2016).
Here, we report the cultivation and characterization of a novel group I.1b Thaumarchoaeta representative, belonging to the Nitrososphaera sister cluster. This representative was enriched from biofilm of rotating biological contactors (RBCs) of a municipal WWTP in Guelph, Canada, where it was first discovered based on DNA and lipid signatures (Sauder et al., 2012). We propose the name Ca. Nitrosocosmicus exaquare G61 for this representative.
Materials and methods
Sampling site
Biofilm for enrichment culture inoculation was collected from the Guelph WWTP in September 2012 from the 8th RBC (‘RBC 8’) of the Southeast (SE) treatment train. For a plant schematic and description of wastewater treatment processes, see Sauder et al. (2012). Biofilm for various experiments was collected at multiple time points; sampling details are summarized in Supplementary Table S1.
Cultivation and experimental incubations
Ca. N. exaquare G61 was cultivated in a growth medium described previously (Krümmel and Harms, 1982; Hatzenpichler et al., 2008), containing (per l): 0.05 g KH2PO4, 0.075 g KCl, 0.05 g MgSO4 × 7H2O, 0.58 g NaCl and 4 g CaCO3. After autoclaving, the medium was supplemented with filter-sterilized NH4Cl (0.5–1 mm), 1 ml selenite-tungstate solution and 1 ml trace element solution (Widdel and Bak, 1992; Könneke et al., 2005). The resulting medium pH was ~8.5.
Culture incubations for generating a growth curve were performed in the described medium supplemented with 1 mm NH4Cl and 0.5 mm sodium pyruvate. At each sampling time, 2 ml culture was removed, pelleted for 10 min at 15 000 × g, and used for genomic DNA extractions. For temperature incubations, cultures were incubated with 0.5 mm NH4Cl at varying temperatures. For testing ammonia and nitrite tolerance, cultures were set-up with varying starting concentrations of NH4Cl and NaNO2, respectively. To test stimulation of Ca. N. exaquare by organic carbon, cells were subcultured (0.1% inoculum) and grown with or without organic carbon in the presence of 0.5 mm NH4Cl. Several organic carbon compounds were tested, including pyruvate, citrate, succinate, malate, acetate (final concentration 0.5 mm), glucose and taurine (0.25 mm), butyrate (0.1 mm), glycerol (0.0007%) and yeast extract (0.01%). All assays above were conducted in triplicate with a 1% inoculum, in the dark and without shaking.
Inhibitor assays on WWTP biofilm
Biofilm and wastewater samples were obtained from the Guelph WWTP in April and December 2015 and were stored on ice until returned to the laboratory (~1 h). Incubations were performed in 125 ml glass serum bottles using 20 ml volumes. Incubations consisted of 2% (w/v) biofilm suspensions in 0.22-μm filtered RBC influent amended with 1 mm ammonium chloride. Flasks were supplemented with inhibitors as appropriate, including 6 μm acetylene (aqueous), 10 μm allylthiourea (ATU), 8 μm octyne (aqueous) and both 200 and 400 μm PTIO. Incubations were performed in triplicate, in the dark, without shaking.
Water chemistry measurements
Ammonia and nitrite concentrations were measured colourimetrically according to previously published protocols using Nessler’s reagent (Meseguer-Lloret et al., 2002) and Griess reagent (Miranda et al., 2001), respectively. Absorbance values were measured at 550 nm (nitrite) and 450 nm (ammonia) using a Filtermax F5 Multi-Mode Microplate Reader (Molecular Devices, Sunnyvale, CA, USA). All technical measurements were performed in duplicate. Substrate concentrations were determined by comparison to standards using SoftMax Pro 6.4 (Molecular Devices). Note that for reported ammonia concentrations, values refer to total ammonia (NH4++NH3), unless otherwise indicated.
DNA extractions and quantitative PCR
Genomic DNA extractions were performed using the PowerSoil DNA Isolation Kit (for biofilm) or the Ultraclean Microbial DNA Isolation Kit (for laboratory cultures), according to the manufacturer’s instructions (MO BIO, Carlsbad, CA, USA). Beadbeating was performed using a FastPrep-24 (MP Biomedicals, Santa Ana, CA, USA) at 5.5 m s−1 for 45 s. Quantification of thaumarchaeotal and bacterial 16S rRNA genes was performed using primers 771F and 957R (Ochsenreiter et al., 2003) and 341F and 518R (Muyzer et al., 1993), respectively. AOB amoA genes were quantified using primers amoA1F and amoA2R (Rotthauwe et al., 1997). All qPCR amplifications used SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) and were performed using technical duplicates on a CFX96 system (Bio-Rad). All efficiencies were >80% and R2 values were >0.99. For growth curves, Ca. N. exaquare cell numbers were estimated based on measured 16S rRNA gene copies (assuming one chromosome per cell), divided in half to account for two 16S rRNA gene copies per genome. Generation time was estimated from the slope of the natural log-transformed thaumarchaeotal cell numbers during exponential growth.
Phylogenetic analyses
Ca. N. exaquare amoA gene sequences were compared with cultivated AOA representatives and environmental sequences obtained from GenBank. Global alignment of sequences was performed using MUSCLE (Edgar, 2004). Evolutionary histories were inferred using the maximum likelihood method based on the general time reversible model of sequence evolution. A Gamma distribution was used to model evolutionary rate differences among sites. Bootstrap testing was conducted with 500 replicates. All alignments and phylogenetic analyses were conducted in MEGA6 (Tamura et al., 2013).
Microscopy
For scanning electron microscopy (SEM), cells were applied to a silicon wafer attached to an aluminum stub with conductive carbon tape, then imaged (unfixed and unstained) using a LEO 1550 field-emission scanning electron microscope (Zeiss, Oberkochen, DE, USA) with an InLens SE detector, an EHT of 7 kV and a working distance of 10.3 mm.
For catalyzed reporter deposition-fluorescence in situ microscopy (CARD-FISH), samples were fixed and processed as described previously (Ishii et al., 2004; Mussmann et al., 2011), with modifications. Briefly, cells were permeabilized with proteinase K (15 μg ml−1) for 10 min at room temperature, followed by proteinase K inactivation by 0.01 m HCl for 20 min. To inactivate endogenous peroxidases, slides were treated with 0.3% H2O2 in 100% methanol for 30 min (enrichment cultures) or overnight (biofilm). Probes Arch915 (Stahl and Amann, 1991) or Thaum726 (Beam, 2015) were used for detecting thaumarchaeotes, with formamide concentrations of 10% and 25%, respectively. Thaum726 was used with competitor probes thaum726_compA and thaum726_compB (Supplementary Table S2). For AOB and Nitrospira, previously published probe mixes were used (Supplementary Table S2). DAIME image analysis software (Daims et al., 2006) was used for quantitative FISH (AOB and nitrite oxidizing bacteria (NOB)) and quantitative CARD-FISH (AOA) analyses.
MAR-FISH
Incubations of Ca. N. exaquare were prepared in 5 ml volumes in 50 ml tissue culture flasks. Cells were incubated with 0.5 mm NH4Cl in HEPES-buffered freshwater medium (FWM; Tourna et al., 2011), with calcium carbonate excluded to minimize unlabeled inorganic carbon availability. Each flask was amended with 10 μCi 14C-bicarbonate (Hanke Laboratory Products, Vienna, Austria). Flasks were incubated at 28 °C for 24 h in the dark, without shaking. For biofilm incubations, biofilm from SE RBC 1 and 8 was collected and chilled until experimental set-up. Biofilm suspensions were diluted 1:5 in RBC influent, and pre-incubated for 3 h with 0.05 mm NH4Cl at ambient temperature. Biofilm suspensions were then aliquoted into 5 ml volumes amended with 0.5 mm NH4Cl and 10 μCi 14C-bicarbonate (in duplicate). Flasks were incubated at ambient temperature in the dark, without shaking, and biomass was removed and fixed after 6 and 20 h of incubation. Biomass was fixed with 4% paraformaldehyde and CARD-FISH was performed as described above; MAR was performed as described previously (Lee et al., 1999; Mussmann et al., 2011). Enrichment culture samples were exposed for 12 days, and biofilm culture samples were exposed for 9, 12 and 15 days. Samples were imaged on a Leica TCS SP8 confocal laser scanning microscope (CLSM). When necessary, MAR signals were recorded with a color-CCD camera (DFC 450, Leica Microsystem, Wetzlar, Germany), attached to the CLSM, to differentiate between black silver grains and brown biofilm material.
Sequencing, genome assembly and genome annotation
Genomic DNA for sequencing was extracted from Ca. N. exaquare using the PowerSoil DNA Isolation Kit (MO BIO Laboratories). Enrichment cultures containing either no organic carbon or supplemented with 0.5 mm taurine were extracted separately to generate metagenomes suitable for differential abundance binning.
Genomic DNA was prepared for sequencing using the TruSeq PCR-free kit (Illumina, San Diego, CA, USA) using alternative nebulizer fragmentation, gel-free size selection and a 550 bp target insert size. A mate-pair library was prepared with the Nextera Mate Pair Sample Preparation Kit (Illumina) and sequenced (2 × 301 bases) using MiSeq Reagent Kit v3 (Illumina). Paired-end FASTQ reads were imported to CLC Genomics Workbench version 7.0 (CLC Bio, Qiagen, Hilden, Germany) and trimmed using a minimum Phred score of 20 and length of 50 bases. Paired-end reads were assembled using the CLC de novo assembly algorithm, using a kmer length of 63 and a minimum scaffold length of 1 kb. Mate-pair reads were trimmed using NextClip (Leggett et al., 2014) and only reads in class A were used for mapping.
Metagenome binning and data generation was conducted as described previously (Albertsen et al., 2013) using the mmgenome R package and scripts (http://madsalbertsen.github.io/mmgenome/). The genome was manually scaffolded using paired-end and mate-pair connections aided by visualization in Circos (Krzywinski et al., 2009). Gaps were closed using GapFiller (Boetzer and Pirovano, 2012) and manually through inspections of read alignments in CLC Genomics Workbench.
The assembled genome was annotated using both Integrated Microbial Genomes Expert Review (IMG ER; Markowitz et al., 2009) and the MicroScope platform for microbial genome annotation (MaGe; Vallenet et al., 2009). Locus tags are based on MaGe annotations. Comparative analysis of MetaCyc degradation, utilization and assimilation pathways were generated automatically in MaGe and updated manually to remove incorrect automatic assignments. The full genome sequence of Ca. N. exaquare G61 has been deposited in GenBank (accession CP017922) and associated annotations are publicly available in both IMG ER (ID 2603880166) and MicroScope (#U7DNPY). Summary data and genome accession numbers for associated enrichment culture bacteria are summarized in Supplementary Table S3.
Results
Ca. N. exaquare enrichment culture
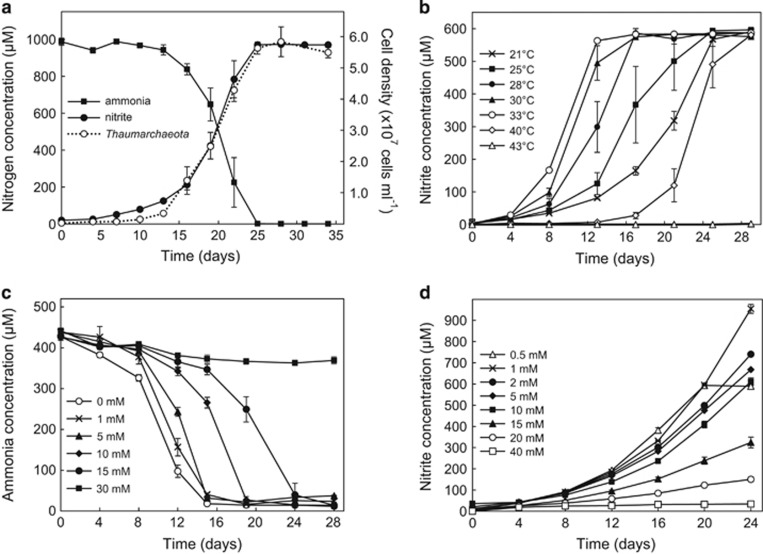
Ca. N. exaquare has been growing in enrichment culture for over three years and, based on quantitative PCR of thaumarchaeotal and bacterial 16S rRNA genes, comprises approximately 99% of cells present. Ca. N. exaquare depletes ammonia and produces nitrite at near-stoichiometric levels (Figure 1a), with an associated generation time of 51.7 h, and a cell density of approximately 5.6 × 107 cells ml−1 (after oxidizing 1 mm NH4Cl). Ca. N. exaquare grows over a broad temperature range, with optimal ammonia-oxidizing activity at 33 °C and complete inhibition at 43 °C (Figure 1b). Nitrite-free medium results in the fastest ammonia-oxidizing activity by Ca. N. exaquare, and activity slows as initial nitrite concentrations increase (Figure 1c). Despite an increased lag time, Ca. N. exaquare cells fully oxidized 0.5 mm NH4Cl in the presence of up to 15 mm nitrite. Optimal activity was observed at ammonia concentrations of 0.5–1 mm NH4Cl, but initiation of ammonia-oxidizing activity persisted up to 20 mm ammonium (Figure 1d). At 15 mM NH4Cl, Ca. N. exaquare oxidized all supplied ammonia to nitrite, although an incubation of approximately six months was required (data not shown). Ca. N. exaquare cells could be revived following short-term and long-term storage at 4 °C and cryopreservation at −80 °C (Supplementary Figures S1A and B).
Figure 1.
Growth and activity of Ca. N. exaquare. (a) Growth curve, indicating conversion of 1 mm ammonia to nitrite and corresponding increase in thaumarchaeotal cell numbers. Cell numbers were estimated by thaumarchaeotal 16S rRNA gene copy numbers, halved to account for two 16S rRNA gene copies per genome. (b) Ca. N. exaquare nitrite production at varying incubation temperatures. (c) Depletion of 0.5 mm ammonia by Ca N. exaquare at varying initial nitrite concentrations. Starting nitrite concentrations are indicated in the panel legend. For this panel, ammonia depletion is shown due to high background levels of nitrite. (d) Ca. N. exaquare nitrite production at varying starting ammonia concentrations. Starting ammonia concentrations are indicated in the panel legend. Note that nitrite production plateaus in the 0.5 mm condition due to depletion of supplied ammonia. All incubations were prepared from a 1% inoculum from an actively growing enrichment culture and were performed in the dark, without shaking. Unless otherwise indicated, the incubation temperature was 30 °C. Error bars indicate the standard error of the mean for biological triplicates. Error bars not seen are contained within the symbols.
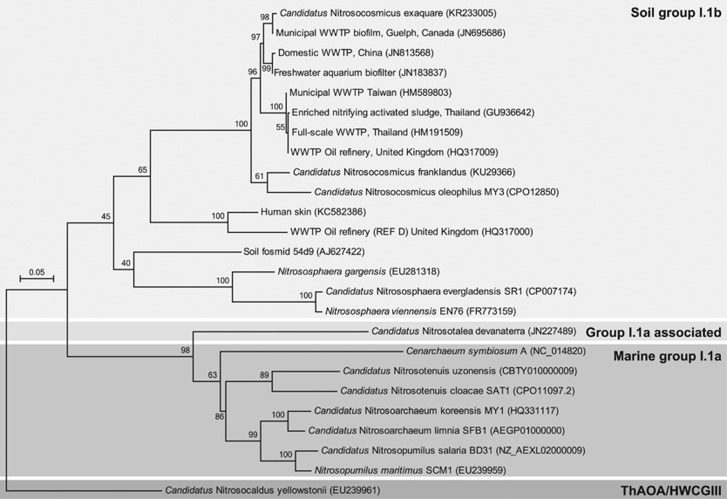
On the basis of amoA (Figure 2) and 16S rRNA (Supplementary Figure S2) gene sequences, Ca. N. exaquare belongs to the soil group I.1b Thaumarchaeota cluster, specifically in the Nitrososphaera sister cluster, and is related to Ca. N. franklandus (Lehtovirta-Morley et al., 2016) and Ca. N. oleophilus (Jung et al., 2016). In addition, Ca. N. exaquare gene sequences are closely related to environmental sequences originating from several municipal and industrial WWTPs. Ca. N. exaquare has sequence identities of >99% to amoA and 16S rRNA gene sequences obtained from the original Guelph WWTP biofilm.
Figure 2.
Phylogenetic affiliations of amoA gene sequences for Ca.N. exaquare and other thaumarchaeotal representatives, inferred using the maximum likelihood method based on the general time reversible model. The tree is drawn to scale, with branch lengths measured as the number of substitutions per site. Bootstrap values are located above branches and are based on 500 replicates. Only bootstrap values >50% are indicated on the tree. The scale bar represents 5% nucleotide divergence.
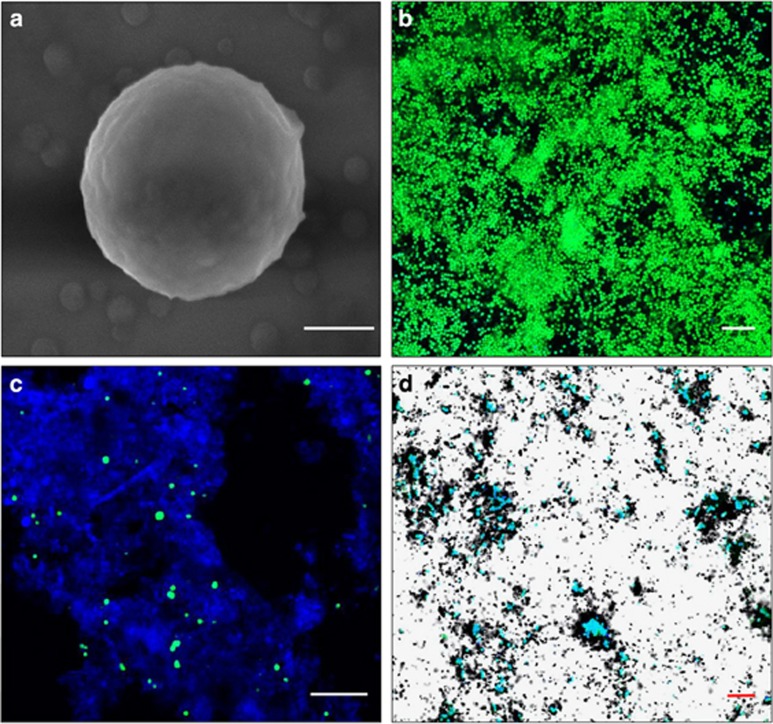
Ca. N. exaquare cells are coccoid and approximately 1.3 μm in diameter (Figures 3a and b). Cells up to ~2 μm in diameter were observed, with large cells often having two discrete regions of nucleic acid (Supplementary Figure S3A). Cells often appeared in pairs or groups, which may be covered in an extracellular matrix (Supplementary Figures S3B and C). Thaumarchaeota in RBC biofilm samples were also coccoid with a diameter of 1–2 μm (Figure 3c; Supplementary Figure S3D). Ca. N. exaquare cells in enrichment culture assimilated 14C-bicarbonate coincident with ammonia-oxidizing activity, as indicated by microautoradiographic signal in association with CARD-FISH labeling of thaumarchaeotal cells (Figure 3d).
Figure 3.
Micrographs of Ca.N. exaquare cells. (a) Scanning electron micrograph with a 400-nm scale bar (measured cell diameter is 1.3 μm). (b) CARD-FISH image of cells in enrichment culture. (c) CARD-FISH image of thaumarchaeotal cells in RBC biofilm from the Guelph WWTP. (d) CARD-FISH combined with MAR of Ca. N. exaquare enrichment culture cells labeled with 14C-bicarbonate. CARD-FISH images in b–d were obtained with probe thaum726 applied together with two competitor probes (compA, compB; see Supplementary Table S2) and FITC-labeled tyramides (green). In these panels, DAPI was applied as a nucleic acid stain (blue) and scale bars are 10 μm.
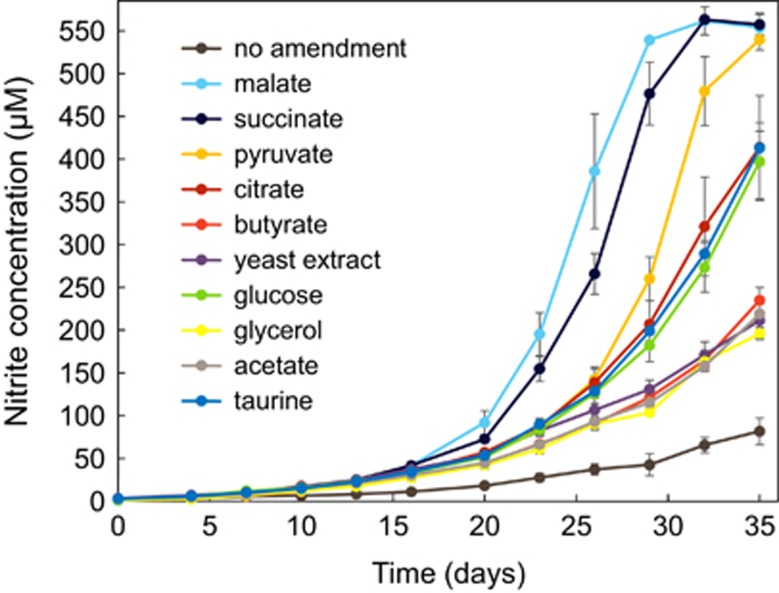
Despite incorporation of inorganic carbon, slowing of growth and activity of Ca. N. exaquare cells was observed as the culture became more highly enriched, a pattern reported previously for N. viennensis (Tourna et al., 2011). Amendment of the growth medium with various sources of organic carbon stimulated growth of Ca. N. exaquare in comparison to a no-organic carbon control (Figure 4). All tested organic carbon compounds stimulated growth, with tricarboxylic acid cycle intermediates malate and succinate providing the most stimulation, followed by pyruvate.
Figure 4.
Growth of Ca.N. exaquare amended with organic carbon and incubated with 0.5 mM NH4Cl at 28 °C. All conditions shown in this figure were performed using the same inoculum and medium batch, and were set-up on the same day. At the time of inoculation, the proportion of Ca. N. exaquare in the mixed culture was ~85%, as measured by quantitative PCR for thaumarchaeotal and bacterial 16S rRNA genes. Error bars indicate the standard error of the mean for biological triplicates. Error bars not seen are contained within symbols.
Ca. N. exaquare genome
The genome of Ca. N. exaquare was sequenced and assembled using metagenomic binning (Supplementary Figure S4), resulting in one circular contiguous sequence of 2.99 Mbps (Supplementary Figure S5), with very low sequence variation of 478 putative SNPs across the entire genome. The genome has a G+C content of 33.9%, 3162 predicted protein-coding sequences, and a coding density of 77.2%. It encodes 39 tRNA genes, one 5S rRNA gene, and two identical copies each of 16S and 23S rRNA genes (Table 1). Synthesis of several vitamins is encoded, including cobalamin (vitamin B12), consistent with a previous study linking marine cobalamin production with Thaumarchaeota (Doxey et al., 2015). Ca. N. exaquare encodes transporters for amino acids and di/oligopeptides, as well as two sodium-dependent dicarboxylate transporters (A4241_1584, 2720). Several genes encoding enzymes associated with detoxification of reactive oxygen species (ROS) are present in the genome, including catalase (A4241_2297), peroxidase (A4241_2636), superoxide dismutase (A4241_1350), alkyl hydroperoxide reductase/peroxiredoxins and thioredoxins (Table 1). Ca. N. exaquare has several genes related to ammonia oxidation, including amoA, amoB, and three copies of amoC, in an arrangement that differs from both AOB and other AOA representatives (Supplementary Figure S6). In addition, Ca. N. exaquare encodes urea transporters and urease enzyme subunits and accessory proteins (Table 1), and produces nitrite from urea in enrichment culture (data not shown). Unlike other I.1b Thaumarchaeota, Ca. N. exaquare does not encode genes associated with chemotaxis or flagellar synthesis (Table 1).
Table 1. Genome features of Ca. N. exaquare G61 and other selected members of the Thaumarchaeota.
| Genome features | Nitrosocosmicus exaquare G61 | Nitrososphaera gargensis Ga9-2a | Nitrosophaera viennensis EN76b | Nitrososphaera evergladensis SR1c | Nitrosopumilus maritimus SCM1d |
|---|---|---|---|---|---|
| Cluster | I.1b | I.1b | I.1b | I.1b | I.1a |
| Genome size (Mb) | 2.99 | 2.83 | 2.53 | 2.95 | 1.6 |
| Contigs | 1 | 1 | 1 | 1 | 1 |
| GC (%) | 33.9 | 48.4 | 52.7 | 50.14 | 34.2 |
| Total genes | 3206 | 3609 | 3167 | 3548 | 1847 |
| Protein-coding genes | 3162 | 3566 | 3027 | 3505 | 1796 |
| Coding density (%) | 77.2 | 81.5 | 87.3 | 83.6 | 90.8 |
| 16-23S rRNA | 2 | 1 | 1 | 1 | 1 |
| 5S rRNA | 1 | 1 | 1 | 1 | 1 |
| tRNA | 39 | 40 | 39 | 39 | 44 |
| CRISPR loci | 2 | 1 | 2 | 1 | − |
| Cell division | FtsZ, Cdv | FtsZ, Cdv | FtsZ, Cdv | FtsZ, Cdv | FtsZ, Cdv |
| Motility/chemotaxis | − | + | + | + | − |
| Carbon fixation | 3HP/4HB | 3HP/4HB | 3HP/4HB | 3HP/4HB | 3HP/4HB |
| Ammonium transporters | 1 | 3 | 3 | 3 | 2 |
| NirK | 1 | 1 | 1 | 1 | 1 |
| Urease and urea transport | + | + | + | + | − |
| Cyanate lyase | − | + | − | − | − |
| Coenzyme F420 | + | + | + | + | + |
| Vitamin B12 (cobalamin) synthesis | + | + | + | + | + |
| Polyhydroxyalkanoate synthesis | + | + | + | + | − |
| Dicarboxylate transporter (SdcS) | 2 | − | − | − | 1 |
| Amino acid transporters | + | + | + | + | − |
| Dipeptide/oligopeptide transporters | + | + | + | + | + |
| Catalase (Mn-containing) | 1 | 1e | 0 | 1 | 0 |
| Peroxidase | 1 | 0 | 0 | 0 | 0 |
| Superoxide dismutase | 1 | 1 | 1 | 1 | 2 |
| Alkyl hydroperoxide reductases | + | + | + | + | + |
| Thioredoxins | + | + | + | + | + |
Truncated gene.
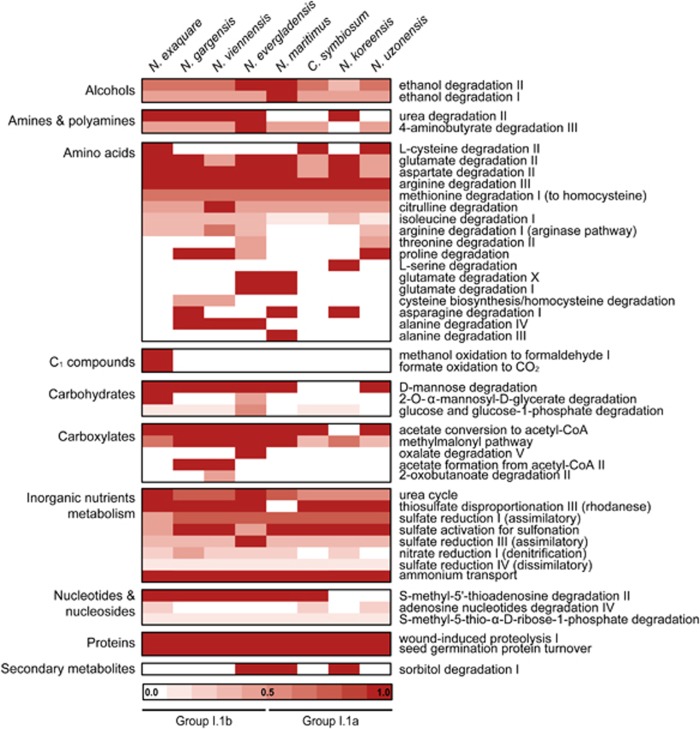
There was little synteny between Ca. N. exaquare and N. maritimus (Supplementary Figure S7A), N. gargensis (Supplementary Figure S7B) or N. viennensis (Supplementary Figure S7C). In comparison, N. gargensis and N. viennensis have several syntenic regions (Supplementary Figure S7D). Average amino acid identities (AAIs) with other AOA were very low, and ranged from 46% with N. maritimus to 53% with N. gargensis (Supplementary Table S4). MetaCyc degradation pathways encoded by Ca. N. exaquare demonstrate metabolic similarity with other AOA representatives, especially group I.1b Thaumarchaeota (Figure 5). For example, all AOA representatives share pathways associated with proteolysis and degradation of amino acids. Few pathways involved in carbohydrate metabolism were identified, with the only unique MetaCyc pathway involving degradation of 2-O-α-mannosyl-d-glycerate. Automated annotation of the Ca. N. exaquare genome also suggested pathways associated with one-carbon (C1) compound utilization, including methanol oxidation to formaldehyde, and formate oxidation to CO2 (Figure 5). In addition, Ca. N. exaquare encodes two copies of S-(hydroxymethyl)glutathione (A4241_3046, 3091), which oxidizes formaldehyde in the presence of glutathione.
Figure 5.
Comparative analysis of MetaCyc degradation pathways of Ca.N. exaquare and other selected Thaumarchaeota. The heat map was generated using the MicroScope platform and includes all MetaCyc pathways under the category ‘degradation, utilization and assimilation’.
Guelph WWTP biofilm
Inhibitors used for biofilm activity assays were first tested on Ca. N. exaquare and N. europaea. Oxidation of 0.5 mm NH4Cl by Ca. N. exaquare was only inhibited by relatively high concentrations of ATU (⩾100 μm; Supplementary Figure S8A) and was not inhibited by octyne at any tested concentration (up to 30 μm; Supplementary Figure S8B). Conversely, PTIO was strongly inhibitory at 30 μm, with total inhibition observed at 100 μm (Supplementary Figure S8C), consistent with previous observations for this organism (Sauder et al., 2016). For N. europaea, 10 μm ATU and 8 μm octyne completely inhibited the oxidation of 0.5 mm NH4Cl, whereas inhibition was not observed by 200 or 400 μm PTIO (Supplementary Figure S9).
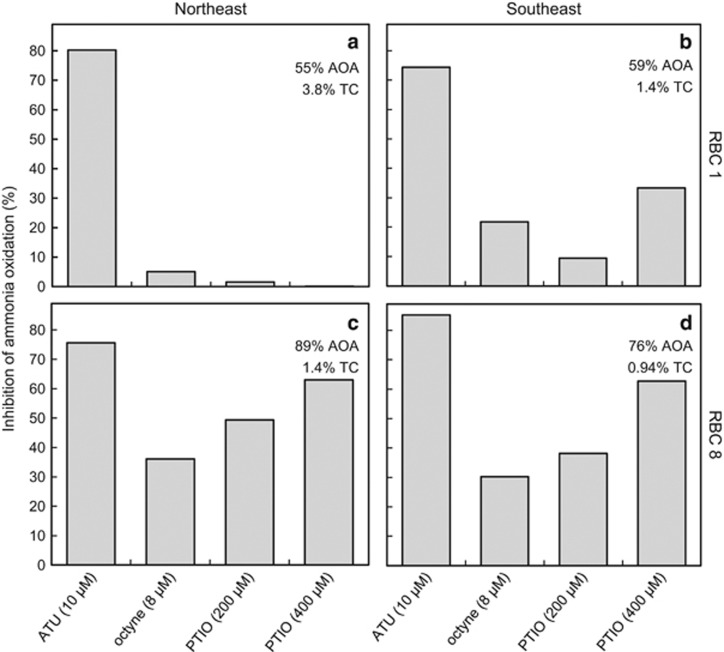
Biofilm samples obtained from Guelph RBCs (December 2015) oxidized 1 mm NH4Cl within 2–4 days (Supplementary Figure S10). Acetylene control flasks showed no ammonia depletion over the incubation period. In fact, an increase in ammonia concentration was observed, likely due to mineralization. Compared with no-inhibitor controls, ATU inhibited between 75% and 85% of ammonia oxidation after 42 h of incubation (Figure 6). Octyne amendment resulted in less inhibition of ammonia oxidation (5–36%). Addition of 200 μm PTIO resulted in little inhibition in RBC 1 biofilm (2–9%) with more inhibition in RBC 8 samples (38–49%). With 400 μm PTIO, patterns were similar, but inhibition was stronger in RBC 8 biofilm samples (63%). In April 2015, similar activity experiments were performed with Guelph RBC biofilm and inhibitors, and demonstrated that ammonia-oxidizing activity in all tested RBCs was partially inhibited by the addition of 200 μm PTIO and strongly inhibited by 10 μm ATU (Supplementary Figure S11).
Figure 6.
Ammonia oxidation by RBC biofilm in the presence of nitrification inhibitors. Biofilm samples were incubated in wastewater supplemented with 1 mm NH4Cl. Percent inhibition of ammonia oxidation is compared with a no-inhibitor control after 42 h of incubation. See Supplementary Figure S10 at ammonia concentrations for all time points. % AOA depicts the proportion of AOA amoA gene of amoA genes of AOA and AOB detected by qPCR in these samples. %TC represents the relative abundance of AOA to the total community (TC), as measured by qPCR of total bacterial plus thaumarchaeal 16S rRNA genes (Supplementary Table S5).
In RBC 1 and 8 biofilm samples, Thaumarchaeota comprised 55–60% and 76–89% of the total putative ammonia oxidizers, respectively (Figure 6; Supplementary Table S5). Although the proportion of thaumarchaeota was higher in RBC 8, the absolute abundance of AOA-associated genes per ng genomic DNA was approximately 3-fold higher in genomic DNA from RBC 1 biofilm. The absolute abundance of AOB was also higher in RBC 1 biofilm compared with the corresponding RBC 8 samples (Supplementary Table S5). Overall, AOA contributed between 0.9% and 3.8% of the total 16S rRNA genes measured in RBC biofilm samples (Figure 6). Quantitative FISH performed for SE biofilm samples (at an earlier time point) provided similar results, with AOA comprising 4.2% (RBC 1) and 2.4% (RBC 8) of the total DAPI stained biomass. In comparison, AOB comprised 1% (RBC 1) and 0.6% (RBC 8) of the total biomass.
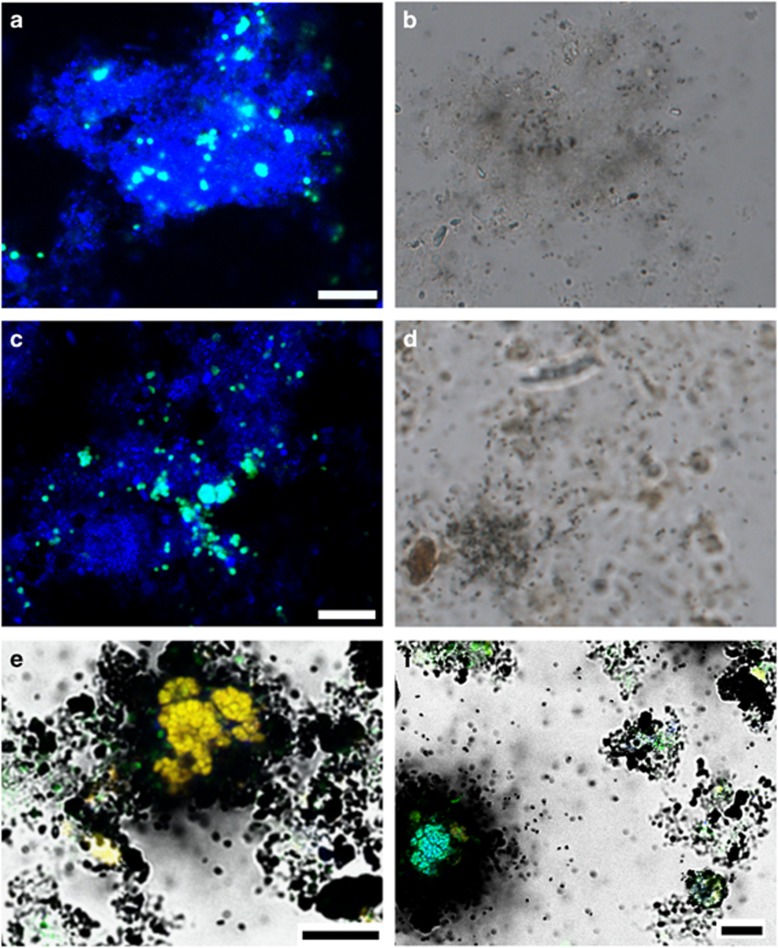
In CARD-FISH-MAR experiments, the thaumarchaeotes in SE RBC 1 and RBC 8 did not show incorporation of 14C-labeled bicarbonate in the presence of 0.5 mm ammonia after incubation for 6 or 20 h, using MAR exposure times of 9, 12 and 15 days (Figure 7). MAR results were negative for many thaumarchaeotal cells, but no statement could be made for some cells located in proximity to AOB or Nitrospira. In contrast, positive MAR signals were obtained for many AOB and Nitrospira microcolonies in both RBC 1 and 8 (Figure 7). In RBC 1, following 6 h and 20 h of incubation, 59% and 71% of AOB colonies, and 14% and 36% of Nitrospira colonies, were MAR-positive, respectively. In RBC 8, following 6 and 20 h of incubation, 46% and 86% of AOB colonies, and 8% and 16% of the Nitrospira microcolonies were MAR-positive, respectively. In no-ammonia control experiments, no MAR-positive AOB nor NOB microcolonies were detected in RBC 8. In RBC 1, up to 10% of the AOB and 7% of the Nitrospira microcolonies showed a positive MAR signal, which was most likely the result of autotrophy associated with ammonia mineralization. Control experiments with dead biofilm samples resulted in no detectable MAR-positive cells, excluding chemographic effects.
Figure 7.
CARD-FISH-MAR for thaumarchaeotes in RBC 1 (a and b) and RBC 8 (c and d) biofilm. FISH-MAR for AOB (orange/yellow) and Nitrospira (blue) (e and f; both RBC 8). Green signals in e and f originate from the EUB338 probe mix targeting most bacteria. Microautoradiographic images were taken with the CCD black/white camera integrated in the CLSM (e and f) or with an external CCD color camera (DFC 450, Leica Microsystem, Wetzlar, Germany; b and d). All scale bars represent 10 μm.
Discussion
Little is known about the metabolism and activity of Thaumarchaeota detected in WWTPs, despite the importance of these environments to human and environmental health. Here, we report the cultivation of a Thaumarchaeota representative originating from a municipal WWTP, which oxidizes ammonia, fixes inorganic carbon, and possesses a genomic repertoire consistent with chemolithoautotrophy. The genus name is based on the related organisms Ca. N. franklandus (Lehtovirta-Morley et al., 2016) and Ca. N. oleophilus (Jung et al., 2016), and the species name ‘exaquare’ (latin for ‘water running out’ or ‘sewage’) reflects its wastewater origin.
Ca. N. exaquare produces nitrite from ammonia at near-stoichiometric values (Figure 1a), and thaumarchaeotal cell numbers follow nitrite production closely, providing evidence that energy for cell growth is derived from the oxidation of ammonia to nitrite. The 51.7 h generation time of Ca. N. exaquare is similar to that originally reported for N. viennensis (46 h), although a shorter generation time of 27.5 h was later reported (Stieglmeier et al., 2014). Ca. N. exaquare is mesophilic, with optimal growth observed at 33 °C (Figure 1b). No growth was observed for Ca. N. exaquare above 40 °C, in contrast to N. gargensis, which grows optimally at 46 °C (Hatzenpichler et al., 2008) and N. viennensis, which can tolerate temperatures of at least 47 °C (Tourna et al., 2011).
Ca. N. exaquare can withstand relatively high concentrations of both ammonia and nitrite (Figures 1c and d). Ammonia oxidation proceeded in the presence of up to 15 mm NaNO2, with complete inhibition observed at 30 mm. N. viennensis oxidizes ammonia with little inhibition at 10 mm NaNO2, but ammonia oxidation ceased if ~3.5 mm nitrification-derived nitrite accumulated (Tourna et al., 2011). In contrast, Ca. N. exaquare fully oxidizes at least 15 mm NH4Cl, indicating that it may be better able to tolerate nitrite or other ammonia oxidation intermediates. Initiation of ammonia oxidation can be achieved with ammonia concentrations of up to 20 mm, with complete inhibition not observed until 30 mm (Figure 1d). For the growth conditions used (that is, pH 8, 30 °C), 20 mm NH4Cl is equivalent to 1.49 mm un-ionized ammonia (NH3). For comparison, reported inhibitory concentrations of un-ionized ammonia are only 18–27 μm, <9 μm and 0.51–0.75 μm for N. maritimus, Ca. N. devanaterra and N. viennensis, respectively (see Hatzenpichler, 2012 for a review). High tolerance to ammonia is perhaps unsurprising given that Ca. N. exaquare originates from a municipal WWTP, where ammonia concentrations would be higher than in most naturally occurring soil or aquatic environments. Niche partitioning occurs between AOA and AOB based on ammonia availability (Erguder et al., 2009; Jia and Conrad, 2009; Martens-Habbena et al., 2009; Schleper, 2010; Verhamme et al., 2011; Sauder et al., 2012), but given the high diversity of AOA and their global distribution across diverse environments, it is also possible that niche partitioning based on ammonia concentrations also occurs within the Thaumarchaeota. Similarly, nitrite has been suggested as a major driver of niche partitioning for nitrite-oxidizers from the genus Nitrospira (Maixner et al., 2006).
Ca. N. exaquare is the first reported representative of the Nitrososphaera sister cluster originating from a WWTP. Both amoA (Figure 2) and 16S rRNA (Supplementary Figure S2) gene sequences clustered with other Thaumarchaeota from WWTPs, both industrial and municipal, as well as other waste-related environments such as landfills and landfill-contaminated soils. Most detected thaumarchaeotal sequences from WWTPs affiliate with the I.1b soil group, often in the Nitrososphaera sister cluster (Mussmann et al., 2011; Sauder et al., 2012; Gao et al., 2013; Limpiyakorn et al., 2013). These engineered environments represent comparatively nutrient-rich habitats, characterized by relatively high levels of organic carbon and ammonia. Combined with the observed high tolerance to ammonia and nitrite, this clustering could reflect an adaptation of Ca. N. exaquare-like Thaumarchaeota to high nutrient environments.
With cell sizes up to 2 μm in diameter, Ca. N. exaquare is the largest reported member of the Thaumarchaeota. Most observed cells were ~1.3 μm (Figure 3a), which is substantially larger than group I.1a Thaumarchaeota (for example, N. maritimus cells are 0.2 μm × 0.7 μm; Könneke et al., 2005). Group I.1b Thaumarchaeota are larger, including N. gargensis and N. viennensis, which have cell diameters of ~0.9 μm and 0.5–0.8 μm, respectively (Hatzenpichler et al., 2008; Tourna et al., 2011). Although coccoid morphologies have been reported for all group I.1b Thaumarchaoeta, Ca. N. exaquare cells appear smoothly spherical (Figure 2a), whereas N. viennensis cells are irregular, with concave areas that appear collapsed into the cell (Tourna et al., 2011). The cell size and morphology of Ca. N. exaquare closely resemble thaumarchaeotal cells previously detected by CARD-FISH in wastewater sludge samples from industrial WWTPs (Mussmann et al., 2011).
Ca. N. exaquare incorporated bicarbonate into biomass in association with ammonia oxidation (Figure 3d) and encodes the 3-hydroxypropionate/4-hydroxybutyrate (3HP/4HB) carbon fixation pathway (Table 1), which is used by all known Thaumarchaeota (Berg et al., 2007; Berg, 2011). Moreover, it has grown in enrichment culture for several years without any externally supplied organic carbon. These data indicate that Ca. N. exaquare combines ammonia oxidation with autotrophic carbon fixation, as expected for a classical ammonia-oxidizing microorganism. Despite this, Ca. N. exaquare is strongly stimulated by the addition of organic carbon (Figure 4), which may indicate a mixotrophic metabolism, or an indirect benefit. A variety of organic carbon sources accelerated ammonia-oxidizing activity by Ca. N. exaquare, with malate and succinate resulting in the highest level of stimulation (Figure 4). Ca. N. exaquare may be able to incorporate these metabolic intermediates directly into its tricarboxylic acid cycle, which could provide reducing power or precursors for biomolecule synthesis. However, given the wide variety of stimulatory compounds (for example, glycerol, yeast extract, butyrate), it is likely that not all organic carbon sources directly stimulate growth, but instead provide indirect benefits via remaining heterotrophic bacteria. A mixotrophic lifestyle would be consistent with previous environmental observations. For example, marine archaea assimilate amino acids (Ouverney and Fuhrman, 2000), and radiocarbon analyses of the membrane lipids of pelagic marine Thaumarchaeota indicate that communities are composed of combination of autotrophs and heterotrophs, or a single mixotrophic population (Ingalls et al., 2006). In addition, N. viennensis and marine thaumarchaeotal strains require pyruvate or α-ketoglutaric acid for optimal growth (Tourna et al., 2011; Qin et al., 2014), although the mechanism of action of these compounds is detoxification of ROS (Kim et al., 2016). Although ROS detoxification was not demonstrated with succinate, it is possible that this compound stimulates growth of heterotrophs that in turn detoxify ROS and thereby encourage growth of AOA. However, Ca. N. exaquare encodes a variety of genes that may confer protection from ROS: in addition to several genes shared among many Thaumarchaeota (for example, superoxide dismutase, alkyl hydroperoxide reductase), Ca. N. exaquare encodes a peroxidase, which is unique among sequenced thaumarchaeotal genomes, and a manganese-dependent catalase, which is also present in Ca. N. evergladensis (Table 1).
The genome of Ca. N. exaquare encodes two gene copies of a sodium-dependent dicarboxylate transporter (SdcS; Table 1), which transports succinate, malate and fumarate (Hall and Pajor, 2005, 2007). SdcS-type dicarboxylate transporters are also encoded in the genomes of Ca. N. evergladensis (Zhalnina et al., 2014) and Ca. N. uzonensis (Lebedeva et al., 2013). Expression of this transporter could provide an explanation for the observed stimulation of Ca. N. exaquare by succinate and malate (Figure 4). In addition to being a tricarboxylic acid cycle intermediate, succinate is a central compound in the 3HB/4HP cycle (Berg, 2011) and could feed directly into this carbon fixation pathway. Labeling studies with Metallosphaera medulla, which also uses the 3HP/4HP cycle (Berg et al., 2007), demonstrated that the majority of anabolic precursors are derived from succinate (Estelmann et al., 2011). Given the presence of this transporter and the strong stimulatory effects of succinate and malate, C4 compounds may have an important role in supplementing Ca. N. exaquare metabolism.
At 2.99 Mbps, Ca. N. exaquare encodes the largest reported AOA genome, and shares several features with Group I.1b soil Thaumarchaeota (Table 1). Interestingly, Ca. N. exaquare has a G+C content (33.9%) that is lower than other Group I.1b Thaumarchaeota (~50%), but comparable to group I.1a Thaumarchaeota. The genome encodes all key components for ammonia oxidation and bicarbonate fixation pathways, supporting its role as a chemolithoautotrophic ammonia oxidizer. In addition, Ca. N. exaquare has a similar metabolic profile to other AOA (Figure 5), with few genes associated with carbohydrate catabolism. An encoded pathway for degradation of mannosylglycerate was identified as unique among thaumarchaeotal genomes, but most likely relates to osmostic regulation, which has been suggested previously (Spang et al., 2012; Zhalnina et al., 2012). Ca. N. exaquare also encodes genes associated with C1 metabolism, including formate dehydrogenase and glutathione-dependent formaldehyde dehydrogenase. Several autotrophic NOB can oxidize or assimilate formate (Malavolta et al., 1962; Van Gool and Laudelout, 1966; Gruber-Dorninger et al., 2015; Koch et al., 2015). However, these encoded enzymes could be used for detoxification, and further work is necessary to assess whether C1 substrates could supplement autotrophic metabolism.
AOA have been detected and quantified in several WWTPs, but only one study has assessed the relative contributions of ammonia-oxidizing prokaryotes to ammonia oxidation in WWTPs. Mussman et al. (2011) detected Thaumarchaeota in industrial WWTPs treating oil refinery waste but found no evidence for bicarbonate fixation, despite active expression of amoA genes. These Thaumarchaeota are phylogenetically related (Figure 2) and morphologically similar to Ca. N. exaquare, which oxidizes ammonia, assimilates bicarbonate and encodes a genome supporting chemolithoautotrophic metabolism. However, ammonia monooxygenase substrate promiscuity has been reported (Pester et al., 2011), and different growth conditions elicit different physiological responses, so the role of Ca. N. exaquare in situ is likely more complex in natural environments.
Incubations of Guelph WWTP RBC biofilm with differential inhibitors indicated that ATU was highly inhibitory, octyne had little effect and PTIO was partially inhibitory. More inhibition by PTIO was observed in RBC 8 biofilms compared with RBC 1 of the same treatment train, suggesting that a larger proportion of the ammonia-oxidizing activity results from AOA. This is supported by qPCR data demonstrating that thaumarchaeotes comprise a higher proportion of the putative ammonia-oxidizing prokaryotes in RBC 8 compared with RBC 1 (Figure 6; Supplementary Table S5). Octyne and ATU are specific inhibitors of AOB (Hatzenpichler et al., 2008; Shen et al., 2013; Taylor et al., 2013) and results obtained from these compounds should ideally be similar, but were inconsistent in this study. Several advantages have been reported for octyne (Taylor et al., 2013), but it has not been used previously with samples from a WWTP environment, and may have been degraded by biofilm microorganisms. Two PTIO concentrations were included because lower concentrations may be insufficient in environmental samples due to production of nitric oxide from non-nitrification processes (for example, denitrification), but higher concentrations might result in inhibition of some AOB. For example, PTIO concentrations of 400 μm are partially inhibitory to N. multiformis (Shen et al., 2013), although not to N. europaea (Supplementary Figure S9).
Although questions remain regarding the efficacy of octyne at inhibiting AOB in this biofilm, and whether AOB were also inhibited using 400 μm PTIO, the observed inhibition of ammonia-oxidizing activity by 200 μm PTIO suggests that Thaumarchaeota contribute to ammonia-oxidizing activity of the biofilm. The previously reported relationship between ammonia concentration and thaumarchaeotal abundance in this biofilm (Sauder et al., 2012) supports the role of Ca. N. exaquare-like AOA as ammonia oxidizers in situ. Although this work only considers AOA and AOB, completely nitrifying Nitrospira organisms (that is, comammox bacteria; van Kessel et al., 2015; Daims et al., 2015), with unknown sensitivities to these inhibitors, could be contributing to nitrification activity in the biofilm.
The CARD-MAR-FISH data from the RBC biofilm samples indicated that when supplied with ammonia, both AOB and Nitrospira are MAR-positive, whereas there was no evidence for bicarbonate fixation by Thaumarchaeota (Figure 7). This suggests that the detected Thaumarchaeota cells were either predominantly relying on an alternative metabolism, or that they were oxidizing ammonia for energy but assimilating a carbon source other than bicarbonate. Similarly, related Thaumarchaeota in another WWTP did not assimilate inorganic carbon in the presence of ammonia (Mussmann et al., 2011). Most observed AOB showed strong MAR signals, which is consistent with the strongly inhibitory effect of ATU. Positive MAR signals were observed for some Nitrospira microcolonies (Figure 7), which could have arisen from either nitrite-oxidizing or comammox activity.
Ca. N. exaquare is the first group I.1b Thaumarchaeota representative cultivated from a WWTP, and clusters phylogenetically with AOA originating from wastewater environments. The laboratory activity and genetic complement of Ca. N. exaquare suggest that it is a classic ammonia-oxidizing microorganism, which may be stimulated by organic carbon. In the wastewater biofilm from which it originates, both qPCR and qFISH indicate that Thaumarchaeota consistently outnumber AOB. However, the metabolic role played in situ by Ca. N. exaquare appears to be more complex than strictly chemolithoautotrophic nitrification, and it is possible that the relatively high abundance of Thaumarchaeota could be explained by growth on other substrates present in the biofilm. Further work is needed to elucidate the contributions of Ca. N. exaquare-like Thaumarchaeota to ammonia oxidation in this system and to assess the in situ potential for autotrophic or mixotrophic metabolisms.
Acknowledgments
We thank the Guelph WWTP staff for access to the facility as well as their enthusiastic support of our study. We thank Jake Beam for providing probe sequences, and Richard Sparling, Barbara Butler and Markus Schmid for helpful suggestions. This research was supported by a European Research Council Advanced Grant (294343) to MW, a Vanier Canada Graduate Scholarship to LAS and a Discovery Grant to JDN, both provided by the National Sciences and Engineering Research Council of Canada (NSERC).
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. (2013). Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31: 533–538. [DOI] [PubMed] [Google Scholar]
- Bagchi S, Vlaeminck SE, Sauder LA, Mosquera M, Neufeld JD, Boon N. (2014). Temporal and spatial stability of ammonia-oxidizing archaea and bacteria in aquarium biofilters. PLoS One 9: e113515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Sun Q, Wen D, Tang X. (2012). Abundance of ammonia-oxidizing bacteria and archaea in industrial and domestic wastewater treatment systems. FEMS Microbiol Ecol 80: 323–330. [DOI] [PubMed] [Google Scholar]
- Banning NC, Maccarone LD, Fisk LM, Murphy DV. (2015). Ammonia-oxidising bacteria not archaea dominate nitrification activity in semi-arid agricultural soil. Sci Rep 5: 11146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beam JP. (2015) Geobiological Interactions of Archaeal Populations in Acidic and Alkaline Geothermal Springs of Yellowstone National Park. Montana, WY, USA: Montana State University. [Google Scholar]
- Berg IA. (2011). Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol 77: 1925–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IA, Kockelkorn D, Buckel W, Fuchs G. (2007). A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318: 1782–1786. [DOI] [PubMed] [Google Scholar]
- Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. (2011). Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One 6: e16626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Pirovano W. (2012). Toward almost closed genomes with GapFiller. Genome Biol 13: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MN, Briones A, Diana J, Raskin L. (2013). Ammonia-oxidizing archaea and nitrite-oxidizing nitrospiras in the biofilter of a shrimp recirculating aquaculture system. FEMS Microbiol Ecol 83: 17–25. [DOI] [PubMed] [Google Scholar]
- Daims H, Lebedeva E, Pjevac P, Han P, Herbold C, Albertsen M et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 528: 504–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H, Lücker S, Wagner M. (2006). Daime, a novel image analysis program for microbial ecology and biofilm research. Environ Microbiol 8: 200–213. [DOI] [PubMed] [Google Scholar]
- Di HJ, Cameron KC, Shen JP, Winefield CS, O’Callaghan M, Bowatte S et al. (2009). Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci 2: 621–624. [Google Scholar]
- Doxey AC, Kurtz DA, Lynch MD, Sauder LA, Neufeld JD. (2015). Aquatic metagenomes implicate Thaumarchaeota in global cobalamin production. ISME J 9: 461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W. (2009). Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol Rev 33: 855–869. [DOI] [PubMed] [Google Scholar]
- Estelmann S, Hügler M, Eisenreich W, Werner K, Berg IA, Ramos-Vera WH et al. (2011). Labeling and enzyme studies of the central carbon metabolism in Metallosphaera sedula. J Bacteriol 193: 1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J-F, Luo X, Wu G-X, Li T, Peng Y-Z. (2014). Abundance and diversity based on amoA genes of ammonia-oxidizing archaea and bacteria in ten wastewater treatment systems. Appl Microbiol Biotechnol 98: 3339–3354. [DOI] [PubMed] [Google Scholar]
- Gao J-F, Luo X, Wu G-X, Li T, Peng Y-Z. (2013). Quantitative analyses of the composition and abundance of ammonia-oxidizing archaea and ammonia-oxidizing bacteria in eight full-scale biological wastewater treatment plants. Bioresour Technol 138: 285–296. [DOI] [PubMed] [Google Scholar]
- Van Gool A, Laudelout H. (1966). Formate utilization by Nitrobacter winogradskyi. Biochim Biophys Acta 127: 295–301. [DOI] [PubMed] [Google Scholar]
- Gruber-Dorninger C, Pester M, Kitzinger K, Savio DF, Loy A, Rattei T et al. (2015). Functionally relevant diversity of closely related Nitrospira in activated sludge. ISME J 9: 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JA, Pajor AM. (2005). Functional characterization of a Na+-coupled dicarboxylate carrier protein from Staphylococcus aureus. J Bacteriol 187: 5189–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JA, Pajor AM. (2007). Functional reconstitution of SdcS, a Na+-coupled dicarboxylate carrier protein from Staphylococcus aureus. J Bacteriol 189: 880–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM et al. (2006). Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol 4: e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzenpichler R. (2012). Diversity, physiology, and niche differentiation of ammonia-oxidizing archaea. Appl Environ Microbiol 78: 7501–7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzenpichler R, Lebedeva E, Spieck E, Stoecker K, Richter A, Daims H et al. (2008). A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA 105: 2134–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im J, Lee S-W, Bodrossy L, Barcelona MJ, Semrau JD. (2011). Field application of nitrogen and phenylacetylene to mitigate greenhouse gas emissions from landfill cover soils: effects on microbial community structure. Appl Microbiol Biotechnol 89: 189–200. [DOI] [PubMed] [Google Scholar]
- Ingalls AE, Shah SR, Hansman RL, Aluwihare LI, Santos GM, Druffel ERM et al. (2006). Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci USA 103: 6442–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Mussmann M, MacGregor BJ, Amann R. (2004). An improved fluorescence in situ hybridization protocol for the identification of bacteria and archaea in marine sediments. FEMS Microbiol Ecol 50: 203–213. [DOI] [PubMed] [Google Scholar]
- Jia Z, Conrad R. (2009). Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol 11: 1658–1671. [DOI] [PubMed] [Google Scholar]
- Jung M-Y, Kim J-G, Sinninghe Damsté JS, Rijpstra WIC, Madsen EL, Kim S-J et al. (2016). A hydrophobic ammonia-oxidizing archaeon of the Nitrosocosmicus clade isolated from coal tar-contaminated sediment. Environ Microbiol Rep 8: 983–992. [DOI] [PubMed] [Google Scholar]
- Jung M-Y, Park S-J, Min D, Kim J-S, Rijpstra WIC, Sinninghe Damste JS et al. (2011). Enrichment and characterization of an autotrophic ammonia-oxidizing archaeon of mesophilic crenarchaeal Group I.1a from an agricultural soil. Appl Environ Microbiol 77: 8635–8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayee P, Sonthiphand P, Rongsayamanont C, Limpiyakorn T. (2011). Archaeal amoA genes outnumber bacterial amoA genes in municipal wastewater treatment plants in Bangkok. Microb Ecol 62: 776–788. [DOI] [PubMed] [Google Scholar]
- van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJM, Kartal B et al. (2015). Complete nitrification by a single microorganism. Nature 528: 555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-G, Park S-J, Sinninghe Damsté JS, Schouten S, Rijpstra WIC, Jung M-Y et al. (2016). Hydrogen peroxide detoxification is a key mechanism for growth of ammonia-oxidizing archaea. Proc Natl Acad Sci USA 113: 7888–7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch H, Lücker S, Albertsen M, Kitzinger K, Herbold C, Spieck E et al. (2015). Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc Natl Acad Sci USA 112: 11371–11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546. [DOI] [PubMed] [Google Scholar]
- Krümmel A, Harms H. (1982). Effect of organic matter on growth and cell yield of ammonia-oxidizing bacteria. Arch Microbiol 133: 50–54. [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva EV, Hatzenpichler R, Pelletier E, Schuster N, Hauzmayer S, Bulaev A et al. (2013). Enrichment and genome sequence of the group I.1a ammonia-oxidizing archaeon ‘Ca. Nitrosotenuis uzonensis’ representing a clade globally distributed in thermal habitats. PLoS One 8: e80835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Nielsen PH, Andreasen KH, Juretschko S, Nielsen JL, Schleifer K-H et al. (1999). Combination of fluorescent in situ hybridization and microautoradiography-a new tool for structure-function analyses in microbial ecology. Appl Environ Microbiol 65: 1289–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett RM, Clavijo BJ, Clissold L, Clark MD, Caccamo M. (2014). NextClip: an analysis and read preparation tool for Nextera Long Mate Pair libraries. Bioinformatics 30: 566–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtovirta-Morley LE, Ross J, Hink L, Weber EB, Gubry-Rangin C, Thion C et al. (2016). Isolation of ‘Candidatus Nitrosocosmicus franklandus’, a novel ureolytic soil archaeal ammonia oxidiser with tolerance to high ammonia concentration. FEMS Microbiol Ecol 92: fiw057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW et al. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442: 806–809. [DOI] [PubMed] [Google Scholar]
- Li Y, Ding K, Wen X, Zhang B, Shen B, Yang Y. (2016). A novel ammonia-oxidizing archaeon from wastewater treatment plant: Its enrichment, physiological and genomic characteristics. Sci Rep 6: 23747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpiyakorn T, Fürhacker M, Haberl R, Chodanon T, Srithep P, Sonthiphand P. (2013). amoA-encoding archaea in wastewater treatment plants: a review. Appl Microbiol Biotechnol 97: 1425–1439. [DOI] [PubMed] [Google Scholar]
- Limpiyakorn T, Sonthiphand P, Rongsayamanont C, Polprasert C. (2011). Abundance of amoA genes of ammonia-oxidizing archaea and bacteria in activated sludge of full-scale wastewater treatment plants. Bioresour Technol 102: 3694–3701. [DOI] [PubMed] [Google Scholar]
- Maixner F, Noguera DR, Anneser B, Stoecker K, Wegl G, Wagner M et al. (2006). Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ Microbiol 8: 1487–1495. [DOI] [PubMed] [Google Scholar]
- Malavolta E, Delwiche CC, Burge WD. (1962). Formate oxidation by cell-free preparations from Nitrobacter agilis. Biochim Biophys Acta 57: 347–351. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, Mavromatis K, Ivanova NN, Chen I-MA, Chu K, Kyrpides NC. (2009). IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25: 2271–2278. [DOI] [PubMed] [Google Scholar]
- Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461: 976–979. [DOI] [PubMed] [Google Scholar]
- Meseguer-Lloret S, Molins-Legua C, Campins-Falco P. (2002). Ammonium determination in water samples by using OPA-NAC reagent: a comparative study with nessler and ammonium selective electrode methods. Int J Environ Anal Chem 82: 475–489. [Google Scholar]
- Miranda KM, Espey MG, Wink DA. (2001). A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide 5: 62–71. [DOI] [PubMed] [Google Scholar]
- Mosier AC, Lund MB, Francis CA. (2012). Ecophysiology of an ammonia-oxidizing archaeon adapted to low-salinity habitats. Microb Ecol 64: 955–963. [DOI] [PubMed] [Google Scholar]
- Mussmann M, Brito I, Pitcher A, Sinninghe Damste JS, Hatzenpichler R, Richter A et al. (2011). Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc Natl Acad Sci USA 108: 16771–16776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, de Waal EC, Uitterlinden AG. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59: 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C. (2003). Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ Microbiol 5: 787–797. [DOI] [PubMed] [Google Scholar]
- Ouverney CC, Fuhrman JA. (2000). Marine planktonic archaea take up amino acids. Appl Environ Microbiol 66: 4829–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatinszky M, Herbold C, Jehmlich N, Pogoda M, Han P, von Bergen M et al. (2015). Cyanate as an energy source for nitrifiers. Nature 524: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-D, Wells GF, Bae H, Criddle CS, Francis CA. (2006). Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl Environ Microbiol 72: 5643–5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pester M, Rattei T, Flechl S, Gröngröft A, Richter A, Overmann J et al. (2012). amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ Microbiol 14: 525–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pester M, Schleper C, Wagner M. (2011). The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol 14: 300–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AJ, Auerbach AK, Moissl-Eichinger C. (2013). Archaea on human skin. PLoS One 8: e65388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Amin SA, Martens-Habbena W, Walker CB, Urakawa H, Devol AH et al. (2014). Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci USA 111: 12504–12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotthauwe JH, Witzel KP, Liesack W. (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63: 4704–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauder LA, Engel K, Stearns JC, Masella AP, Pawliszyn R, Neufeld JD. (2011). Aquarium nitrification revisited: Thaumarchaeota are the dominant ammonia oxidizers in freshwater aquarium biofilters. PLoS One 6: e23281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauder LA, Peterse F, Schouten S, Neufeld JD. (2012). Low-ammonia niche of ammonia-oxidizing archaea in rotating biological contactors of a municipal wastewater treatment plant. Environ Microbiol 14: 2589–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauder LA, Ross AA, Neufeld JD. (2016). Nitric oxide scavengers differentially inhibit ammonia oxidation in ammonia-oxidizing archaea and bacteria. FEMS Microbiol Lett 363: fnw052. [DOI] [PubMed] [Google Scholar]
- Schleper C. (2010). Ammonia oxidation: different niches for bacteria and archaea? ISME J 4: 1092–1094. [DOI] [PubMed] [Google Scholar]
- Shen T, Stieglmeier M, Dai J, Urich T, Schleper C. (2013). Responses of the terrestrial ammonia-oxidizing archaeon Ca. Nitrososphaera viennensis and the ammonia-oxidizing bacterium Nitrosospira multiformis to nitrification inhibitors. FEMS Microbiol Lett 344: 121–129. [DOI] [PubMed] [Google Scholar]
- Spang A, Poehlein A, Offre P, Zumbrägel S, Haider S, Rychlik N et al. (2012). The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol 14: 3122–3145. [DOI] [PubMed] [Google Scholar]
- Stahl DA, Amann R. (1991). Development and application of nucleic acid probes in bacterial systematics. In: Stackebrandt E, Goodfellow M (eds). Nucleic Acid Techniques in Bacterial Systematics.. Chichester, England: John Wiley & Sons Ltd, pp 205–248. [Google Scholar]
- Sterngren AE, Hallin S, Bengtson P. (2015). Archaeal ammonia oxidizers dominate in numbers, but bacteria drive gross nitrification in N-amended grassland soil. Front Microbiol 6: 1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieglmeier M, Klingl A, Alves RJE, Rittmann SK-MR, Melcher M, Leisch N et al. (2014). Nitrososphaera viennensis gen. nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon from soil and a member of the archaeal phylum Thaumarchaeota. Int J Syst Evol Microbiol 64: 2738–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopnisek N, Gubry-Rangin C, Höfferle S, Nicol GW, Mandic-Mulec I, Prosser JI. (2010). Thaumarchaeal ammonia oxidation in an acidic forest peat soil is not influenced by ammonium amendment. Appl Environ Microbiol 76: 7626–7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AE, Vajrala N, Giguere AT, Gitelman AI, Arp DJ, Myrold DD et al. (2013). Use of aliphatic n-alkynes to discriminate soil nitrification activities of ammonia-oxidizing thaumarchaea and bacteria. Appl Environ Microbiol 79: 6544–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T et al. (2011). Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA 108: 8420–8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet D, Engelen S, Mornico D, Cruveiller S, Fleury L, Lajus A et al. (2009). MicroScope: a platform for microbial genome annotation and comparative genomics. Database 2009: bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhamme DT, Prosser JI, Nicol GW. (2011). Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J 5: 1067–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ et al. (2010). Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA 107: 8818–8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells GF, Park H-D, Yeung C-H, Eggleston B, Francis CA, Criddle CS. (2009). Ammonia-oxidizing communities in a highly aerated full-scale activated sludge bioreactor: betaproteobacterial dynamics and low relative abundance of Crenarchaea. Environ Microbiol 11: 2310–2328. [DOI] [PubMed] [Google Scholar]
- Widdel F, Bak F. (1992). Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Trüper HG (eds). The Prokaryotes, 2nd edn, vol. IV. New York, NY, USA: Springer, pp 352–3378. [Google Scholar]
- Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P et al. (2006). Archaeal nitrification in the ocean. Proc Natl Acad Sci USA 103: 12317–12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N, Otawa K, Nakai Y. (2010). Diversity and abundance of ammonia-oxidizing bacteria and ammonia-oxidizing archaea during cattle manure composting. Microb Ecol 60: 807–815. [DOI] [PubMed] [Google Scholar]
- Yao H, Gao Y, Nicol GW, Campbell CD, Prosser JI, Zhang L et al. (2011). Links between ammonia oxidizer community structure, abundance and nitrification potential in acidic soils. Appl Environ Microbiol 77: 4618–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhalnina K, Dias R, Leonard MT, Dorr de Quadros P, Camargo FAO, Drew JC et al. (2014). Genome sequence of Candidatus Nitrososphaera evergladensis from Group I.1b enriched from everglades soil reveals novel genomic features of the ammonia-oxidizing archaea. PLoS One 9: e101648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhalnina K, de Quadros PD, Camargo FAO, Triplett EW. (2012). Drivers of archaeal ammonia-oxidizing communities in soil. Front Microbiol 3: A210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L-M, Hu H-W, Shen J-P, He J-Z. (2012). Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J 6: 1032–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Jin T, Yan Q, Shao M, Wells G, Criddle CS et al. (2009). Occurrence of ammonia-oxidizing archaea in activated sludges of a laboratory scale reactor and two wastewater treatment plants. J Appl Microbiol 107: 970–977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.