Abstract
Listeria monocytogenes (Lm) is an intracellular Gram-positive bacterium that induces expression of type I interferons (IFN-α/IFN-β) during infection. These cytokines are detrimental to the host during infection by priming leukocytes to undergo Lm-mediated apoptosis. Our previous studies showed that C5aR1−/− and C3aR−/− mice are highly susceptible to Lm infection due to elevated IFN-β–mediated apoptosis of major leukocyte cell populations, including CD4+ and CD8+ T-cells. However, the mechanisms by which C3a and C5a modulate IFN-β expression during Lm infection were not examined in these initial investigations. Accordingly, we report here that C5a and C3a suppress IFN-β production in response to Lm via c-di-AMP, a secondary messenger molecule of Lm, in both J774A.1 macrophage-like cells and in bone marrow-derived dendritic cells (BMDCs). Moreover, C5a and C3a suppress IFN-β production by acting through their respective receptors, as no inhibition was seen in C5aR1−/− or C3aR−/− BMDCs, respectively. C5a and C3a suppress IFN-β production in a manner that is dependent on Bruton’s Tyrosine Kinase (BTK), p38 MAPK, and TANK-Binding Kinase 1 (TBK1), as demonstrated by the individual use of BTK, p38 MAPK, and TBK1 inhibitors. Pre-treatment of cells with C5a and C3a reduced the expression of the IFN-β signaling molecules DDX41, STING, phosphorylated TBK1, and phosphorylated p38 MAPK in WT BMDCs following treatment with c-di-AMP. Collectively, these data demonstrate that C3a and C5a, via direct signaling through their specific receptors, suppress IFN-β expression by modulation of a distinct innate cytosolic surveillance pathway involving DDX41, STING, and other downstream molecular targets of Lm-generated c-di-AMP.
Introduction
Listeria monocytogenes (Lm) is a Gram-positive facultative intracellular pathogen that lives in soil and water and is transmitted to humans via contaminated food. Common sources of infection include uncooked meats and produce, unpasteurized milk and cheeses, and processed meats such as hot dogs and deli meats. Infection with this pathogen, known as listeriosis, can lead to sepsis and meningitis. This bacterium infects mostly older adults (age 65 and over), persons with weakened immune systems (such as those with cancer, HIV/AIDS, diabetes, kidney or liver disease), pregnant women, and their newborns. In the U.S., pregnant women are 10–100 times more likely than others to become infected with Lm, which can lead to miscarriage, stillbirth, and death of the newborn soon after birth (1, 2). According to data compiled from the CDC (www.cdc.gov/listeria/outbreaks), there were 29 outbreaks in the U.S. involving food contaminated with Lm from 2011–2016, which resulted in 363 confirmed cases, 333 hospitalizations, and 70 deaths (21% mortality rate). In a paper published in 2014 (3), which looked at global listeriosis cases reported in 2010, there were 23,150 cases and 5463 deaths (24% mortality rate), and 20.7% of the cases were pregnancy-related.
C3a and C5a are 77-aa and 74-aa peptides, respectively, that are generated during activation of the complement cascade in response to infection and other stimuli. They are known as anaphylatoxins because they cause smooth muscle contraction, histamine release from mast cells, and vasodilation (4). However, C3a and C5a are potent mediators of numerous other biological pro-inflammatory and anti-inflammatory responses. C3a and C5a trigger these biological responses by binding to their specific G protein–coupled receptors, C3aR and C5aR1, respectively (4). C3aR and C5aR1 are expressed on both myeloid and lymphoid cells, as well as on multiple types of non-immune tissue cells (5). In our previous studies, we showed that C3aR (6) and C5aR1 (7) are protective during systemic Lm infection. Compared to WT mice, C3aR−/− mice and C5aR1−/− mice had decreased survival, increased bacterial burdens in the liver and spleen, increased cytokine production, including IFN-β, increased spleen and liver pathology, and increased apoptosis in the spleen. Pre-treatment of the C5aR1−/− mice with an antibody to the type I interferon receptor-1 (IFNAR-1) protected these mice from Lm-induced mortality (7), indicating that excess IFN-β production in the C5aR1−/− mice leads to increased susceptibility to Lm. Production of type I interferons during Lm infection is detrimental to the host (8–10). Through an unknown mechanism, type I interferons prime lymphocytes to undergo apoptosis (9), and mice lacking IFNAR-1 undergo less apoptosis in the spleen and are more resistant to Lm infection (8–10). IFN-β has also been shown to cause death of macrophages when infected with Lm (11). IFN-γ is critical to controlling Lm growth in vivo (12–14), and type I interferons have been shown to decrease the responsiveness of macrophages to IFN-γ during Lm infection (15).
A lot of work has been done in the last several years in an effort to understand how Lm induces type I interferon production. Several pattern recognition receptors and adaptor molecules, such as TLR2, TLR4, TLR9, MyD88, TRIF, TRAM, and the nucleotide-binding domain family of proteins, including receptor interacting protein 2 (10, 16–18), were shown not to be involved in Lm-induced type I interferon production. IFN-β production was, however, found to be dependent on BTK (19), DDX41 (20), STING (21, 22), TBK1 (17), and IRF3 (10, 18) expression. Lm must escape from the phagosome and get into the cytosol of the infected cell to induce IFN-β production, as infection with listeriolysin-deficient Lm does not lead to IFN-β production (8, 16, 17, 23), indicating some cytosolic sensor must be involved in type I interferon production. In 2006, Stetson and Medzhitov discovered that cytosolic delivery of Lm-derived DNA was a strong inducer of type I interferon (24). Subsequently, a cyclic dinucleotide (CDN) from Lm, known as c-di-AMP, was also found to be a powerful inducer of type I interferon (25). DDX41 (20) and STING (26) were identified as cytosolic sensors of CDNs and were found to be required for IFN-β production in response to Lm (20–22, 27). BTK was recently identified as a regulator of DDX41 and STING (19). DDX41 is thought to be the primary sensor of CDNs, with STING acting as a secondary receptor for CDNs (20). Once bound to CDNs, DDX41 associates with STING (20), which binds to TBK1 and phosphorylates IRF3 to induce type I interferon production (21, 28). The absence of either DDX41 (20), STING (20–22, 27), or TBK1 (17) results in defective type I interferon production in response to Lm, demonstrating the importance of all three of these proteins in this signaling pathway (please note that a schematic diagram depicting the CDN-signaling pathway discussed here is displayed as Supplemental Fig. 1).
In our current efforts to understand how C5a and C3a regulate IFN-β production, we demonstrate that pre-treatment of J774A.1 macrophage-like cells or WT BMDCs with C5a or C3a results in a significant reduction in IFN-β production in response to Lm or c-di-AMP. This inhibition is specific for IFN-β, as we saw no significant effect on IL-6, TNF, or MCP-1 expression. We demonstrate that C5aR1, C3aR, BTK, p38 MAPK, and TBK1 are necessary for this inhibitory effect. Finally, we show that pre-treatment of BMDCs with C5a or C3a leads to significant reductions in DDX41, STING, phosphorylated TBK1, and phosphorylated p38 MAPK expression, resulting in decreased IFN-β production in response to Lm and c-di-AMP.
Materials and Methods
Reagents and bacteria
Human C5a and C3a were purchased from Complement Technology. The C5aR agonist (C5aA) (RAARISLGPRSIKAFTE) (29) and C3aR agonist (C3aA) (WWTRRWRGDKLGLAR) (30) were synthesized by GenScript with greater than 95% purity. c-di-AMP, BX795, Wortmannin, U0126, and SB202190 were purchased from Invivogen. LPS from E. coli was purchased from List Labs, and CGI-1746 was purchased from ApexBio. Listeria monocytogenes ATCC strain 13932 (serotype 4b) (MicroBioLogics, Inc.) was used for the infection studies. Bacteria were cultured in Bacto brain heart infusion broth at 37°C to mid-logarithmic phase, harvested by centrifugation, washed once in sterile PBS, and resuspended in sterile PBS.
Mice
The C3aR−/− mice (31) and the C5aR1−/− mice (32) used in these studies were generated in our laboratory and have been described previously. These strains of mice were backcrossed for over ten generations onto the C57BL/6 background. C57BL/6 mice from our own inbred C57BL/6 colony served as wild-type mice. All mice were housed in HEPA-filtered Tecniplast cages in a barrier facility. Bone marrow was collected only from female mice. Institutional guidelines for animal care and welfare were followed.
In vitro experiments
The J774A.1 mouse macrophage cell line was purchased from the ATCC (TIB-67™), and these cells were cultured in DMEM with high glucose and L-glutamine (Lonza BioWhittaker) supplemented with 10% (v/v) heat-inactivated FBS (Gibco) and 1X Penicillin/Streptomycin (HyClone). J774A.1 cells were grown in a loosely adherent manner in non-tissue culture-treated 100 mm petri dishes (Fisher). The night before experiments, the cells were dislodged by pipetting up and down and were plated in a volume of 400 μl at 2 × 105 cells per well in 48 well tissue culture-treated plates (BD Falcon). Mouse bone marrow cells were isolated by flushing each end of the femur with 3 ml of HBSS (Lonza) using a 23 gauge needle and a 10 ml syringe for a total of 6 ml of cells per femur. The cells were kept on ice immediately after harvesting. The bone marrow cells were filtered through a 40 μm cell strainer (BD Falcon), centrifuged at 200 rcf, washed one time with HBSS, and resuspended at a density of 1 × 106 cells/ml in RPMI-1640 (Lonza) supplemented only with 10% (v/v) heat-inactivated FBS, 1X Penicillin/Streptomycin, and 20 ng/ml of recombinant mouse GM-CSF (animal-free) (BioLegend) to select for bone marrow dendritic cells (BMDCs). The BMDCs were plated in a volume of 400 μl at 4 × 105 cells per well in 48 well tissue culture-treated plates (BD Falcon). The media was replaced on days 4 and 6, and the BMDCs were used on day 7. If the cells were going to be infected with Lm, the media did not contain antibiotics beginning on day 6. The BMDCs were maintained in 20 ng/ml GM-CSF throughout the entire experiment.
On the day of the experiment, the appropriate concentration of C5a, C3a, C5aA, or C3aA was added to the cells via total media replacement (with GM-CSF for BMDCs). After 2 h, either c-di-AMP or LPS was added directly to the cells for 4 h or 20 h. In the case of Lm, for J774 studies 107 Lm in 10 μl was added directly to the cells and incubated for 1 h. After 1 h, the media was replaced with warm media containing C5a, C3a, C5aA, or C3aA, and the cells were incubated for 20 h. For BMDC studies, 106 Lm in 10 μl was added directly to the cells for 4 h or 103 Lm in 10 μl was added directly to the cells for 20 h. In some studies, vehicle, CGI-1746, SB202190, BX795, Wortmannin, or U0126 was added to the cells via total media replacement (with GM-CSF) 1 h prior to C5aA or C3aA. The cells were maintained in these inhibitors throughout the entire experiment.
ELISAs
IFN-β was quantitated in cell-free culture supernatants using our own ELISA consisting of recombinant mouse IFN-β as a standard, a polyclonal goat anti-mouse IFN-β used as a capture antibody, a biotinylated hamster anti-mouse IFN-β used as a detection antibody, and HRP Avidin (all from BioLegend) and TMB substrate (Sigma) for signal detection. Mouse IL-6, TNF, and MCP-1 were quantitated in cell-free culture supernatants using ELISA MAX kits from BioLegend. The plates were read using a SpectraMax M2 plate reader (Molecular Devices).
Quantitative PCR
RNA was purified from BMDCs using the RNeasy Plus Mini Kit from Qiagen. cDNA was synthesized from RNA using the High-Capacity cDNA Reverse Transcription Kit from Applied Biosystems. Quantitative PCR was performed using RT2 qPCR Primers for mouse Ifnb and Gapdh (Qiagen), iTaq Universal SYBR Green Supermix (Bio-Rad), and an ABI-7900 thermal cycler. Transcript abundance for Ifnb was normalized to Gapdh for each sample.
Western blots
BMDCs were lysed directly in the 48 well plates using 55 μl of lysis buffer containing 1X Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific), and 20 μl of lysate was electrophoresed on 10% SDS-PAGE reducing gels (BioRad). The gels were transferred to PVDF (BioExpress) and were blocked in 5% (w/v) BSA in TBS/0.1% (v/v) Tween20. The following antibodies used for Western blotting were purchased from Cell Signaling Technology: rabbit anti-STING (D2P2F), rabbit anti-phospho-TBK1 (Ser172) (D52C2), rabbit anti-TBK1 (D1B4), rabbit anti-phospho-p38 MAPK (Thr180/Tyr182) (D3F9), and rabbit polyclonal anti-p38 MAPK. Rabbit anti-DDX41 was purchased from Bethyl Laboratories. Mouse anti-β-actin (clone AC-74) was purchased from Sigma. HRP-conjugated donkey anti-rabbit IgG and donkey anti-mouse IgG were purchased from Jackson Immunoresearch. Clarity Western ECL Substrate (BioRad) was used for detection of HRP on Western blots. OneMinute Plus Western Blot Stripping Buffer from GM Biosciences was used to strip the blots for multiple re-probings. Western blots were scanned using the ChemiDoc Touch Imaging System from BioRad, and protein bands were quantitated using Image Lab software (BioRad). Proteins were normalized either to β-actin, or phosphorylated proteins were normalized to their respective total protein.
Statistical analyses
All statistical analyses were done using GraphPad Prism 5 software. Comparisons were done with the unpaired two-tailed t test, with P values ≤ 0.05 considered significant.
Results
C5a and C3a suppress IFN-β production in J774 cells
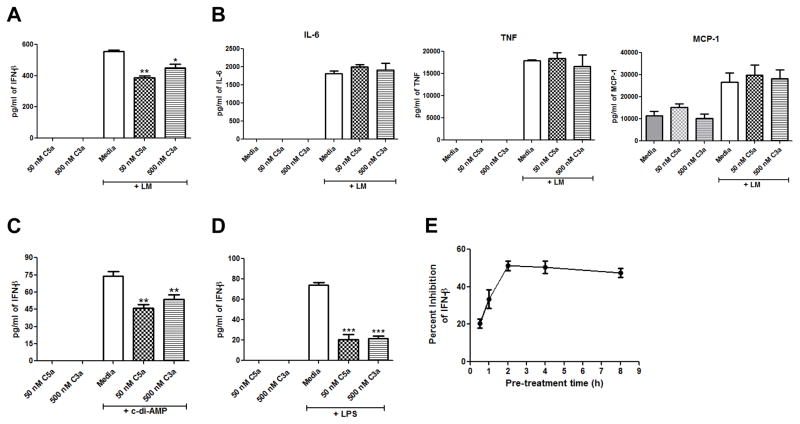
Following 24 hrs of infection with Lm, C5aR1−/− mice (7) and C3aR−/− mice (unpublished data) produce significantly more IFN-β than do WT mice, suggesting that C5a and C3a suppress IFN-β production during Lm infection. To determine if C5a and C3a can suppress IFN-β production in response to Lm in vitro, we pre-treated J774 cells with 50 nM C5a or 500 nM C3a for 2 h and then incubated the cells with 107 Lm as described in Materials and Methods. The concentrations of peptides used in these experiments correspond to the approximate levels of C5a and C3a generated following 10% activation of C5 and C3 in the circulation. As shown in Figure 1A, C5a and C3a suppressed IFN-β production from J774 cells by about 30% in response to Lm. Pre-treating J774 cells with 50 nM C5a and 500 nM C3a together did not increase the amount of inhibition of IFN-β production (data not shown), suggesting that C5a and C3a suppress IFN-β production through the same pathway. To see if C5a and C3a have a broad ability to suppress cytokine production in J774 cells, we also measured IL-6, TNF, and MCP-1 levels in the cell culture supernatants from J774 cells pre-treated with media, C5a, or C3a and then infected with Lm. There was no significant difference in IL-6, TNF, or MCP-1 levels between J774 cells pre-treated with C5a or C3a and those pre-treated with media (Fig. 1B), indicating a specific ability of C5a and C3a to suppress IFN-β production in J774 cells. Lm has been shown to induce IFN-β production through secretion of c-di-AMP (25). To determine if C5a and C3a can suppress IFN-β production in response to c-di-AMP, we pre-treated J774 cells with 50 nM C5a or 500 nM C3a for 2 h and then incubated the cells with 25 μg/ml c-di-AMP overnight for a total of 20 h. As shown in Figure 1C, 50 nM C5a and 500 nM C3a significantly suppressed IFN-β production from J774 cells. Again, pre-treating J774 cells with 50 nM C5a and 500 nM C3a together did not increase the amount of inhibition of IFN-β production (data not shown). To determine if C5a and C3a can also suppress LPS-induced IFN-β production, we pre-treated J774 cells with 50 nM C5a and 500 nM C3a for 2 h and then incubated the cells with 100 ng/ml LPS for 20 h. As shown in Figure 1D, C5a and C3a significantly suppressed IFN-β production in response to LPS so the ability of C5a and C3a to suppress IFN-β production is not specific to only Lm and c-di-AMP. Pre-treatment times with C5a were varied from 30 min to 8 h, and the 2 h pre-treatment time gave the earliest and most inhibitory IFN-β effect (Fig. 1E) so this is the pre-treatment time used for all studies.
Figure 1. C5a and C3a suppress IFN-β production in J774A.1 cells.
J774A.1 cells were pre-treated with either media, 50 nM C5a, or 500 nM C3a for 2 h and then the cells were infected with Lm for 20 h. Cell-free supernatants were used to quantitate (A) IFN-β production or (B) IL-6, TNF-α, and MCP-1 production. J774A.1 cells were pre-treated with either media, 50 nM C5a, or 500 nM C3a for 2 h and then were incubated with (C) 25 μg/ml c-di-AMP or (D) 100 ng/ml LPS for 20 h. IFN-β was quantitated from cell-free supernatants. (E) J774A.1 cells were pre-treated with either media or 50 nM C5a for 30 min, 1 h, 2 h, 4 h, or 8 h and then were incubated with 100 ng/ml LPS for 20 h. IFN-β was quantitated from cell-free supernatants. All data are presented as mean pg/ml ± SEM. These data are pooled from three independent experiments. * P = 0.027; ** P ≤ 0.006; *** P ≤ 0.0001 by t test.
C5a and C3a suppress IFN-β production in bone marrow dendritic cells
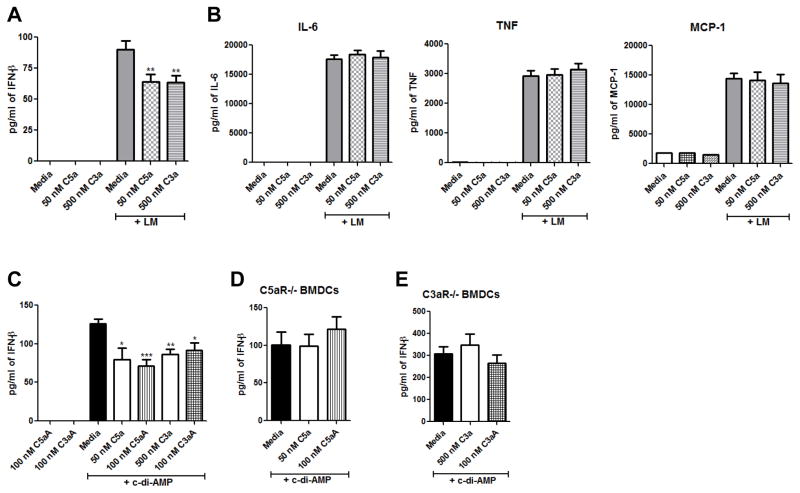
Next, we wanted to determine if our findings in the J774 macrophage cell line would translate to primary mouse cells. Multiple studies have identified dendritic cells as the primary producers of IFN-β during Lm in vivo infection (33, 34). Therefore, we continued our studies using bone marrow derived dendritic cells. WT BMDCs pre-treated with 50 nM C5a or 500 nM C3a for 2 h and then infected with Lm overnight produced 30% less IFN-β than WT BMDCs pre-treated with media (Fig. 2A). WT BMDCs treated with 50 nM C5a or 500 nM C3a, but not Lm, did not produce any IFN-β (Fig. 2A). To see if C5a and C3a have a broad ability to suppress cytokine production in BMDCs, we also measured IL-6, TNF, and MCP-1 levels in the cell culture supernatants from BMDCs pre-treated with media, C5a, or C3a and then infected overnight with Lm. There was no significant difference in IL-6, TNF, or MCP-1 levels between BMDCs pre-treated with C5a or C3a and BMDCs pre-treated with media (Fig. 2B), indicating a specific ability of C5a and C3a to suppress IFN-β production in BMDCs. Peptides (referred to here as C3aA and C5aA) have been developed that exhibit enhanced agonist activity for C3aR (28) and C5aR1 (27), respectively. We tested these peptides to determine if they would suppress IFN-β production more robustly than native purified C3a or C5a. WT BMDCs were pre-treated with either 50 nM C5a, 100 nM C5aA, 500 nM C3a, or 100 nM C3aA for 2 h followed by incubation with 25 μg/ml c-di-AMP overnight. As shown in Fig. 2C, purified C5a and C3a, as well as C5aA and C3aA, all caused about 30% suppression of IFN-β production. BMDCs from C5aR1−/− mice or C3aR−/− mice did not show any suppression in c-di-AMP-induced IFN-β production when pre-treated with C5a or C5aA (Fig. 2D) or C3a or C3aA (Fig. 2E), respectively, indicating that C5a/C5aA and C3a/C3aA are acting through their respective receptors to suppress IFN-β production.
Figure 2. C5a and C3a suppress IFN-β production in BMDCs.
WT BMDCs were pre-treated with either media, 50 nM C5a, or 500 nM C3a for 2 h, and then the cells were infected with Lm overnight. Cell-free supernatants were used to quantitate (A) IFN-β production or (B) IL-6, TNF-α, and MCP-1 production. (C) WT BMDCs were pre-treated with either media, 50 nM C5a, 100 nM C5aA, 500 nM C3a, or 100 nM C3aA for 2 h and then were incubated with 25 μg/ml c-di-AMP for 20 h. IFN-β was quantitated from cell-free supernatants. (D) C5aR−/− BMDCs were pre-treated with either media, 50 nM C5a, or 100 nM C5aA for 2 h and (E) C3aR−/− BMDCs were pre-treated with either media, 500 nM C3a, or 100 nM C3aA for 2 h, and then the cells were incubated with 25 μg/ml c-di-AMP for 20 h. IFN-β was quantitated from cell-free supernatants. All data are presented as mean pg/ml ± SEM. These data are pooled from three independent experiments. * P ≤ 0.017; ** P ≤ 0.009; *** P = 0.0006 by t test.
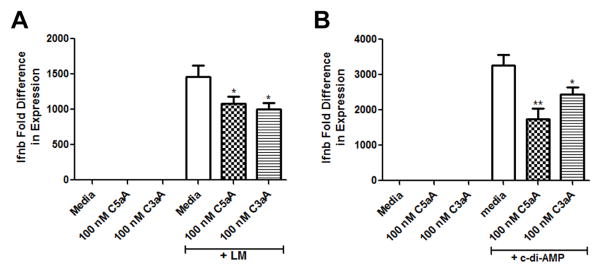
To determine if C3aA and C5aA suppress Ifnb at the message level, BMDCs were pre-treated with media, 100 nM C5aA, or 100 nM C3aA for 2 h and then infected with Lm for 4 h or incubated with c-di-AMP for 4 h. Ifnb message increases early after infection with Lm or stimulation with c-di-AMP (19, 20) so we chose a 4 h time point. As shown in Figure 3, Lm (Fig. 3A) and c-di-AMP (Fig. 3B) caused a dramatic upregulation of Ifnb message in the BMDCs pre-treated with media, but pre-treatment with C5aA or C3aA significantly suppressed Ifnb message induced by both Lm and c-di-AMP. These data demonstrate that C5aA and C3aA suppress IFN-β at both the message and protein levels in BMDCs.
Figure 3. C5aA and C3aA suppress Ifnb message in BMDCs.
WT BMDCs were pre-treated with either media, 100 nM C5aA, or 100 nM C3aA for 2 h and then the cells were (A) infected with Lm or (B) incubated with 25 μg/ml c-di-AMP for 4 h. RNA was harvested from whole cell lysates, and message for Ifnb and Gapdh was quantitated from cDNA. Message for Ifnb was normalized to Gapdh for each sample, and each treatment group was compared to cells treated with only media (set as 1). Data is presented as mean ± SEM. These data are pooled from three independent experiments. * P ≤ 0.048; ** P = 0.003 by t test.
C5aA and C3aA decrease DDX41, STING, TBK1, and p38 MAPK expression
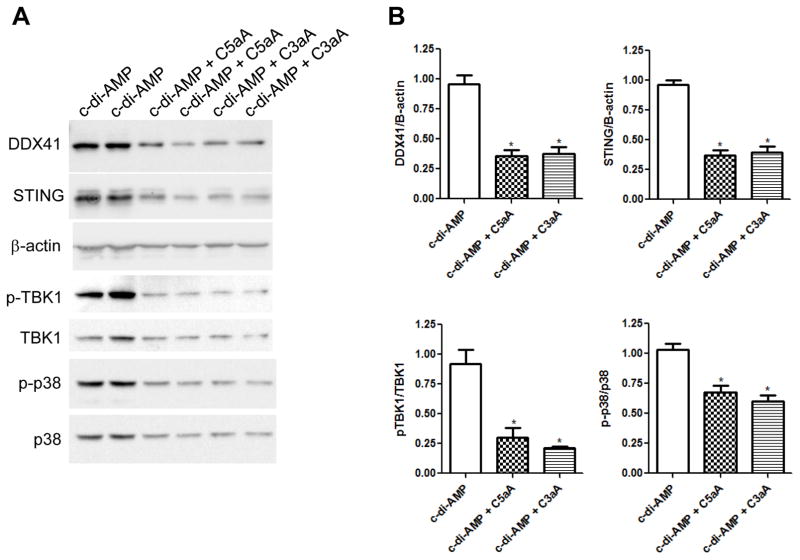
The induction of type I interferons by cytosolic DNA is dependent on a signaling pathway that consists of the helicase DDX41 acting as a sensor of intracellular DNA, forming a complex with the adaptor STING, which then signals to TBK1 and p38 MAPK to activate the type I interferon response (35). To determine if C5a and C3a suppress IFN-β production by reducing the expression of any of these proteins in this signaling pathway, BMDCs were pre-treated with media, C5aA, or C3aA for 2 h and then treated with c-di-AMP for 20 h. BMDCs pre-treated with C5aA or C3aA and then incubated with c-di-AMP showed significantly reduced expression of DDX41, STING, pTBK1/TBK1, and p-p38/p38 MAPK compared to BMDCs pre-treated with media and then c-di-AMP (Fig. 4A and 4B). These data demonstrate that C3a and C5a suppress IFN-β expression by modulation of the DDX41/STING innate cytosolic surveillance pathway involving the downstream targets TBK1 and p38 MAPK.
Figure 4. C5aA and C3aA decrease expression of IFN-β signaling proteins.
(A) Immunoblot analysis of DDX41, STING, β-actin, phosphorylated and total TBK1, and phosphorylated and total p38 MAPK in WT BMDCs that were pre-treated with either media, 100 nM C5aA, or 100 nM C3aA for 2 h followed by incubation with 25 μg/ml c-di-AMP for 20 h. These experiments were performed twice in triplicate, and two of the three cell lysates per treatment group were loaded on the blot. (B) Quantitation of the blots shown in (A) normalized to β-actin for DDX41 and STING (same blots) or the phosphorylated proteins were normalized to the respective total protein (same blots). The data is presented as mean ± SEM. Data are combined from two independent experiments. * P < 0.05 by t test.
BTK, TBK1, and p38 MAPK are necessary for C5aA/C3aA suppression of IFN-β
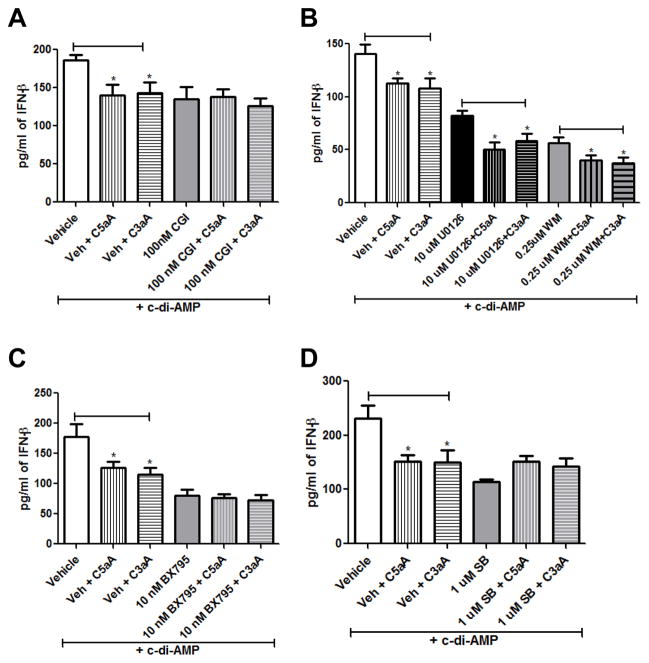
Finally, we wanted to confirm the involvement of TBK1 and p38 MAPK and to determine which additional kinase pathways are important for the ability of C5a and C3a to suppress IFN-β production. Lee et al. (19) demonstrated previously that BTK activates the binding of DDX41 to dsDNA and to STING to initiate IFN-β production. Zhang et al. (35) also demonstrated that Erk1/2, p38 MAPK, and TBK1 became phosphorylated when BMDCs were stimulated with dsDNA, suggesting these kinases are important in the type I interferon response. We also examined the importance of the PI3K/Akt pathway since activation of this pathway has been shown to occur when C5a binds to the C5aR1 and C3a binds to the C3aR in a variety of cell types including dendritic cells (36), T cells (37), mesenchymal stem cells (38), and cancer cells (39). To determine if these kinases are involved in the ability of C5a/C3a to suppress IFN-β production, we used CGI-1746 to inhibit BTK, Wortmannin to inhibit PI3K/Akt, U0126 to inhibit Erk, BX795 to inhibit TBK1, and SB202190 to inhibit p38 MAPK. WT BMDCs were pre-treated with either vehicle or inhibitor for one hour prior to treatment with media or 100 nM C5aA or C3aA for two hours and then c-di-AMP overnight. As shown in Fig. 5A, pre-incubation of the BMDCs in the BTK inhibitor blocked the ability of C5aA and C3aA to suppress IFN-β production, in contrast to cells pre-incubated in vehicle where C5aA and C3aA were able to suppress IFN-β by 30% (Fig. 5A), demonstrating the importance of BTK in C5a/C3a suppression of the type I interferon response. Pre-incubation of the BMDCs in Wortmannin or U0126, however, did not block the ability of C5aA or C3aA to suppress IFN-β production (Fig. 5B), indicating that C5a and C3a do not act through the PI3K/Akt or Erk pathways to inhibit type I interferon. Next, we wanted to confirm that C5a and C3a suppress IFN-β production by acting through the TBK1 signaling pathway. As shown in Fig. 5C, BMDCs treated with vehicle and then C5aA or C3aA produced significantly less IFN-β in response to c-di-AMP than BMDCs treated with vehicle alone. However, BMDCs treated with BX795 and then C5aA or C3aA produced the same amount of IFN-β in response to c-di-AMP as BMDCs treated with BX795 alone, confirming that C5a and C3a act through the TBK1 signaling pathway to suppress IFN-β production. Finally, we wanted to determine the importance of p38 MAPK in the ability of C5a and C3a to suppress IFN-β production. As shown in Fig. 5D, BMDCs treated with vehicle and then C5aA or C3aA, produced significantly less IFN-β in response to c-di-AMP than BMDCs treated with vehicle alone. However, BMDCs treated with SB202190 and then C5aA or C3aA produced the same amount of IFN-β in response to c-di-AMP as BMDCs treated with SB202190 alone, indicating that p38 MAPK is important in the ability of C5a/C3a to suppress IFN-β production. In conclusion, these data demonstrate that C5a and C3a suppress type I interferon production in response to c-di-AMP in BMDCs by decreasing DDX41 and STING expression, which leads to reduced phosphorylation of TBK1 and p38 MAPK, and the ability of C5a and C3a to suppress IFN-β production is dependent on BTK, TBK1, and p38 MAPK activity.
Figure 5. BTK, TBK1, and p38 MAPK are necessary for C5aA and C3aA to suppress IFN-β production.
WT BMDCs were pre-treated with either vehicle, (A) 100 nM CGI-1746 to inhibit BTK, (B) 10 μM U0126 to inhibit Erk, 0.25 μM Wortmannin to inhibit PI3K/Akt, (C) 10 nM BX795 to inhibit TBK1, or (D) 1 μM SB202190 to inhibit p38 MAPK. After 1 h, the cells were treated with either media, 100 nM C5aA, or 100 nM C3aA for 2 h followed by incubation with 25 μg/ml c-di-AMP for 20 h. IFN-β was quantitated from cell-free supernatants. All data are presented as mean pg/ml ± SEM. These data are pooled from three independent experiments. * P < 0.05 by t test.
Discussion
The results of this study show for the first time a critical role for C3a/C3aR and C5a/C5aR1 in regulating IFN-β production in response to Lm and c-di-AMP in both a mouse macrophage-like cell line and in primary mouse BMDCs. Pre-treating J774A.1 cells and WT BMDCs with C5a or C3a resulted in a significant reduction in Lm and c-di-AMP-induced IFN-β production compared to cells that were pre-treated with media (Figs 1 and 2). This IFN-β suppression was specific as C5a and C3a had no significant effect on IL-6, TNF, or MCP-1 production from Lm-infected J774 cells (Fig. 1B) or BMDCs (Fig. 2B). C3a and C5a act through their receptors, C3aR and C5aR1, respectively, because no inhibition was observed in C5aR1−/− or C3aR−/− BMDCs pre-treated with C5a or C3a, respectively (Fig. 2D–E). The ability of C3a and C5a to suppress Ifnb message occurs as early as 4 h after incubation with Lm or c-di-AMP (Fig. 3). Pre-treatment of WT BMDCs with C3aA or C5aA reduced the expression of DDX41, STING, pTBK1, and p-p38 MAPK following incubation with c-di-AMP compared to pre-treatment with media (Fig. 4). C5aA and C3aA were still able to suppress c-di-AMP-induced IFN-β production in the presence of Akt and Erk inhibitors (Fig. 5B), indicating that C5a and C3a do not suppress IFN-β through these pathways. However, C5aA and C3aA were not able to suppress c-di-AMP-induced IFN-β production in the presence of inhibitors for BTK, p38 MAPK, or TBK1, demonstrating the importance of these kinases in this inhibition (Fig. 5).
DDX41 and STING are both necessary for IFN-β production in response to live Lm and c-di-AMP (20–22, 26, 27). DDX41 is thought to be the primary sensor of CDNs, with STING acting as a secondary sensor. Immunofluorescence studies showed that c-di-GMP and DDX41 co-localized about 34% whereas c-di-GMP and STING co-localized about 6%, and c-di-GMP had a much greater binding affinity for DDX41 compared to STING (20). In studies using DDX41-specific shRNA and STING-specific shRNA, DDX41 was necessary for STING to bind c-di-GMP, but STING was not necessary for DDX41 to bind c-di-GMP (20). Immunoprecipitation studies showed that DDX41 and STING interact and localize together, and c-di-AMP and c-di-GMP enhanced the formation of this complex (20). STING then binds to TBK1 to activate the type I interferon response (21). In dendritic cells treated with DDX41-specific shRNA, c-di-AMP- and c-di-GMP-induced activation of STING and TBK1 was dramatically suppressed (20), indicating that DDX41 is upstream of STING and TBK1 and DDX41 expression affects expression of STING and TBK1. Our data in this paper shows that C3a and C5a suppress expression of DDX41, STING, and TBK1 (Fig. 4); therefore, it is likely that C3a and C5a decrease the expression of DDX41 which in turn leads to decreased expression of STING and TBK1.
We found multiple kinases to be involved in the ability of C5aA and C3aA to suppress IFN-β production from both BMDCs (Fig. 5) and from J774A.1 cells (data not shown). Mutations in the Btk gene are responsible for an immune disorder called X-linked agammaglobulinemia (XLA), which is characterized by a lack of circulating B cells as well as an absence of all immunoglobulin classes. Monocytes and macrophages from XLA patients have a defective cytokine response when stimulated with LPS, and LPS has been shown to activate BTK (40). PBMCs from XLA patients also showed decreased phosphorylation of p38 MAPK in response to LPS compared to PBMCs from healthy controls (40). In addition, human and murine B cells treated with the BTK inhibitor, CGI-1746, showed reduced BCR-induced phosphorylation of p38 MAPK (41). These data indicate a necessary role for BTK upstream of p38 MAPK activation. BTK also activates the binding of DDX41 to dsDNA and to STING to initiate the type I interferon response (19). BTK−/− mouse cells produced significantly less Ifnb message compared to WT cells when stimulated with cytosolic DNA or Lm, and these cells also had decreased phosphorylation of TBK1 (19). Mouse dendritic cells in which DDX41 or STING had been knocked down had very low levels of phosphorylated p38 MAPK and TBK1 in response to cytosolic DNA compared to normal cells (35), indicating that DDX41 and STING are required for activation of these MAPKs. In our studies we were able to show that inhibition of BTK kinase activity abrogates the ability of C5a and C3a to suppress IFN-β, which proves that BTK is necessary for C5a and C3a to inhibit IFN-β production.
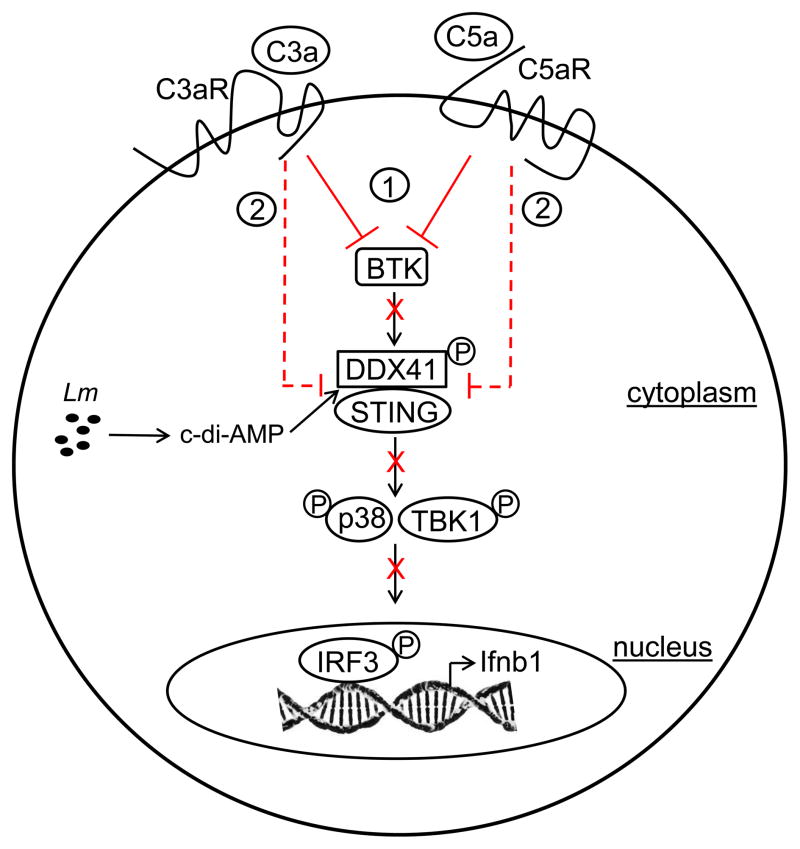
In summary, our findings show for the first time that C3a and C5a on binding and signaling through their respective receptors (C3aR and C5aR1) suppress IFN-β production in Lm-infected cells via modulation of the cyclic dinucleotide-activated cytosolic surveillance pathway involving BTK, DDX41, STING and downstream targets TBK1 and p38 MAPK. Inhibition and expression studies indicated that the impact of C3aR and C5aR1 signaling occurs early in this innate surveillance pathway by 1) inhibiting BTK-mediated activation of DDX41, which is necessary for STING recruitment and CDN-binding, and 2) reduction of DDX41 and STING protein expression, leading to reduced phosphorylation of TBK1 and p38 MAPK. A mechanistic model illustrating how C3a and C5a protect host cells against the detrimental effects of type I interferon expression during Lm infection is depicted in Fig. 6.
Figure 6. Schematic model depicting the C3a- and C5a-mediated suppression of the innate CDN-activated cytosolic surveillance pathway in Lm-infected cells.
During infection Lm- derived CDNs (c-di-AMP) are recognized by the host cell cytosolic pathway by binding to BTK-activated DDX41, which in turn recruits STING. TBK1 and p38 are subsequently recruited and phosphorylated, facilitating activation of IRF3-mediated transcription of Ifnb1. From the data presented here, C3a and C5a, via signaling on binding their respective receptors (C3aR and C5aR1), suppress IFN-β production at two different stages of this innate CDN-activated cytosolic surveillance pathway (shown in red) --- (1) by inhibiting BTK-mediated activation of DDX41 and (2) by reduction of DDX41 and STING protein expression.
Supplementary Material
Acknowledgments
This work was supported by a National Institutes of Health Public Service Grant RO1 AI025011 (to RAW). Support was also provided by the Hans J. Muller-Eberhard and Irma Gigli Distinguished Chair in Immunology.
Abbreviations
- BMDCs
bone marrow-derived dendritic cells
- BTK
Bruton’s Tyrosine Kinase
- C5aR1
C5a receptor 1
- C3aR
C3a receptor
- CDN
cyclic dinucleotide
- DDX41
DEAD-Box Helicase 41
- Lm
Listeria monocytogenes
- MOI
multiplicity of infection
- STING
Stimulator of Interferon Genes
- TBK1
TANK-Binding Kinase 1
References
- 1.Kourtis AP, Read JS, Jamieson DJ. Pregnancy and infection. N Engl J Med. 2014;370:2211–2218. doi: 10.1056/NEJMra1213566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamont RF, Sobel J, Mazaki-Tovi S, Kusanovic JP, Vaisbuch E, Kim SK, Uldbjerg N, Romero R. Listeriosis in human pregnancy: a systematic review. J Perinat Med. 2011;39:227–236. doi: 10.1515/JPM.2011.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maertens de Noordhout C, Devleesschauwer B, Angulo FJ, Verbeke G, Haagsma J, Kirk M, Havelaar A, Speybroeck N. The global burden of listeriosis: a systematic review and meta-analysis. Lancet Infect Dis. 2014;14:1073–1082. doi: 10.1016/S1473-3099(14)70870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wetsel RA, Kildsgaard J, Haviland DL. Complement anaphylatoxins (C3a, C4a, C5a) and their receptors (C3aR, C5aR/CD88) as therapeutic targets in inflammation. In: Lambris JD, Holers VM, editors. Contemporary Immunology: Therapeutic Interventions in the Complement System. Humana Press; Totowa, NJ: 2000. pp. 113–154. [Google Scholar]
- 5.Klos A, Tenner AJ, Johswich KO, Ager RR, Reis ES, Kohl J. The role of the anaphylatoxins in health and disease. Mol Immunol. 2009;46:2753–2766. doi: 10.1016/j.molimm.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mueller-Ortiz SL, Morales JE, Wetsel RA. The receptor for the complement C3a anaphylatoxin (C3aR) provides host protection against Listeria monocytogenes-induced apoptosis. J Immunol. 2014;193:1278–1289. doi: 10.4049/jimmunol.1302787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calame DG, Mueller-Ortiz SL, Morales JE, Wetsel RA. The C5a anaphylatoxin receptor (C5aR1) protects against Listeria monocytogenes infection by inhibiting type 1 IFN expression. J Immunol. 2014;193:5099–5107. doi: 10.4049/jimmunol.1401750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Auerbuch V, Brockstedt DG, Meyer-Morse N, O’Riordan M, Portnoy DA. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J Exp Med. 2004;200:527–533. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrero JA, Calderon B, Unanue ER. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J Exp Med. 2004;200:535–540. doi: 10.1084/jem.20040769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, Zarnegar B, Perry AK, Nguyen BO, Lane TF, Taniguchi T, Miller JF, Cheng G. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J Exp Med. 2004;200:437–445. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stockinger S, Materna T, Stoiber D, Bayr L, Steinborn R, Kolbe T, Unger H, Chakraborty T, Levy DE, Muller M, Decker T. Production of type I IFN sensitizes macrophages to cell death induced by Listeria monocytogenes. J Immunol. 2002;169:6522–6529. doi: 10.4049/jimmunol.169.11.6522. [DOI] [PubMed] [Google Scholar]
- 12.Buchmeier NA, Schreiber RD. Requirement of endogenous interferon-gamma production for resolution of Listeria monocytogenes infection. Proc Natl Acad Sci USA. 1985;82:7404–7408. doi: 10.1073/pnas.82.21.7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harty JT, Bevan MJ. Specific immunity to Listeria monocytogenes in the absence of IFN gamma. Immunity. 1995;3:109–117. doi: 10.1016/1074-7613(95)90163-9. [DOI] [PubMed] [Google Scholar]
- 14.Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, Vilcek J, Zinkernagel RM, Aguet M. Immune response in mice that lack the interferon-gamma receptor. Science. 1993;259:1742–1745. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- 15.Rayamajhi M, Humann J, Penheiter K, Andreasen K, Lenz LL. Induction of IFN-alphabeta enables Listeria monocytogenes to suppress macrophage activation by IFN-gamma. J Exp Med. 2010;207:327–337. doi: 10.1084/jem.20091746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCaffrey RL, Fawcett P, O’Riordan M, Lee KD, Havell EA, Brown PO, Portnoy DA. A specific gene expression program triggered by Gram-positive bacteria in the cytosol. Proc Natl Acad Sci USA. 2004;101:11386–11391. doi: 10.1073/pnas.0403215101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Connell RM, Vaidya SA, Perry AK, Saha SK, Dempsey PW, Cheng G. Immune activation of type I IFNs by Listeria monocytogenes occurs independently of TLR4, TLR2, and receptor interacting protein 2 but involves TNFR-associated NF kappa B kinase-binding kinase 1. J Immunol. 2005;174:1602–1607. doi: 10.4049/jimmunol.174.3.1602. [DOI] [PubMed] [Google Scholar]
- 18.Stockinger S, Reutterer B, Schaljo B, Schellack C, Brunner S, Materna T, Yamamoto M, Akira S, Taniguchi T, Murray PJ, Muller M, Decker T. IFN regulatory factor 3-dependent induction of type I IFNs by intracellular bacteria is mediated by a TLR- and Nod2-independent mechanism. J Immunol. 2004;173:7416–7425. doi: 10.4049/jimmunol.173.12.7416. [DOI] [PubMed] [Google Scholar]
- 19.Lee KG, Kim SS, Kui L, Voon DC, Mauduit M, Bist P, Bi X, Pereira NA, Liu C, Sukumaran B, Renia L, Ito Y, Lam KP. Bruton’s tyrosine kinase phosphorylates DDX41 and activates its binding of dsDNA and STING to initiate type 1 interferon response. Cell Reports. 2015;10:1055–1065. doi: 10.1016/j.celrep.2015.01.039. [DOI] [PubMed] [Google Scholar]
- 20.Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, Liu ZJ, Modlin RL, Liu YJ, Cheng G. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol. 2012;13:1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Riordan M, Yi CH, Gonzales R, Lee KD, Portnoy DA. Innate recognition of bacteria by a macrophage cytosolic surveillance pathway. Proc Natl Acad Sci USA. 2002;99:13861–13866. doi: 10.1073/pnas.202476699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen K, Prabakaran T, Laustsen A, Jorgensen SE, Rahbaek SH, Jensen SB, Nielsen R, Leber JH, Decker T, Horan KA, Jakobsen MR, Paludan SR. Listeria monocytogenes induces IFNbeta expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J. 2014;33:1654–1666. doi: 10.15252/embj.201488029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baranyi L, Campbell W, Okada H. Antisense homology boxes in C5a receptor and C5a anaphylatoxin: a new method for identification of potentially active peptides. J Immunol. 1996;157:4591–4601. [PubMed] [Google Scholar]
- 30.Bellows-Peterson ML, Fung HK, Floudas CA, Kieslich CA, Zhang L, Morikis D, Wareham KJ, Monk PN, Hawksworth OA, Woodruff TM. De novo peptide design with C3a receptor agonist and antagonist activities: theoretical predictions and experimental validation. J Med Chem. 2012;55:4159–4168. doi: 10.1021/jm201609k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kildsgaard J, Hollmann TJ, Matthews KW, Bian K, Murad F, Wetsel RA. Cutting edge: targeted disruption of the C3a receptor gene demonstrates a novel protective anti-inflammatory role for C3a in endotoxin-shock. J Immunol. 2000;165:5406–5409. doi: 10.4049/jimmunol.165.10.5406. [DOI] [PubMed] [Google Scholar]
- 32.Hollmann TJ, Mueller-Ortiz SL, Braun MC, Wetsel RA. Disruption of the C5a receptor gene increases resistance to acute Gram-negative bacteremia and endotoxic shock: opposing roles of C3a and C5a. Mol Immunol. 2008;45:1907–1915. doi: 10.1016/j.molimm.2007.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dresing P, Borkens S, Kocur M, Kropp S, Scheu S. A fluorescence reporter model defines “Tip-DCs” as the cellular source of interferon beta in murine listeriosis. PloS One. 2010;5:e15567. doi: 10.1371/journal.pone.0015567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stockinger S, Kastner R, Kernbauer E, Pilz A, Westermayer S, Reutterer B, Soulat D, Stengl G, Vogl C, Frenz T, Waibler Z, Taniguchi T, Rulicke T, Kalinke U, Muller M, Decker T. Characterization of the interferon-producing cell in mice infected with Listeria monocytogenes. PLoS Path. 2009;5:e1000355. doi: 10.1371/journal.ppat.1000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li K, Fazekasova H, Wang N, Peng Q, Sacks SH, Lombardi G, Zhou W. Functional modulation of human monocytes derived DCs by anaphylatoxins C3a and C5a. Immunobiology. 2012;217:65–73. doi: 10.1016/j.imbio.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strainic MG, Liu J, Huang D, An F, Lalli PN, Muqim N, Shapiro VS, Dubyak GR, Heeger PS, Medof ME. Locally produced complement fragments C5a and C3a provide both costimulatory and survival signals to naive CD4+ T cells. Immunity. 2008;28:425–435. doi: 10.1016/j.immuni.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schraufstatter IU, Discipio RG, Zhao M, Khaldoyanidi SK. C3a and C5a are chemotactic factors for human mesenchymal stem cells, which cause prolonged ERK1/2 phosphorylation. J Immunol. 2009;182:3827–3836. doi: 10.4049/jimmunol.0803055. [DOI] [PubMed] [Google Scholar]
- 39.Cho MS, Vasquez HG, Rupaimoole R, Pradeep S, Wu S, Zand B, Han HD, Rodriguez-Aguayo C, Bottsford-Miller J, Huang J, Miyake T, Choi HJ, Dalton HJ, Ivan C, Baggerly K, Lopez-Berestein G, Sood AK, Afshar-Kharghan V. Autocrine effects of tumor-derived complement. Cell Reports. 2014;6:1085–1095. doi: 10.1016/j.celrep.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horwood NJ, Mahon T, McDaid JP, Campbell J, Mano H, Brennan FM, Webster D, Foxwell BM. Bruton’s tyrosine kinase is required for lipopolysaccharide-induced tumor necrosis factor alpha production. J Exp Med. 2003;197:1603–1611. doi: 10.1084/jem.20021845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Paolo JA, Huang T, Balazs M, Barbosa J, Barck KH, Bravo BJ, Carano RA, Darrow J, Davies DR, DeForge LE, Diehl L, Ferrando R, Gallion SL, Giannetti AM, Gribling P, Hurez V, Hymowitz SG, Jones R, Kropf JE, Lee WP, Maciejewski PM, Mitchell SA, Rong H, Staker BL, Whitney JA, Yeh S, Young WB, Yu C, Zhang J, Reif K, Currie KS. Specific Btk inhibition suppresses B cell- and myeloid cell-mediated arthritis. Nat Chem Biol. 2011;7:41–50. doi: 10.1038/nchembio.481. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.