Abstract
Background
Oral squamous cell carcinoma (OSCC), which is the most common head and neck cancer, accounts for 1%–2% of all human malignancies and is characterized by poor prognosis and reduced survival rates. WNT1-inducible signaling pathway protein 1 (WISP1), a cysteine-rich protein belonging to the Cyr61, CTGF, Nov (CCN) family of matricellular proteins, has many developmental functions and may be involved in carcinogenesis. This study investigated WISP1 single-nucleotide polymorphisms (SNPs) to elucidate OSCC susceptibility and clinicopathologic characteristics.
Methodology/Principal findings
Real-time polymerase chain reaction was used to analyze 6 SNPs of WISP1 in 900 OSCC patients and 1200 cancer-free controls. The results showed that WISP1 rs2929970 polymorphism carriers with at least one G allele were susceptible to OSCC. Moreover, compared with smokers, non-smoker patients with higher frequencies of WISP1 rs2929970 (AG + GG) variants had a late stage (stages III and IV) and a large tumor size. In addition, OSCC patients who were betel quid chewers and carried WISP1 rs16893344 (CT + TT) variants had a low risk of lymph node metastasis.
Conclusion
Our results demonstrate that a joint effect of WISP1 rs2929970 with smoking as well as WISP1 rs16893344 with betel nut chewing causally contributes to the occurrence of OSCC. WISP1 polymorphism may serve as a marker or a therapeutic target in OSCC.
Introduction
Oral squamous cell carcinoma (OSCC), which is the most common head and neck cancer, accounts for 1%–2% of all human malignancies. OSCC accounts for more than 90% of oral malignancies [1]. OSCC is characterized by poor prognosis and reduced survival rates [2]. Despite improved imaging techniques and advances in surgery, chemotherapy, and radiation, the prognosis and mortality of OSCC remain poor, with a 5-year survival rate of approximately 50% [3–6].
The Cyr61, CTGF, Nov (CCN) family is a group of 6 secreted proteins that regulates adhesion and migration or functions as growth factors that modulate cell proliferation and differentiation [7]. WNT1-inducible signaling pathway protein 1 (WISP1) is a cysteine-rich protein belonging to the CCN family of matricellular proteins, and WISP1 has many developmental functions [8, 9]. Increasing evidence suggests that WISP1 is involved in carcinogenesis [10]. Research on colon cancer revealed that high WISP1 expression is associated with apoptosis, invasion, and poor prognosis [11]. In esophageal squamous cell carcinoma, WISP1 has been found to be an independent prognostic factor for poor overall survival and has been confirmed to mediate resistance to radiotherapy [12, 13]. In OSCC, WISP1 expression is regulated by methylation, and WISP1 hypomethylation contributes to lymph node (LN) metastasis [14]. Moreover, a previous study indicated that WISP1 enhances the migration of OSCC cells by increasing intercellular adhesion molecule-1 (ICAM-1) expression through the αvβ3 integrin receptor and apoptosis signal-regulating kinase 1 (ASK1), c-Jun N-terminal kinase (JNK)/p38, and activator protein-1 (AP-1) signal transduction pathways [15]. WISP1 may also promote OSCC angiogenesis through vascular endothelial growth factor (VEGF)-A expression [16]. Thus, WISP1 can act as an oncogene and may be a promising therapeutic target in OSCC.
Single-nucleotide polymorphism (SNP) is the most common genetic variant in DNA expression, and the expression of a specific gene may be affected or regulated by its genetic variations [17–21]. Previous data have demonstrated the possible role of WISP1 SNPs in various cancers or diseases [22–26]. For example, in postmenopausal Japanese women, the rs2929970 SNP in the WISP1 3′-UTR region was suggested to be associated with spinal osteoarthritis [24]. WISP1 rs2929970 was also suggested to be correlated with hypertension in men, and men carrying the G allele of rs2929970 had higher blood pressure [26]. Moreover, WISP1 rs2929973 was associated with lung function in asthmatic children [25]. In lung cancer, a series of WISP1 genetic polymorphisms, such as rs16893344, rs2977530, rs2977537, rs62514004, rs11778573, rs2977536, and rs2977549, were significantly associated with lung cancer susceptibility or the chemotherapy response [22]. However, little is known about the effects of WISP1 gene variants on the predisposition to OSCC. Therefore, we hypothesized that polymorphisms of the WISP1 gene may play an essential role in OSCC. In this study, we aimed to investigate the WISP1 SNPs of rs62514004, rs16893344, rs2977530, rs2977537, rs2929970 and rs2929973, which are located in the promoter or the 3′-UTR region, to elucidate their contribution to OSCC susceptibility and clinicopathologic characteristics.
Materials and methods
Subjects and specimen collection
We enrolled 900 male patients with OSCC from Chung Shan Medical University Hospital in Taichung and Changhua Christian Hospital in Changhua, Taiwan, from 2007 to 2016. For the control group, we selected 1200 healthy male individuals with no self-reported history of cancer at any site from Taiwan Biobank. Each subject completed a questionnaire on demographic characteristics, betel quid chewing, tobacco use, and alcohol consumption, and medical histories. The Institutional Review Board of Chung Shan Medical University Hospital approved this study (CSMUH No: CS13214-1), and informed written consent was obtained from each participant.
Selection of WISP1 polymorphisms
A total of six SNPs in WISP1 were selected from the International HapMap Project data for this study. The previous studies have reported the effect of WISP1 genetic polymorphisms on human cancer susceptibility [22, 23, 27]. Thus, we included the SNP rs62514004, rs2929970 and rs2929973 in the promoter region and 3' untranslated region, respectively. Three SNPs (rs16893344, rs2977530 and rs2977537) which locate in introns of WISP1 were selected in this study since these SNPs were suggested to be significantly associated with lung cancer susceptibility or chemotherapy response in Chinese population [22].
DNA extraction and WISP1 genotyping
Genomic DNA from OSCC group was isolated from peripheral blood using the QIAamp DNA blood mini kit and used as the template for polymerase chain reaction as described previously [28, 29]. Allelic discrimination of the rs62514004, rs16893344, rs2977530, rs2977537, rs2929970 and rs2929973 of WISP1 gene was assessed with the ABI StepOne Real-Time PCR System (Applied Biosystems, Foster City, CA, USA), and analyzed with SDS vers. 3.0 software using the TaqMan assay.
Bioinformatics analysis
The stem-loop portion of the miRNA-miRNA duplex structure of pre-miRNAs was identified by miRNA target prediction using the MicroRNA.org resource. Models of the miRNA-target duplex were determined using the RNAhybrid web tool on Bielefeld Bioinformatics Server. Boxplot chart showing the differential expressions of miR-99a-5p in 420 OSCC patients and 43 normal controls, as taken from the Pan-Cancer dataset [30].
Statistical analysis
The Mann-Whitney U test was used to compare differences in the distribution of age and demographic characteristics between the controls and OSCC patients. The common haplotypes were estimated by PHASE version 2.1. ORs with 95% CIs were estimated using logistic regression models. AORs with 95% CIs were used to assess association between genotype frequencies with OSCC risk and clinical factors. p values less than 0.05 were considered significant. The data were analysed with SPSS 12.0 statistical software (SPSS Inc., Chicago, IL, USA).
Results
The statistical analysis of demographic characteristics is shown in Table 1. We analyzed the demographic characteristics of sample specimens and observed that 16.6% (199/1200) of controls and 78.6% (707/900) of OSCC patients had the habit of betel quid chewing. Moreover, 53.0% (636/1200) of controls and 88.9% (800/900) of OSCC patients were smokers. In addition, 19.7% (237/1200) of controls and 53.0% (480/900) of OSCC patients consumed alcohol. Significant distributional differences were observed in betel quid chewing (p < 0.001), cigarette smoking (p < 0.001), and alcohol consumption (p < 0.001) between controls and OSCC patients.
Table 1. The distributions of demographical characteristics in 1200 controls and 900 male patients with oral cancer.
| Variable | Controls (N = 1200) | Patients (N = 900) | p value |
|---|---|---|---|
| Age (yrs) | 53.91 ± 10.02 | 55.08 ± 11.09 | |
| <55 | 566 (47.2%) | 440 (48.9%) | p = 0.434 |
| ≥ 55 | 634 (52.8%) | 460 (51.1%) | |
| Betel quid chewing | |||
| No | 1001 (83.4%) | 193 (21.4%) | |
| Yes | 199 (16.6%) | 707 (78.6%) | p <0.001* |
| Cigarette smoking | |||
| No | 564 (47.0%) | 100 (11.1%) | |
| Yes | 636 (53.0%) | 800 (88.9%) | p <0.001* |
| Alcohol drinking | |||
| No | 963 (80.3%) | 420 (46.7%) | |
| Yes | 237 (19.7%) | 480 (53.3%) | p <0.001* |
| Stage | |||
| I+II | 444 (49.3%) | ||
| III+IV | 456 (50.7%) | ||
| Tumor T status | |||
| T1+T2 | 519 (57.7%) | ||
| T3+T4 | 381 (42.3%) | ||
| Lymph node status | |||
| N0 | 606 (67.3%) | ||
| N1+N2+N3 | 294 (32.7%) | ||
| Metastasis | |||
| M0 | 890 (98.9%) | ||
| M1 | 10 (1.1%) | ||
| Cell differentiation | |||
| Well differentiated | 127 (14.1%) | ||
| Moderately or poorly differentiated | 773 (85.9%) |
Mann-Whitney U test was used between healthy controls and patients with oral cancer.
* p value < 0.05 as statistically significant.
The genotype distributions and associations between OSCC and the WISP1 genetic polymorphisms are shown in Table 2. The WISP1 rs62514004, rs16893344, rs2977530, rs2977537, rs2929970 and rs2929973 genetic polymorphisms exhibited the highest distribution frequency in both control and OSCC patients homozygous for AA, homozygous for CC, heterozygous for AG, heterozygous for GA, heterozygous for AG and heterozygous for TG, respectively. In these controls, the frequencies of WISP1 rs62514004, rs16893344, rs2977530, rs2977537, rs2929970 and rs2929973 were in the Hardy-Weinberg equilibrium (p = 0.442, χ2 value: 0.590; p = 0.957, χ2 value: 0.003; p = 0.132, χ2 value: 2.274; p = 0.290, χ2 value: 1.119; p = 0.490, χ2 value: 0.476; and p = 0.314, χ2 value: 1.014, respectively). After adjustment for several variables, no significant differences were observed between OSCC patients and control group in the rs62514004, rs16893344, rs2977530, rs2977537 and rs2929973 polymorphisms of the WISP1 gene. However, patients with WISP1 polymorphic rs2929970 GG genotypes exhibited a higher risk of OSCC than the corresponding WT homozygous patients (OR [odds ratios] = 1.463, 95% confidence interval [CI] = 1.045–2.048, p = 0.027). Moreover, a similar result was observed in patients with WISP1 polymorphic rs2929970 AG + GG genotypes (OR = 1.298, 95% CI = 1.026–1.642, p = 0.030).
Table 2. Odds ratio (OR) and 95% confidence interval (CI) of oral cancer associated with WISP1 genotypic frequencies.
| Variable | Controls (N = 1200) n (%) | Patients (N = 900) n (%) | OR (95% CI) | AOR (95% CI) |
|---|---|---|---|---|
| rs62514004 | ||||
| AA | 926 (77.2%) | 707 (78.6%) | 1.00 | 1.00 |
| AG | 253 (21.1%) | 180 (20.0%) | 0.932 (0.752–1.155) | 0.956 (0.726–1.259) |
| GG | 21 (1.7%) | 13 (1.4%) | 0.811 (0.403–1.630) | 0.682 (0.276–1.687) |
| AG+GG | 274 (22.8%) | 193 (21.4%) | 0.923 (0.749–1.137) | 0.943 (0.714–1.211) |
| rs16893344 | ||||
| CC | 877 (73.1%) | 662 (73.6%) | 1.00 | 1.00 |
| CT | 298 (24.8%) | 222 (24.7%) | 0.987 (0.807–1.207) | 0.956 (0.737–1.239) |
| TT | 25 (2.1%) | 16 (1.8%) | 0.848 (0.449–1.601) | 0.795 (0.349–1.812) |
| CT+TT | 323 (26.9%) | 238 (26.4%) | 0.976 (0.803–1.187) | 0.943 (0.733–1.214) |
| rs2977530 | ||||
| AA | 337 (28.1%) | 263 (29.2%) | 1.00 | 1.00 |
| AG | 573 (47.8%) | 435 (48.3%) | 0.973 (0.793–1.193) | 0.840 (0.645–1.093) |
| GG | 290 (24.1%) | 202 (22.5%) | 0.893 (0.701–1.136) | 0.810 (0.592–1.108) |
| AG+GG | 863 (71.9%) | 637 (70.8%) | 0.946 (0.781–1.145) | 0.830 (0.648–1.063) |
| rs2977537 | ||||
| GG | 330 (27.5%) | 233 (25.9%) | 1.00 | 1.00 |
| GA | 581 (48.4%) | 435 (48.3%) | 1.060 (0.861–1.307) | 1.033 (0.789–1.353) |
| AA | 289 (24.1%) | 232 (25.8%) | 1.137 (0.894–1.447) | 1.191 (0.873–1.625) |
| GA+AA | 870 (72.5%) | 667 (74.1%) | 1.086 (0.893–1.321) | 1.085 (0.842–1.397) |
| rs2929970 | ||||
| AA | 439 (36.6%) | 302 (33.6%) | 1.00 | 1.00 |
| AG | 583 (48.6%) | 443 (49.2%) | 1.105 (0.912–1.338) | 1.248 (0.973–1.600) |
| GG | 178 (14.8%) | 155 (17.2%) | 1.266 (0.976–1.642) | 1.463 (1.045–2.048)* |
| AG+GG | 761 (63.4%) | 598 (66.4%) | 1.142 (0.953–1.370) | 1.298 (1.026–1.642)* |
| rs2929973 | ||||
| TT | 503 (41.9%) | 395 (43.9%) | 1.00 | 1.00 |
| TG | 560 (46.7%) | 391 (43.4%) | 0.889 (0.739–1.069) | 0.976 (0.769–1.238) |
| GG | 137 (11.4%) | 114 (12.7%) | 1.060 (0.800–1.404) | 1.133 (0.789–1.627) |
| TG+GG | 697 (58.1%) | 505 (56.1%) | 0.923 (0.775–1.099) | 1.007 (0.804–1.262) |
The odds ratio (OR) with their 95% confidence intervals were estimated by logistic regression models.
The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for betel nut chewing, alcohol and tobacco consumption.
The interactive effects between environmental risk factors and the WISP1 genetic polymorphisms are shown in Table 3. To reduce the possible interference of confounding variables, we used adjusted ORs (AORs) with 95% CIs that were estimated using multiple logistic regression models after controlling for betel nut chewing and alcohol consumption in each comparison. Among 664 non-smokers, those with WISP1 rs2977530 AG + GG genotypes exhibited a 0.598 fold-lower risk of OSCC (95% CI = 0.364–0.980, p = 0.041) (Table 3) but no difference was observed among 1436 smokers (Table A in S1 File).
Table 3. Odds ratio (OR) and 95% confidence interval (CI) of oral cancer associated with WISP1 genotypic frequencies in non-smoker.
| Variable | Controls (N = 564) n (%) | Patients (N = 100) n (%) | OR (95% CI) | AOR (95% CI) |
|---|---|---|---|---|
| rs62514004 | ||||
| AA | 444 (78.7%) | 78 (78.0%) | 1.00 | 1.00 |
| AG | 110 (19.5%) | 22 (22.0%) | 1.138 (0.679–1.909) | 1.096 (0.610–1.968) |
| GG | 10 (1.8%) | 0 (0%) | --- | --- |
| AG+GG | 120 (21.3%) | 22 (22.0%) | 1.044 (0.624–1.745) | 1.014 (0.567–1.814) |
| rs16893344 | ||||
| CC | 411 (72.9%) | 75 (75.0%) | 1.00 | 1.00 |
| CT | 141 (25.0%) | 24 (24.0%) | 0.933 (0.567–1.535) | 0.784 (0.441–1.397) |
| TT | 12 (2.1%) | 1 (1.0%) | 0.457 (0.059–3.564) | 0.172 (0.016–1.894) |
| CT+TT | 153 (27.1%) | 25 (25.0%) | 0.895 (0.549–1.460) | 0.720 (0.407–1.273) |
| rs2977530 | ||||
| AA | 165 (29.3%) | 41 (41.0%) | 1.00 | 1.00 |
| AG | 261 (46.3%) | 40 (40.0%) | 0.617 (0.383–0.994)* | 0.585 (0.340–1.006) |
| GG | 138 (24.4%) | 19 (19.0%) | 0.554 (0.307–0.999)* | 0.623 (0.325–1.197) |
| AG+GG | 399 (70.7%) | 59 (59.0%) | 0.595 (0.384–0.922)* | 0.598 (0.364–0.980)* |
| rs2977537 | ||||
| GG | 153 (27.1%) | 27 (27.0%) | 1.00 | 1.00 |
| GA | 274 (48.6%) | 41 (41.0%) | 0.848 (0.502–1.433) | 0.879 (0.489–1.578) |
| AA | 137 (24.3%) | 32 (32.0%) | 1.324 (0.755–2.321) | 1.180 (0.622–2.237) |
| GA+AA | 411 (72.9%) | 73 (73.0%) | 1.006 (0.623–1.625) | 0.982 (0.573–1.684) |
| rs2929970 | ||||
| AA | 203 (36.0%) | 31 (31.0%) | 1.00 | 1.00 |
| AG | 271 (48.0%) | 50 (50.0%) | 1.208 (0.745–1.960) | 1.127 (0.657–1.932) |
| GG | 90 (16.0%) | 19 (19.0%) | 1.382 (0.742–2.577) | 1.120 (0.550–2.284) |
| AG+GG | 361 (64.0%) | 69 (69.0%) | 1.252 (0.792–1.977) | 1.125 (0.675–1.875) |
| rs2929973 | ||||
| TT | 233 (41.3%) | 46 (46.0%) | 1.00 | 1.00 |
| TG | 263 (46.6%) | 43 (43.0%) | 0.828 (0.527–1.301) | 0.810 (0.487–1.347) |
| GG | 68 (12.1%) | 11 (11.0%) | 0.819 (0.402–1.668) | 0.695 (0.307–1.571) |
| TG+GG | 331 (58.7%) | 54 (54.0%) | 0.826 (0.539–1.267) | 0.785 (0.485–1.272) |
The odds ratio (OR) with their 95% confidence intervals were estimated by logistic regression models.
The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for betel nut chewing and alcohol.
We used Haploview software and the PHASE program to analyze the common haplotypes. As shown in Table 4, compared with the reference group G-A-T (WISP1 rs2977537/rs2929970/rs2929973), carriers with G-G-T or A-G-T had 1.857-fold (95% CI 1.374–2.510) and 2.048-fold (95% CI 1.217–3.448) significantly increased risks of OSCC (Table 4).
Table 4. Frequencies of WISP1 haplotypes in OSCC patients and control subjects.
| Haplotype block | Controls | Patients | |||
|---|---|---|---|---|---|
| rs2977537 G/A | rs2929970 A/G | rs2929973 T/G | n = 2400 | n = 1800 | OR (95% CI) |
| G | A | T | 852 (35.5%) | 599 (33.3%) | 1.000 (reference) |
| A | A | T | 604 (25.2%) | 435 (24.2%) | 1.024 (0.872–1.204) |
| A | G | G | 530 (22.1%) | 422 (23.4%) | 1.133 (0.960–1.336) |
| G | G | G | 299 (12.5%) | 184 (10.2%) | 0.875 (0.709–1.081) |
| G | G | T | 85 (3.5%) | 111 (6.2%) | 1.857 (1.374–2.510)a |
| A | G | T | 25 (1.0%) | 36 (2.0%) | 2.048 (1.217–3.448)b |
| G | A | G | 5 (0.2%) | 7 (0.4%) | 1.991 (0.629–6.304) |
| A | A | G | 0 (0.0%) | 6 (0.3%) | - |
a p< 0.001
b p = 0.007
To clarify the role of the WISP1 genetic polymorphisms in OSCC clinicopathologic statuses, such as clinical stage, tumor size, LN metastasis, distant metastasis, and cell differentiation, the distribution frequency of clinical statuses and WISP1 genotype frequencies in OSCC patients were estimated. The rs62514004, rs2977530, rs2977537 and rs2929973 genetic polymorphisms showed no significant association with the clinicopathologic statuses. However, among 707 OSCC patients who were betel quid chewers, those carrying the polymorphic rs16893344 gene had a lower risk of LN metastasis (OR = 0.674, 95% CI = 0.465–0.979, p = 0.038) than those carrying the rs16893344 WT gene, but no difference was observed in clinical stage, tumor size, distant metastasis, or cell differentiation (Table 5). However, no significant differences were observed among 193 non-betel quid chewers (Table B in S1 File). Among 100 non-smoker OSCC patients, those carrying the polymorphic rs2929970 gene had a higher risk of late-stage (OR = 2.428, 95% CI = 0.998–5.909, p = 0.048) and a larger tumor size (OR = 2.965, 95% CI = 1.129–7.789, p = 0.024) than those carrying the rs2929970 WT gene, but no difference was observed in LN metastasis and cell differentiation (Table 6). Moreover, Among 800 smoker OSCC patients, no difference was observed in stage, tumor size, LN metastasis, distant metastasis and cell differentiation (Table C in S1 File).
Table 5. Clinical statuses and WISP1 rs16893344 genotype frequencies in oral cancer among 707 betel quid chewers.
| Variable | WISP1 rs16893344 (betel quid chewers) | |||
|---|---|---|---|---|
| CC (n = 518) n (%) | CT+TT (n = 189) n (%) | OR (95% CI) | p value | |
| Clinical Stage | ||||
| Stage I/II | 251 (48.5%) | 98 (51.9%) | 1.00 | p = 0.424 |
| Stage III/IV | 267 (51.5%) | 91 (48.1%) | 0.873 (0.625–1.218) | |
| Tumor size | ||||
| ≤T2 | 292 (56.4%) | 105 (55.6%) | 1.00 | p = 0.847 |
| > T2 | 226 (43.6%) | 84 (44.4%) | 1.034 (0.739–1.445) | |
| Lymph node metastasis | ||||
| No | 341 (65.8%) | 140 (74.1%) | 1.00 | p = 0.038* |
| Yes | 177 (34.2%) | 49 (25.9%) | 0.674 (0.465–0.979) | |
| Distant metastasis | ||||
| No | 512 (98.8%) | 187 (98.9%) | 1.00 | p = 0.911 |
| Yes | 6 (1.2%) | 2 (1.1%) | 0.913 (0.183–4.561) | |
| Cell differentiation | ||||
| well | 73 (14.1%) | 32 (16.9%) | 1.00 | p = 0.348 |
| Moderate/poor | 445 (85.9%) | 157 (83.1%) | 0.805 (0.511–1.267) | |
* p<0.05
Table 6. Clinical statuses and WISP1 rs2929970 genotype frequencies in oral cancer among 100 non-smoker.
| Variable | WISP1 rs2929970 | |||
|---|---|---|---|---|
| AA (n = 31) n (%) | AG+GG (n = 69) n (%) | OR (95% CI) | p value | |
| Clinical Stage | ||||
| Stage I/II | 21 (67.7%) | 32 (46.4%) | 1.00 | p = 0.048* |
| Stage III/IV | 10 (32.3%) | 37 (53.6%) | 2.428 (0.998–5.909) | |
| Tumor size | ||||
| ≤T2 | 24 (77.4%) | 37 (53.6%) | 1.00 | p = 0.024* |
| > T2 | 7 (22.6%) | 32 (46.4%) | 2.965 (1.129–7.789) | |
| Lymph node metastasis | ||||
| No | 21 (67.7%) | 44 (63.8%) | 1.00 | p = 0.700 |
| Yes | 10 (32.3%) | 25 (36.2%) | 1.193 (0.486–2.932) | |
| Cell differentiation | ||||
| well | 4 (12.9%) | 5 (7.2%) | 1.00 | p = 0.361 |
| Moderate/poor | 27 (87.1%) | 64 (92.8%) | 1.896 (0.473–7.610) | |
* p<0.05
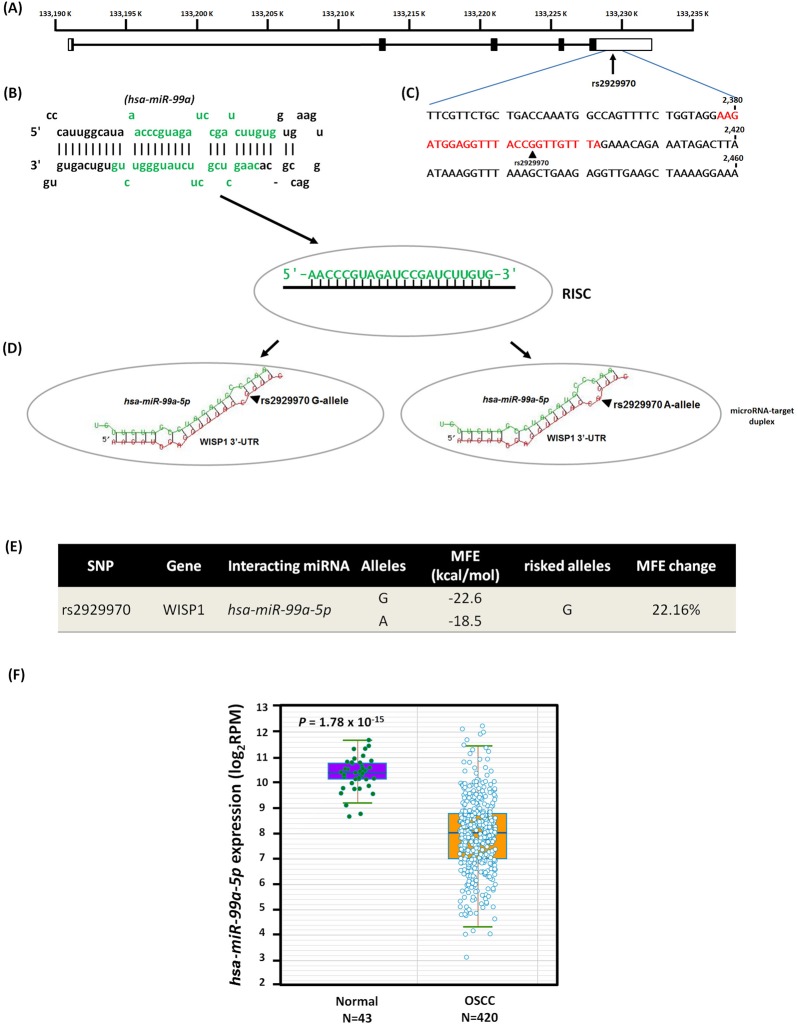
As shown in Fig 1A, the 3′-untranslated region (3′-UTR) of the WISP1 gene is 5.0 kb long and might be among the region’s most sensitive to microRNA (miRNA) epigenetic regulation (Fig 1A). The miRNA hsa-miR-99a (miRBase ID: MI0003190, Fig 1B) shares binding site complementarily with rs2929970 in the 3′-UTR region (Fig 1C and 1D). In addition, compared with the [A]-allele, the OSCC-associated risk [G]-allele creates a slight kink in the WISP1 mRNA structure, which results in a less negative free energy state and less stable hybridization [MFE (minimum free energy) changes: 22.16%, from −22.6 to −18.5 kcal/mol] (Fig 1E). Furthermore, starBase analysis revealed a significant difference in hsa-miR-99a expression in 420 OSCC patients and 43 subjects from the Pan-Cancer data set [30] (Fig 1F) (p = 1.78 × 10−15).
Fig 1. Binding site polymorphism from SNP rs2929970 [G/A] in human WISP1 3’-UTR mRNA with microRNA hsa-miR-99a-5p to decrease oral cancer susceptibility among Taiwan HNSCC population.
(A) Exons of WISP1 are shown by the filled boxes from the chromosome positions (chr.13, reference genome GRCh37.p13). (B) The stem-loop portion of miRNA-miRNA duplex structure on pre-miRNAs (hsa-miR-99a; miRBase ID: MI0003190) was identified by microRNA target prediction on MicroRNA.org resource. The hsa-miR-99a-5p sequence marked by green fonts. (C) Sequence of the human WISP1 3’-UTR region and number shown the positions of mRNA (NM_003882). Predicted hsa-miR-99a-5p binding site with SNP rs2929970 was highlighted by color red fonts. (D) The models of microRNA-target duplex were determined using the RNAhybrid web tool on the Bielefeld Bioinformatics Server. RISC, RNA-induced silencing complex, arrows indicate the locus of rs2929970. (E) The SNP rs2929970 A-allele reduces the free binding energy (MFE, minimum free energy; change: 22.16%). (F) Boxplot chart counting the differential expressions of microRNA hsa-miR-99a-5p in the 420 OSCC patients and 43 normal from Pan-Cancer dataset.
Discussion
In this study, we revealed the correlations between WISP1 SNPs and OSCC. Previous studies have indicated the correlations of risk factors such as alcohol consumption, betel quid chewing, and smoking with oral cancer carcinogenesis [31–33]. In our study, we confirmed that betel quid chewing, cigarette smoking, and alcohol consumption are associated with OSCC (Table 1). However, the associations of these risk factors with WISP1 regulation have yet to be extensively investigated. Smoking is one of the crucial risk factors for OSCC [31, 33]. In a pilot study, whole genome expression profiling of Indian patients with tobacco chewing-associated oral cancers implicated that WISP1 is one of the representative apoptosis-related deregulated genes in oral cancer [34]. Chen et al. revealed that WISP1 was overexpressed in non–small cell lung carcinoma (NSCLC) samples compared with their normal lung tissue counterparts, implicating that WISP1 might act as an oncoprotein in NSCLC [10]. However, WISP1 expression was not associated with clinical parameters such as family history, metastasis, smoking history, tuberculosis, gender, tumor type, and tumor size in NSCLC individuals [10]. WISP1 mRNA expression levels were higher in both lung and head and neck tumor tissues compared with their normal tissue counterparts [35], and smoking history was inconsistent. Therefore, it can be assumed that smoking has a limited ability to induce WISP1 overexpression.
Recent studies have suggested the vital role of WISP1 in cancer [35–37]. However, the WISP1 SNPs contributing to cancer progression have yet to be extensively investigated. In the present study, we found that the WISP1 SNP of rs2929970 was associated with OSCC risk (Table 2); rs2929970 is located in the 3′ untranslated region within a region of splicing variation [38]. Our present study confirmed the association of the clinically examined rs2929970 in the WISP1 3′-UTR region with OSCC risk; this association is most likely attributed to a putative hsa-miRNA-99a binding site (Fig 1). This finding suggests that the SNP corresponding to the RNA bulge region may affect the binding strength of specific miRNA/target duplexes, resulting in low minimum free energy and modulating mRNA stability.
A previous study indicated that the rs2929970 SNP may be associated with spinal osteoarthritis in postmenopausal Japanese women [24]. Yamada et al. [26] indicated that WISP1 rs2929970 was associated with hypertension in men carrying the G allele, and the men carrying this polymorphism had higher blood pressure. These results demonstrate the different effects of WISP1 rs2929970 expression in different diseases. Such inconsistent results for WISP1 rs2929970 expression may be due to different ethnicities or diseases; therefore, different genotype distributions may be observed. Although the detailed mechanism of WISP1 rs2929970 remains unclear, the WISP1 rs2929970 polymorphism certainly plays a role in cancers or diseases.
In the present study, we determined that nonsmoker controls and OSCC patients with WISP1 polymorphic rs2977530 AG + GG genotypes had a low risk of OSCC (AOR = 0.598, Table 3). Chen et al. indicated that lung cancer patients carrying the A alleles of WISP1 rs2977530 polymorphisms may have an increased risk of lung cancer [22]. However, in that study, the smoking status of controls was not adjusted [22]. Consistent with this result, our data reveal that the G alleles of WISP1 rs2977530 were associated with a low risk of OSCC. We also analyzed the WISP1 SNP of rs1689334 and showed that 707 OSCC betel quid chewers carrying WISP1 rs16893344 SNP CT + TT genotypes exhibited a low risk of LN metastasis (OR = 0.674, p = 0.038, Table 5). However, Chen et al. [22] indicated that lung cancer patients carrying the T allele of the WISP1 rs16893344 polymorphism may have an increased risk of lung cancer. Tao et al. also showed that WISP1 rs16893344 C > T polymorphisms significantly increased myocardial infarction risk [39]. These results demonstrate that the various WISP1 SNPs may be expressed in different cancers and diseases. Moreover, a previous study suggested that WISP1 expression is regulated by methylation, and WISP1 hypomethylation contributes to LN metastasis in OSCC [14]. The WISP1 expression is correlated with the DNA methylation of its promoter, and reduced methylation levels are correlated with increased WISP1 expression [14]. Therefore, although the interaction of betel quid chewing with WISP1 expression and the functions of WISP1 rs16893344 have not been extensively investigated, the WISP1 SNP of rs16893344 may contribute to changes that influence WISP1 gene transcription.
We analyzed the correlations of WISP1 SNP expression with the clinical statues of OSCC patients. We observed that among 100 nonsmoker OSCC patients, those carrying WISP1 rs2929970 AG + GG genotypes had later stage OSCC and a larger tumor size (Table 6). Because smoking is a well-known risk factor for OSCC [31, 33], this result in nonsmoker OSCC patients implicated the pivotal role of the WISP1 SNP of rs2929970 in cancer progression and WISP1 regulation. A previous study showed that WISP1 binds to αvβ3 integrin and causes the activation of the ASK1, JNK/p38, and AP-1 pathways, which upregulate ICAM-1 expression and promote the migration of human OSCC cells [15], and tumor-secreted WISP1 promotes angiogenesis through VEGF-A expression and increased angiogenesis-related tumor growth [16]. Although the mechanism and regulation of WISP1 SNPs in cancer progression have not been extensively investigated, the WISP1 SNPs of rs16893344 and rs2929970 might be involved in WISP1 regulation through WISP1-induced ICAM-1 upregulation and VEGF-A expression. The WISP1 SNPs of rs16893344 and rs2929970 may interfere with or enhance the binding activity of WISP1 to αvβ3 integrin, triggering the regulation of the ASK1, JNK/p38, and AP-1 pathways and VEGF-A expression, leading to a more favorable or poorer prognosis. Moreover, Mercer et al. also reports that, in a rat model of alcohol-induced liver disease, chronic alcohol consumption can significant upregulation in WISP1 expression [40]. Approximately half of OSCC patients (53.3%) in our study consumed alcohol (Table 1), these OSCC patients may exhibit a higher level of serum WISP1, and alcohol consumption may exert a synergistic effect on WISP1 upregulation. However, the WISP1 SNPs contributing to cancer progression and WISP1 regulation require further investigation to elucidate their detailed mechanisms.
In conclusion, WISP1 SNPs are correlated with OSCC. The WISP1 SNP of rs2929970 is associated with OSCC susceptibility, and rs2929970 A/G polymorphisms may be correlated with a worse prognosis of OSCC, such as later stage OSCC or larger tumor size. WISP1 rs2929970 may serve as a marker or a therapeutic target in OSCC.
Supporting information
Table A. Odds ratio (OR) and 95% confidence interval (CI) of oral cancer associated with WISP1 genotypic frequencies in smoker. Table B. Clinical statuses and WISP1 rs16893344 genotype frequencies in oral cancer among 193 non-betel quid chewers. Table C. Clinical statuses and WISP1 rs2929970 genotype frequencies in oral cancer among 800 smoker.
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by CSH-2017-E-001-Y2 from Chung Shan Medical University Hospital Taiwan, respectively. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral oncology. 2009;45(4–5):309–16. doi: 10.1016/j.oraloncology.2008.06.002 [DOI] [PubMed] [Google Scholar]
- 2.Kim SY, Nam SY, Choi SH, Cho KJ, Roh JL. Prognostic value of lymph node density in node-positive patients with oral squamous cell carcinoma. Annals of surgical oncology. 2011;18(8):2310–7. doi: 10.1245/s10434-011-1614-6 [DOI] [PubMed] [Google Scholar]
- 3.Haddad RI, Shin DM. Recent advances in head and neck cancer. The New England journal of medicine. 2008;359(11):1143–54. doi: 10.1056/NEJMra0707975 [DOI] [PubMed] [Google Scholar]
- 4.Rogers SN, Brown JS, Woolgar JA, Lowe D, Magennis P, Shaw RJ, et al. Survival following primary surgery for oral cancer. Oral oncology. 2009;45(3):201–11. doi: 10.1016/j.oraloncology.2008.05.008 [DOI] [PubMed] [Google Scholar]
- 5.Su SC, Lin CW, Liu YF, Fan WL, Chen MK, Yu CP, et al. Exome Sequencing of Oral Squamous Cell Carcinoma Reveals Molecular Subgroups and Novel Therapeutic Opportunities. Theranostics. 2017;7(5):1088–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng HL, Liu YF, Su CW, Su SC, Chen MK, Yang SF, et al. Functional genetic variant in the Kozak sequence of WW domain-containing oxidoreductase (WWOX) gene is associated with oral cancer risk. Oncotarget. 2016;7(43):69384–96. Epub 2016/09/23. doi: 10.18632/oncotarget.12082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inkson CA, Ono M, Kuznetsov SA, Fisher LW, Robey PG, Young MF. TGF-beta1 and WISP-1/CCN-4 can regulate each other's activity to cooperatively control osteoblast function. Journal of cellular biochemistry. 2008;104(5):1865–78. PubMed Central PMCID: PMC2729692. doi: 10.1002/jcb.21754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holbourn KP, Acharya KR, Perbal B. The CCN family of proteins: structure-function relationships. Trends in biochemical sciences. 2008;33(10):461–73. PubMed Central PMCID: PMC2683937. doi: 10.1016/j.tibs.2008.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perbal B. NOV (nephroblastoma overexpressed) and the CCN family of genes: structural and functional issues. Molecular pathology: MP. 2001;54(2):57–79. PubMed Central PMCID: PMC1187006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen PP, Li WJ, Wang Y, Zhao S, Li DY, Feng LY, et al. Expression of Cyr61, CTGF, and WISP-1 correlates with clinical features of lung cancer. PloS one. 2007;2(6):e534 PubMed Central PMCID: PMC1888724. doi: 10.1371/journal.pone.0000534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu J, Long Z, Cai H, Du C, Liu X, Yu S, et al. High expression of WISP1 in colon cancer is associated with apoptosis, invasion and poor prognosis. Oncotarget. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Luo H, Hu Z, Peng J, Jiang Z, Song T, et al. Targeting WISP1 to sensitize esophageal squamous cell carcinoma to irradiation. Oncotarget. 2015;6(8):6218–34. PubMed Central PMCID: PMC4467433. doi: 10.18632/oncotarget.3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagai Y, Watanabe M, Ishikawa S, Karashima R, Kurashige J, Iwagami S, et al. Clinical significance of Wnt-induced secreted protein-1 (WISP-1/CCN4) in esophageal squamous cell carcinoma. Anticancer research. 2011;31(3):991–7. [PubMed] [Google Scholar]
- 14.Clausen MJ, Melchers LJ, Mastik MF, Slagter-Menkema L, Groen HJ, van der Laan BF, et al. Identification and validation of WISP1 as an epigenetic regulator of metastasis in oral squamous cell carcinoma. Genes, chromosomes & cancer. 2016;55(1):45–59. [DOI] [PubMed] [Google Scholar]
- 15.Chuang JY, Chang AC, Chiang IP, Tsai MH, Tang CH. Apoptosis signal-regulating kinase 1 is involved in WISP-1-promoted cell motility in human oral squamous cell carcinoma cells. PloS one. 2013;8(10):e78022 PubMed Central PMCID: PMC3804520. doi: 10.1371/journal.pone.0078022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuang JY, Chen PC, Tsao CW, Chang AC, Lein MY, Lin CC, et al. WISP-1 a novel angiogenic regulator of the CCN family promotes oral squamous cell carcinoma angiogenesis through VEGF-A expression. Oncotarget. 2015;6(6):4239–52. PubMed Central PMCID: PMC4414186. doi: 10.18632/oncotarget.2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM. Silent polymorphisms speak: how they affect pharmacogenomics and the treatment of cancer. Cancer research. 2007;67(20):9609–12. doi: 10.1158/0008-5472.CAN-07-2377 [DOI] [PubMed] [Google Scholar]
- 18.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430(7001):743–7. PubMed Central PMCID: PMC2966974. doi: 10.1038/nature02797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chou CH, Chou YE, Chuang CY, Yang SF, Lin CW. Combined effect of genetic polymorphisms of AURKA and environmental factors on oral cancer development in Taiwan. PloS one. 2017;12(2):e0171583 Epub 2017/02/06. PubMed Central PMCID: PMCPMC5289639. doi: 10.1371/journal.pone.0171583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung TT, Pan MS, Kuo CL, Wong RH, Lin CW, Chen MK, et al. Impact of RECK gene polymorphisms and environmental factors on oral cancer susceptibility and clinicopathologic characteristics in Taiwan. Carcinogenesis. 2011;32(7):1063–8. Epub 2011/05/14. doi: 10.1093/carcin/bgr083 [DOI] [PubMed] [Google Scholar]
- 21.Su CW, Huang YW, Chen MK, Su SC, Yang SF, Lin CW. Polymorphisms and Plasma Levels of Tissue Inhibitor of Metalloproteinase-3: Impact on Genetic Susceptibility and Clinical Outcome of Oral Cancer. Medicine. 2015;94(46):e2092 Epub 2015/11/19. PubMed Central PMCID: PMCPMC4652830. doi: 10.1097/MD.0000000000002092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Yin JY, Li XP, Wang Y, Zheng Y, Qian CY, et al. Association of Wnt-Inducible Signaling Pathway Protein 1 Genetic Polymorphisms With Lung Cancer Susceptibility and Platinum-Based Chemotherapy Response. Clinical lung cancer. 2015;16(4):298–304 e1-2. Epub 2015/02/07. doi: 10.1016/j.cllc.2014.12.008 [DOI] [PubMed] [Google Scholar]
- 23.Frank B, Hoffmeister M, Klopp N, Illig T, Chang-Claude J, Brenner H. Single nucleotide polymorphisms in Wnt signaling and cell death pathway genes and susceptibility to colorectal cancer. Carcinogenesis. 2010;31(8):1381–6. Epub 2010/04/21. doi: 10.1093/carcin/bgq082 [DOI] [PubMed] [Google Scholar]
- 24.Urano T, Narusawa K, Shiraki M, Usui T, Sasaki N, Hosoi T, et al. Association of a single nucleotide polymorphism in the WISP1 gene with spinal osteoarthritis in postmenopausal Japanese women. Journal of bone and mineral metabolism. 2007;25(4):253–8. Epub 2007/06/27. doi: 10.1007/s00774-007-0757-9 [DOI] [PubMed] [Google Scholar]
- 25.Wang SH, Xu F, Dang HX, Yang L. Genetic variations in the Wnt signaling pathway affect lung function in asthma patients. Genetics and molecular research: GMR. 2013;12(2):1829–33. Epub 2013/01/15. doi: 10.4238/2013.January.4.1 [DOI] [PubMed] [Google Scholar]
- 26.Yamada Y, Ando F, Shimokata H. Association of polymorphisms of SORBS1, GCK and WISP1 with hypertension in community-dwelling Japanese individuals. Hypertension research: official journal of the Japanese Society of Hypertension. 2009;32(5):325–31. Epub 2009/03/14. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Yin J, Li X, Wang Y, Zheng Y, Qian C, et al. WISP1 polymorphisms contribute to platinum-based chemotherapy toxicity in lung cancer patients. International journal of molecular sciences. 2014;15(11):21011–27. Epub 2014/11/19. PubMed Central PMCID: PMCPMC4264209. doi: 10.3390/ijms151121011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su SC, Hsieh MJ, Liu YF, Chou YE, Lin CW, Yang SF. ADAMTS14 Gene Polymorphism and Environmental Risk in the Development of Oral Cancer. PloS one. 2016;11(7):e0159585 Epub 2016/07/28. PubMed Central PMCID: PMCPMC4962993. doi: 10.1371/journal.pone.0159585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho HY, Lin CW, Chien MH, Reiter RJ, Su SC, Hsieh YH, et al. Melatonin suppresses TPA-induced metastasis by downregulating matrix metalloproteinase-9 expression through JNK/SP-1 signaling in nasopharyngeal carcinoma. Journal of pineal research. 2016;61(4):479–92. Epub 2016/10/26. doi: 10.1111/jpi.12365 [DOI] [PubMed] [Google Scholar]
- 30.Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nature genetics. 2013;45(10):1113–20. Epub 2013/09/28. PubMed Central PMCID: PMCPMC3919969. doi: 10.1038/ng.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ko YC, Huang YL, Lee CH, Chen MJ, Lin LM, Tsai CC. Betel quid chewing, cigarette smoking and alcohol consumption related to oral cancer in Taiwan. Journal of oral pathology & medicine: official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. 1995;24(10):450–3. [DOI] [PubMed] [Google Scholar]
- 32.Lu CT, Yen YY, Ho CS, Ko YC, Tsai CC, Hsieh CC, et al. A case-control study of oral cancer in Changhua County, Taiwan. Journal of oral pathology & medicine: official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. 1996;25(5):245–8. [DOI] [PubMed] [Google Scholar]
- 33.Yen TT, Lin WD, Wang CP, Wang CC, Liu SA. The association of smoking, alcoholic consumption, betel quid chewing and oral cavity cancer: a cohort study. European archives of oto-rhino-laryngology: official journal of the European Federation of Oto-Rhino-Laryngological Societies. 2008;265(11):1403–7. [DOI] [PubMed] [Google Scholar]
- 34.Chakrabarti S, Multani S, Dabholkar J, Saranath D. Whole genome expression profiling in chewing-tobacco-associated oral cancers: a pilot study. Medical oncology. 2015;32(3):60 doi: 10.1007/s12032-015-0483-4 [DOI] [PubMed] [Google Scholar]
- 35.Gurbuz I, Chiquet-Ehrismann R. CCN4/WISP1 (WNT1 inducible signaling pathway protein 1): a focus on its role in cancer. The international journal of biochemistry & cell biology. 2015;62:142–6. [DOI] [PubMed] [Google Scholar]
- 36.Jun JI, Lau LF. Taking aim at the extracellular matrix: CCN proteins as emerging therapeutic targets. Nature reviews Drug discovery. 2011;10(12):945–63. PubMed Central PMCID: PMC3663145. doi: 10.1038/nrd3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Ye L, Owen S, Weeks HP, Zhang Z, Jiang WG. Emerging role of CCN family proteins in tumorigenesis and cancer metastasis (Review). International journal of molecular medicine. 2015;36(6):1451–63. PubMed Central PMCID: PMC4678164. doi: 10.3892/ijmm.2015.2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez-Torres J, Hernandez-Diaz C, Espinosa-Morales R, Camacho-Galindo J, Galindo-Sevilla Ndel C, Lopez-Macay A, et al. Polymorphic variation of hypoxia inducible factor-1 A (HIF1A) gene might contribute to the development of knee osteoarthritis: a pilot study. BMC musculoskeletal disorders. 2015;16:218 PubMed Central PMCID: PMC4546180. doi: 10.1186/s12891-015-0678-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tao J, Wang YT, Abudoukelimu M, Yang YN, Li XM, Xie X, et al. Association of genetic variations in the Wnt signaling pathway genes with myocardial infarction susceptibility in Chinese Han population. Oncotarget. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mercer KE, Hennings L, Sharma N, Lai K, Cleves MA, Wynne RA, et al. Alcohol consumption promotes diethylnitrosamine-induced hepatocarcinogenesis in male mice through activation of the Wnt/beta-catenin signaling pathway. Cancer prevention research (Philadelphia, Pa). 2014;7(7):675–85. Epub 2014/04/30. PubMed Central PMCID: PMCPMC4082466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A. Odds ratio (OR) and 95% confidence interval (CI) of oral cancer associated with WISP1 genotypic frequencies in smoker. Table B. Clinical statuses and WISP1 rs16893344 genotype frequencies in oral cancer among 193 non-betel quid chewers. Table C. Clinical statuses and WISP1 rs2929970 genotype frequencies in oral cancer among 800 smoker.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.