SUMMARY
Mitochondrial fission mediated by the GTPase dynamin-related protein-1 (Drp1) is an attractive drug target in numerous maladies that range from heart disease to neurodegenerative disorders. The compound mdivi-1 is widely reported to inhibit Drp1-dependent fission, elongate mitochondria, and mitigate brain injury. Here, we show that mdivi-1 reversibly inhibits mitochondrial Complex I-dependent O2 consumption and reverse electron transfer-mediated reactive oxygen species (ROS) production at concentrations (e.g. 50 μM) used to target mitochondrial fission. Respiratory inhibition is rescued by bypassing Complex I using yeast NADH dehydrogenase Ndi1. Unexpectedly, respiratory impairment by mdivi-1 occurs without mitochondrial elongation, is not mimicked by Drp1 deletion, and is observed in Drp1-deficient fibroblasts. In addition, mdivi-1 poorly inhibits recombinant Drp1 GTPase activity (Ki>1.2 mM). Overall, results suggest that mdivi-1 is not a specific Drp1 inhibitor. The ability of mdivi-1 to reversibly inhibit Complex I and modify mitochondrial ROS production may contribute to effects observed in disease models.
eTOC
Bordt, Clerc et al. show that the putative Drp1 inhibitor mdivi-1 reversibly inhibits mitochondrial complex I without impairing Drp1 GTPase activity or lengthening mitochondria. Mdivi-1 attenuates mitochondrial reactive oxygen species production under conditions relevant to ischemia/reperfusion injury. These mechanisms may provide an alternative explanation for some of mdivi-1’s in vivo effects.
INTRODUCTION
Mitochondrial fission-fusion events occur physiologically and are involved in the segregation and elimination of damaged mitochondrial elements by autophagy (Twig et al., 2008). Basal Drp1-dependent mitochondrial fission is required for mitochondrial trafficking to synapses, mitochondrial quality control, and brain development (Ishihara et al., 2009; Wakabayashi et al., 2009; Kageyama et al., 2014). However, mitochondrial fragmentation also occurs simultaneous to cytochrome c release during programmed cell death (Frank et al., 2001). The fission GTPase Drp1 promotes Bax-dependent cytochrome c redistribution from mitochondria to the cytoplasm (Frank et al., 2001), an event which initiates activation of caspase protease executioners. Therefore, Drp1 is a drug target in numerous degenerative diseases that involve aberrant mitochondrial fission and/or disrupted membrane integrity.
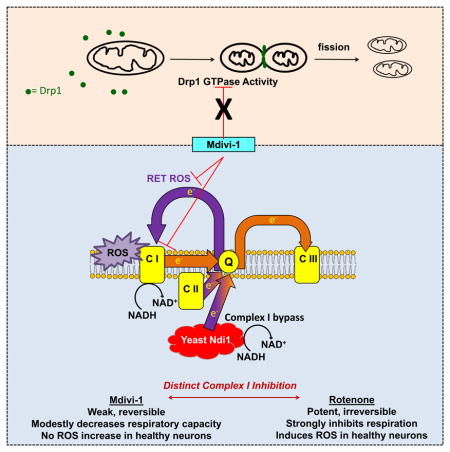
Mdivi-1 is a quinazolinone derivative identified as a mitochondrial fission inhibitor in a chemical library screen for compounds that influence mitochondrial morphology in yeast (Cassidy-Stone et al., 2008). Mdivi-1 impaired the GTPase activity of Dnm1, the yeast homologue of the mammalian fission factor Drp1 (Cassidy-Stone et al., 2008). However, mdivi-1 failed to inhibit the GTPase activity of recombinant human Drp1 in the same study. Human Drp1 was less active than its yeast homologue and incapable of self-assembly, leading to speculation that the human protein was folded incorrectly. Nevertheless, mdivi-1 caused elongation of mammalian mitochondria in COS cells within an hour (Cassidy-Stone et al., 2008) and lengthened mitochondria in several additional studies (Rosdah et al., 2016). Consequently, mdivi-1 is widely considered to be a small molecule inhibitor of mitochondrial fission that specifically targets Drp1.
Mdivi-1 crosses the blood-brain barrier and is protective in several preclinical disease animal models that include heart and brain ischemia-reperfusion injury (Grohm et al., 2012; Ong et al., 2010), traumatic brain injury (Wu et al., 2016), and Parkinson’s disease (Rappold et al., 2014). Mdivi-1 also blocks pro-apoptotic Bax-dependent cytochrome c release from isolated mitochondria (Cassidy-Stone et al., 2008) and attenuates neural cell death in vitro and in vivo (Grohm et al., 2012), consistent with the possibility that Drp1 is a bona fide therapeutic drug target. However, because Drp1 is an essential regulator of mitochondrial fission under normal conditions, it is important to determine whether its inhibition by mdivi-1 impacts cellular bioenergetics over the short or long term, and if so, whether the effects of mdivi-1 are directly due to blocking Drp1 activity.
Here, we set out to test the hypothesis that pharmacological inhibition of Drp1 by mdivi-1 leads to impaired mitochondrial bioenergetics. We predicted that mdivi-1 would rapidly elongate mitochondria and cause Drp1-dependent changes in mitochondrial respiration. Unexpectedly, mdivi-1 treatment failed to lengthen mitochondria in neurons, wild type (WT) or Drp1 knockout (KO) immortalized mouse embryonic fibroblasts (MEFs), or COS-7 cells. Mdivi-1 also poorly antagonized recombinant human Drp1 GTPase activity. However, mdivi-1 rapidly and reversibly inhibited electron transport chain (ETC) Complex I-dependent O2 consumption by cells in a Drp1-independent fashion. In addition, mdivi-1 attenuated Complex I-dependent reverse electron transfer (RET)-mediated reactive oxygen species (ROS) production by brain mitochondria oxidizing succinate. Collectively, these results establish mitochondrial Complex I as a previously unknown target of mdivi-1 action and suggest a re-evaluation of prior studies attributing the effects of mdivi-1 exclusively to inhibition of Drp1.
RESULTS
Mdivi-1 Inhibits Complex I-dependent Mitochondrial O2 Consumption
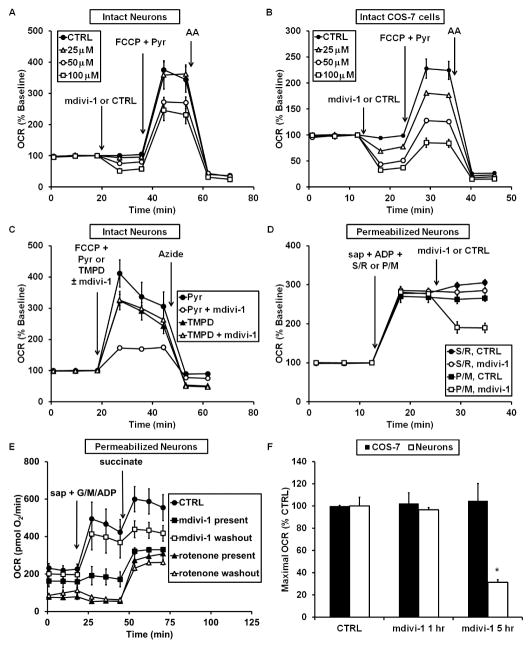
To determine the impact of mdivi-1 on bioenergetics, we first measured neuronal O2 consumption rate (OCR) prior to and immediately following injection of mdivi-1 (25–100 μM). Mdivi-1 caused significant inhibition of basal respiration in primary cortical neurons at concentrations of 50 and 100 μM. Maximal respiration, measured after addition of the uncoupler carbonyl cyanide-p-trifluoromethyoxyphenylhydrazone (FCCP), was also impaired (Figure 1A). In COS-7 cells, where the effects of mdivi-1 on mitochondrial morphology were initially reported, mdivi-1 (25–100 μM) even more robustly inhibited basal and maximal respiration (Figure 1B). Injection of the Complex III inhibitor antimycin A (AA) confirmed that the mdivi-1 effect on cellular O2 consumption was on the mitochondrial ETC as mdivi-1 failed to alter antimycin-A insensitive non-mitochondrial O2 consumption (Figures 1A and 1B).
Figure 1. Mdivi-1 reversibly inhibits basal and maximal respiration at Complex I.
(A) OCR traces for neurons or (B) COS-7 cells receiving mdivi-1 or DMSO vehicle (CTRL), FCCP (3 μM for neurons, 2 μM for COS-7) plus pyruvate (Pyr, 10 mM), and antimycin A (AA, 1 μM). Traces are mean ± SD from 3 wells and are representative of 4 independent experiments (2–3 wells each). (C) FCCP together with pyruvate or together with TMPD (0.4 mM) plus ascorbate (0.4 mM) were injected along with mdivi-1 (100 μM) or CTRL while measuring OCR. The Complex IV inhibitor azide (5 mM) was then injected. (D) Neurons were permeabilized by saponin (sap, 25 μg/ml), and OCR was stimulated by 1 mM ADP in the presence of pyruvate and malate (P/M, 5 mM each) or succinate (S, 5 mM) in the presence of rotenone (R, 0.5 μM). Mdivi-1 (50 μM) or vehicle control was then injected. (E) Neurons were treated with rotenone or mdivi-1 (50 μM) for 1 hr prior to OCR measurements. Rotenone or mdivi-1 was then either left on for the duration of the assay (“present”), or washed out and replaced with drug-free aCSF (“washout”). Following permeabilization by sap and addition of ADP with glutamate and malate (G/M, 5 mM each), 5 mM succinate was added. (F) Neurons or COS-7 cells were treated with DMSO or mdivi-1 (50 μM) for 1 or 5 hr. The drug was then washed out and maximal OCR was determined. See also Figure S1.
To investigate where in the ETC respiratory inhibition occurred, we first tested whether mdivi-1 inhibits Complex IV. Mdivi-1 was added to intact neurons either together with FCCP and pyruvate or together with FCCP and a combination of the cell-permeable artificial electron donor N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD), ascorbate, and antimycin A. TMPD, which is reduced by ascorbate, donates electrons to cytochrome c-Complex IV, bypassing upstream components of the ETC (Packer and Mustafa, 1966). Mdivi-1 inhibited the respiration of neurons metabolizing glucose and pyruvate but did not inhibit TMPD/ascorbate-dependent O2 consumption by Complex IV (Figure 1C), indicating that mdivi-1 impairs respiration upstream of Complex IV.
To further investigate the mechanism of mdivi-1-mediated respiratory inhibition, the neuronal plasma membrane was selectively permeabilized with saponin and mitochondria within permeabilized cells were supplied with substrates specific for Complex I or Complex II (Clerc and Polster, 2012). Mdivi-1-mediated respiratory inhibition was observed in the presence of the Complex I substrates pyruvate and malate (Figure 1D) or glutamate and malate (see Figure 1E) but not in the presence of the Complex II substrate succinate (Figure 1D). Mdivi-1 inhibition of Complex I-dependent respiration could be rescued by washing out the drug after 1 hour, in contrast to irreversible Complex I inhibition mediated by rotenone (Figure 1E), indicating that mdivi-1 is a reversible inhibitor. Succinate stimulated respiration in both rotenone-treated and mdivi-1-treated cells (Figure 1E), confirming that decreased OCR was primarily due to inhibition of Complex I rather than due to cell death or downstream ETC inhibition. In response to chronic treatment with mdivi-1 for 5 hr, respiratory inhibition in COS-7 cells remained fully reversible, however it became irreversible in neurons (Figure 1F), suggesting sustained respiratory alterations in the more oxidative phosphorylation-reliant neurons.
We use bovine serum albumin (BSA) in our artificial cerebrospinal fluid (aCSF) assay medium as a surrogate for extracellular protein (Clerc and Polster, 2012). We found that respiratory inhibition by mdivi-1 was not observed in the absence of BSA in XF24 assays (Figure S1A), possibly due to binding of the hydrophobic mdivi-1 to the polystyrene assay plates. To further confirm the ability of mdivi-1 to impair respiration, a polarographic Clark O2 electrode in an acrylic-walled chamber was used to measure O2 consumption by isolated brain mitochondria in the absence of BSA. Similar to results with permeabilized cells, mdivi-1 rapidly inhibited the Complex I-dependent (Figure S1B) but not Complex II-dependent (Figure S1C) respiration of isolated mitochondria. Therefore, BSA is not required for mdivi-1 to inhibit respiration.
Mdivi-1 Fails to Elongate Mitochondria or Inhibit Drp1 GTPase Activity
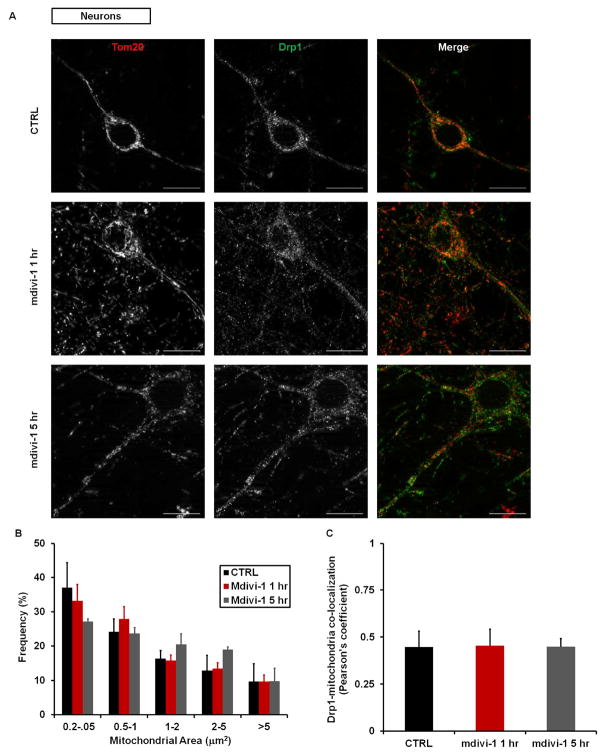
To test whether attenuation of OCR by mdivi-1 is associated with elongation of mitochondria, we used immunofluorescence microscopy for Tom20 to measure mitochondrial size in cortical neurons treated with mdivi-1 (50 μM) for 1 or 5 hr (Figure 2A). We did not observe a significant difference in size in neurons treated with mdivi-1 compared to vehicle control at either time point (Figure 2B). We also failed to observe a significant effect of mdivi-1 exposure on mitochondrial morphology in vehicle or staurosporine-treated COS-7 cells, with mitochondria visualized at multiple time points by three different methods [MitoTracker Red staining (Figures S2A–S2C), cytochrome c immunofluorescence (Figure S2D) or mitochondrially targeted green fluorescent protein (mito-GFP, Movies S1 and S2)]. Staurosporine caused robust mitochondrial fragmentation as reported (Frank et al., 2001).
Figure 2. Mdivi-1 does not alter mitochondrial size in neuronal processes.
(A) Representative immunofluorescence of Tom20 (red) and Drp1 (green) in neurons following treatment with DMSO vehicle (CTRL) or mdivi-1 (75 μM) for 1 or 5 hr (scale bars, 10 μm). Extrasomal Tom20 and Drp1 fluorescence is primarily localized to a meshwork of neuronal processes. (B) Frequency of binned mitochondrial areas for the treatments described in (A). (C) Pearson’s coefficient for Drp1 co-localized with mitochondrial Tom20 in neurons treated with CTRL or 75 μM mdivi-1. Results are mean ± SD, where ~800–1000 mitochondria were counted per treatment group per experiment, and 3 separate experiments were conducted. See also Figure S2.
A lack of mitochondrial elongation despite a significant effect on bioenergetics within the same time frame was unexpected, raising the possibility that inhibition of OCR by mdivi-1 is not due to an effect on Drp1. Furthermore, we found only limited co-localization of Drp1 with mitochondria in neurons with or without mdivi-1 treatment (Figures 2A and 2C), consistent with the primarily cytoplasmic Drp1 distribution in healthy cells (Frank et al., 2001). Consequently, we re-evaluated whether mdivi-1 can directly antagonize mammalian Drp1 GTPase activity. Using recombinant human Drp1 that exhibits assembly-stimulated GTPase activity (Chang et al., 2010), we found that mdivi-1 poorly inhibited Drp1 GTPase activity (Ki>1.2 mM, Figure 3) at concentrations that cause significant attenuation of basal and maximal respiration (see Figure 1). As expected, robust inhibition of Drp1 GTPase activity was achieved using the non-hydrolyzable GTP analogue GTP-γ-S. As an additional control, the previously reported inhibition of yeast Dnm1 by mdivi-1 (Cassidy-Stone et al., 2008) was confirmed (Figure S3).
Figure 3. Mdivi-1 is a poor inhibitor of human Drp1 GTPase activity.
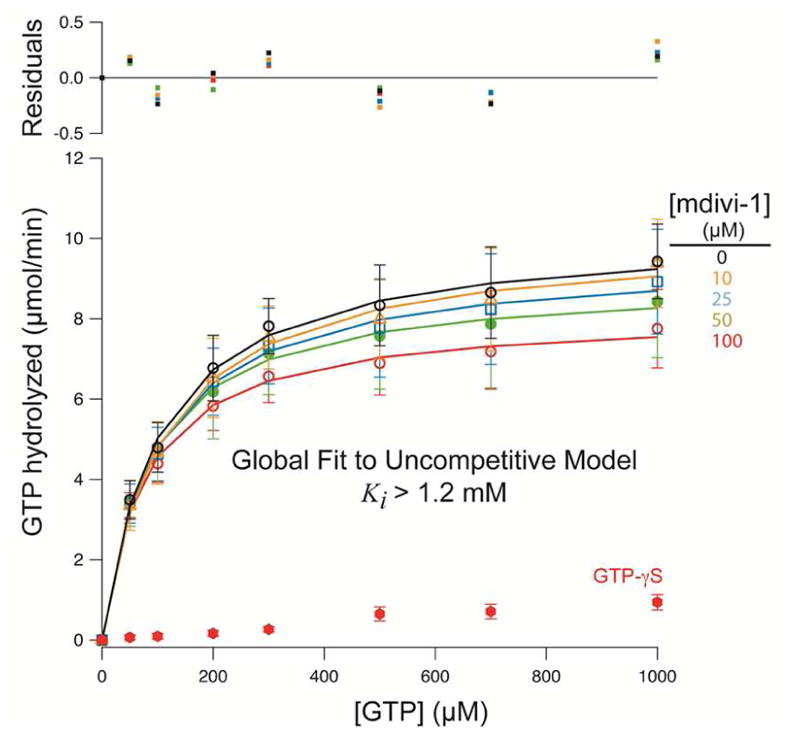
Substrate kinetics of recombinant human Drp1 was measured at 0–100 μM mdivi-1. Data were globally fit to an uncompetitive model yielding a Ki > 1.2mM. The data and SEM are for 3 independent preparations of Drp1. Residuals to the fit are shown above the graph. See also Figure S3.
Mdivi-1 Inhibits Complex I-dependent Respiration in the Absence of Drp1
When Drp1 is recruited to mitochondria it resides on the mitochondrial outer membrane (Frank et al., 2001). To test whether a mitochondrial outer membrane or intermembrane space target is required for mdivi-1 to attenuate Complex I-dependent respiration, we permeabilized the mitochondrial outer membrane in neurons, as well as the plasma membrane, by adding a 20-fold higher saponin concentration compared to that used earlier (Figures S4A and S4B). Mdivi-1 impaired respiration to the same extent whether the mitochondrial outer membrane was intact or permeabilized (Figures S4C and S4D), indicating that the mdivi-1 target likely resides in the mitochondrial inner membrane or matrix rather than the outer membrane or intermembrane space.
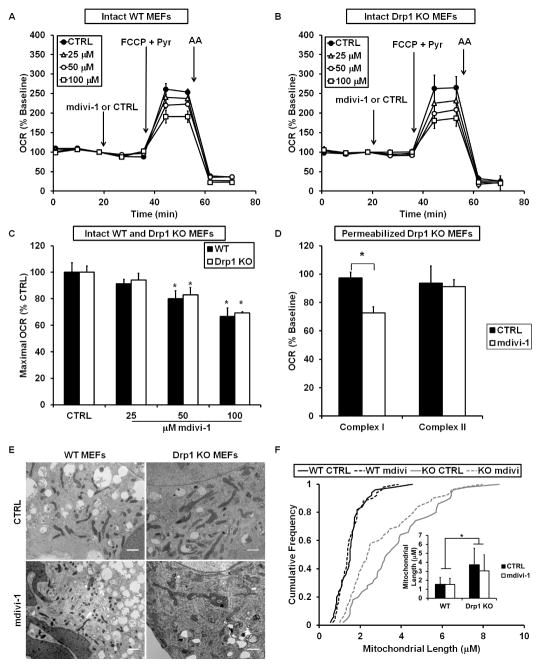
To directly test for involvement of the mitochondrial fission protein Drp1 in mdivi-1-mediated respiratory inhibition, mdivi-1 (25–100 μM) was added to immortalized WT and Drp1 KO MEFs. As was the case for neurons, mdivi-1 inhibited the maximal respiratory stimulation induced by uncoupler in both WT and Drp1 KO MEFs (Figures 4A–4C). In contrast to the impairment induced by mdivi-1, knockout of Drp1 in MEFs did not significantly alter respiration stimulated by the uncoupler FCCP (Figures S5A and S5B), similar to previous findings (Kageyama et al., 2014). Hence, not only did the bioenergetic response of mitochondria to mdivi-1 not require Drp1, but elimination of the putative protein target of mdivi-1, Drp1, did not recapitulate the effect of the drug on mitochondrial OCR. Consistent with the specific effect of mdivi-1 on Complex I-dependent respiration in neurons, mdivi-1 significantly inhibited Complex I-linked but not Complex II-linked respiration in permeabilized Drp1 knockout MEFs (Figure 4D).
Figure 4. Mdivi-1-induced respiratory inhibition is not mimicked by Drp1 KO or dependent upon Drp1 expression.
WT (A) or Drp1 KO (B) MEFs were treated with mdivi-1 (25–100 μM) or vehicle followed by FCCP (3 μM) plus pyruvate (10 mM) and antimycin A (AA, 1 μM) while OCR was measured. Traces here and below are mean ± SD of 3 wells and representative of at least 3 experiments. (C) Maximal OCR calculated immediately following FCCP plus pyruvate addition, expressed as a percentage of the maximal rate in vehicle-treated cells (mean ± SD, n=3). (D) Drp1 KO MEFs were permeabilized with 5 μg/mL saponin, along with ADP (1 mM) and either pyruvate and malate (5 mM each) or succinate (5 mM) in the presence of rotenone (0.5 μM). MEFs were then treated with 50 μM mdivi-1 or vehicle (CTRL). OCR is expressed as a percentage of OCR prior to drug injection. Results are mean ± SD, n=3. * p<0.05 compared to control. (E) Electron micrographs of WT or Drp1 KO MEFs treated with CTRL or mdivi-1 (50 μM) for 16 min. Scale bars = 1 μm. (F) Cumulative frequency distribution plot for mitochondrial length in WT or Drp1 KO MEFs ± mdivi-1. Bar graph inset is mean mitochondrial length ± SD. Results are from 50 mitochondria from 2 different experiments. See also Figures S4–S6.
Complex I inhibitors decrease the rate of cell proliferation in medium depleted of pyruvate (Sullivan et al., 2015). To further test whether mdivi-1 behaves like a Complex I inhibitor even in the absence of Drp1, we measured MEF proliferation in medium lacking pyruvate. Consistent with the possibility of Drp1-independent Complex I inhibition by mdivi-1, mdivi-1 slowed cell proliferation in both WT and Drp1 KO MEFs (Figures S5C and S5D).
Next, we examined whether mdivi-1 treatment, Drp1 KO, or a combination of the two had an effect on mitochondrial morphology in MEFs. Electron micrographs of MEFs revealed significantly elongated mitochondria in Drp1 KO cells compared to WT cells (Figures 4E and 4F), consistent with impaired mitochondrial fission in the absence of Drp1. However, mdivi-1 failed to elongate WT or Drp1 KO mitochondria within the same time frame (16 min) that it attenuated O2 consumption. Thus, mdivi-1 did not mimic the effect of deleting its suggested target, Drp1, on mitochondrial morphology at a time when functional effects on respiration were already apparent.
We also considered the possibility that mdivi-1 impairs mitochondrial Complex I-dependent respiration by inhibiting a yet to be discovered mammalian homologue of the yeast GTPase Dnm1. Complex I-dependent respiration by permeabilized neurons was measured either in the presence of GTPγS, a non-hydrolyzable GTP analogue that antagonizes all GTPases, including Drp1 (see Figure 3), or dynasore, a general dynamin inhibitor also reported to inhibit Drp1 (Macia et al., 2006). In contrast to mdivi-1, neither GTPγS (Figure S6A) nor dynasore (Figures S6B and S6C) impaired Complex I-dependent respiration.
Mdivi-1 Impairs Complex I-dependent NADH Oxidation by the Electron Transport Chain
Having established that suppression of mitochondrial respiration by mdivi-1 is independent of Drp1 or other GTPases, we investigated whether mdivi-1 is a direct inhibitor of Complex I. We measured Complex I activity in detergent-solubilized brain mitochondria by monitoring the Complex I-dependent reduction of the artificial electron acceptor 2,6-dichloroindophenol (DCIP). DCIP reduction was almost entirely abolished by the Complex I inhibitor rotenone whereas 50 μM mdivi-1 did not alter the reduction of DCIP (Figure S7A). Surprisingly, 100 μM mdivi-1 stimulated Complex I-dependent DCIP reduction (Figures S7A and S7B). DCIP reduction in the presence of 100 μM mdivi-1 remained fully rotenone-sensitive (Figure S7B). These results indicate that the mdivi-1-mediated Complex I inhibition observed in cells likely occurs by a different mechanism than that induced by rotenone and may require an intact inner mitochondrial membrane, and/or matrix components not present in detergent-solubilized mitochondria.
One possibility is that mdivi-1 impedes Complex I-dependent respiration upstream of Complex I by interfering with the supply of NADH substrate, for example, by inhibiting mitochondrial dehydrogenase enzymes. To test this possibility, we permeabilized neurons with saponin, added the pore-forming peptide alamethicin to porate the mitochondrial inner membrane, and then measured O2 consumption in the presence of exogenous NADH and cytochrome c. Exogenous NADH bypasses matrix dehydrogenases in this assay and O2 consumption measures Complex I–III–IV linked activity (Ji et al., 2012). OCR was impaired by the Complex I inhibitor rotenone or by the Complex IV inhibitor azide but not by the Complex II inhibitor 2-thenoyltrifluoroacetone (TTFA), validating the linked activity assay using the Seahorse XF24-based method (Figure S7C). Like rotenone, mdivi-1 inhibited NADH-supported OCR (Figure S7D), suggesting that mdivi-1 inhibits Complex I downstream of matrix dehydrogenases, possibly by interacting with the Complex I enzyme itself.
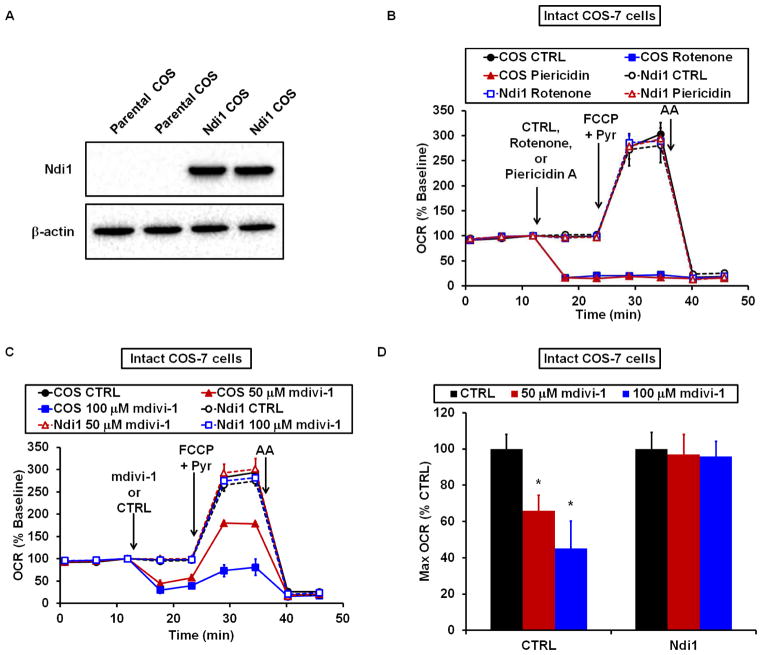
To specifically evaluate whether respiratory inhibition by mdivi-1 occurs at Complex I in intact cells, we ectopically expressed the gene encoding the internal NADH dehydrogenase (Ndi1) enzyme from Saccharomyces cerevisiae in COS-7 cells. Ndi1 is a single protein that can functionally substitute for Complex I in the ETC while exhibiting resistance to mammalian inhibitors and regulatory mechanisms (Seo et al., 1998). The respiration of COS-7 cells expressing Ndi1 protein (Figure 5A) was not impaired by the Complex I inhibitors rotenone or piericidin A (Figure 5B), indicating complete functional substitution for Complex I by Ndi1. The respiration of Ndi1-expressing cells was similarly insensitive to mdivi-1 treatment (Figures 5C and 5D), providing strong evidence that inhibition of the ETC by mdivi-1 occurs at Complex I.
Figure 5. Yeast Ndi1 NADH dehydrogenase prevents respiratory inhibition by mdivi-1.
(A) Ndi1 and β-actin protein levels in COS-7 cells (COS) transfected with the NDI1 gene and selected for NDI1 expression compared to parental COS. (B) COS or NDI1-transfected COS (Ndi1) were treated with vehicle (CTRL), rotenone (1 μM), or piericidin A (100 nM), followed by FCCP (2 μM) plus pyruvate (10 mM), and then antimycin A (1 μM) while OCR was measured. (C) OCR traces for Ndi1-COS-7 or parental COS-7 cells treated with DMSO or mdivi-1, followed by the drugs in (B). Traces in (B) and (C) are mean ± SD of 3 wells and representative of 3 experiments. (D) Bar graph showing quantification of traces in (C). See also Figure S7.
Mdivi-1 Preferentially Attenuates Complex I ROS Produced by Reverse Electron Transfer
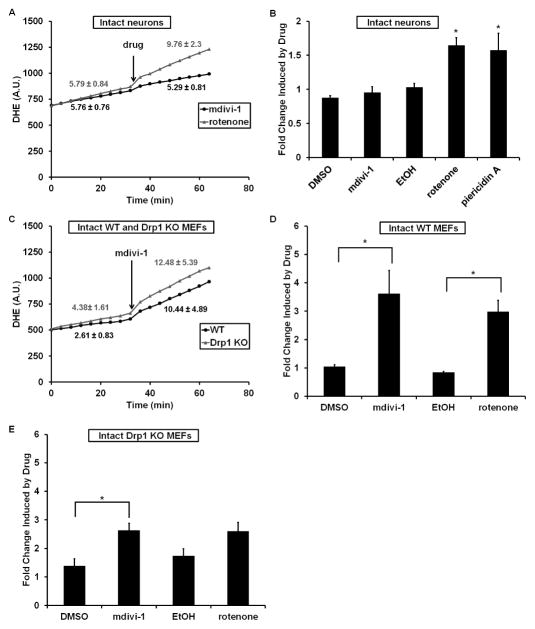
To further understand the mdivi-1 modulation of Complex I activity, we investigated its effect on ROS. Classical Complex I inhibitors like rotenone stimulate ROS production when mitochondria oxidize Complex I-linked substrates (Chinta et al., 2009; Yadava and Nicholls, 2007). As expected, complete inhibition of Complex I by rotenone led to an increase in ROS-dependent oxidation of the fluorescent dye dihydroethidium (DHE) in both neurons and MEFs (Figure 6). The effect of piericidin A on neurons was indistinguishable from rotenone (Figure 6B). Interestingly, partial inhibition of respiration by 50 μM mdivi-1 was insufficient to elevate ROS levels in neurons (Figures 6A and 6B), but increased ROS levels in WT and Drp1 KO MEFs (Figures 6C–6E).
Figure 6. Mdivi-1 increases ROS in WT and Drp1 KO fibroblasts but not neurons.
(A) DHE fluorescence (arbitrary units, A.U.) was measured over time in neurons. Rotenone (1 μM) or mdivi-1 (50 μM) was added as indicated (drug). Numbers are rates (A.U./min, mean ± SEM) before and after drug addition. (B) The rate of DHE fluorescence change following drug addition was divided by the basal rate to determine fold change. Piericidin A was added at 0.5 μM. DMSO was the vehicle for mdivi-1 and EtOH was the vehicle for rotenone and piericidin A. (C) DHE fluorescence measured over time in WT or Drp1 KO MEFs. Mdivi-1 was 50 μM. Numbers are rates before and after mdivi-1 addition. (D) and (E) The rate of DHE fluorescence change following drug addition was calculated in either WT (D) or Drp1 KO (E) MEFs as in (B). All data in bar graphs are mean ± SEM, n=5. * p<0.05.
Similar to findings with intact neurons, mdivi-1 failed to stimulate ROS emission from isolated brain mitochondria oxidizing Complex I substrates in the absence of ADP (Figures 7A and 7B). ADP significantly reduced the rate of ROS emission in both the absence and presence of mdivi-1, consistent with the oxidized shift in NADH/NAD+ ratio that accompanies phosphorylating respiration (Starkov and Fiskum, 2003). However, ROS emission following ADP addition was significantly higher when mdivi-1 was present. Notably, the mdivi-1 augmentation of ROS release during phosphorylating respiration was several-fold lower than that observed when rotenone was added (Figures 7A and B).
Figure 7. Mdivi-1 modulates ROS production by brain mitochondria as predicted by Complex I inhibition.
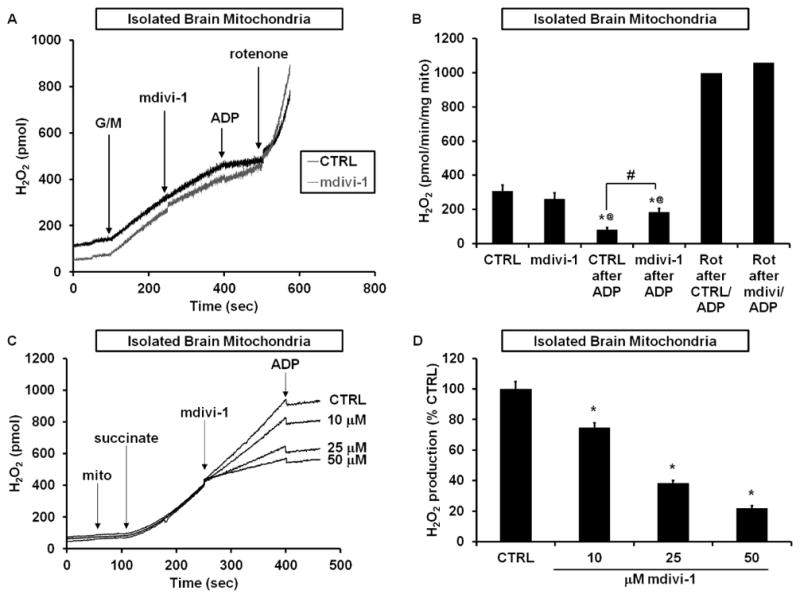
(A) Representative traces depicting H2O2 production by isolated brain mitochondria oxidizing glutamate and malate (G/M, 5 mM each) before and after addition of DMSO (CTRL) or mdivi-1 (50 μM), followed by addition of ADP (2 mM), and then rotenone (1 μM). (B) Quantification of the data in (A) (mean ± SD, n=5). * p<0.05 compared to control, @ p<0.05 compared to the respective condition with no ADP, # p<0.05 for mdivi-1 rate after ADP compared to control rate after ADP. Each rotenone (Rot) bar is the mean of 2 experiments. (C) Representative traces depicting RET-mediated H2O2 production by brain mitochondria incubated with succinate, followed by addition of CTRL or mdivi-1, and then ADP (2 mM). (D) Quantification of the data in (C) (mean ± SD, n=3). * p<0.05 compared to control.
When well-coupled brain mitochondria oxidize succinate rather than Complex I-linked substrates, ROS are produced from the quinone-binding (Q) site of Complex I (Andreyev et al., 2005). Rotenone and similar Complex I inhibitors attenuate this so-called reverse electron transfer (RET) ROS production (Tretter et al., 2007; Chinta et al., 2009). Like rotenone, mdivi-1 caused a significant and dose-dependent attenuation of RET-mediated ROS production (Figures 7C and 7D). Subsequent addition of ADP to slightly decrease the electrochemical proton gradient by stimulating proton flux through the ATP synthase largely abolished ROS emission by brain mitochondria oxidizing succinate. This finding is consistent with the high protonmotive force requirement to drive reverse electron transfer from ubiquinone to Complex I (Votyakova and Reynolds, 2001; Tretter et al., 2007) and further supports the Complex I origin of succinate-stimulated ROS under our experimental conditions.
DISCUSSION
Since its identification from a chemical library screen in 2008, mdivi-1 has been used prevalently as a selective inhibitor of the mammalian mitochondrial fission protein Drp1. Here, we showed that mdivi-1 inhibits mitochondrial Complex I-dependent respiration in mammalian neurons, fibroblasts, and kidney cells at concentrations reported to antagonize mitochondrial fission. Drp1 was not required for mdivi-1 to inhibit respiration, and knockout of Drp1 or general dynamin GTPase inhibition did not mimic the effect of mdivi-1 on mitochondrial OCR. Furthermore, mdivi-1 modified cell proliferation and mitochondrial ROS in a manner consistent with Complex I inhibition, and the yeast Complex I functional equivalent Ndi1 rescued the respiration of mammalian cells treated with mdivi-1. Consequently, we conclude that the effects of mdivi-1 on mitochondria are not specific to Drp1.
Whether mdivi-1 can directly inhibit Drp1 in mammalian cells remains unclear. Elegant work indicates that mdivi-1 impairs yeast Dnm1 GTPase activity, likely via an allosteric binding mechanism that prohibits Dnm1 self-assembly (Cassidy-Stone et al., 2008). However, because mdivi-1 was initially tested without effect on recombinant Drp1 protein that could not self-assemble, Drp1 antagonism in mammalian cells was inferred from the drug’s effect on mitochondrial morphology and its ability to inhibit the yeast homologue. Here, we showed that the GTPase activity of recombinant human Drp1 protein capable of self-assembly (Chang et al., 2010) was impaired with very poor potency (Ki>1.2 mM). No inhibition was observed at concentrations (e.g. 50 μM) reported to elongate mitochondria.
Our studies do not exclude the possibility that in cells mdivi-1 targets Drp1-dependent fission steps other than GTP hydrolysis or, alternatively, inhibits Drp1 activity by indirect mechanisms. However, we found that despite rapid effects on Complex I-dependent respiration, mdivi-1 did not significantly alter mitochondrial size. Therefore, the suppression of respiration by mdivi-1 does not require changes in the mitochondrial network by either Drp1-dependent or Drp1-independent mechanisms.
Interestingly, Berman et al. observed that in primary cortical neurons ~25% of mitochondria underwent a fission event within 15 minutes (Berman et al., 2009), suggesting that the majority of mitochondria should have undergone fission after one hour. Nevertheless, we saw no effect of mdivi-1 on cortical neuron mitochondrial size at this time point, or after a longer, five hour incubation. In addition, we failed to see an effect of mdivi-1 on COS-7 cell mitochondrial morphology following incubations of 1, 2, 6, or 24 hours, and mdivi-1 did not prevent mitochondrial fragmentation induced by staurosporine. We measured mitochondrial morphology in cells using multiple methods, including those used in prior studies with mdivi-1: TOM20 or cytochrome c immunofluorescence, Mito-GFP fluorescence, MitoTracker Red staining, and electron microscopy. A subtle difference in experimental conditions compared to published studies may have prevented us from seeing an effect of mdivi-1 on mitochondrial morphology.
Since the initial description of the anti-mitochondrial fission effect of mdivi-1, the evidence that mdivi-1 inhibits mammalian Drp1 is correlative; i.e. data showing that mdivi-1 affects mitochondrial size in the same manner as Drp1 siRNA or K38A dominant negative Drp1 (Rosdah et al., 2016). To our knowledge, there are no convincing demonstrations that mdivi-1 directly inhibits Drp1. One study showed a minor ~25% decrease in cellular GTPase activity ascribed to Drp1 following a 24 hour treatment with 50 μM mdivi-1 (Manczak and Reddy, 2015). Notably, mitochondrial fusion proteins Mfn1, Mfn2 and Opa1 were all significantly upregulated at this time point, suggesting that mdivi-1 may elongate mitochondria in some cells by Drp1-independent mechanisms.
The major finding of our study is the identification of mdivi-1 as a reversible inhibitor of Complex I. Unexpectedly, mdivi-1 stimulated rather than inhibited Complex I-catalyzed electron transfer to an artificial electron donor in detergent-solubilized mitochondria, suggesting that the integrity of the lipid bilayer and/or other mitochondrial components influence how mdivi-1 interacts with Complex I. Nevertheless, mdivi-1 acts as a Complex I inhibitor in cells since it impaired respiration, which was rescued by Ndi1 expression, and it triggered ROS accumulation in MEFs.
The well-characterized Complex I inhibitor rotenone causes parkinsonian neurodegeneration in rodents (Sherer et al., 2003). In contrast, mdivi-1 has little to no reported in vivo toxicity and is instead neuroprotective in several animal models, including mouse models of Parkinson’s disease (Rappold et al., 2014). The toxicity of rotenone was linked to the induction of oxidative stress (Sherer et al., 2003). We found that mdivi-1 is a weak Complex I inhibitor compared to rotenone, and 50 μM mdivi-1 generated almost no ROS from isolated brain mitochondria either in the absence or presence of ADP, compared to >4 fold stimulation by a saturating concentration of rotenone (Figure 7B). Strikingly, mdivi-1 failed to elevate ROS in intact neurons, which may partly explain its lack of in vivo brain toxicity. Importantly, in contrast to chronic mdivi-1 treatment (Rappold et al., 2014), conditional knockout of Drp1 in dopaminergic neurons caused degeneration associated with axonal mitochondria loss (Berthet et al., 2014). The absence of dopaminergic neurodegeneration following several days of mdivi-1 administration suggests that mdivi-1 is not a potent antagonist of Drp1 in vivo.
We further investigated the ability of mdivi-1 to modulate Complex I-dependent ROS production by measuring ROS released by brain mitochondria incubated with succinate. Succinate accumulates during ischemia and is oxidized rapidly during reperfusion, producing ROS by reverse electron transfer that contribute to injury (Chouchani et al., 2014; Brand et al., 2016). RET ROS is also thought to play a role in Alzheimer’s disease (Zhang et al., 2015) and lifespan (Lambert et al., 2007; Scialo et al., 2016). We found that mdivi-1 dose dependently inhibits ROS produced by RET. Whereas mdivi-1 barely elevated ROS by brain mitochondria oxidizing Complex I substrates in the presence of ADP, mdivi-1 was effective as a RET inhibitor, with ~80% ROS inhibition at 50 μM mdivi-1. Interestingly, 25 μM mdivi-1 had no effect on respiratory capacity in cortical neurons, yet still inhibited succinate-driven ROS production by ~60%. Notably, multiple reports indicate that mdivi-1 mitigates oxidative stress in animals and suggest that this effect is due to Drp1 inhibition (Liu et al., 2015; Sharp et al., 2015). However, conditional knockout of Drp1 in the cerebellum led to increased rather than decreased oxidative stress (Kageyama et al., 2012). Because Drp1 ablation alters oxidative stress in vivo, it is problematic to use KO mice to evaluate the ability of mdivi-1 to alter ROS in the absence of Drp1.
Overall, our results raise the possibility that mdivi-1 is a relatively unusual Complex I inhibitor that is not only weak and reversible, but has the ability to attenuate pathological ROS production at the Complex I Q site with limited impact to ROS in healthy neurons. Interestingly, metformin, a drug widely prescribed for the treatment of Type II diabetes, partially inhibits Complex I (El-Mir et al., 2000) and RET ROS (Batandier et al., 2006), but, in contrast to mdivi-1, is reported to exacerbate toxicity in a mouse model of Parkinson’s disease (Ismaiel et al., 2016). The translational potential of mdivi-1 is supported by the success of metformin in humans while its potentially different mode of action may increase its utility in some disorders. Nevertheless, it is important to note that because the structure of mdivi-1 contains a thiophenol, it likely has multiple cellular targets and additional experiments will be needed to demonstrate the specificity of its effects.
Drugs that prevent mitochondrial dysfunction are highly sought. Because we find that mdivi-1 influences multiple aspects of mitochondrial function—respiration and ROS—even in the absence of Drp1, it has limited utility in studies aiming to demonstrate a specific role for Drp1-dependent fission in biological processes. However, its ability to target several aspects of mitochondrial dysfunction, particularly succinate-driven RET ROS and cytochrome c release (Cassidy-Stone et al., 2008), make it an attractive therapeutic drug candidate for numerous diseases.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Brian M. Polster (bpolster@anes.umm.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Rat Primary Cortical Neurons
All procedures were in accordance with the NIH Guide for the Care and Use of Laboratory Animals and were approved by the University of Maryland Institutional Animal Care and Use Committee. Embryonic day 18 rat cortices were used to prepare primary cortical neurons according to established procedures (Yakovlev et al., 2001). Male and female rat cortices were mixed together for this study. Cortices were isolated, placed in Hank’s Balanced Salt Solution (HBSS), and meninges were removed. The cortices were then gently mixed by passing through a 5 ml pipette. Tissue was minced and dissociated with 1800 U/ml trypsin (Sigma-Aldrich, St. Louis, MO) at 37°C for 15 minutes. DNase I (200 U/ml, Sigma-Aldrich) was then added to the suspension and mixed by gentle inversion. This tissue mixture was centrifuged at 1,000 × g for 10 minutes, suspended in 2 ml of Neurobasal medium (Thermo Fisher, Waltham, MA) that included 10% fetal bovine serum (Sigma-Aldrich), 1x Gem21 (Gemini Bio-Products, Broderick, CA), 1x Glutamax (Thermo Fisher), and 100 IU/ml penicillin with 100 μg/ml streptomycin (Gemini Bio-Products), and then filtered through a 40 μm filter. Cells were seeded at a density of 0.8 × 105 cells/well (0.32 cm2) in V7 microplates (Agilent Technologies, Santa Clara, CA) and maintained in a humidified atmosphere of 95% air/5% CO2 at 37ºC. Glial proliferation was inhibited by addition of cytosine arabinofuranoside (5 μM) after 4 days in vitro (DIV). Neurons were used for experiments at DIV 10–14.
Cell Line Culture and Transfection
WT and Drp1 KO MEFs (Wakabayashi et al., 2009) that were spontaneously immortalized by serial passage (Kageyama et al., 2012) were cultured in Iscove’s Modified Dulbecco’s Medium supplemented with 10% FBS and 100 μg/ml primocin (InvivoGen, San Diego, CA). WT and Drp1 KO cell lines were authenticated by western blot for Drp1. COS-7 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS, penicillin (100 IU/ml), and streptomycin (100 μg/ml). COS-7 cells were transfected with the NDI1-puromycin construct (see below for description) using Lipofectamine 2000 (Thermo Fisher). At 24 hr post-transfection, cells were treated with 3 μg/ml puromycin for 4 days to select for transfected cells. Ndi1 expression was confirmed by immunoblot.
METHOD DETAILS
XF24 Microplate-based Respirometry
O2 consumption measurements from intact and permeabilized cells were performed using an XF24 Extracellular Flux Analyzer (Agilent Technologies) (Clerc and Polster, 2012). Artificial cerebrospinal fluid (aCSF) assay medium consisted of 120 mM NaCl, 3.5 mM KCl, 1.3 mM CaCl2, 0.4 mM KH2PO4, 1 mM MgCl2, 5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 15 mM glucose, and 4 mg/ml fatty acid free bovine serum albumin (BSA), pH 7.4. Cells were incubated in an aCSF volume of 0.675 ml in a CO2-free incubator at 37°C for one hour prior to assays to allow temperature and pH equilibration. Cells were then loaded into the instrument and further equilibrated for 15 min by three 3 min mix, 2 min wait cycles prior to measurements. Compounds of interest prepared in assay medium (75 μl) were pre-loaded into reagent delivery chambers a, b, c, and d at 10X, 11X, 12X, and 13X the final working concentration, respectively. Saponin and pyruvate were prepared fresh from powder for each individual experiment. The molecular identity of mdivi-1 from two different commercial sources (Sigma-Aldrich, St. Louis, MO and Enzo Life Sciences, Farmingdale, NY) was verified by the NMR and Mass Spectrometry facilities at the Medical College of Wisconsin (Milwaukee, WI). O2 consumption rate (OCR) measurements were made and drugs were injected sequentially as described in figure legends. For permeabilized cell assays, saponin (25 μg/ml unless otherwise indicated) was co-injected with 3.6 mM K2HPO4, 1 mM ADP, 5 mM EGTA, and the indicated mitochondrial substrate(s) to initiate permeabilization and ADP-stimulated respiration in aCSF assay medium. Note that saponin should be titrated for every individual lot obtained as the optimal concentration of saponin depends on source and purity. Substrate combinations for complex I-linked respiration consisted of 5 mM pyruvate plus 5 mM malate or 5 mM glutamate plus 5 mM malate. Succinate (5 mM) in combination with rotenone (0.5 μM) was used to assay complex II-dependent respiration. The ATP synthase inhibitor oligomycin (0.3 μg/ml) was used to measure OCR in the absence of oxidative phosphorylation, the protonophore carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP, 2–3 μM) was added to measure uncoupled respiration, and the complex III inhibitor antimycin A (1 μM) was used to inhibit O2 consumption by the mitochondrial electron transport chain.
Oxygen Consumption by Isolated Mitochondria
Sprague-Dawley rat non-synaptosomal forebrain mitochondria were isolated and purified on a Percoll™ gradient (Sims, 1990; Kristian and Fiskum, 2004). Following decapitation, the olfactory bulb and cerebellum were removed and the rest of the brain was placed in ice-cold mitochondrial isolation buffer (75 mM mannitol, 225 mM sucrose, 1 mM EGTA, 5 mM HEPES, pH 7.4) at 4°C. All tissue was minced with scissors in mitochondrial isolation buffer, and homogenized by hand with 10 up and down strokes. This homogenate was then centrifuged for 3 minutes at 1,330 × g at 4°C. Following supernatant removal, the pellet was resuspended in mitochondrial isolation buffer and again centrifuged at 1,330 × g at 4°C. Supernatant was then centrifuged at 4°C for 10 minutes at 21,200 × g. The resultant pellet was resuspended in 15% Percoll (100% Percoll stock consisted of 225 mM sucrose, 1 mM EGTA, 75 mM mannitol, 5 mM HEPES at pH 7.4; 100% Percoll stock was diluted with mitochondrial isolation buffer to obtain final concentrations). This 15% Percoll/tissue solution was layered into a two-step discontinuous gradient of 1.5 ml of 40% Percoll and 3.7 ml of 24% Percoll. This gradient was centrifuged at 4°C for 8 minutes at 30,700 × g. The mitochondrial fraction, which was at the interface of the bottom two layers, was taken and diluted 1:4 in mitochondrial isolation buffer, after which it was centrifuged at 4°C for 10 minutes at 16,700 × g. The resultant pellet was resuspended in 0.5 ml of 10 mg/ml BSA (made up in mitochondrial isolation buffer) and diluted 1:10 in mitochondrial isolation buffer. This mixture was centrifuged at 4°C for 10 minutes at 6,900 × g. The final mitochondrial pellet was then resuspended in 100 μl of mitochondrial isolation buffer lacking EGTA. Oxygen consumption was measured polarographically with a Clark-type oxygen electrode (Hansatech Instruments, obtained through PP Systems, Amesbury, MA) (Chinta et al., 2009). Mitochondria (0.5 mg/ml) were added to medium containing 125 mM KCl, 20 mM HEPES, 2 mM K2HPO4, pH 7.0 at 37°C. Complex I-dependent oxygen consumption was measured in the presence of glutamate and malate (5 mM each) and MgCl2 (1 mM). Complex II-dependent oxygen consumption was measured in the presence of succinate (5 mM), the complex I inhibitor rotenone (2 μM), and MgCl2 (1 mM). State 3 (phosphorylating) respiration was initiated by addition of ADP (1 mM).
Immunofluorescence and Mitochondrial Size
Primary cortical neurons were treated with mdivi-1 (75 μM) or vehicle for 60 min, after which they were fixed for 20 minutes in pre-warmed 4% formaldehyde, diluted 1:4 in PBS from stock (Thermo Fisher). Cells were then washed 3× with PBS, and permeabilized with 0.15% Trixton-X-100 in PBS for 20 minutes. Following 2× washing with PBS, cells were blocked with 7.5% BSA in PBS for 45 minutes. Cells were then immunostained with primary antibody in 7.5% BSA for 90 minutes. Primary antibodies used were anti-Tom20 (FL-145, Santa Cruz Biotechnology, Dallas, TX, 1:2,000) and anti-Drp1 (Clone 8/DLP1, BD Transduction Laboratories, San Jose, CA, 1:500). Following 2× washing with PBS, secondary antibodies were incubated for 60 minutes in 7.5% BSA. Secondary antibodies used were anti-rabbit IgG Alexa Fluor 594 (ThermoFisher Scientific, 1:250) and anti-mouse IgG Alexa Fluor 488 (ThermoFisher Scientific, 1:250). Cells were then washed 2× with PBS and left in PBS. At least eight random images for each condition were captured with a Zeiss ApoTome- and AxioCamMRm Rev.3 camera-equipped AxioObserver Z1 inverted microscope with a 100x/1.4 Plan-Apochromat objective lens. Tom20 images were converted to 32-bit grayscale format using ImageJ software, inverted, and analyzed with the same threshold settings in each experiment. Mitochondria were selected and analyzed for area in neuronal processes by a blinded observer using the particle analysis function. Quantification of mitochondrial size specifically in neuronal processes allowed for unambiguous identification of individual mitochondria. The restriction of mitochondrial width by the width of the neuronal processes was an additional advantage that enabled the use of mitochondrial area as a reasonable measure of mitochondrial length. Mitochondria were then binned for analysis of the neuronal mitochondrial population.
Analysis of Mitochondrial Morphology
COS-7 cells were grown in 2-well chamber slides (model 1 German borosilicate; Lab-Tek, Thermo Fisher). Cells were treated with mdivi-1 (50 μM), staurosporine (1 μM), or mdivi-1 plus staurosporine in phenol red-free cell culture medium (“DMEM”) or aCSF for a total of 2 hours. Some cells were treated with mdivi-1 (50 μM) or DMSO vehicle in DMEM for 24 hours. MitoTracker Red CMXRos (20 nM) was added 30 minutes prior to imaging to label mitochondria for analysis of morphology. Alternatively, cells were treated with mdivi-1 (50 μM) or DMSO vehicle for 1 or 6 hours, fixed, and immunostained for cytochrome c for morphology analysis. Immunostaining and analysis were performed as described above; primary antibody for cytochrome c was mouse monoclonal anti-cytochrome c (Clone 6H2.B4, BD Biosciences, San Jose, CA, 1:500). Images were acquired with a Zeiss AxioObserver Z1 fluorescence microscope, equipped with a 100X/1.45 a-Plan-FLUAR objective lens (Zeiss MicroImaging, Thornwood, NJ). Mitochondrial morphologies in cells were scored using blinded cell counting. Cells were divided into three categories based on mitochondrial morphology: “Normal”, “Fragmented,” and “Elongated” (Xu et al., 2016). The data represent at least 50 cell counts per condition.
Live Cell Time-Lapse Imaging
Cells were grown in 2-well chambered coverslides and transfected with 0.375 μg of mitochondrial matrix-localized green fluorescent protein (mito-GFP) construct using Lipofectamine 2000 (Thermo Fisher). Cells with mito-GFP labelled mitochondria were imaged at ~24hr post transfection in imaging medium containing phenol red-free DMEM, supplemented with 10% heat-inactivated FBS, 2 mM GlutaMAX, 1 mM sodium pyruvate, non-essential amino acids, 100 U/ml penicillin, 100 μg/ml streptomycin, and 25 mM HEPES (pH 7.4). Vehicle (DMSO)- or mdivi-1-treated cells were imaged for 65 minutes at room temperature. Sixty-five minutes was selected because mdivi-1 was reported to elongate mitochondria in COS cells within 60 minutes (Cassidy-Stone et al., 2008). The image acquisition interval was set at 5 minutes, producing a total of 13 images per experiment. Each image consisted of 6 z-sections taken with an interval of 0.5 μm. Images were acquired using a Nikon TI-E inverted microscope, equipped with CFI60 Plan Apochromat Lambda 100X Oil Immersion Objective Lens (N.A. 1.45), Perfect Focus module, monochrome Zyla sCMOS 5.5 Megapixel camera, and NIS-Elements imaging software. After acquisition, maximum intensity projections were obtained using NIS-Elements software. Image cropping and global adjustments to brightness and contrast and time lapse movie formatting were performed using ImageJ software (National Institutes of Health, Bethesda, MD).
Purification of Recombinant Drp1 and Dnm1
Human Drp1 (isoform 1) was expressed and purified as a His-tagged fusion construct as previously described (Cahill et al., 2015). Specifically, plasmid encoding Drp1 isoform 1 was transformed into Escherichia coli BL21 (DE3). Protein expression was induced with 0.5 mM isopropyl 1-thio-β-D-galactopyranoside for 12–16 hours and then affinity chromatography using Ni2+ Sepharose High Performance beads (GE Healthcare, Pittsburgh, PA) was employed to isolate His6-tagged Drp1. Following washing, resin-bound Drp1 was eluted with buffer containing 20 mM HEPES, pH 7.4, 400 mM NaCl, 5 mM MgCl2, and 500 mM imidazole. Fractions containing Drp1 were then pooled, dialyzed overnight at 4°C into buffer containing 20 mM HEPES, pH 7.4, 5 mM MgCl2, and 500 mM NaCl, and then concentrated. Purified protein was frozen in 0.5 ml aliquots using a dry ice and ethanol bath. Frozen aliquots were stored at −80°C. No loss of activity compared to fresh protein was observed if frozen aliquots were used within 12 hours of thawing.
Yeast Dnm1 was expressed and purified as a maltose binding protein (MBP) fusion construct as previously described (Koppenol-Raab et al., 2016). Briefly, MBP-Dnm1 was cloned into a modified pEG(KT) backbone by homologous recombination in yeast. The glutathione S-transferase (GST) open reading frame present in pEG(KT) (Mitchell et al., 1993) was removed by digesting the vector with SacI followed by gel purification of the pEG(KT) backbone lacking GST. The full-length MBP-Dnm1 open reading frame was PCR amplified using pMalc2xDnm1 (Wells et al., 2007) as a template and with homology to pEG(KT) at the 5′ and 3′ ends. The MBP-Dnm1 PCR fragment and SacI-digested pEG(KT) were transformed into SEY6210 yeast using the LiAc method (Schiestl et al., 1989). Circularized plasmids were recovered by plasmid rescue, and successful recombination of MBP-Dnm1 into the pEG(KT) backbone was verified by sequencing.
Protease-deficient DDY1810 cells (MATa, leu23,trp1, ura3-52, prb1-1122, pep4-3, pre1-451) (Shang et al., 2003) were transformed with the pEGKT-MBPDnm1 plasmid using the LiAc method (Schiestl et al., 1989). For large scale purification, plasmid-containing cells were grown in 50 ml synthetic complete media lacking uracil and with 2% glucose for 36–48 hrs at 30°C, shaking at 200 rpm until reaching OD 600 of 2. Cultures were then back-diluted (25 ml into 1 l) into synthetic complete medium lacking uracil and leucine, with 2% raffinose and 2× tryptophan. Cells were grown (~24h) at 26°C, shaking at 250 rpm, and expression was induced by the addition of 10 g yeast extract, 20 g Bacto-tryptone and 100 ml 20% w/v galactose. Cells were grown overnight at 26°C with shaking. Harvested cells were then washed in sterile water, resuspended to 80 ml in sterile water with AEBSF (0.1 mM final) and 10 ml-aliquotted into conical tubes. The cells were stored at −80°C. Four aliquots at a time were thawed on ice, and buffer A (50mM sodium phosphate pH 7.4, 500mM NaCl, 2mM DTT and 5mM MgCl2) was added to an 80 ml total volume. One tablet of EDTA-free protease inhibitors (Roche Applied Science) and PMSF (2 mM final) were added. Cells were lysed by 5–6 passes on an Emulsiflex C3 Homogenizer (Avestin, Ottawa, CA) at 21,000 psi. DNase was added at 1 μg/ml, and the lysates were assessed by microscopy for lysis. The lysate was then clarified by centrifugation for 45 min at 15,000 rpm. The resulting soluble MBP-Dnm1 was then purified as described (Wells et al., 2007). TEV protease was added to pooled fractions from the elution at 2:1 molar ratio (TEV:MBP-Dnm1) for 24 hours at 4°C to cleave the N-terminal MBP tag. Cleaved Dnm1 was then run on a SE-75 sizing column (GE Healthcare, Pittsburgh PA). Sample purity was assessed by Coomassie-stained SDS-Page gel and found to be at least 90% pure.
GTPase Activity Assay
Drp1 activity was measured in the absence and presence of mdivi-1 using a continuous, coupled GTPase assay (Ingerman and Nunnari, 2005). GTPase activity was determined in a 96-well format using 150μl of GTPase reaction buffer (25mM HEPES, 25mM PIPES, 7.5mM KCl, 5mM MgCl2, 1mM phosphoenolpyruvate, 20 units/ml pyruvate kinase/lactate dehydrogenase, 600μM NADH, pH 7.0), which was placed into a 96-well Corning Costar flat, clear bottom plate (Sigma-Aldrich). The oxidation of NADH was measured at A340 for 40 minutes at 25°C using a Molecular Devices (Sunnyvale, CA) Flexstation 3 Multi-Detection Reader with Integrated Fluid Transfer. Reactions were run using 5 μM Drp1, 150 mM NaCl, and 5% DMSO at indicated GTP and mdivi-1 concentrations. GTPase assays were started by the addition of GTP to the desired concentration. Global analysis of the data was fit using Igor Pro to the following equation that models uncompetitive inhibition: Vo = (Vmax*[GTP])/(KM+(1+(I/Ki))*[GTP]). Data from three independent preparations of enzyme are reported with standard errors of the mean and are representative of similar observations conducted with multiple preparations at different concentrations of Drp1.
Purified Dnm1 from yeast (10 μM) was incubated for 30 min on ice with 1 mM GTP and mdivi-1 at the indicated concentrations. Samples (160 μl) were placed in 96-well plates, and the reactions were started by addition of 40 μl of a 5× master mix, including 125 mM HEPES pH 7.0, 125 mM PIPES pH 7.0, 5 mM MgCl2, 37.5 mM KCl, 5 mM phosphoenolpyruvate, 100U/ml pyruvate kinase/lactate dehydrogenase, and 1.5 mM NADH. Reactions (150 μl) were transferred with a multichannel pipette to a new plate, and absorbance at 340 nm was recorded for 40 min at 30 sec intervals on a Spectra Pro X. Absorbance was plotted vs. time, and the linear portions of the plot fit to a line. The slope (3A340/sec) was converted to activity in nmol/min with the following equation: activity = (3A340/sec * 60 sec/min) * (0.001 l) * (19 nmol/mol) / (6220 M−1cm−1) / (0.41 cm). A buffer control for each GTP concentration was done and the background rate subtracted from the activity.
Cell Proliferation Assay
WT or Drp1 KO mouse embryonic fibroblasts were plated in DMEM cell culture medium without pyruvate on 6-well plates and allowed 24 hours to attach and recover, after which they were treated with vehicle (DMSO) or mdivi-1 (25 or 50 μM). At 0, 24, or 48 hours following treatment, cells were fixed with 4% formaldehyde and stained with 1 μg/ml Hoechst 33342 (Thermo Fisher). Images were taken at 20X using an EVOS FL Auto (Thermo Fisher) fluorescence microscope, with an excitation filter of 357/44 nm and an emission filter of 447/60 nm. At least 5 images were acquired per treatment group per experiment, with at least 3 experiments performed. The number of Hoechst-stained cells was quantified at each time point and expressed as a percentage of cells present at 0 hours.
Electron Microscopy
Drp 1 WT and KO cells were plated on 10 mm diameter glass coverslips at 2 × 104 cells/coverslip and cultured for 24 hours. Cells were then treated with mdivi-1 (50 μM) or DMSO vehicle control for 16 min and fixed in Trump’s Fixative (McDowell and Trump, 1976), post-fixed in 1% osmium tetroxide, dehydrated in graded alcohols, enbloc stained with uranyl acetate, embedded in epoxy resin, sectioned (50–100 nm) on an ultramicrotome, stained with lead acetate and examined in a Tecnai G12 electron microscope. Mitochondrial profile lengths were measured using a Calculated Industries Scale Master Pro Digital Plan measuring device.
Complex I Activity Assay
Mdivi-1 intrinsic fluorescence interferes with Complex I assays that are based on NADH autofluorescence. Therefore, Complex I activity was measured using 2,6-dichloroindophenol (DCIP) as a terminal electron acceptor. Decylubiquinone reduced by Complex I delivers electrons to DCIP which is followed spectrophotometrically at 600 nm. An incubation volume of 1 ml containing 25 mM potassium phosphate (pH 7.8), 3.5 g/l BSA, 120 μM DCIP, 140 μM decylubiquinone, 1 μM antimycin A, and 0.4 mM NADH was used. Rat brain mitochondria (50 μg) were pre-incubated at 37°C in 1 ml of assay buffer without NADH. After 3 min, 0.4 mM NADH was added and absorbance at 600 nm was measured. Drug was added at 4 min and then 4 min of additional measurements were made.
Ndi1-expressing Construct
Yeast genomic DNA was extracted from wild type Saccharomyces cerevisiae (strain FY833) using Yeast DNA Extraction Kit (Thermo Scientific) according to the manufacturer’s instructions. The NDI1 gene was PCR-amplified from yeast genomic DNA using two primers 5′ AAAGAATTCGCCACCATGCTATCGAAGAATTTGTATAG 3′ and 5′ AAAGGTACCGTTAATCCTTTAAAAAAGTCTC 3′. After digestion with EcoRI and KpnI, PCR products were ligated into the pEGFP-N1 expression vector (Clontech, Mountain View, CA) and confirmed by DNA sequencing. NDI1 was sub-cloned from pEGFP-N1 vector into a pSMPUW base plasmid with a puromycin resistance gene (Cell BioLabs, San Diego, CA) by the University of Maryland, Baltimore Recombinant Virus Core.
Immunoblotting
NDI1-transfected or parental COS-7 cells were lysed by sonication in sodium dodecyl sulfate and protease inhibitor cocktail (Millipore, Billerica, MA). SDS-PAGE and immunodetection for Ndi1 (1:1000, graciously provided by Takao Yagi (The Scripps Research Institute, La Jolla, CA)) and β-actin (1:5000; Sigma-Aldrich) were performed as described (Sharma et al., 2011). In detail, 30 μg of protein were loaded on Novex WedgeWell 4–20% Tris-Glycine Mini Gels (Thermo Fisher), and run at 200V for 45 minutes, after which they were transferred to PVDF membranes. Blots were then washed 3× 10 minutes in PBS, following by blocking in 5% BSA in PBS-T (20× stock from ThermoFisher was diluted 1:20 in H2O). Primary antibodies were then incubated in 5% BSA in PBS-T overnight at 4°C. The next morning, blots were washed 3× 10 minutes in PBS-T, following by incubation with secondary antibodies at 1:10,000 dilution for 1 hour in 5% BSA in PBS-T. Finally, blots were washed 3× 10 minutes in PBS-T, after which they were placed in PBS. Proteins were visualized using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher) and imaged with a ChemiDoc MP system (Bio-Rad, Hercules, CA).
Measurement of Intracellular ROS
Rat cortical neurons (2 × 105/well, DIV10–14) or MEFs (variable densities to achieve ~70% confluence at the time of imaging) were plated on borosilicate 4-well chamber slides. On the day of imaging, DHE (Thermo Fisher) was diluted to 2 mM in aCSF, and then incubated with Dowex cation exchange beads (Sigma-Aldrich) for 30 min to remove oxidized DHE products. Post-bead incubation, DHE was diluted in aCSF to a final concentration of 5 μM for MEFs and of 1 μM for neurons. Cells were incubated at 37°C for 1 hour to allow DHE uptake and then imaged at 20X using an EVOS FL Auto fluorescence microscope, with an excitation filter of 531/40 nm and an emission filter of 593/40 nm. Images were acquired at 4 min intervals and fluorescence intensity over time was analyzed in blinded fashion using ImageJ software.
H2O2 Detection Using Amplex® UltraRed
Guinea pig brain mitochondria were isolated on a discontinuous Percoll™ gradient as described for oxygen consumption measurements. Mitochondria (0.1 mg/ml) were added to medium containing 125 mM KCl, 20 mM HEPES, 2 mM K2HPO4, 1 mM MgCl2, 0.1 mM EGTA, 0.025% fatty acid-free BSA, 1 μM Amplex® UltraRed, and 2.5 U/ml horseradish peroxidase, pH 7.0 at 37°C. Dye was excited at 550 nm, and emission was measured at 585 nm using a fluorescence spectrophotometer (Tretter et al., 2007). Traces were calibrated in pmol H2O2 for each experiment using known concentrations of freshly prepared H2O2.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical parameters, including the exact value of n, dispersion and precision measures (mean ± SD or mean ± SE), and statistical significance are reported in the Figures and Figure Legends. Statistical analyses were performed using SigmaPlot 12.0 (Systat Software, Inc., San Jose, CA). OCR and electron microscopy length measurements comparing treatments and genotype (Figures 4C and 4F, respectively) were evaluated by two-way analysis of variance (ANOVA) with Tukey’s post-hoc analysis, while all other measurements were analyzed by one-way ANOVA with Tukey’s post-hoc analysis. p < 0.05 was considered to be statistically significant.
Supplementary Material
Highlights.
Mdivi-1 does not impair Drp1 GTPase activity or acutely elongate mitochondria
Mdivi-1 reversibly inhibits respiration at mitochondrial complex I
Mdivi-1 inhibits reverse electron transfer reactive oxygen species (ROS) production
Effects of mdivi-1 on respiration and ROS are independent of Drp1
Acknowledgments
The authors thank Ru-ching Hsia, Ph.D. of the University of Maryland Dental School Core Imaging Facility for her assistance in collecting the electron microscopic images. They also thank Takao Yagi (The Scripps Research Institute, San Diego, CA) for the generous gift of the Ndi1 antibody. This research was supported by NINDS Grants R01NS064978, R01NS085165, and R21NS096538 (B.M.P.), by NICHD Grant P01HD016596 and by NIGMS Grants R01GM089853 (H.S.) and R01GM067180 (R.B.H.), OTKA (NK 81983, V.A.), Hungarian Academy of Sciences MTA TKI 02001 (V.A.), and Hungarian Brain Research Program - Grant No. KTIA_13_NAP-A-III/6 (V.A.).
Footnotes
AUTHOR CONTRIBUTIONS
E.A.B, P.C., B.A.R., E.C., S.X.G., T.C.F., T.K., S.U., A.M., and M.K. performed and/or designed and analyzed bioenergetics and imaging experiments. E.A.B., L.T., and V. A-V. conducted ROS measurements. A.J.S. designed the electron microscopy study. H.S. provided Drp1 knockout MEFs and conceptual advice. K.I. cloned NDI1 into the pEGPF-N1 vector. A.K. and N.Y. helped design the linked Complex I–III–IV activity assay. N.W.K., L.K.P., and R.B.H. conducted and analyzed GTPase assays. E.A.B., P.C., R.B.H., and B.M.P. designed the study and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andreyev AY, Kushnareva YE, Starkov AA. Mitochondrial metabolism of reactive oxygen species. Biochemistry (Mosc ) 2005;70:200–214. doi: 10.1007/s10541-005-0102-7. [DOI] [PubMed] [Google Scholar]
- Batandier C, Guigas B, Detaille D, El-Mir MY, Fontaine E, Rigoulet M, Leverve XM. The ROS production induced by a reverse-electron flux at respiratory-chain complex 1 is hampered by metformin. J Bioenerg Biomembr. 2006;38:33–42. doi: 10.1007/s10863-006-9003-8. [DOI] [PubMed] [Google Scholar]
- Berman SB, Chen YB, Qi B, McCaffery JM, Rucker EB, III, Goebbels S, Nave KA, Arnold BA, Jonas EA, Pineda FJ, Hardwick JM. Bcl-x L increases mitochondrial fission, fusion, and biomass in neurons. J Cell Biol. 2009;184:707–719. doi: 10.1083/jcb.200809060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet A, Margolis EB, Zhang J, Hsieh I, Zhang J, Hnasko TS, Ahmad J, Edwards RH, Sesaki H, Huang EJ, Nakamura K. Loss of mitochondrial fission depletes axonal mitochondria in midbrain dopamine neurons. J Neurosci. 2014;34:14304–14317. doi: 10.1523/JNEUROSCI.0930-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand MD, Goncalves RL, Orr AL, Vargas L, Gerencser AA, Borch JM, Wang YT, Melov S, Turk CN, Matzen JT, Dardov VJ, Petrassi HM, Meeusen SL, Perevoshchikova IV, Jasper H, Brookes PS, Ainscow EK. Suppressors of Superoxide-H2O2 Production at Site IQ of Mitochondrial Complex I Protect against Stem Cell Hyperplasia and Ischemia-Reperfusion Injury. Cell Metab. 2016;24:582–592. doi: 10.1016/j.cmet.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill TJ, Leo V, Kelly M, Stockenhuber A, Kennedy NW, Bao L, Cereghetti G, Harper AR, Czibik G, Lao C, Bellahcene M, Steeples V, Ghaffari S, Yavari A, Mayer A, Poulton J, Ferguson DJ, Scorrano L, Hettiarachchi NT, Peers C, Boyle J, Hill RB, Simmons A, Watkins H, Dear TN, Ashrafian H. Resistance of Dynamin-related Protein 1 Oligomers to Disassembly Impairs Mitophagy, Resulting in Myocardial Inflammation and Heart Failure. J Biol Chem. 2015;290:25907–25919. doi: 10.1074/jbc.M115.665695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, Kurth MJ, Shaw JT, Hinshaw JE, Green DR, Nunnari J. Chemical inhibition of the mitochondrial division dynamin reveals its role in bax/bak-dependent mitochondrial outer membrane permeabilization. Dev Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CR, Manlandro CM, Arnoult D, Stadler J, Posey AE, Hill RB, Blackstone C. A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division. J Biol Chem. 2010;285:32494–32503. doi: 10.1074/jbc.M110.142430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinta SJ, Rane A, Yadava N, Andersen JK, Nicholls DG, Polster BM. Reactive oxygen species regulation by AIF- and complex I-depleted brain mitochondria. Free Radic Biol Med. 2009;46:939–947. doi: 10.1016/j.freeradbiomed.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchani ET, Pell VR, Gaude E, Aksentijevic D, Sundier SY, Robb EL, Logan A, Nadtochiy SM, Ord EN, Smith AC, Eyassu F, Shirley R, Hu CH, Dare AJ, James AM, Rogatti S, Hartley RC, Eaton S, Costa AS, Brookes PS, Davidson SM, Duchen MR, Saeb-Parsy K, Shattock MJ, Robinson AJ, Work LM, Frezza C, Krieg T, Murphy MP. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515:431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc P, Ge SX, Hwang H, Waddell J, Roelofs BA, Karbowski M, Sesaki H, Polster BM. Drp1 is dispensable for apoptotic cytochrome c release in primed MCF10A and fibroblast cells but affects Bcl-2 antagonist-induced respiratory changes. Br J Pharmacol. 2014;171:1988–1999. doi: 10.1111/bph.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc P, Polster BM. Investigation of mitochondrial dysfunction by sequential microplate-based respiration measurements from intact and permeabilized neurons. PLoS ONE. 2012;7:e34465. doi: 10.1371/journal.pone.0034465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Mir MY, Nogueira V, Fontaine E, Averet N, Rigoulet M, Leverve X. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J Biol Chem. 2000;275:223–228. doi: 10.1074/jbc.275.1.223. [DOI] [PubMed] [Google Scholar]
- Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- Grohm J, Kim SW, Mamrak U, Tobaben S, Cassidy-Stone A, Nunnari J, Plesnila N, Culmsee C. Inhibition of Drp1 provides neuroprotection in vitro and in vivo. Cell Death Differ. 2012;19:1446–1458. doi: 10.1038/cdd.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerman E, Nunnari J. A continuous, regenerative coupled GTPase assay for dynamin-related proteins. Methods Enzymol. 2005;404:611–619. doi: 10.1016/S0076-6879(05)04053-X. [DOI] [PubMed] [Google Scholar]
- Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, Otera H, Nakanishi Y, Nonaka I, Goto Y, Taguchi N, Morinaga H, Maeda M, Takayanagi R, Yokota S, Mihara K. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol. 2009;11:958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- Ismaiel AA, Espinosa-Oliva AM, Santiago M, Garcia-Quintanilla A, Oliva-Martin MJ, Herrera AJ, Venero JL, de Pablos RM. Metformin, besides exhibiting strong in vivo anti-inflammatory properties, increases mptp-induced damage to the nigrostriatal dopaminergic system. Toxicol Appl Pharmacol. 2016;298:19–30. doi: 10.1016/j.taap.2016.03.004. [DOI] [PubMed] [Google Scholar]
- Ji F, Sharpley MS, Derbeneva O, Alves LS, Qian P, Wang Y, Chalkia D, Lvova M, Xu J, Yao W, Simon M, Platt J, Xu S, Angelin A, Davila A, Huang T, Wang PH, Chuang LM, Moore LG, Qian G, Wallace DC. Mitochondrial DNA variant associated with Leber hereditary optic neuropathy and high-altitude Tibetans. Proc Natl Acad Sci U S A. 2012;109:7391–7396. doi: 10.1073/pnas.1202484109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y, Hoshijima M, Seo K, Bedja D, Sysa-Shah P, Andrabi SA, Chen W, Hoke A, Dawson VL, Dawson TM, Gabrielson K, Kass DA, Iijima M, Sesaki H. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 2014;33:2798–2813. doi: 10.15252/embj.201488658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y, Zhang Z, Roda R, Fukaya M, Wakabayashi J, Wakabayashi N, Kensler TW, Reddy PH, Iijima M, Sesaki H. Mitochondrial division ensures the survival of postmitotic neurons by suppressing oxidative damage. J Cell Biol. 2012;197:535–551. doi: 10.1083/jcb.201110034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol-Raab M, Harwig MC, Posey AE, Egner JM, MacKenzie KR, Hill RB. A Targeted Mutation Identified through pKa Measurements Indicates a Postrecruitment Role for Fis1 in Yeast Mitochondrial Fission. J Biol Chem. 2016;291:20329–20344. doi: 10.1074/jbc.M116.724005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristian T, Fiskum G. A fluorescence-based technique for screening compounds that protect against damage to brain mitochondria. Brain Res Brain Res Protoc. 2004;13:176–182. doi: 10.1016/j.brainresprot.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Lambert AJ, Boysen HM, Buckingham JA, Yang T, Podlutsky A, Austad SN, Kunz TH, Buffenstein R, Brand MD. Low rates of hydrogen peroxide production by isolated heart mitochondria associate with long maximum lifespan in vertebrate homeotherms. Aging Cell. 2007;6:607–618. doi: 10.1111/j.1474-9726.2007.00312.x. [DOI] [PubMed] [Google Scholar]
- Liu JM, Yi Z, Liu SZ, Chang JH, Dang XB, Li QY, Zhang YL. The mitochondrial division inhibitor mdivi-1 attenuates spinal cord ischemia-reperfusion injury both in vitro and in vivo: Involvement of BK channels. Brain Res. 2015;1619:155–165. doi: 10.1016/j.brainres.2015.03.033. [DOI] [PubMed] [Google Scholar]
- Ma D, Taneja TK, Hagen BM, Kim BY, Ortega B, Lederer WJ, Welling PA. Golgi export of the Kir2.1 channel is driven by a trafficking signal located within its tertiary structure. Cell. 2011;145:1102–1115. doi: 10.1016/j.cell.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Manczak M, Reddy PH. Mitochondrial division inhibitor 1 protects against mutant huntingtin-induced abnormal mitochondrial dynamics and neuronal damage in Huntington’s disease. Hum Mol Genet. 2015;24:7308–7325. doi: 10.1093/hmg/ddv429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell EM, Trump BF. Histologic fixatives suitable for diagnostic light and electron microscopy. Arch Pathol Lab Med. 1976;100:405–414. [PubMed] [Google Scholar]
- Mitchell DA, Marshall TK, Deschenes RJ. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–722. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- Ong SB, Subrayan S, Lim SY, Yellon DM, Davidson SM, Hausenloy DJ. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation. 2010;121:2012–2022. doi: 10.1161/CIRCULATIONAHA.109.906610. [DOI] [PubMed] [Google Scholar]
- Packer L, Mustafa MG. Pathways of electron flow established by tetramethylphenylenediamine in mitochondria and ascites tumor cells. Biochim Biophys Acta. 1966;113:1–12. doi: 10.1016/s0926-6593(66)80115-7. [DOI] [PubMed] [Google Scholar]
- Rappold PM, Cui M, Grima JC, Fan RZ, de Mesy-Bentley KL, Chen L, Zhuang X, Bowers WJ, Tieu K. Drp1 inhibition attenuates neurotoxicity and dopamine release deficits in vivo. Nat Commun. 2014;5:5244. doi: 10.1038/ncomms6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosdah AA, Holien K, Delbridge LM, Dusting GJ, Lim SY. Mitochondrial fission - a drug target for cytoprotection or cytodestruction? Pharmacol Res Perspect. 2016;4:e00235. doi: 10.1002/prp2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Scialo F, Sriram A, Fernandez-Ayala D, Gubina N, Lohmus M, Nelson G, Logan A, Cooper HM, Navas P, Enriquez JA, Murphy MP, Sanz A. Mitochondrial ROS Produced via Reverse Electron Transport Extend Animal Lifespan. Cell Metab. 2016;23:725–734. doi: 10.1016/j.cmet.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo BB, Kitajima-Ihara T, Chan EK, Scheffler IE, Matsuno-Yagi A, Yagi T. Molecular remedy of complex I defects: rotenone-insensitive internal NADH-quinone oxidoreductase of Saccharomyces cerevisiae mitochondria restores the NADH oxidase activity of complex I-deficient mammalian cells. Proc Natl Acad Sci U S A. 1998;95:9167–9171. doi: 10.1073/pnas.95.16.9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang C, Hazbun TR, Cheeseman IM, Aranda J, Fields S, Drubin DG, Barnes G. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol Biol Cell. 2003;14:3342–3355. doi: 10.1091/mbc.E02-11-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma LK, Fang H, Liu J, Vartak R, Deng J, Bai Y. Mitochondrial respiratory complex I dysfunction promotes tumorigenesis through ROS alteration and AKT activation. Hum Mol Genet. 2011;20:4605–4616. doi: 10.1093/hmg/ddr395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp WW, Beiser DG, Fang YH, Han M, Piao L, Varughese J, Archer SL. Inhibition of the mitochondrial fission protein dynamin-related protein 1 improves survival in a murine cardiac arrest model. Crit Care Med. 2015;43:e38–e47. doi: 10.1097/CCM.0000000000000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer TB, Betarbet R, Testa CM, Seo BB, Richardson JR, Kim JH, Miller GW, Yagi T, Matsuno-Yagi A, Greenamyre JT. Mechanism of toxicity in rotenone models of Parkinson’s disease. J Neurosci. 2003;23:10756–10764. doi: 10.1523/JNEUROSCI.23-34-10756.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims NR. Rapid isolation of metabolically active mitochondria from rat brain and subregions using Percoll density gradient centrifugation. J Neurochem. 1990;55:698–707. doi: 10.1111/j.1471-4159.1990.tb04189.x. [DOI] [PubMed] [Google Scholar]
- Starkov AA, Fiskum G. Regulation of brain mitochondrial H2O2 production by membrane potential and NAD(P)H redox state. J Neurochem. 2003;86:1101–1107. doi: 10.1046/j.1471-4159.2003.01908.x. [DOI] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162:552–563. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter L, Adam-Vizi V. Moderate dependence of ROS formation on DeltaPsim in isolated brain mitochondria supported by NADH-linked substrates. Neurochem Res. 2007;32:569–575. doi: 10.1007/s11064-006-9130-y. [DOI] [PubMed] [Google Scholar]
- Tretter L, Mayer-Takacs D, Adam-Vizi V. The effect of bovine serum albumin on the membrane potential and reactive oxygen species generation in succinate-supported isolated brain mitochondria. Neurochem Int. 2007;50:139–147. doi: 10.1016/j.neuint.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votyakova TV, Reynolds IJ. DeltaPsi(m)-Dependent and -independent production of reactive oxygen species by rat brain mitochondria. J Neurochem. 2001;79:266–277. doi: 10.1046/j.1471-4159.2001.00548.x. [DOI] [PubMed] [Google Scholar]
- Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, Sesaki H. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186:805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RC, Picton LK, Williams SC, Tan FJ, Hill RB. Direct binding of the dynamin-like GTPase, Dnm1, to mitochondrial dynamics protein Fis1 is negatively regulated by the Fis1 N-terminal arm. J Biol Chem. 2007;282:33769–33775. doi: 10.1074/jbc.M700807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Xia SX, Li QQ, Gao Y, Shen X, Ma L, Zhang MY, Wang T, Li YS, Wang ZF, Luo CL, Tao LY. Mitochondrial division inhibitor 1 (Mdivi-1) offers neuroprotection through diminishing cell death and improving functional outcome in a mouse model of traumatic brain injury. Brain Res. 2016;1630:134–143. doi: 10.1016/j.brainres.2015.11.016. [DOI] [PubMed] [Google Scholar]
- Xu S, Cherok E, Das S, Li S, Roelofs BA, Ge SX, Polster BM, Boyman L, Lederer WJ, Wang C, Karbowski M. Mitochondrial E3 ubiquitin ligase MARCH5 controls mitochondrial fission and cell sensitivity to stress-induced apoptosis through regulation of MiD49 protein. Mol Biol Cell. 2016;27:349–359. doi: 10.1091/mbc.E15-09-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadava N, Nicholls DG. Spare respiratory capacity rather than oxidative stress regulates glutamate excitotoxicity after partial respiratory inhibition of mitochondrial complex I with rotenone. J Neurosci. 2007;27:7310–7317. doi: 10.1523/JNEUROSCI.0212-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovlev AG, Ota K, Wang G, Movsesyan V, Bao WL, Yoshihara K, Faden AI. Differential expression of apoptotic protease-activating factor-1 and caspase-3 genes and susceptibility to apoptosis during brain development and after traumatic brain injury. J Neurosci. 2001;21:7439–7446. doi: 10.1523/JNEUROSCI.21-19-07439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang S, Maezawa I, Trushin S, Minhas P, Pinto M, Jin LW, Prasain K, Nguyen TD, Yamazaki Y, Kanekiyo T, Bu G, Gateno B, Chang KO, Nath KA, Nemutlu E, Dzeja P, Pang YP, Hua DH, Trushina E. Modulation of mitochondrial complex I activity averts cognitive decline in multiple animal models of familial Alzheimer’s Disease. EBioMedicine. 2015;2:294–305. doi: 10.1016/j.ebiom.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.