Abstract
Metaplastic breast carcinoma is an aggressive form of invasive breast cancer with histological evidence of epithelial to mesenchymal transition (EMT). However, the defining molecular events are unknown. Here we show that CCN6 (WISP3), a secreted matricellular protein of the CCN (CYR61/CTGF/NOV) family, is significantly down regulated in clinical samples of human spindle cell metaplastic breast carcinoma. We generated a mouse model of mammary epithelial-specific Ccn6 deletion by developing a floxed Ccn6 mouse which was bred with an MMTV-Cre mouse. Ccn6fl/fl; MMTV-Cre mice displayed severe defects in ductal branching and abnormal age-related involution compared to littermate controls. Ccn6fl/fl ;MMTV-Cre mice developed invasive high grade mammary carcinomas with bona fide EMT, histologically similar to human metaplastic breast carcinomas. Global gene expression profiling of Ccn6fl/fl mammary carcinomas and comparison of orthologous genes with a human metaplastic carcinoma signature revealed a significant overlap of 87 genes (p=5×10−11). Among the shared deregulated genes between mouse and human are important regulators of epithelial morphogenesis including Cdh1, Ck19, Cldn3 and 4, Ddr1, and Wnt10a. These results document a causal role for Ccn6 deletion in the pathogenesis of metaplastic carcinomas with histological and molecular similarities with human disease. We provide a platform to study new targets in the diagnosis and treatment of human metaplastic carcinomas, and a new disease relevant model in which to test new treatment strategies.
Keywords: CCN6, WISP3, matricellular, microenvironment, metaplastic carcinoma, triple negative, breast cancer
INTRODUCTION
Metaplastic breast carcinomas constitute approximately 1% of invasive carcinomas and are characterized by histological evidence of epithelial to mesenchymal transition (EMT) towards spindle, squamous, and less frequently heterologous elements including chondroid and osseous differentiation (Oberman 1987). Metaplastic carcinomas are a subtype of triple negative breast cancer (TNBC) poorly responsive to chemotherapy with exhibit high propensity for distant metastasis (Bae et al 2011, Luini et al 2007). Recent comprehensive genomic analyses revealed that metaplastic carcinomas belong to the mesenchymal-like subtype of TNBC, with a distinct transcriptional profile than other invasive breast carcinomas (Burstein et al 2015, Hennessy et al 2009, Weigelt et al 2015). However, the molecular determinants of metaplastic carcinomas needed to inform effective treatments are still incompletely understood.
Metaplasia is the reversible change in which one adult cell type is replaced by another adult cell type. Epithelial cells may undergo metaplasia through EMT, which during tumorigenesis results in the acquisition of molecular and phenotypic changes resulting in spindle morphology and dysfunctional cell-cell adhesion, leading to invasion and metastasis (Hugo et al 2007). Of all human cancers, metaplastic carcinomas provide the clearest evidence of morphological and molecular EMT with deregulation of genes involved in cell adhesion and low expression of claudins (Burstein et al 2015, Hennessy et al 2009, Weigelt et al 2015).
CCN6 (WISP3) is a secreted matricellular protein of the CCN family, which includes 6 members that play regulatory rather than structural roles in embryonic development, cell attachment, and growth (Jun and Lau 2011). CCN proteins act in a cell and tissue-specific manner primarily through direct binding to cell surface receptors including integrins and by modulating the effect of extracellular growth factors on epithelial cells (Huang et al 2008, Lau 2011, Lorenzatti et al 2011, Pal et al 2012a, Zhang 2005). We have shown that CCN6 is secreted by ductal epithelial cells in the breast, and that CCN6 downregulation in nontumorigenic breast cells is a robust inducer of EMT and is sufficient to confer growth factor independent survival and resistance to anoikis, a property of metastatic cancer cells (Huang et al 2008, Huang et al 2010, Lorenzatti et al 2011, Pal et al 2012a, Pal et al 2012b, Zhang 2005). Despite this knowledge, a direct role of CCN6 in breast tumorigenesis and the underlying mechanisms have remained elusive in part due to the lack of a physiologically relevant genetically engineered mouse model.
RESULTS
CCN6 protein is reduced in human metaplastic carcinomas
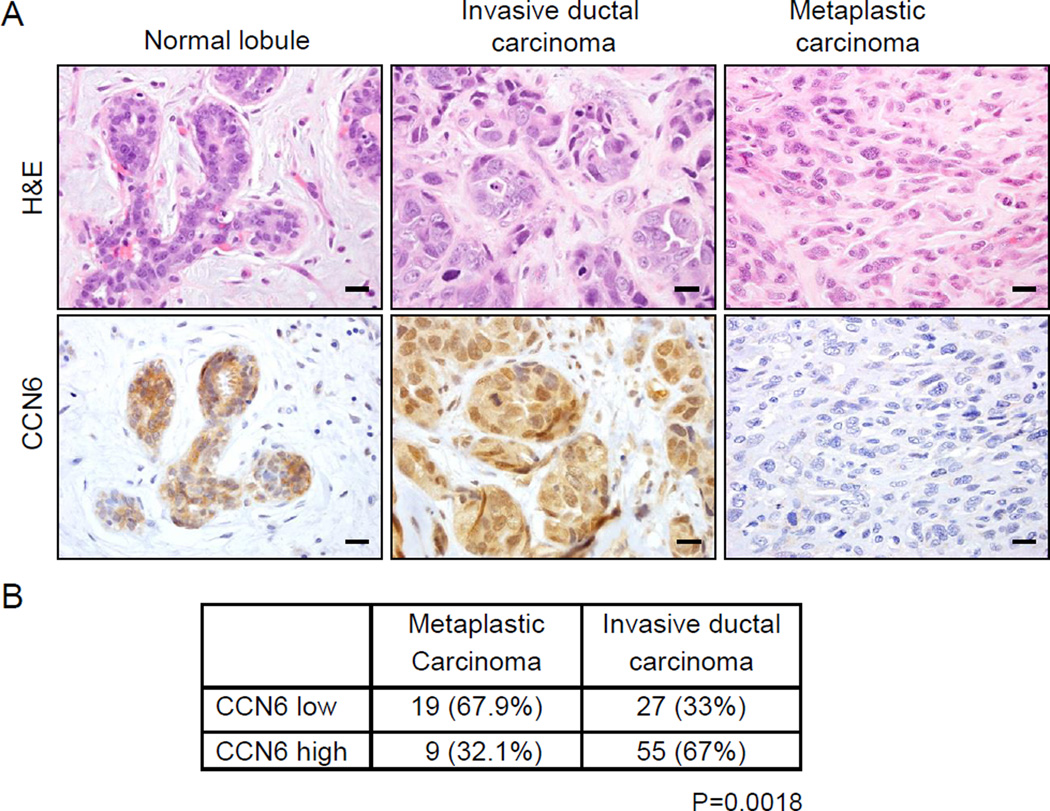
Based on our previous studies showing that CCN6 regulates the transition between epithelial and mesenchymal states in breast cells, we set out to investigate CCN6 expression in breast tissues including invasive carcinomas and metaplastic carcinomas. Immunohistochemical evaluation revealed that CCN6 level is significantly reduced in spindle metaplastic carcinomas compared to normal breast and invasive ductal carcinomas (Fig.1A). All normal breast lobules examined, and 67% of invasive ductal carcinomas maintained high levels of CCN6, and 33% had low levels. This distribution was shifted in metaplastic carcinomas, among which 67.9% had low CCN6 expression (Fig.1B).
Fig. 1. CCN6 protein is reduced in human spindle metaplastic carcinomas of the breast compared to invasive ductal carcinoma and normal breast lobules.
A. images of a normal breast lobule, an invasive ductal carcinoma with no special features, and a spindle metaplastic carcinoma stained with hematoxylin and eosin (H&E) and CCN6 immunostaining (magnification 400x). B. Invasive metaplastic and ductal carcinomas with high or low CCN6 protein. Chi square test P=0.0018. (Scale bars, 20 µm.)
Mammary epithelial specific Ccn6 deletion causes abnormal mammary gland development
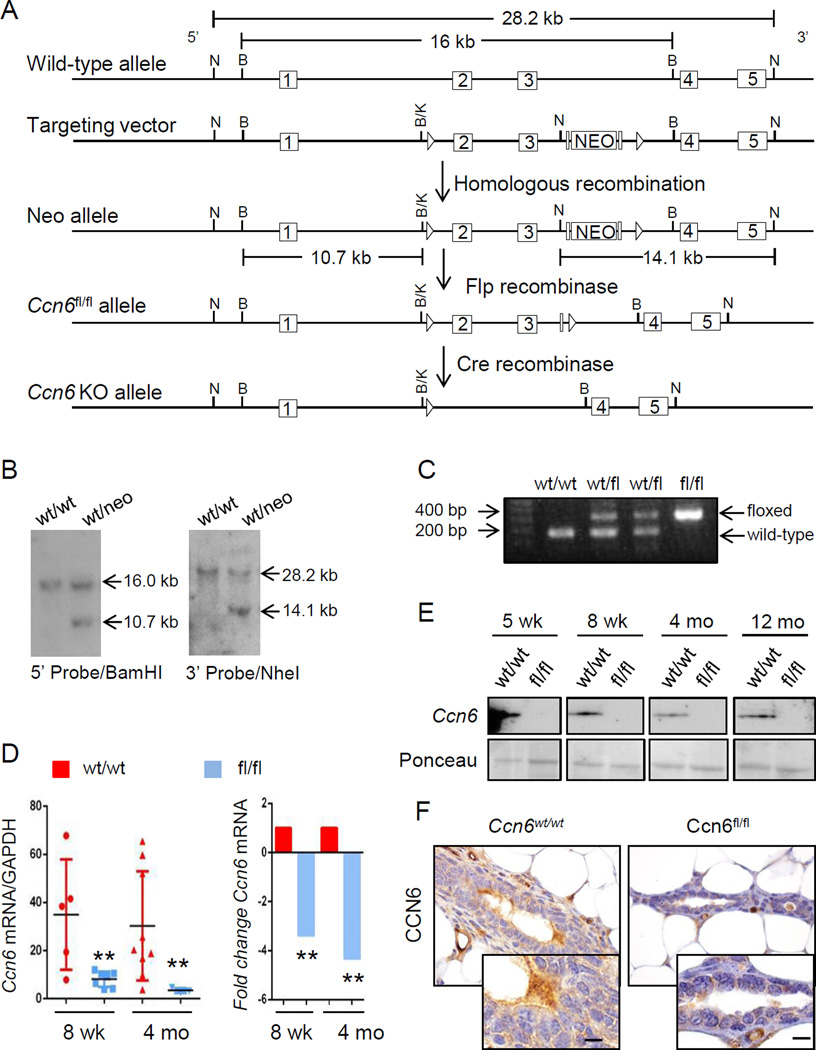
To directly investigate whether loss of Ccn6 in vivo triggers the development of carcinomas that recapitulate human metaplastic carcinomas, we generated a new model of epithelial cell specific Ccn6 deletion in the mammary gland using Cre/loxp-mediated recombination (Fig.2A-B). We specifically inactivated the Ccn6 gene in the mammary epithelium, by intercrossing the floxed Ccn6 mice with the MMTV-Cre mice. The offspring were genotyped using primers specific for various Ccn6 alleles (i.e. floxed, wild type, and deleted) and with primers specific for Cre (Fig.2C). We confirmed the presence of Ccn6 deletion in the mammary glands by quantitative RT-PCR and Western blots using whole mammary gland protein extracts, and by immunohistochemistry in mammary gland tissue samples (Fig.2D-F).
Fig. 2. Conditional deletion of Ccn6 in the mammary epithelium using the Cre-Lox system.
A. Mouse Ccn6 containing exons 2 and 3. Pictured: wild type allele; targeted allele after homologous recombination in ES cells; the floxed allele in which exons 2 and 3 are flanked by loxP sites and the neomycin cassette has been excised; the allele in which exons 2 and 3 have been deleted in Ccn6fl/fl;MMTV-Cre mice. 5’ homology arm (~5.6 kb), 3’ homology arm (~3.7 kb) and cKO region (~4.5 kb) are marked. B. Embryonic stem cell clones were screened for recombination of the targeting vector at the 5′ (BamHI) or 3′ (NheI) ends. C. Example of genotyping results demonstrating amplification of the Ccn6wt and Ccn6fl/fl alleles. D. Relative expression of Ccn6 at the mRNA level as assessed by qt-PCR in 8 week-old mice and 4 month old mice (n≥3 mice per genotype per timepoint; p<0.01 for 8 weeks old, p<0.05 for 4 months old). E. Immunoblot for Ccn6 in representative samples from different timepoints in Ccn6fl/fl;MMTV-Cre and littermate control (Ccn6wt/wt;MMTV-Cre) mice. F. Immunohistochemistry showing the expression of Ccn6 protein in 4 month-old mice mammary glands (at least 3 mice were stained from each genotype). (Scale bars, 20 µm.)
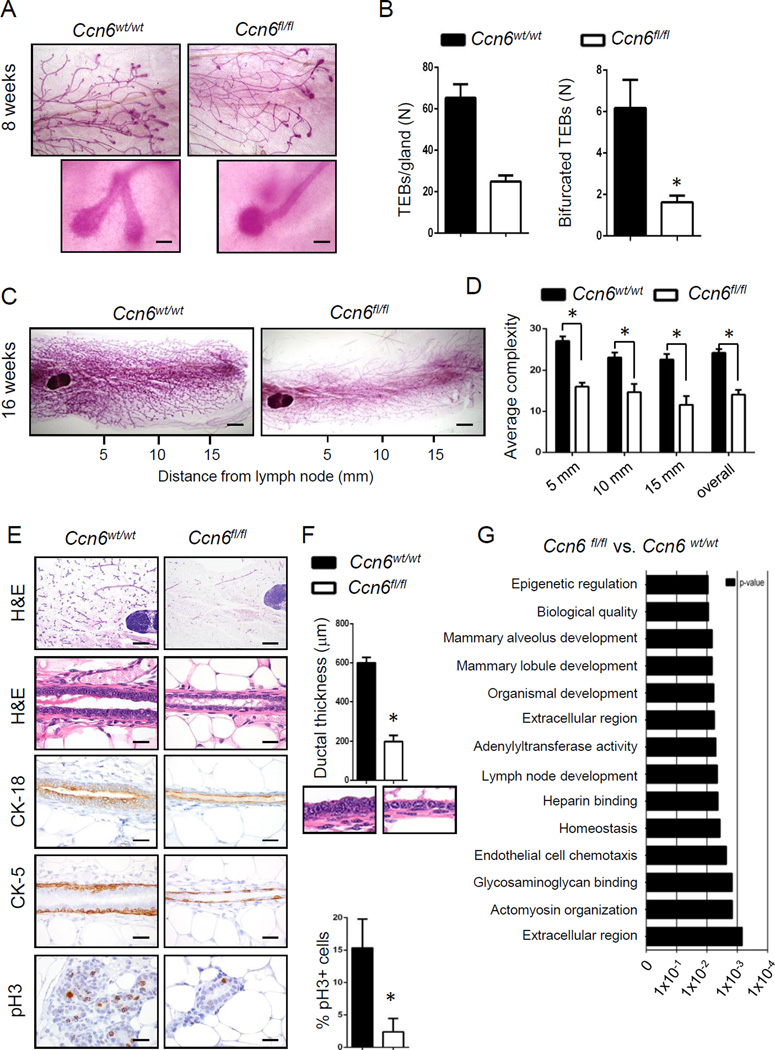
Our initial characterization studies show that pre-pubertal (5 week old) and pubertal (8 week old) Ccn6fl/fl;MMTV-Cre mice exhibit reduced numbers of TEBs and of bifurcated TEBs compared with controls (Fig.3A-B, Supplementary Fig.1A-D). At 16 weeks adult virgin Ccn6fl/fl;MMTV-Cre mice display a significant reduction in mammary gland complexity in Carmine alum-stained whole mounts and histopathology compared to littermate controls (Fig.3C-D, Supplementary Fig.2A-C). Ccn6 deletion resulted in a hypoplastic ductal epithelium with significantly reduced ductal thickness. The ductal epithelial cells were flat compared to the tall columnar cells present in the control ducts (Fig.3E-F). The ductal epithelial hypoplasia of Ccn6fl/fl;MMTV-Cre mice was highlighted by immunostaining with CK-18 and CK-5 marking luminal and basal cells, respectively. Consistent with the hypoplastic ductal phenotype of Ccn6fl/fl;MMTV-Cre mammary glands we detected reduced mitosis identified by pH3 immunostaining compared to controls (Fig.3E). Despite these abnormalities, Ccn6fl/fl;MMTV-Cre dams were able to nurse their litters, and glands underwent post lactational involution similar to controls (Fig.3A-C).
Fig. 3. Mammary epithelial cell specific Ccn6 knockout leads to defects in branching and ductal epithelial hypoplasia.
A. Representative pictures of the distal end of invading ducts in pubertal Ccn6wt/wt;MMTV-Cre and Ccn6fl/fl;MMTV-Cre mice, 2x magnification. Inset of a normal bifurcated duct giving rise to two TEBs in Ccn6wt/wt;MMTV-Cre, compared to a duct that failed to normally bifurcate in an Ccn6fl/fl;MMTV-Cre mouse (20x magnification). B. Quantification of the average number of TEBs and bifurcated TEBs of Ccn6fl/fl;MMTV-Cre and littermate controls. C. Representative mammary whole mounts from 4 month-old Ccn6fl/fl;MMTV-Cre and Ccn6wt/wt;MMTV-Cre mice. D. Quantification of the number of ducts and acini at 5, 10 and 15 mm from the mammary lymph node. E. Representative images of Ccn6fl/fl;MMTV-Cre and Ccn6wt/wt;MMTV-Cre glands stained with H&E and with antibodies against the indicated proteins. F. Ductal epithelial thickness was quantified using image J. G. Gene ontology (GO) analysis of Ccn6fl/fl;MMTV-Cre and Ccn6wt/wt;MMTV-Cre 8 week-old mice mammary glands. For all experiments quantification was performed blindly to genotype by two independent investigators with results reported as average ± SEM, n= at least 3 mice per genotype, in triplicate. Two tailed Student’s t test *P<0.05. (Scale bars, 20 µm.)
To assess the global impact of CCN6 ablation on mammary epithelial cell development in vivo in an unbiased fashion, isolated RNA from 8 week old virgin Ccn6fl/fl;MMTV-Cre mammary glands and littermate controls was subjected to transcriptional profiling. Ccn6 deletion significantly altered the expression of ~180 unique transcripts with marked effects noted in gene ontologies associated with extracellular signaling, matrix-associated functions, and mammary gland lobule development including Bmp8a, Acta1, Cxcl13, and Dmbt1 (Fig.3G, Supplementary Fig.4A-C). Together, the results indicate that Ccn6 is required for branching morphogenesis and proper development of the mammary ductal epithelium in virgin mice.
Deletion of Ccn6 in the mammary epithelium results in abnormal age-related involution
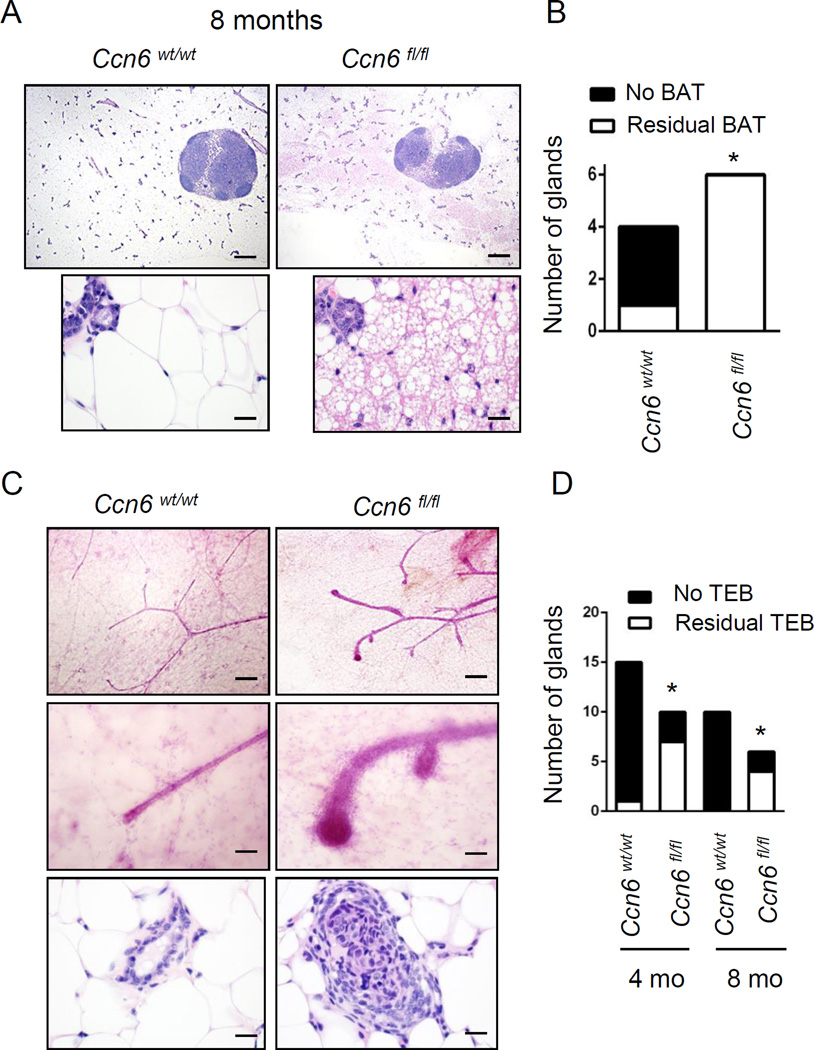
In humans, delayed age-related mammary gland involution has been associated with increased risk for breast cancer (Milanese et al 2006, Radisky and Hartmann 2009). We found evidence of abnormal age-related involution in virgin Ccn6fl/fl;MMTV-Cre mice compared to controls. As expected, while at 8 months of age 1 of 4 (25%) control mice had foci of brown adipose tissue (BAT), all Ccn6fl/fl;MMTV-Cre mice examined had residual BAT in the mammary glands (Fig.4A-B). Likewise, while at 8 months of age there were no residual TEBs in virgin control mice, TEBs persisted in 4 of 6 (66.7%) virgin Ccn6fl/fl;MMTV-Cre mice (Fig.4C-D). Thus, Ccn6 expression is required for proper age-related involution of the virgin murine mammary gland.
Fig. 4. Ccn6 KO mice exhibit residual persistent brown adipose tissue (BAT) and terminal end bud (TEB)-like structures at 8 months of age.
A. Histological H&E sections from 8 month-old virgin Ccn6wt/wt;MMTV-Cre and Ccn6fl/fl;MMTV-Cre mice. Note the presence of brown adipose tissue (BAT) in Ccn6fl/fl;MMTV-Cre mice. B. Quantification of the prevalence of BAT in the mammary whole mounts of virgin mice. C. Mammary whole mounts of 8 month-old Ccn6fl/fl;MMTV-Cre and Ccn6wt/wt;MMTV-Cre mice show the persistence of TEB-like structures in Ccn6fl/fl;MMTV-Cre mice. D. Bars show the prevalence of TEB-like structures, n=at least 3 mice per genotype. Two-tailed Fischer’s exact tests were used for all statistical analyses. *P<0.05.
Ccn6 is a breast tumor suppressor in mice
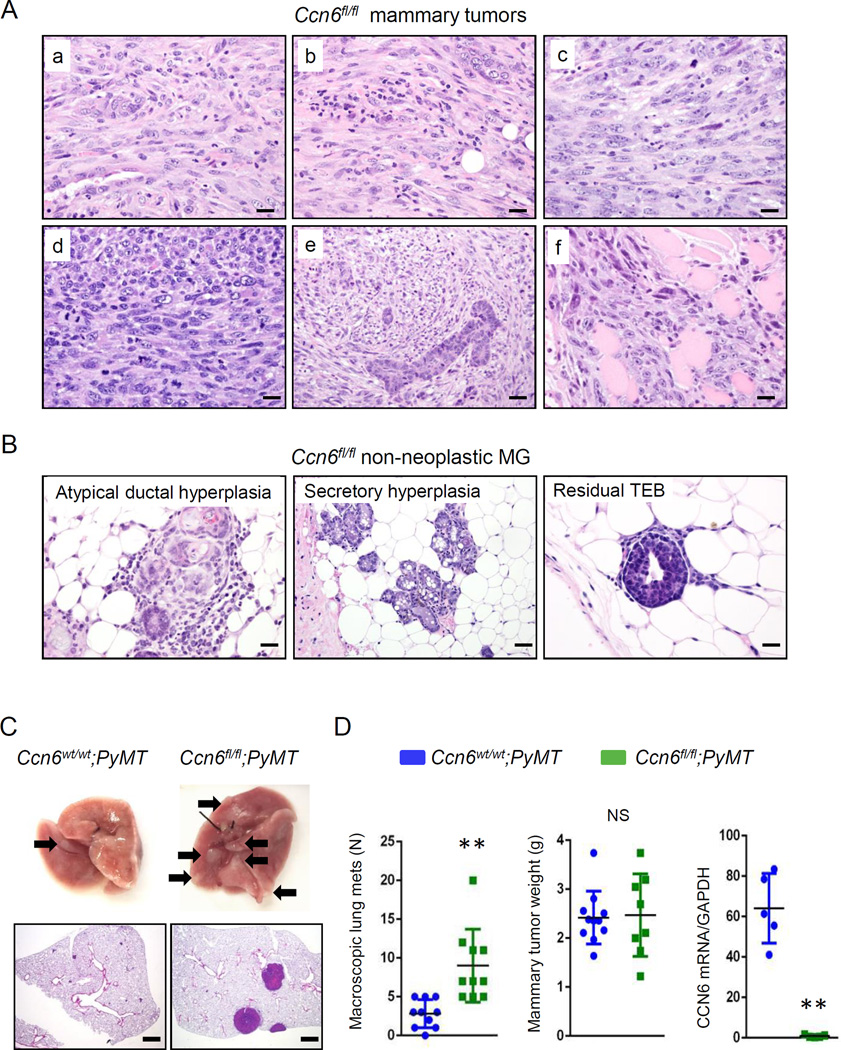
Thirteen of 18 (72.2%) Ccn6fl/fl;MMTV-Cre mice formed mammary carcinomas (age range 10–21 months, mean 15.5 months), compared to 3 of 24 (12.5%) of the Ccn6wt/wt;MMTV-Cre (Fisher’s exact test P < 0.005). The mammary carcinomas of Ccn6fl/fl;MMTV-Cre exhibit mitotically active elongated cells of high histological grade, remarkably similar to the spindle subtype of human metaplastic carcinomas (Fig.5Aa-f). Ccn6fl/fl;MMTV-Cre mice tumors were highly invasive into the surrounding mammary tissues and skeletal muscle (Fig.5Aa-f). The non-neoplastic mammary gland tissues of Ccn6fl/fl;MMTV-Cre mice display a variety of histological abnormalities including persistent TEBs, secretory hyperplasia, and atypical ductal hyperplasia similar to the human counterpart (Fig.5B, Table 1).
Fig. 5. Ccn6 deletion induces mammary tumors morphologically similar to human spindle metaplastic carcinomas and enhances distant metastasis.
A. Histological sections of Ccn6fl/fl;MMTV-Cre mammary tumors (a-f) showing a strikingly spindle cell morphology and high tumor grade. In (f) the carcinoma infiltrates skeletal muscle. 400x magnification. B. Histological sections of non-neoplastic mammary glands of Ccn6fl/fl;MMTV-Cre mice adjacent to invasive carcinomas showing atypical ductal hyperplasia, secretory hyperplasia and residual TEBs. C. Representative images of fresh lungs of Ccn6fl/fl;PyMT and Ccn6wt/wt;PyMT mice. Arrows indicate grossly identified metastasis. Below are histological sections of the lungs showing metastatic nodules, 100x magnification. D. Number of macroscopic lung metastasis and primary tumor volume in Ccn6fl/fl;PyMT and Ccn6wt/wt;PyMT mice. Shown is also CCN6 mRNA expression by quantitative RT-PCR in primary mammary tumors. **P<0.005, two-tailed Student’s t test. (Scale bars, 50 µm.)
Table 1.
Histopathological features of 13 mammary carcinomas from Ccn6fl/fl virgin female mice.
| Tumor | Age (months) |
Histology | Tumor grade |
EMT | Metastasis | Non-neoplastic mammary gland |
|---|---|---|---|---|---|---|
| 1 | 10 | Spindle with necrosis |
3 | Yes | - | Atypical hyperplasia |
| 2 | 12 | Spindle | 3 | Yes | Soft tissue | Atypia, residual TEBs |
| 3 | 13 | Spindle | 3 | Yes | - | Secretory hyperplasia |
| 4 | 13 | Glandular and spindle |
3 | Yes | - | Hyperplasia |
| 5 | 14 | Spindle | 3 | Yes | Lung | Atypia |
| 6 | 14 | Spindle, squamous and glandular |
3 | Yes | Lung | Secretory hyperplasia |
| 7 | 15 | Spindle | 3 | Yes | - | Secretory hyperplasia and atypia |
| 8 | 16 | Spindle with necrosis |
3 | Yes | Lung | Secretory hyperplasia and atypia |
| 9 | 16 | Spindle | 3 | Yes | Lung | Severe atypia |
| 10 | 18 | Spindle and nested |
3 | Yes | - | Secretory hyperplasia |
| 11 | 19 | Spindle | 3 | Yes | - | Secretory hyperplasia |
| 12 | 21 | Spindle | 3 | Yes | Lung | Secretory hyperplasia, atypia |
| 13 | 21 | Spindle | 3 | Yes | - | Atypia, residual TEBs |
Distant metastasis developed in 6 of 13 (46.2%) tumor bearing Ccn6fl/f;MMTV-Cre mice, 5 to the lungs, and 1 to the soft tissues of the neck, compared to none of the Ccn6wt/wt;MMTV-Cre control mice. To investigate the effect of Ccn6 deletion in metastasis in greater detail, we crossed Ccn6fl/fl;MMTV-Cre mice with polyoma middle-T antigen (PyMT)-MMTV mice. PyMT-MMTV mice uniformly develop multifocal mammary tumors with a high incidence of pulmonary metastasis (Guy et al 1992). Despite no differences in primary mammary tumor volume, female virgin Ccn6fl/fl;MMTV-Cre/PyMT mice showed a significant increase in the number of grossly evident lung metastasis compared to Ccn6wt/wt;MMTV-Cre/PyMT mice (Fig.5C-D).
Mammary carcinomas of Ccn6fl/fl;MMTV-Cre mice share an 87-gene signature with human metaplastic breast carcinomas
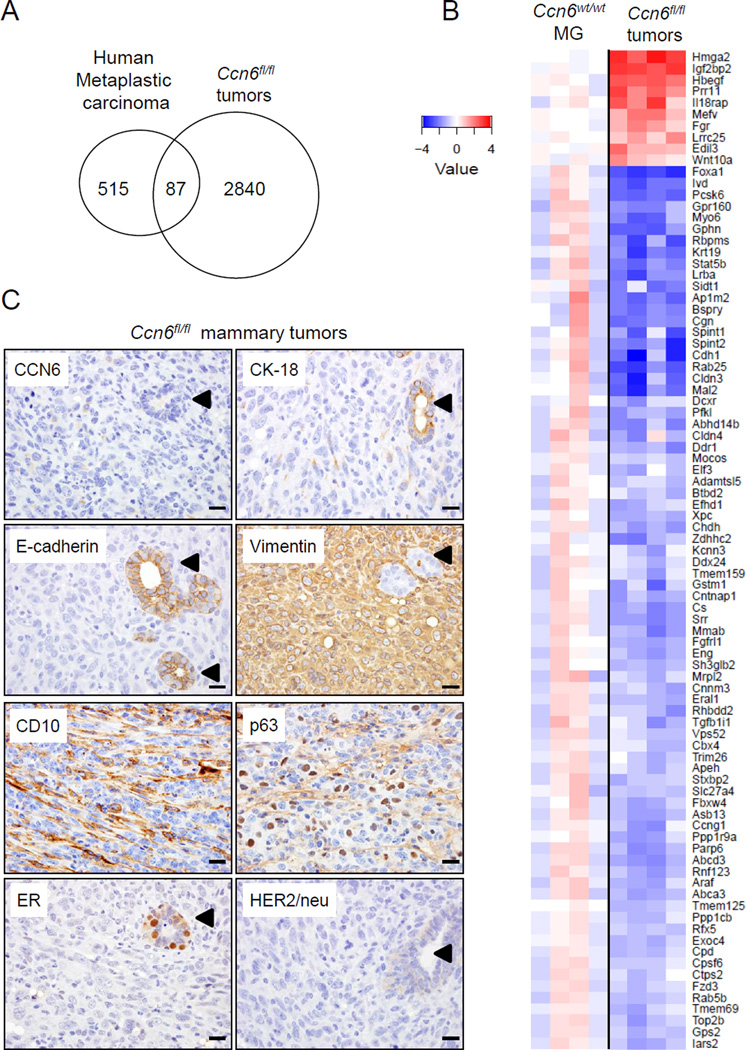
We next sought to examine whether Ccn6fl/fl;MMTV-Cre mammary carcinomas recapitulate human metaplastic carcinoma phenotypes as defined by gene expression patterns. Transcriptional profiling of Ccn6fl/fl;MMTV-Cre mammary carcinomas using Affymetrix revealed 5,354 significantly deregulated gene transcripts compared to age-matched wild-type mammary gland controls. We included genes with a fold change of 2 or greater and an adjusted p-value of 0.05 or less, using a false discovery rate of 0.05 or less. We compared the gene profile of Ccn6fl/fl;MMTV-Cre tumors with a published human metaplastic breast carcinoma signature derived by comparing gene expression profiles of metaplastic carcinomas with profiles of all other breast cancers (Hennessy et al 2009). By directly comparing the relative expression ratio of orthologous genes between human and mouse, we found that Ccn6fl/fl;MMTV-Cre tumors exhibit similarities with the gene expression activity of human metaplastic carcinomas. Of the 602 orthologous genes in the human metaplastic carcinoma signature, there were 87 (14.4%) shared genes concordantly up or down regulated with human metaplastic carcinomas (Fisher’s exact test p=5×10−11, Fig.6A-B).
Fig. 6. Mammary carcinomas in Ccn6fl/fl;MMTV-Cre mice share an 87 gene signature with human metaplastic carcinomas.
A. Venn diagram showing the overlapping 87 orthologous genes between Ccn6fl/fl;MMTV-Cre tumors and a previously reported profile of human high grade metaplastic carcinomas (Hennessy et al 2009) (p= 5×10−11 for the overlap). Of the 2,930 orthologous genes with at least 2-fold up or down regulation in Ccn6fl/fl;MMTV-Cre tumors compared to Ccn6wt/wt;MMTV-Cre mammary glands, 87 showed significant overlap with the human metaplastic carcinoma signature (Hennessy et al 2009). B. Heatmap of the 87 overlapping orthologous genes. C. Representative images Ccn6fl/fl;MMTV-Cre mice mammary carcinomas immunostained with anti-Ccn6 and clinically used markers of human metaplastic carcinoma. Arrowheads point to a normal gland entrapped by the spindle metaplastic carcinoma. 400x magnification. (Scale bars, 50 µm.)
In Gene Ontology analyses, the 87 shared orthologous genes fell into several functional groups. The top five enriched biological processes were morphogenesis of an epithelium, embryo development, lateral plasma membrane, cellular amino acid metabolic process, and very long chain fatty acid catabolic process (Table 2, Suppl. Fig.5). Ccn6fl/fl;MMTV-Cre tumors exhibited downregulated expression of Cldn3, Cldn5, Cdh1, Krt19, previously reported in human metaplastic carcinomas (Burstein et al 2015, Hennessy et al 2009, Weigelt et al 2015). Significantly, Ddr1, Fzd3, Elf3, Stat5b, and Foxa1 are among the shared downregulated genes, while Wnt10a, Hmga2, Hbegf, and Edil3 are among shared upregulated genes (Fig.6B). While these genes are important for normal epithelial homeostasis and have been implicated in carcinogenesis, they have not been previously considered in the pathogenesis of metaplastic carcinoma.
Table 2.
Summary of selected genes significantly deregulated in Ccn6fl/fl mammary tumors and in human metaplastic carcinomas. *Average gene expression ratios (log2-transformed) between the human and mouse tumors.
| Unigene |
Gene symbol |
Fold Change |
||||
|---|---|---|---|---|---|---|
| Human | Mouse | Human | Mouse | Description | Human | Mouse |
| Morphogenesis of an epithelium | ||||||
| Hs.631988 | Mm.5021 | DDR1 | Ddr1 | Discoidin domain receptor family, member 1 | 0.46 | 0.32 |
| Hs.461086 | Mm.35605 | CDH1 | Cdh1 | Cadherin 1 | 0.24 | 0.13 |
| Hs.76753 | Mm.225297 | ENG | Eng | Endoglin | 0.60 | 0.33 |
| Hs.40735 | Mm.214687 | FZD3 | Fzd3 | Frizzled homolog 3 (Drosophila) | 0.49 | 0.47 |
| Hs.799 | Mm.289681 | HBEGF | Hbegf | Heparin-binding EGF-like growth factor | 1.93 | 5.13 |
| Hs.163484 | Mm.4578 | FOXA1 | Foxa1 | Forkhead box A1 | 0.05 | 0.10 |
| Hs.233950 | Mm.104955 | SPINT1 | Spint1 | Serine protease inhibitor, Kunitz type 1 | 0.62 | 0.24 |
| Hs.513530 | Mm.3248 | TGFB1I1 | Tgfb1i1 | TGF beta 1 induced transcript 1 | 0.60 | 0.39 |
| Embyo development | ||||||
| Hs.603657 | Mm.441439 | ELF3 | Elf3 | E74-like factor 3 | 0.25 | 0.43 |
| Hs.654568 | Mm.439699 | KRT19 | Krt19 | Keratin 19 | 0.30 | 0.17 |
| Hs.149387 | Mm.4040 | MYO6 | Myo6 | Myosin VI | 0.35 | 0.19 |
| Hs.498494 | Mm.294007 | PCSK6 | Pcsk6 | Proprotein convertase subtilisin/kexin type 6 | 0.64 | 0.15 |
| Lateral plasma membrane | ||||||
| Hs.647023 | Mm.158662 | CLDN3 | Cldn3 | Claudin 3 | 0.28 | 0.16 |
| Hs.647036 | Mm.7339 | CLDN4 | Cldn4 | Claudin 4 | 0.43 | 0.35 |
| Cellular amino acid metabolic process | ||||||
| Hs.301961 | Mm.37199 | GSTM1 | Gstm1 | Glutathione S-transferase, mu 1 | 0.51 | 0.31 |
| Hs.595276 | Mm.34064 | STAT5B | Stat5b | Signal trans. & activator of transcription 5B | 0.62 | 0.19 |
| Hs.126688 | Mm.259916 | CHDH | Chdh | Choline dehydrogenase | 0.42 | 0.33 |
| Hs.461954 | Mm.131443 | SRR | Srr | Serine racemase | 0.65 | 0.25 |
| Hs.262823 | Mm.331142 | IARS2 | Iars2 | Isoleucine-tRNA synthetase 2, mitochondrial | 0.52 | 0.37 |
| Hs.227049 | Mm.2065 | CTPS2 | Ctps2 | Cytidine -triphosphate synthase 2 5’ | 0.57 | 0.50 |
| Hs.513646 | Mm.6635 | IVD | Ivd | Isovaleryl coenzyme A dehydrogenase | 0.61 | 0.16 |
| Very long-chain fatty acid catabolic process | ||||||
| Hs.700576 | Mm.399042 | ABCD3 | Abcd3 | ATP-binding cassette, sub-family D, member 3 | 0.38 | 0.27 |
| Hs.656699 | Mm.330113 | SLC27A4 | Slc27a4 | Solute carrier family 27, member 4 | 0.53 | 0.41 |
In daily pathology practice, the diagnosis of metaplastic carcinoma is supported by detection of a combination of epithelial and mesenchymal proteins (Koker and Kleer 2004, Rakha et al 2015, Reis-Filho et al 2005, Rungta and Kleer 2012, Weigelt et al 2015). Ccn6fl/f;MMTV-Cre mice tumors had a protein expression profile similar to human metaplastic carcinoma characterized by negative ER and HER-2/neu expression, reduced expression of epithelial markers E-cadherin and cytokeratin-18, and increased expression of the mesenchymal markers vimentin, and the human metaplastic carcinoma markers CD10 and p63 (Koker and Kleer 2004, Reis-Filho et al 2003, Reis-Filho and Schmitt 2003) (Fig.6C). Taken together, these data document that Ccn6 deletion in the mammary gland epithelium is sufficient to induce carcinomas with histological, immunophenotypic, and transcriptomic features that recapitulate human spindle metaplastic carcinomas.
DISCUSSION
Metaplastic carcinomas, and in particular those with spindle histopathology, are less responsive to chemotherapy than other TNBC (Hennessy et al 2006, Rakha et al 2015). Elucidation of the molecular underpinnings of this unusual but aggressive type of breast cancer is necessary to develop effective therapeutic strategies. Our previous identification of Ccn6 frame shift mutations in human metaplastic carcinoma tissues (Hayes et al 2008) coupled with the observation that CCN6 expression is reduced or absent in clinical samples of metaplastic carcinoma raised the suspicion that Ccn6 may be causally involved in their pathogenesis rather than just serve as a marker for these tumors. Our results demonstrate that genetic ablation of Ccn6 in the mammary epithelium of mice induces the development of invasive tumors that recapitulate human metaplastic spindle carcinomas on their histopathology, transcriptional activity, and biological behavior.
Ample mechanistic and functional data support the importance of the process of EMT in human tumorigenesis, however, direct identification of cancer cells undergoing EMT in clinical cancer tissue samples has been challenging (Anwar and Kleer 2013). Metaplastic carcinomas of the breast and other organs are the only human malignancies with overt EMT consisting of spindle cells and mesenchymal-like components, loss of epithelial and gain of mesenchymal marker proteins, and a distinctive transcriptional profile rich in genes involved in EMT (Burstein et al 2015, Hennessy et al 2009, Weigelt et al 2015).
CCN proteins are secreted extracellular matrix associated proteins that regulate stroma-epithelial cross-talk to influence angiogenesis, cell proliferation, invasion, adhesion and other important cell functions in a context-dependent manner (Huang et al 2008, Lau 2011, Lorenzatti et al 2011, Pal et al 2012a, Zhang 2005). CCN6 mutations were reported in patients with progressive pseudorheumatoid dysplasia, and deregulated expression of CCN6 was found in colon and breast cancer (Hurvitz et al 1999, Pennica et al 1998). However, global Ccn6 deletion in transgenic mice revealed no discernible skeletal phenotype (Kutz et al 2005). In breast cancer, we have demonstrated that CCN6 regulates the transition between epithelial and mesenchymal states in vivo and in vitro (Huang et al 2008, Huang et al 2016, Kleer et al 2007, Lorenzatti et al 2011). CCN6 knockdown in nontumorigenic breast cells induces a spindle and invasive phenotype with inhibited expression of E-cadherin and cytokeratin, and upregulation of vimentin and EMT transcription factors (Huang et al 2008, Lorenzatti et al 2011). In contrast, CCN6 overexpression in breast cancer cells reduces invasion and metastasis in xenograft models, and leads to a mesenchymal to epithelial transition (MET) (Huang et al 2016, Pal et al 2012a). Despite these data, proof that Ccn6 deficiency results in mammary tumor development and the relevance of these data to human metaplastic carcinomas was unknown. The mammary epithelial cell-specific Ccn6 knockout model developed here provides direct demonstration of an essential role for CCN6 in the pathogenesis of mammary carcinomas with overt EMT.
During mammary gland development, pre-pubertal and nulliparous adult Ccn6fl/fl;MMTV-Cre mice exhibit fewer mammary ducts and a hypoplastic breast epithelium with defective developmental patterns and reduced mitosis compared to littermate controls. The restricted development of the mammary glands of Ccn6fl/fl;MMTV-Cre mice highlights a role for Ccn6 in mammary gland morphogenesis and may have implications for tumorigenesis. A similar phenotype has been reported after deletion of classical tumor suppressors including Brca1 and Brca2 (Ferguson et al 2012, McAllister et al 2002, Xu et al 1999). Because Ccn6-related tumorigenesis is characterized by an initial growth disadvantage it is not unexpected that tumor formation occurs after a long latency, albeit with high frequency, as observed in Brca1 conditional knockout mice (Xu et al 1999).
The developmental delay in pre-pubertal Ccn6-deficient mice and the propensity to develop carcinomas in adult life may be linked to the deregulation of gene targets. Transcriptional profiling of Ccn6 deleted versus Ccn6 wild type pre-pubertal mammary glands uncovered a complex range of deregulated targets, especially those associated with mammary epithelial development. We noted a significant downregulation of Dmbt1, a glycoprotein of the scavenger receptor cysteine-rich (SRCR) family, with roles in immunity and cancer (Mollenhauer et al 1997). Dmbt1 was identified as a modifier of susceptibility to mammary tumors in Trp53+/− BALB/c breast cancer model (Blackburn et al 2007), and Dmbt1 polymorphisms are associated with increased risk of breast cancer (Tchatchou et al 2010).
In contrast with invasive ductal or lobular carcinomas of the breast, metaplastic carcinomas often lack an associated ductal carcinoma in situ component, and its precursors have not been identified in tissue samples (Rungta and Kleer 2012). The conditional Ccn6 knockout mice generated here shed light into the histological abnormalities that precede the development of spindle cell metaplastic carcinomas. Virgin Ccn6fl/fl;MMTV-Cre mammary glands failed to involute properly with age, as evidenced by abnormal persistence of residual TEBs and brown adipose tissue at 8 months of age, in contrast with the mammary glands of age-matched littermate control mice. Of note, defective age-related involution with persistent BAT has been reported in mammary glands from adult Brca1 mutant mice prior to development of poorly differentiated carcinomas (Jones et al 2011). These data are remarkable in light of recent studies showing that defective age-related involution in human mammary glands is significantly associated with increased risk for breast cancer development (Milanese et al 2006, Radisky and Hartmann 2009). Postmenopausal women with delayed age-related mammary gland involution have a 3-fold increased risk of breast cancer compared with women with complete involution (Milanese et al 2006, Radisky and Hartmann 2009). Collectively, our model provides insights into the role of Ccn6 loss during preneoplastic progression and may have clinical implications for preventative strategies.
Our transcriptional profiling studies show that Ccn6fl/fl;MMTV-Cre tumors share an 87-gene signature with human metaplastic carcinomas (Hennessy et al 2009). Among the 87 shared genes (10 up- and 77 are downregulated) are several that have been previously studied as markers of human metaplastic carcinoma including downregulated expression of Cdh1, Cldn3, Cldn4, and Krt19 (Hennessy et al 2009, Koker and Kleer 2004, Rungta and Kleer 2012, Zhang et al 2012). Also significantly overlapping with human metaplastic carcinomas are novel genes, which have not been previously studied in this context. Among the 3 top uregulated genes are Hmga2, Igfbp2, and Hbegf. Hmga2 (high mobility group AT-hook 2) was previously reported to promote EMT and metastasis by activating the TGFβ receptor II signaling (Morishita et al 2013). Igf2bp2 (p62/IMP2), a member of the family of insulin-like growth factor 2 mRNA binding proteins, promotes breast cancer cell migration and to reduce adhesion in TNBC cells (Li et al 2015). Hbegf (heparin binding EGF-like growth factor) induces breast cancer intravasation and metastasis (Zhou et al 2014). Among the most significantly downregulated genes shared between Ccn6fl/fl;MMTV-Cre tumors and human metaplastic carcinomas are Foxa1, Spint1 and 2, and Ddr1. Foxa1, an important regulator of the ERα and the androgen receptor, whose silencing increases invasion and induces an aggressive, basal-like breast cancer phenotype (Bernardo and Keri 2012). Spint 1 and 2 (hepatocyte growth factor activation inhibitors HAI-1 and HAI-2) are potent matriptase inhibitors reducing invasion and metastasis in TNBC (Parr and Jiang 2006). The Discoidin Domain Receptor 1 (Ddr1), a collagen-binding receptor tyrosine kinase, is associated with worse survival in women with TNBC (Toy et al 2015). Elucidation the functional significance and underlying mechanism of these novel genes which were previously unconsidered in the context of metaplastic carcinoma, may lead to new diagnostic, prognostic, and therapeutic targets.
In conclusion, data presented herein establish that mammary epithelial cell loss of Ccn6 triggers the development of spindle metaplastic carcinomas recapitulating human disease. We believe that this mouse model will be useful in understanding how Ccn6 serves its breast cancer suppression function.
MATERIALS AND METHODS
Construction of the targeting vector and generation of floxed Ccn6 mice
All animal experiments followed procedures approved by the UCUCA of the University of Michigan, protocol UCUCA #PRO00005009. Mice with mammary epithelial-cell specific Ccn6 deletion were generated using the Cre-Lox recombination system. The mouse chromosome 10 sequence of mWisp3 gene was retrieved from the Ensembl database and used as reference. Using genomic DNA from FVB mice as a template, the 5’ homology arm (~5.6 kb), 3’ homology arm (~3.7 kb) and the conditional knockout region (~4.5 kb, containing exons 2 and 3) of the Ccn6 allele were generated by PCR using high fidelity Taq DNA polymerase. These fragments were cloned in the LoxFtNwCD or pCR4.0 vector; aside from the homology arms, the final vector also contained LoxP sequences flanking the conditional KO region (~4.5 kb), Frt sequences flanking the Neo expression cassette (for positive selection of the electroporated ES cells), and a DTA expression cassette (for negative selection of the ES cells). The final vector was confirmed by both restriction digestion and end sequencing analysis. For southern blot analysis and identification of the ES positive for homologous recombination with single neo integration, 5’ and 3’ external probes were generated by PCR. 30 µg of NotI-linearized targeting vector DNA was electroporated into FVB ES cells and selected with 200 µg/ml G418 (Gibco, #10131-035). The primary ES cell screening was performed with 3’ PCR and distal LoxP PCR. Five potential targeted clones (A9, B7, B8, H1 and H6) were identified from one plate, which were expanded for further analysis. Upon completion of the ES clone expansion, additional Southern confirmation analysis was performed. Based on this analysis, two out of the five expanded clones (B7 and B8) were confirmed for homologous recombination with single neo integration. Flp electroporation was performed on these clones, and subsequent Neo deleted clones were identified and confirmed by PCR upon expansion. Confirmed Neo-deleted ES cell clones were injected into mouse C57BL/6NTac blastocysts and breeding of the male chimeras to wild-type FVB females resulted in germline transmission. Heterozygous Ccn6fl/wt mice were crossed with MMTV-Cre line A mice on an FVB background (kind gift of Dr. Stephen Weiss). Generation of the targeting vector, cloning, electroporation into embryonic stem cells, and breeding of chimeras to F1 generation mice was carried out by Taconic Biosciences, Inc.
Ccn6fl/fl;MMTV-Cre mice were intercrossed with MMTV-PyMT mice (FVB/n) to generate breeding cohorts. After weaning, female mice were examined twice a week for the development of mammary tumors by palpation. Tumors were then measured once per week until tumors reached 1 cm3, after which tumors were measured twice per week until mice were sacrificed. Mice were sacrificed when tumors reached 2 cm3, to control for tumor size when considering the effect of Ccn6 loss on metastasis. All mammary tumors were excised, weighed, and collected for histology and RNA isolation as described elsewhere in the methods. Lungs were inflated with 10% NBF, macrometastases were counted immediately by eye, and then lobes were separated and placed in a cassette for histology.
Genotyping
Mice and embryos were genotyped by PCR analysis of genomic DNA from mouse tail samples. Isolation of genomic DNA was done using the DNeasy Blood and Tissue Kit (Qiagen, #69506) according to manufacturer’s instructions. Primers used to genotype CCN6 alleles were P1, 5’-TTC AAA ATT GTG GGA ATA GCT CCA GTA TT-3’; P2 5’-CCA TTG ATA CTG GTT GAG AAC ACA GTG AG-3’. Amplification with these primers results in a 196-bp fragment from the CCN6WT allele and a 344-bp fragment from the CCN6fl allele after neo deletion. Primers used to genotype CRE alleles were P1, 5’-GGT TCT GAT CTG AGC TCT GAG TG-3’; P2 5’-CAT CAC TCG TTG CAT CGA CCG G-3’. PCR was performed for 35 cycles of 94°C (30 sec), 56°C (90 sec), and 72°C (60 sec) for amplification of Ccn6 and Cre alleles.
Animal studies
All procedures were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at the University of Michigan (UCUCA #PRO00005009). Mice were housed in standard conditions, at 23°C, with a 14/10 h light/dark cycle, and food and water supplied ad libitum. We observed no difference among any of the control mice genotypes when considering histological and morphological phenotype, immunoblots or qPCR data; therefore the wild-type control groups presented here consist of a mixture of all of the genotypes.
Mice were euthanized and the left # 4 inguinal mammary gland was excised, spread onto a microscope slide and immediately fixed in Carnoy’s fixative for 5 hours. Subsequently, the tissue was washed in graded alcohol solutions (70, 50, 25%; 15 min each) and finally in ddH2O for 5 min. Staining was carried out overnight in carmine alum stain. The tissue was then dehydrated in graded alcohol solutions (70, 80, 95, and 100%; 15 min each), cleared in xylene overnight, and mounted using Permount. Whole mounts were observed under a Leica MZFL III Stereo/Dissecting microscope (Leica Microsystems GmbH, Wetzlar, Germany), and digital images were recorded using an Olympus DP-70 digital camera. A portion of the mammary gland was placed in a cassette and fixed in 10% neutral buffered formalin for 48 h at room temperature. After fixation, tissues were transferred to 70% ethanol, embedded in paraffin, sectioned and stained with H&E for routine histological examination, or left unstained for later immunohistochemistry.
Microarray analysis and statistics
RNA was isolated from either 8 week old knockout and control mouse mammary glands (n=4 for each group), or mammary tumors from Ccn6fl/fl;MMTV-Cre mice and age matched Ccn6wt/wt;MMTV-Cre control mammary glands (n=4 for each group). Isolated RNA was processed on a Mouse Gene ST 2.1 strip assay at the University of Michigan Microarray Core using the Affy Plus Kit (Affymetrix). For statistical analyses, robust multi-array average (RMA) was used to fit log2 expression values to probesets using the oligo package of bioconductor (Irizarry et al 2003). Linear models were fit using the limma package of bioconductor to identify differentially expressed probesets (Smyth 2004). Arrays were weighted based on a gene-by-gene update algorithm designed to downweight genes deemed less reproducible (Ritchie et al 2006). P-values were adjusted using false discovery rate (FDR) (Benjamini and Hochberg 1995). Comparison to human metaplastic carcinoma was done using data from a custom Agilent array of 12 patient samples developed at the University of North Carolina, GEO# GSE10885 (Hennessy et al 2009). The mouse Affymetrix array data were compared to the human Agilent array data to identify the matching gene symbols between the arrays; further, a list of the gene symbols that were significantly deregulated in the same direction in both arrays was compiled. Statistical analysis of the resulting contingency table was done using a Fisher’s exact test.
Immunoblots
Whole mammary tissue sample (50 mg) was weighed from the most distal portion of the #4 inguinal mammary gland and lysed in RIPA lysis buffer (Thermo-Fisher Scientific) with protease and phosphatase (Thermo-Fisher Scientific) inhibitors at 100x dilution in Precellys 2mL tissue homogenizing mixed beads kit (Cayman Chemical) using a Precellys homogenizer at 6500 rpm (2 cycles of 15 sec motion-15 sec rest). Immunoblots were carried out with 40µg of total protein. Membranes were blocked in TBS-T (Bio-Rad, with 0.05% Tween20) with 4% milk (Bio-Rad, #170-6404) and incubated with primary antibodies in TBS-T at 4°C overnight. Membranes were stained with Ponceau-S solution (Sigma-Aldrich) to confirm equal loading. Anti-CCN6 (Santa Cruz Biotechnology, #SC-25443) was the primary antibody.
Immunohistochemical studies
Immunohistochemistry of human breast cancer tissue samples was performed in 110 cases of invasive breast carcinoma comprising 82 invasive ductal and 28 metaplastic carcinomas obtained with University of Michigan IRB approval (HUM00050330). Hematoxylin and eosin stained samples were histologically evaluated and 5 µ thick sections were immunostained using anti-CCN6 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) following published protocols (Huang et al 2008). CCN6 expression was evaluated as either low or high based on intensity of staining and percentage of cells staining (Huang et al 2008).
To immunostain mouse mammary glands formalin-fixed, paraffin-embedded sections were cut at 5 µm and rehydrated with water. Heat induced epitope retrieval was performed with FLEX TRS High pH Retrieval Buffer (9.01) for CCN6, for 20 minutes (Dako, Carpinteria, CA). Immunohistochemical staining was performed using Peroxidase for 5 min to quench endogenous peroxidases, followed by incubation for 60 minutes with one of the following antibodies: CCN6 (Santa Cruz Biotechnology), CK-18 (Abcam), E-cadherin (BD Biosciences), Vimentin (BD Biosciences), CD10 (BD Biosciences), p63 (Santa Cruz Biotechnology), ERα (Santa Cruz Biotechnology), and HER-2/neu (Santa Cruz Biotechnology). The EnVision + Rabbit HRP System was used for detection (DakoCytomation). DAB chromagen was then applied for 10 minutes. Slides were counterstained with hematoxylin for 5 seconds and then dehydrated and coverslipped. For quantification, three representative areas from three Ccn6fl/fl;MMTV-Cre and three Ccn6wt/wt;MMTV-Cre mice were counted for epithelial cells that stained positive for the protein of interest and calculated as a percentage of the total number of epithelial cells; all counts were done blind to genotype by two separate investigators.
Statistics
Data are expressed as mean ± SD. All experiments were repeated at least 3 times with similar results. The 2-tailed Student’s t test was performed to determine the probability of statistically significant difference (P values) and recorded in figure legends. A P value less than 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH) grant R01CA125577 and R01CA107469 (C.G.K), F30CA196084 (T.A.), R25GM086262 (PREP program, C.A-G), and the University of Michigan’s Cancer Center Support grant 5 P30 CA46592. We thank Dr. Stephen Weiss for the MMTV-Cre line A mice in FVB background, and members of the Kleer lab for critical review of the manuscript.
Footnotes
CONFLICT OF INTEREST: NONE
References
- Anwar TE, Kleer CG. Tissue-based identification of stem cells and epithelial-to-mesenchymal transition in breast cancer. Hum Pathol. 2013;44:1457–1464. doi: 10.1016/j.humpath.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae SY, Lee SK, Koo MY, Hur SM, Choi MY, Cho DH, et al. The prognoses of metaplastic breast cancer patients compared to those of triple-negative breast cancer patients. Breast Cancer Res Treat. 2011;126:471–478. doi: 10.1007/s10549-011-1359-8. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach in multiple testing. Journal of Royal Statistical Society. 1995 Series B 57. [Google Scholar]
- Bernardo GM, Keri RA. FOXA1: a transcription factor with parallel functions in development and cancer. Biosci Rep. 2012;32:113–130. doi: 10.1042/BSR20110046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn AC, Hill LZ, Roberts AL, Wang J, Aud D, Jung J, et al. Genetic mapping in mice identifies DMBT1 as a candidate modifier of mammary tumors and breast cancer risk. Am J Pathol. 2007;170:2030–2041. doi: 10.2353/ajpath.2007.060512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21:1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BW, Gao X, Kil H, Lee J, Benavides F, Abba MC, et al. Conditional Wwox deletion in mouse mammary gland by means of two Cre recombinase approaches. PLoS One. 2012;7:e36618. doi: 10.1371/journal.pone.0036618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes MJ, Thomas D, Emmons A, Giordano TJ, Kleer CG. Genetic changes of Wnt pathway genes are common events in metaplastic carcinomas of the breast. Clin Cancer Res. 2008;14:4038–4044. doi: 10.1158/1078-0432.CCR-07-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy BT, Giordano S, Broglio K, Duan Z, Trent J, Buchholz TA, et al. Biphasic metaplastic sarcomatoid carcinoma of the breast. Ann Oncol. 2006;17:605–613. doi: 10.1093/annonc/mdl006. [DOI] [PubMed] [Google Scholar]
- Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhang Y, Varambally S, Chinnaiyan AM, Banerjee M, Merajver SD, et al. Inhibition of CCN6 (Wnt-1-induced signaling protein 3) down-regulates E-cadherin in the breast epithelium through induction of snail and ZEB1. Am J Pathol. 2008;172:893–904. doi: 10.2353/ajpath.2008.070899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Gonzalez ME, Toy KA, Banerjee M, Kleer CG. Blockade of CCN6 (WISP3) activates growth factor-independent survival and resistance to anoikis in human mammary epithelial cells. Cancer Res. 2010;70:3340–3350. doi: 10.1158/0008-5472.CAN-09-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Martin EE, Burman B, Gonzalez ME, Kleer CG. The matricellular protein ccn6 (wisp3) decreases notch1 and suppresses breast cancer initiating cells. Oncotarget. 2016 doi: 10.18632/oncotarget.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED, et al. Epithelial--mesenchymal and mesenchymal--epithelial transitions in carcinoma progression. J Cell Physiol. 2007;213:374–383. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- Hurvitz JR, Suwairi WM, Van Hul W, El-Shanti H, Superti-Furga A, Roudier J, et al. Mutations in the CCN gene family member WISP3 cause progressive pseudorheumatoid dysplasia. Nat Genet. 1999;23:94–98. doi: 10.1038/12699. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jones LP, Buelto D, Tago E, Owusu-Boaitey KE. Abnormal Mammary Adipose Tissue Environment of Brca1 Mutant Mice Show a Persistent Deposition of Highly Vascularized Multilocular Adipocytes. J Cancer Sci Ther. 2011 doi: 10.4172/1948-5956.s2-004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JI, Lau LF. Taking aim at the extracellular matrix: CCN proteins as emerging therapeutic targets. Nat Rev Drug Discov. 2011;10:945–963. doi: 10.1038/nrd3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleer CG, Zhang Y, Merajver SD. CCN6 (WISP3) as a new regulator of the epithelial phenotype in breast cancer. Cells Tissues Organs. 2007;185:95–99. doi: 10.1159/000101308. [DOI] [PubMed] [Google Scholar]
- Koker MM, Kleer CG. p63 expression in breast cancer: a highly sensitive and specific marker of metaplastic carcinoma. Am J Surg Pathol. 2004;28:1506–1512. doi: 10.1097/01.pas.0000138183.97366.fd. [DOI] [PubMed] [Google Scholar]
- Kutz WE, Gong Y, Warman ML. WISP3, the gene responsible for the human skeletal disease progressive pseudorheumatoid dysplasia, is not essential for skeletal function in mice. Mol Cell Biol. 2005;25:414–421. doi: 10.1128/MCB.25.1.414-421.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau LF. CCN1/CYR61: the very model of a modern matricellular protein. Cell Mol Life Sci. 2011;68:3149–3163. doi: 10.1007/s00018-011-0778-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Francia G, Zhang JY. p62/IMP2 stimulates cell migration and reduces cell adhesion in breast cancer. Oncotarget. 2015;6:32656–32668. doi: 10.18632/oncotarget.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzatti G, Huang W, Pal A, Cabanillas AM, Kleer CG. CCN6 (WISP3) decreases ZEB1-mediated EMT and invasion by attenuation of IGF-1 receptor signaling in breast cancer. J Cell Sci. 2011;124:1752–1758. doi: 10.1242/jcs.084194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luini A, Aguilar M, Gatti G, Fasani R, Botteri E, Brito JA, et al. Metaplastic carcinoma of the breast, an unusual disease with worse prognosis: the experience of the European Institute of Oncology and review of the literature. Breast Cancer Res Treat. 2007;101:349–353. doi: 10.1007/s10549-006-9301-1. [DOI] [PubMed] [Google Scholar]
- McAllister KA, Bennett LM, Houle CD, Ward T, Malphurs J, Collins NK, et al. Cancer susceptibility of mice with a homozygous deletion in the COOH-terminal domain of the Brca2 gene. Cancer Res. 2002;62:990–994. [PubMed] [Google Scholar]
- Milanese TR, Hartmann LC, Sellers TA, Frost MH, Vierkant RA, Maloney SD, et al. Age-related lobular involution and risk of breast cancer. J Natl Cancer Inst. 2006;98:1600–1607. doi: 10.1093/jnci/djj439. [DOI] [PubMed] [Google Scholar]
- Mollenhauer J, Wiemann S, Scheurlen W, Korn B, Hayashi Y, Wilgenbus KK, et al. DMBT1, a new member of the SRCR superfamily, on chromosome 10q25.3-26.1 is deleted in malignant brain tumours. Nat Genet. 1997;17:32–39. doi: 10.1038/ng0997-32. [DOI] [PubMed] [Google Scholar]
- Morishita A, Zaidi MR, Mitoro A, Sankarasharma D, Szabolcs M, Okada Y, et al. HMGA2 is a driver of tumor metastasis. Cancer Res. 2013;73:4289–4299. doi: 10.1158/0008-5472.CAN-12-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberman HA. Metaplastic carcinoma of the breast. A clinicopathologic study of 29 patients. Am J Surg Pathol. 1987;11:918–929. doi: 10.1097/00000478-198712000-00002. [DOI] [PubMed] [Google Scholar]
- Pal A, Huang W, Li X, Toy KA, Nikolovska-Coleska Z, Kleer CG. CCN6 modulates BMP signaling via the Smad-independent TAK1/p38 pathway, acting to suppress metastasis of breast cancer. Cancer Res. 2012a;72:4818–4828. doi: 10.1158/0008-5472.CAN-12-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal A, Huang W, Toy KA, Kleer CG. CCN6 knockdown disrupts acinar organization of breast cells in three-dimensional cultures through up-regulation of type III TGF-beta receptor. Neoplasia. 2012b;14:1067–1074. doi: 10.1593/neo.121322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr C, Jiang WG. Hepatocyte growth factor activation inhibitors (HAI-1 and HAI-2) regulate HGF-induced invasion of human breast cancer cells. Int J Cancer. 2006;119:1176–1183. doi: 10.1002/ijc.21881. [DOI] [PubMed] [Google Scholar]
- Pennica D, Swanson TA, Welsh JW, Roy MA, Lawrence DA, Lee J, et al. WISP genes are members of the connective tissue growth factor family that are up-regulated in wnt-1-transformed cells and aberrantly expressed in human colon tumors. Proc Natl Acad Sci U S A. 1998;95:14717–14722. doi: 10.1073/pnas.95.25.14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisky DC, Hartmann LC. Mammary involution and breast cancer risk: transgenic models and clinical studies. J Mammary Gland Biol Neoplasia. 2009;14:181–191. doi: 10.1007/s10911-009-9123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakha EA, Tan PH, Varga Z, Tse GM, Shaaban AM, Climent F, et al. Prognostic factors in metaplastic carcinoma of the breast: a multi-institutional study. Br J Cancer. 2015;112:283–289. doi: 10.1038/bjc.2014.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis-Filho JS, Milanezi F, Paredes J, Silva P, Pereira EM, Maeda SA, et al. Novel and classic myoepithelial/stem cell markers in metaplastic carcinomas of the breast. Appl Immunohistochem Mol Morphol. 2003;11:1–8. doi: 10.1097/00129039-200303000-00001. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Schmitt FC. p63 expression in sarcomatoid/metaplastic carcinomas of the breast. Histopathology. 2003;42:94–95. doi: 10.1046/j.1365-2559.2003.01513_2.x. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Milanezi F, Carvalho S, Simpson PT, Steele D, Savage K, et al. Metaplastic breast carcinomas exhibit EGFR, but not HER2, gene amplification and overexpression: immunohistochemical and chromogenic in situ hybridization analysis. Breast Cancer Res. 2005;7:R1028–R1035. doi: 10.1186/bcr1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Diyagama D, Neilson J, van Laar R, Dobrovic A, Holloway A, et al. Empirical array quality weights in the analysis of microarray data. BMC Bioinformatics. 2006;7:261. doi: 10.1186/1471-2105-7-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rungta S, Kleer CG. Metaplastic carcinomas of the breast: diagnostic challenges and new translational insights. Arch Pathol Lab Med. 2012;136:896–900. doi: 10.5858/arpa.2012-0166-CR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Tchatchou S, Riedel A, Lyer S, Schmutzhard J, Strobel-Freidekind O, Gronert-Sum S, et al. Identification of a DMBT1 polymorphism associated with increased breast cancer risk and decreased promoter activity. Hum Mutat. 2010;31:60–66. doi: 10.1002/humu.21134. [DOI] [PubMed] [Google Scholar]
- Toy KA, Valiathan RR, Nunez F, Kidwell KM, Gonzalez ME, Fridman R, et al. Tyrosine kinase discoidin domain receptors DDR1 and DDR2 are coordinately deregulated in triple-negative breast cancer. Breast Cancer Res Treat. 2015;150:9–18. doi: 10.1007/s10549-015-3285-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B, Ng CK, Shen R, Popova T, Schizas M, Natrajan R, et al. Metaplastic breast carcinomas display genomic and transcriptomic heterogeneity [corrected] Mod Pathol. 2015;28:340–351. doi: 10.1038/modpathol.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Toy KA, Kleer CG. Metaplastic breast carcinomas are enriched in markers of tumor-initiating cells and epithelial to mesenchymal transition. Mod Pathol. 2012;25:178–184. doi: 10.1038/modpathol.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pan Q, Zhong H, Merajver SD, Kleer CG. Inhibition of CCN6 (WISP3) expression promotes neoplastic progression and enhances the effects of insulin-like growth factor-1 on breast epithelial cells. Breast Cancer Research. 2005;7:R1080–R1089. doi: 10.1186/bcr1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZN, Sharma VP, Beaty BT, Roh-Johnson M, Peterson EA, Van Rooijen N, et al. Autocrine HBEGF expression promotes breast cancer intravasation, metastasis and macrophage-independent invasion in vivo. Oncogene. 2014;33:3784–3793. doi: 10.1038/onc.2013.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.