Abstract
Salmonella enterica is a facultative intracellular bacterium that is the leading cause of food borne illnesses in humans. The cytokine IFN-γ has well-established antibacterial properties against Salmonella and other intracellular microbes, for example its capacity to activate macrophages, promote phagocytosis, and destroy phagocytosed microbes by free radical-driven toxification of phagosomes. But IFN-γ induces the expression of hundreds of uncharacterized genes, suggesting that this cytokine deploys additional antimicrobial strategies that await discovery. Recently, one such mechanism, mediated by a family of IFN-inducible small GTPases called Guanylate Binding Proteins (GBPs) has been uncovered. GBPs were shown to facilitate the pyroptotic clearance of Salmonella from infected macrophages by rupturing the protective intracellular vacuole this microbe forms around itself. Once this protective vacuole is lost, exposed Salmonella activates pyroptosis, which destroys the infected cell. In this review, we summarize such emerging roles for IFN-γ in restricting Salmonella pathogenesis.
Keywords: Interferon-γ, Salmonella, GBPs, pyroptosis, inflammasomes
Introduction
Salmonella enterica is a facultative intracellular bacterium implicated in a variety of illnesses, from gastric disorders to cancer, and is the leading cause of food borne illnesses in humans1,2. S. enterica infects over 90 million people worldwide, including 1.2 million annually in the United States3. There are many serovars of S. enterica, but this review will primarily focus on Typhi and Typhimurium, as these are the best-characterized in terms of their interactions with host cells in culture in and in vivo. Both serovars infect epithelial cells of the small intestine but differ in the nature and magnitude of illnesses they cause. S. Typhi is a major health concern in developing countries, where it infects over 21 million people each year, primarily via the fecal-oral route following ingestion of contaminated food and, more commonly, drinking water4, 5. S. Typhi is efficient in evading the host immune response, and causes a systemic severe disease, called typhoid fever, that is often fatal6. S. Typhimurium is also spread through contaminated food and drinking water, but is efficiently controlled by host immunity and typically only causes self-limiting gastroenteritis in humans7. Notably, S. Typhimurium is pathogenic in mice, where infection leads to systemic disease, and promotes many of the same symptoms as S. Typhi infection does in humans8. In contrast, S. Typhi is unable to infect or cause disease in mice, for which reason S. Typhimurium is often used as an experimental model of systemic salmonellosis9,10. S. Typhimurium can also be used as a model to study Salmonella-induced gastroenteritis, if mice are first treated with streptomycin to alter their gut microbiota11. Thus, infecting mice with S. Typhimurium can either induce a typhoid fever-like disease or colitis-like disease depending on the mouse model used, which allows for broad study of Salmonella pathogenesis. In this review, we will summarize new insights into innate host defense against S. Typhimurium induced fever or colitis, with a focus on emerging roles for IFN-γ in controlling Salmonella pathogenesis.
The life cycle of Salmonella in mammalian hosts
Upon ingestion, Salmonella travels through the stomach into the small intestine, where it begins its pathogenesis cycle. The initial cells targeted by Salmonella are those of the intestinal epithelium12. Invasion of intestinal epithelial cells is mediated by a chromosomal genetic island called Salmonella Pathogenicity-Island 1 (SPI-1)13–16. SPI-1 encodes a Type III secretion system (T3SS) that injects dozens of effector proteins into the cytosol of epithelial cells17. A subset of these effectors, most notably the Sop proteins SopB, SopE, and SopE2, remodel host actin, allowing ingress of Salmonella into the host cytosol18–21. In parallel, SipA and SipC inhibit actin depolymerization, thus maintaining the sustained polymerization required to move plasma membrane around the bacterium until it is fully engulfed22–24. Once Salmonella is engulfed, another SPI-1 protein, SptP, deactivates the actin polymerization proteins Cdc42 and Rac1, allowing for the closure of the membrane “ruffle” and formation of a vacuole around the bacterium. This vacuole is called the Salmonella containing vacuole, or SCV25.
Although most Salmonella remain enclosed in the SCV, occasional bacteria are able to escape and replicate freely in the cytosol of non-phagocytic cells, eventually disseminating through the intestinal lining into the underlying lamina propina, which houses macrophages, dendritic cells and polymorphonuclear cells that can phagocytose the bacterium26. Salmonella can also invade and disseminate through M cells into Peyer’s patches, where it is phagocytosed by dendritic cells (DCs). DCs then carry the bacterium to mesenteric lymph nodes, allowing spread to distal organs such as the liver and spleen, via lymphatic ducts. Alternatively, Salmonella can be phagocytosed by CD18+ monocytes in the intestinal lumen and spread to distal organs through the bloodstream without having to first travel to mesenteric lymph nodes27.
Normally, the acidic environment of the phagocyte is toxic to bacteria, but Salmonella has evolved to combat this environment through the use of another plasmid-encoded pathogenicity island, SPI-228. SPI-2 induces the manipulation of vesicle trafficking to prevent the phagocyte SCV from rupturing or fusing with the lysosome29. SPI-2 encoded proteins of the Sif family are essential for maintenance of SCV integrity. Of these, SifA is best studied, and interacts with host adaptor proteins Pleckstrin homology domain containing, family M member 1 (PLEKHM1), Rab7, and Homotypic fusion and protein sorting protein (HOPS)30. These proteins normally act to stabilize the interaction between autophagosomes and lysosomes to help degrade damaged proteins or pathogens in mitophagy and xenophagy, respectively31. However, Salmonella has co-opted these proteins to stabilize the SCV by preventing lysosomal fusion as well as by maintaining SCV pH balance and optimal membrane dynamics30, 31. Like SifA, the T3SS protein SseJ also cooperates in these activities32. SseJ and SifA also interact with an adaptor protein called SifA kinesin interacting protein (SKIP), which binds to microtubule-based motor kinesin-1, recruiting it to the vacuole, and promoting efficient interaction between the SCV and the microtubule network of the cell32. SCV forming proteins aid retention of early endosome markers, including Rab5, transferrin receptor (TnfR) and early endosome associated antigen 1 (EEA1) on the SCV, which prevent the SCV from fusing with lysosomes33.
These and other immune-evasion mechanisms enable the SCV in phagocytes to serve not only as a replicative niche for Salmonella, but also as a protective barrier against cytosolic anti-microbial effector mechanisms29.
Salmonella clearance in non-phagocytic cells
Within the SCV, Salmonella remains well-contained12, but Salmonella that escape the SCV and enter the cytosol of non-phagocytic cells, replicate well in this environment and eventually lyse the infected cell, allowing spread to surrounding cells34. The SCV in non-phagocytic cells thus protects the host by limiting the spread of Salmonella to uninfected neighboring cells, and such cells have in place mechanisms that maintain the integrity of the SCV34, 35.
A key mechanism by which the non-phagocytic cell maintains SCV integrity involves the host kinase TBK1. Although TBK1 is best known for its role in antiviral innate immunity, where it functions as an IRF3/7 kinase and is a pivotal player in the induction of type I IFNs36, recent studies have revealed a distinct function for this protein in regulating the SCV37. TBK1 regulates levels of aquaporin-1 (AQP1), which controls the osmotic balance of the SCV and prevents it from expanding or collapsing38. TBK1 deficient MEFs, which cannot form a stable SCV, are thus hypersensitive to Salmonella replication, as are cells overexpressing AQP139.
In cases where Salmonella is able to escape the SCV and enter the cytosol, it can be ubiquitinated and targeted for degradation, via the host cell’s autophagy machinery. Ubiquitination of Salmonella is mediated by an E3 ubiquitin ligase called Leucine Rich Repeat and Sterile Alpha Motif Containing 1 (LRSAM1)40. Nuclear dot protein 52 kDa (NDP52) and its paralog TAX1BP1 recognize ubiquitinated Salmonella and bind to myosin VI, allowing for autophagosome formation around the bacterium39, 41. Like NDP52, the adaptor proteins optineurin and p62 (also called SQSTM1), which are activated by TBK1-mediated phosphorylation, can recognize poly-ubiquitinated Salmonella42. Upon activation, these proteins bind to the autophagosome marker LC3, linking the autophagosome machinery to ubiquitinated Salmonella and targeting the bacterium for degradation43. Like LRSAM1, Parkin can also ubiquitinate Salmonella, leading to autophagic clearance, although how LRSAM1- and Parkin-driven ubiquitination mechanisms differ from each other await elucidation44. Autophagosomes engulf and subsequently degrade the bacterium, controlling, to some extent, Salmonella infection in the cytosol of cells. It is noteworthy, however, that cytosolic mechanisms for control of Salmonella in non-phagocytic cells may not be very efficient, as TBK1-deficent cells, or cells infected with mutant SifA Salmonella that cannot form an SCV and directly enter the cytosol of cells, display increased Salmonella replication compared to controls45. Interestingly, while phagocytic cells produce high levels of antibacterial proteins, such as defensins, that combat Salmonella, these proteins are produced at much lower levels in non-phagocytic cells, providing an explanation for the differences in cytosolic replication between the two cell types27.
IFN-γ and control of Salmonella in the phagocyte SCV
When Salmonella infects a macrophage, it “hides” in SCVs. This allows the bacterium to both survive the harsh environment of the macrophage and replicate without interference from cytosolic antimicrobial factors46. However, at least two mechanisms are in place to eliminate Salmonella from the macrophage, both of which require IFN-γ at crucial steps to mediate bacterial clearance. First, IFN-γ activates transcription of a group of proteins called Guanylate Binding Proteins (GBPs) that localize to the SCV, inducing its lysis and subsequently releasing Salmonella into the cytosol47. Second, IFN-γ “primes” the macrophage through induction of reactive oxygen species, which activates the NLRP3 inflammasome, leading to pro-inflammatory cell death and eventual clearance of the bacterium48. Each of these anti-Salmonella strategies will be discussed next.
Recently, an IFN-γ-driven mechanism has come to light that ruptures the macrophage SCV, preventing Salmonella from hiding within the protective confines of this vacuole. The drivers of release of Salmonella into the cytosol are a group of small interferon-inducible GTPases called Guanylate Binding Proteins (GBPs)49. There are 11 genes encoding GBPs in mice and seven in humans, of which, at least five (GBPs 1,2,3,5,7) are critical for Salmonella clearance50. GBPs are diffusely cytosolic proteins that are recruited to the SCV in infected macrophages. Recruitment of GBPs to the SCV requires dimerization and subsequent prenylation of these proteins, the latter step of which is mediated by the interferon-stimulated gene Immunity-related GTPase family M protein 1 (IRGM1), a GTPase51. Upon sensing ubiquitinated bacterial structures, IRGM1 activates farnesyltransferase-I and geranylgeranyltransferase-I that prenylate GBPs on conserved CaaX sequences in their C-termini52. This prenylation step induces their localization to the Salmonella-containing vacuole53. Once at the SCV, GBPs insert into and puncture the integrity of the vacuolar membrane, rupturing the SCV and releasing Salmonella into the cytosol54. GBPs also participate in directly activating the NLRP3 inflammasome in response to both Salmonella and Chlamydia55, 56. Further, GBPs recruit the interferon-inducible protein IRGB10 to intracellular bacteria, which then mediates the release of bacterial ligands for detection by inflammasome sensors57.
IFN-γ is critical to the function of GBPs at multiple distinct steps in this process. First, IFN-γ is required for the induction of GBP-encoding genes. IFN-γ induces expression of GBPs in a two-step mechanism: IFN-γ-mediated JAK/STAT1 signaling first induces rapid expression of the transcription factor IRF-1, which then transactivates GBP-encoding genes58, 59. Second, IFN-γ is necessary for activating GBPs in the cytoplasm, as well as for localizing these GTPases to the SCV60. Details of how IFN-γ promotes the activation of GBPs and how IRGM1 mediated prenylation sites recruit GBPs to the SCV are currently unclear. Notably, simply overexpressing GBPs does not mimic their induction by IFN-γ, demonstrating the requirement for IFN-γ signaling at additional post-transcriptional steps in the biology of these intriguing enzymes61.
Besides facilitating the rupture of SCVs, IFN-γ promotes the destruction of Salmonella and other intracellular microbes by inducing the production of reactive oxygen species (ROS) and reactive nitrogen species (RNS) in phagocytes62. ROS, besides being directly toxic to Salmonella, can also promote Salmonella clearance by other means. For example, ROS promotes activation of NF-κB, a transcription factor that induces expression of a variety of anti-bacterial immune factors, including NLRP363. ROS can also initiate autophagy to control Salmonella in phagocytic cells. This process is mediated by the enzyme NOX2, which recruits LC3 to phagosomes by sensing ROS mediated cellular damage64.
Pyroptosis and clearance of Salmonella-infected cells
Upon rupture of the SCV, Salmonella is released into the cytosol of macrophages, which can trigger pyroptosis, a rapid pro-inflammatory form of cell death. The rupturing of membranes and the release of IL-1β from the cell, producing robust inflammation, characterize pyroptosis65. Activation of caspase-1 (the driver of IL-1β release) and secretion of IL-1β are mediated by cytoplasmic signaling complexes called inflammasomes66. All canonical inflammasomes contain Nod-like (NLR) proteins, but differ in the pathogen-associated molecular patterns (PAMPs) they recognize67. Salmonella is detected by two canonical inflammasomes, NLRC4 and NLRP368,69. Notably, NLRC4 and NLRP3 interact with each other and may co-operate to activate caspase-1 in response to Salmonella infection70. The NLRC4 inflammasome senses the PAMP flagellin, as well as components of the Salmonella T3SS, specifically, the SPI-1-encoded needle proteins PrgJ and PrgI, to activate caspase-1 and trigger pyroptosis71. What triggers NLRP3 inflammasome activation during Salmonella infection is less known, although extracellular ATP and citrate are required for this process72, 73,74.
Pyroptosis can be induced non-canonically, through a mechanism reliant on recognition of Salmonella LPS by caspase-11. In this pathway, caspase-11 directly senses cytosolic LPS as a signal to release IL-1α, mediating cell death without need for NLRC4 and NLRP375. Notably, caspase-11 is also needed for caspase-1 induced cleavage of IL-1β by canonical inflammasomes76. Once activated, caspase-1/11 then cleaves a protein called gasdermin D. Upon cleavage, the N-terminus of gasdermin D translocates to the membrane, triggering rupture of the cell and release of IL-1β77–79.
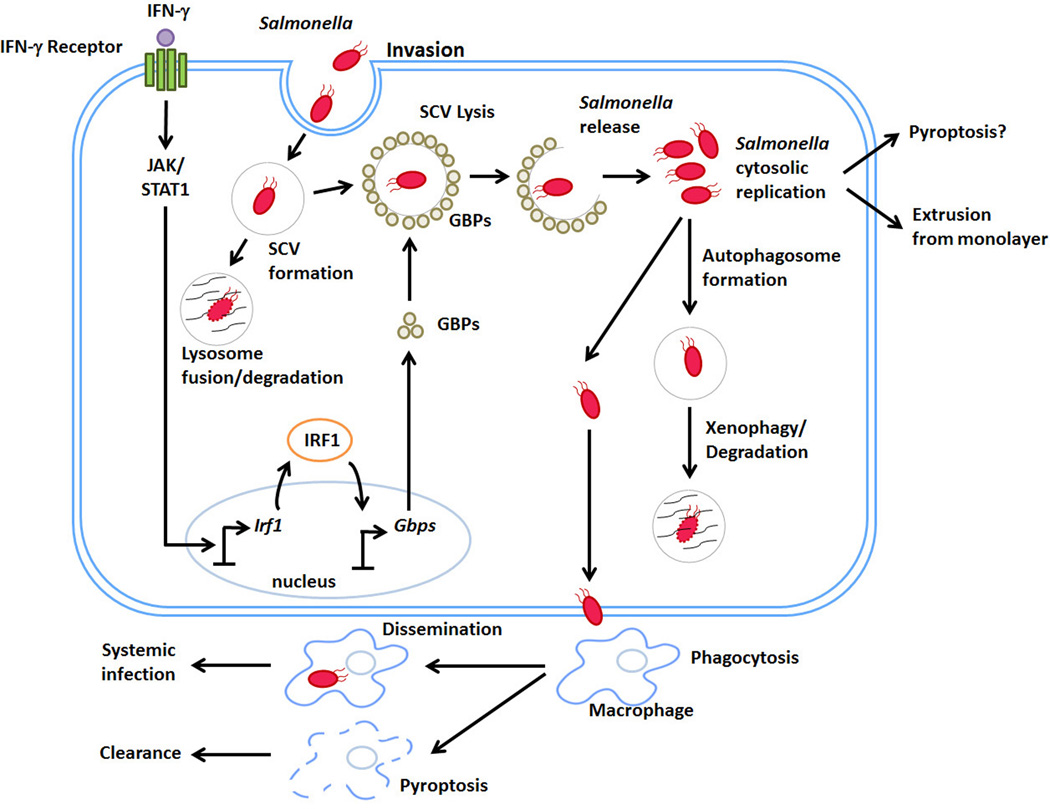
Figure 1. IFN-γ control of Salmonella life cycle in non-phagocytic cells.
Upon ingestion, Salmonella travels to the small intestine where it invades non-phagocytic cells through use of a TTSS encoded by the pathogenicity island SPI-1. After invasion, a Salmonella-containing vacuole (SCV) forms around the bacterium, from which is eventually cleared by fusion with lysosomes. Following exposure to IFN-γ, a family IFN-inducible GTPases called GBPs are transcribed by a two-step mechanism involving JAK/STAT1 dependent induction of the transcription factor IRF1. IRF1 then drives GBP expression. Once synthesized, GBPs lyse the SCV, releasing Salmonella into the cytosol of the cell. Salmonella can replicate in the host cytosol, and is often cleared by autophagosome-mediated mechanisms. In some cases where Salmonella hyper-replicates in the cytosol, the infected cell is extruded from the monolayer. A few bacteria can escape these processes and disseminate into the underlying lamina propina, where they are phagocytosed by macrophages.
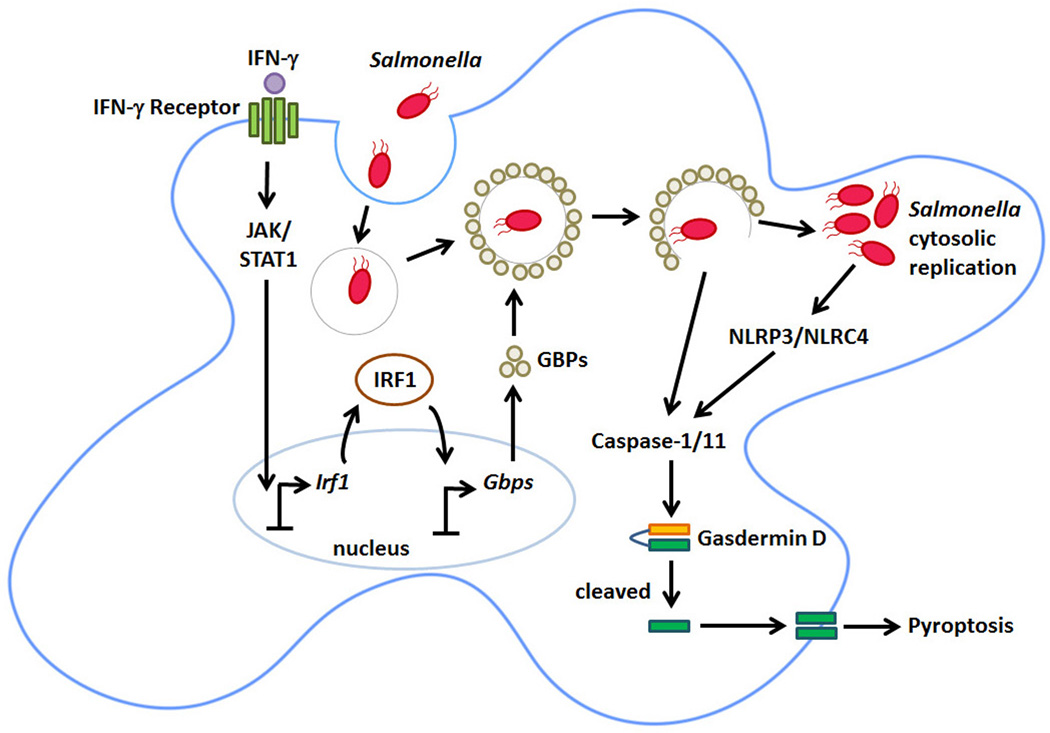
Figure 2. IFN-γ control of Salmonella lifecycle in phagocytic cells.
Salmonella is phagocytosed by the macrophage and immediately forms an SCV to survive in the unfavorable acidic environment of the cell. IFN-γ leads to JAK/STAT1 dependent IRF1 induction and transcription of GBPs, which localize to the SCV, lysing it. Upon lysis, Salmonella is released into the cytosol of the macrophage where the bacterium is recognized by the inflammasome sensors NLRC4 and NLRP3, which activate caspase-1/11. In parallel, cytosolic LPS can be directly sensed by caspase-11. Once activated, these caspases cleave Gasdermin D. The N-terminal cleavage product of Gasdermin D then localizes to and punctures holes in the plasma membrane, triggering pyroptotic death.
Outstanding Questions.
Although much is known about how Salmonella induces disease, there are still multiple questions that remain to be answered. First, the molecular underpinnings of GBP activation leading to rupture of the SCV are unclear. Second, the role of IFN-γ in clearance of Salmonella in non-phagocytic cells is relatively unexplored. Recent studies demonstrate that epithelial cells infected with Salmonella manifest caspase-11 (caspase-4 in humans) cleavage and extrusion of infected cells from the intestinal epithelial monolayer35. Whether IFN-γ-mediated induction of GBPs is responsible for such clearance, or whether other ISGs are required to lyse the SCV and trigger caspase-11 activation remains to be seen. Finally, mechanisms by which inflammasomes are negatively-regulated to prevent their inopportune activation, or to terminate the inflammatory response, remain relatively unexplored.
Highlights.
IFN-γ is important in controlling Salmonella replication and spread in phagocytes
IFN-γ induces expression of Guanylate Binding Proteins (GBPs)
GBPs promote Salmonella clearance by rupturing its protective intracellular vacuole
Salmonella thus exposed triggers pyroptosis of the host phagocyte
Acknowledgments
We thank Shoko Nogusa for help with figure preparation. Work in the Balachandran lab is supported by NIH grants CA168621, CA190542, and AI113469. Additional funds were provided by NIH Cancer Center Support Grant P30CA006927 and an appropriation from the Commonwealth of Pennsylvania.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pham OH, McSorley SJ. Protective host immune responses to Salmonella infection. Future Microbiol. 2015;10:101–110. doi: 10.2217/fmb.14.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scanu T, Spaapen RM, Bakker JM, Pratap CB, Wu LE, Hofland I, Broeks A, Shukla VK, Kumar M, Janssen H, Song JY, Neefjes-Borst EA, te Riele H, Holden DW, Nath G, Neefjes J. Salmonella Manipulation of Host Signaling Pathways Provokes Cellular Transformation Associated with Gallbladder Carcinoma. Cell Host Microbe. 2015;17:763–774. doi: 10.1016/j.chom.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Schwille-Kiuntke J, Unverdorben A, Weimer K, Schlarb AA, Gulewitsch MD, Ellert U, Enck P. Bacterial infections in childhood: A risk factor for gastrointestinal and other diseases? United European Gastroenterol J. 2015;3:31–38. doi: 10.1177/2050640614558346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gunn JS, Marshall JM, Baker S, Dongol S, Charles RC, Ryan ET. Salmonella chronic carriage: epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol. 2014;22:648–655. doi: 10.1016/j.tim.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bula-Rudas FJ, Rathore MH, Maraqa NF. Salmonella Infections in Childhood. Adv Pediatr. 2015;62:29–58. doi: 10.1016/j.yapd.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Dougan G, Baker S. Salmonella enterica serovar Typhi and the pathogenesis of typhoid fever. Annu Rev Microbiol. 2014;68:317–336. doi: 10.1146/annurev-micro-091313-103739. [DOI] [PubMed] [Google Scholar]
- 7.Broz P, Ohlson MB, Monack DM. Innate immune response to Salmonella typhimurium, a model enteric pathogen. Gut Microbes. 2012;3:62–70. doi: 10.4161/gmic.19141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng L, Song J, Gao X, Wang J, Yu H, Chen X, Varki N, Naito-Matsui Y, Galan JE, Varki A. Host adaptation of a bacterial toxin from the human pathogen Salmonella Typhi. Cell. 2014;159:1290–1299. doi: 10.1016/j.cell.2014.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song J, Willinger T, Rongvaux A, Eynon EE, Stevens S, Manz MG, Flavell RA, Galan JE. A mouse model for the human pathogen Salmonella typhi. Cell Host Microbe. 2010;8:369–376. doi: 10.1016/j.chom.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Brien AD, Rosenstreich DL, Taylor BA. Control of natural resistance to Salmonella typhimurium and Leishmania donovani in mice by closely linked but distinct genetic loci. Nature. 1980;287:440–442. doi: 10.1038/287440a0. [DOI] [PubMed] [Google Scholar]
- 11.Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Jong HK, Parry CM, van der Poll T, Wiersinga WJ. Host-pathogen interaction in invasive Salmonellosis. PLoS Pathog. 2012;8:e1002933. doi: 10.1371/journal.ppat.1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruno VM, Hannemann S, Lara-Tejero M, Flavell RA, Kleinstein SH, Galan JE. Salmonella Typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PLoS Pathog. 2009;5:e1000538. doi: 10.1371/journal.ppat.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee CA, Jones BD, Falkow S. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc Natl Acad Sci U S A. 1992;89:1847–1851. doi: 10.1073/pnas.89.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lara-Tejero M, Galan JE. Salmonella enterica serovar typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect Immun. 2009;77:2635–2642. doi: 10.1128/IAI.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen-Wester I, Hensel M. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 2001;3:549–559. doi: 10.1016/s1286-4579(01)01411-3. [DOI] [PubMed] [Google Scholar]
- 17.Que F, Wu S, Huang R. Salmonella pathogenicity island 1(SPI-1) at work. Curr Microbiol. 2013;66:582–587. doi: 10.1007/s00284-013-0307-8. [DOI] [PubMed] [Google Scholar]
- 18.Galan JE, Zhou D. Striking a balance: modulation of the actin cytoskeleton by Salmonella. Proc Natl Acad Sci U S A. 2000;97:8754–8761. doi: 10.1073/pnas.97.16.8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norris FA, Wilson MP, Wallis TS, Galyov EE, Majerus PW. SopB, a protein required for virulence of Salmonella dublin, is an inositol phosphate phosphatase. Proc Natl Acad Sci U S A. 1998;95:14057–14059. doi: 10.1073/pnas.95.24.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardt WD, Chen LM, Schuebel KE, Bustelo XR, Galan JE. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 1998;93:815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 21.Stender S, Friebel A, Linder S, Rohde M, Mirold S, Hardt WD. Identification of SopE2 from Salmonella typhimurium, a conserved guanine nucleotide exchange factor for Cdc42 of the host cell. Mol Microbiol. 2000;36:1206–1221. doi: 10.1046/j.1365-2958.2000.01933.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhou D, Mooseker MS, Galan JE. Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science. 1999;283:2092–2095. doi: 10.1126/science.283.5410.2092. [DOI] [PubMed] [Google Scholar]
- 23.Zhou D, Mooseker MS, Galan JE. An invasion-associated Salmonella protein modulates the actin-bundling activity of plastin. Proc Natl Acad Sci U S A. 1999;96:10176–10181. doi: 10.1073/pnas.96.18.10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayward RD, Koronakis V. Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 1999;18:4926–4934. doi: 10.1093/emboj/18.18.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Humphreys D, Hume PJ, Koronakis V. The Salmonella effector SptP dephosphorylates host AAA+ ATPase VCP to promote development of its intracellular replicative niche. Cell Host Microbe. 2009;5:225–233. doi: 10.1016/j.chom.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knodler LA. Salmonella enterica: living a double life in epithelial cells. Curr Opin Microbiol. 2015;23:23–31. doi: 10.1016/j.mib.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 27.Watson KG, Holden DW. Dynamics of growth and dissemination of Salmonella in vivo. Cell Microbiol. 2010;12:1389–1397. doi: 10.1111/j.1462-5822.2010.01511.x. [DOI] [PubMed] [Google Scholar]
- 28.Srikanth CV, Mercado-Lubo R, Hallstrom K, McCormick BA. Salmonella effector proteins and host-cell responses. Cell Mol Life Sci. 2011;68:3687–3697. doi: 10.1007/s00018-011-0841-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Figueira R, Holden DW. Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology. 2012;158:1147–1161. doi: 10.1099/mic.0.058115-0. [DOI] [PubMed] [Google Scholar]
- 30.McEwan DG, Richter B, Claudi B, Wigge C, Wild P, Farhan H, McGourty K, Coxon FP, Franz-Wachtel M, Perdu B, Akutsu M, Habermann A, Kirchof A, Helfrich MH, Odgren PR, Van Hul W, Frangakis AS, Rajalingam K, Macek B, Holden DW, Bumann D, Dikic I. PLEKHM1 regulates Salmonella-containing vacuole biogenesis and infection. Cell Host Microbe. 2015;17:58–71. doi: 10.1016/j.chom.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 31.McEwan DG, Dikic I. PLEKHM1: Adapting to life at the lysosome. Autophagy. 2015;11:720–722. doi: 10.1080/15548627.2015.1034419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolodziejek AM, Miller SI. Salmonella modulation of the phagosome membrane, role of SseJ. Cell Microbiol. 2015;17:333–341. doi: 10.1111/cmi.12420. [DOI] [PubMed] [Google Scholar]
- 33.Steele-Mortimer O. The Salmonella-containing vacuole: moving with the times. Curr Opin Microbiol. 2008;11:38–45. doi: 10.1016/j.mib.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malik-Kale P, Winfree S, Steele-Mortimer O. The bimodal lifestyle of intracellular Salmonella in epithelial cells: replication in the cytosol obscures defects in vacuolar replication. PLoS One. 2012;7:e38732. doi: 10.1371/journal.pone.0038732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knodler LA, Crowley SM, Sham HP, Yang H, Wrande M, Ma C, Ernst RK, Steele-Mortimer O, Celli J, Vallance BA. Noncanonical inflammasome activation of caspase-4/caspase-11 mediates epithelial defenses against enteric bacterial pathogens. Cell Host Microbe. 2014;16:249–256. doi: 10.1016/j.chom.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikeda F, Hecker CM, Rozenknop A, Nordmeier RD, Rogov V, Hofmann K, Akira S, Dotsch V, Dikic I. Involvement of the ubiquitin-like domain of TBK1/IKK-i kinases in regulation of IFN-inducible genes. EMBO J. 2007;26:3451–3462. doi: 10.1038/sj.emboj.7601773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radtke AL, Delbridge LM, Balachandran S, Barber GN, O'Riordan MX. TBK1 protects vacuolar integrity during intracellular bacterial infection. PLoS Pathog. 2007;3:e29. doi: 10.1371/journal.ppat.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radtke AL, O'Riordan MX. Homeostatic maintenance of pathogen-containing vacuoles requires TBK1-dependent regulation of aquaporin-1. Cell Microbiol. 2008;10:2197–2207. doi: 10.1111/j.1462-5822.2008.01199.x. [DOI] [PubMed] [Google Scholar]
- 39.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 40.Huett A, Heath RJ, Begun J, Sassi SO, Baxt LA, Vyas JM, Goldberg MB, Xavier RJ. The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella Typhimurium. Cell Host Microbe. 2012;12:778–790. doi: 10.1016/j.chom.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tumbarello DA, Manna PT, Allen M, Bycroft M, Arden SD, Kendrick-Jones J, Buss F. The Autophagy Receptor TAX1BP1 and the Molecular Motor Myosin VI Are Required for Clearance of Salmonella Typhimurium by Autophagy. PLoS Pathog. 2015;11:e1005174. doi: 10.1371/journal.ppat.1005174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 43.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dotsch V, Bumann D, Dikic I. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, Cox JS. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512–516. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brumell JH, Rosenberger CM, Gotto GT, Marcus SL, Finlay BB. SifA permits survival and replication of Salmonella typhimurium in murine macrophages. Cell Microbiol. 2001;3:75–84. doi: 10.1046/j.1462-5822.2001.00087.x. [DOI] [PubMed] [Google Scholar]
- 46.Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, Holden DW. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meunier E, Dick MS, Dreier RF, Schurmann N, Kenzelmann Broz D, Warming S, Roose-Girma M, Bumann D, Kayagaki N, Takeda K, Yamamoto M, Broz P. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509:366–370. doi: 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- 48.Heid ME, Keyel PA, Kamga C, Shiva S, Watkins SC, Salter RD. Mitochondrial reactive oxygen species induces NLRP3-dependent lysosomal damage and inflammasome activation. J Immunol. 2013;191:5230–5238. doi: 10.4049/jimmunol.1301490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD. A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332:717–721. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- 50.Shenoy AR, Kim BH, Choi HP, Matsuzawa T, Tiwari S, MacMicking JD. Emerging themes in IFN-gamma-induced macrophage immunity by the p47 and p65 GTPase families. Immunobiology. 2007;212:771–784. doi: 10.1016/j.imbio.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Traver MK, Henry SC, Cantillana V, Oliver T, Hunn JP, Howard JC, Beer S, Pfeffer K, Coers J, Taylor GA. Immunity-related GTPase M (IRGM) proteins influence the localization of guanylate-binding protein 2 (GBP2) by modulating macroautophagy. J Biol Chem. 2011;286:30471–30480. doi: 10.1074/jbc.M111.251967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Britzen-Laurent N, Bauer M, Berton V, Fischer N, Syguda A, Reipschlager S, Naschberger E, Herrmann C, Sturzl M. Intracellular trafficking of guanylate-binding proteins is regulated by heterodimerization in a hierarchical manner. PLoS One. 2010;5:e14246. doi: 10.1371/journal.pone.0014246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vestal DJ, Jeyaratnam JA. The guanylate-binding proteins: emerging insights into the biochemical properties and functions of this family of large interferon-induced guanosine triphosphatase. J Interferon Cytokine Res. 2011;31:89–97. doi: 10.1089/jir.2010.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pilla DM, Hagar JA, Haldar AK, Mason AK, Degrandi D, Pfeffer K, Ernst RK, Yamamoto M, Miao EA, Coers J. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc Natl Acad Sci U S A. 2014;111:6046–6051. doi: 10.1073/pnas.1321700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shenoy AR, Wellington DA, Kumar P, Kassa H, Booth CJ, Cresswell P, MacMicking JD. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 2012;336:481–485. doi: 10.1126/science.1217141. [DOI] [PubMed] [Google Scholar]
- 56.Finethy R, Jorgensen I, Haldar AK, de Zoete MR, Strowig T, Flavell RA, Yamamoto M, Nagarajan UM, Miao EA, Coers J. Guanylate binding proteins enable rapid activation of canonical and noncanonical inflammasomes in Chlamydia-infected macrophages. Infect Immun. 2015;83:4740–4749. doi: 10.1128/IAI.00856-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Man SM, Karki R, Sasai M, Place DE, Kesavardhana S, Temirov J, Frase S, Zhu Q, Malireddi RK, Kuriakose T, Peters JL, Neale G, Brown SA, Yamamoto M, Kanneganti TD. IRGB10 Liberates Bacterial Ligands for Sensing by the AIM2 and Caspase-11-NLRP3 Inflammasomes. Cell. 2016;167:382 e17–396 e17. doi: 10.1016/j.cell.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Briken V, Ruffner H, Schultz U, Schwarz A, Reis LF, Strehlow I, Decker T, Staeheli P. Interferon regulatory factor 1 is required for mouse Gbp gene activation by gamma interferon. Mol Cell Biol. 1995;15:975–982. doi: 10.1128/mcb.15.2.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Man SM, Karki R, Malireddi RK, Neale G, Vogel P, Yamamoto M, Lamkanfi M, Kanneganti TD. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat Immunol. 2015;16:467–475. doi: 10.1038/ni.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meunier EDM, Dreier RF, Schürmann N, Kenzelmann Broz D, Warming S, Roose-Girma M, Bumann D, Kayagaki N, Takeda K, Yamamoto M, Broz P. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509:366–370. doi: 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- 61.Yamamoto M, Okuyama M, Ma JS, Kimura T, Kamiyama N, Saiga H, Ohshima J, Sasai M, Kayama H, Okamoto T, Huang DC, Soldati-Favre D, Horie K, Takeda J, Takeda K. A cluster of interferon-gamma-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity. 2012;37:302–313. doi: 10.1016/j.immuni.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 62.Mastroeni P. Immunity to systemic Salmonella infections. Curr Mol Med. 2002;2 doi: 10.2174/1566524023362492. [DOI] [PubMed] [Google Scholar]
- 63.Morgan MJ, Liu ZG. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res. 2011;21:103–115. doi: 10.1038/cr.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007;17:422–427. doi: 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 65.LaRock CN, Cookson BT. Burning down the house: cellular actions during pyroptosis. PLoS Pathog. 2013;9:e1003793. doi: 10.1371/journal.ppat.1003793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharma D, Kanneganti TD. The cell biology of inflammasomes: Mechanisms of inflammasome activation and regulation. J Cell Biol. 2016;213:617–629. doi: 10.1083/jcb.201602089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Storek KM, Monack DM. Bacterial recognition pathways that lead to inflammasome activation. Immunol Rev. 2015;265:112–129. doi: 10.1111/imr.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miao EA, Warren SE. Innate immune detection of bacterial virulence factors via the NLRC4 inflammasome. J Clin Immunol. 2010;30:502–506. doi: 10.1007/s10875-010-9386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Broz P, Newton K, Lamkanfi M, Mariathasan S, Dixit VM, Monack DM. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J Exp Med. 2010;207:1745–1755. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qu Y, Misaghi S, Newton K, Maltzman A, Izrael-Tomasevic A, Arnott D, Dixit VM. NLRP3 recruitment by NLRC4 during Salmonella infection. J Exp Med. 2016;213:877–885. doi: 10.1084/jem.20132234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, Liu L, Shao F. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 72.Franceschini A, Capece M, Chiozzi P, Falzoni S, Sanz JM, Sarti AC, Bonora M, Pinton P, Di Virgilio F. The P2X7 receptor directly interacts with the NLRP3 inflammasome scaffold protein. FASEB J. 2015;29:2450–2461. doi: 10.1096/fj.14-268714. [DOI] [PubMed] [Google Scholar]
- 73.Gombault A, Baron L, Couillin I. ATP release and purinergic signaling in NLRP3 inflammasome activation. Front Immunol. 2012;3:414. doi: 10.3389/fimmu.2012.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wynosky-Dolfi MA, Snyder AG, Philip NH, Doonan PJ, Poffenberger MC, Avizonis D, Zwack EE, Riblett AM, Hu B, Strowig T, Flavell RA, Jones RG, Freedman BD, Brodsky IE. Oxidative metabolism enables Salmonella evasion of the NLRP3 inflammasome. J Exp Med. 2014;211:653–668. doi: 10.1084/jem.20130627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 76.Broz P, Ruby T, Belhocine K, Bouley DM, Kayagaki N, Dixit VM, Monack DM. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature. 2012;490:288–291. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kayagaki N, Stowe IB, Lee BL, O'Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, Liu PS, Lill JR, Li H, Wu J, Kummerfeld S, Zhang J, Lee WP, Snipas SJ, Salvesen GS, Morris LX, Fitzgerald L, Zhang Y, Bertram EM, Goodnow CC, Dixit VM. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 78.Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, Sun H, Wang DC, Shao F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016 doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- 79.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 80.Pereira M, Tourlomousis P, Wright J, T PM, Bryant CE. CARD9 negatively regulates NLRP3-induced IL-1beta production on Salmonella infection of macrophages. Nat Commun. 2016;7:12874. doi: 10.1038/ncomms12874. [DOI] [PMC free article] [PubMed] [Google Scholar]