Summary
Increasing grain yield and improving grain quality are two important goals for rice breeding. A better understanding of the factors that contribute to the overall grain quantity and nutritional quality of rice will lay the foundation for developing new breeding strategies. RAG2 is a member of 14‐to‐16‐kDa α‐amylase/trypsin inhibitors in rice, which belong to the albumin of seed storage proteins. We found that RAG2 was specifically expressed in ripening seed and its transcription peak was between 14 and 21 days after flowering. Grain size and 1000‐grain weight were obviously increased in RAG2‐overexpressed lines compared with wild type, and grain size was reduced in RAG2‐suppressed lines. In addition, the major storage substances of the seeds differed significantly in RAG2‐overexpressed and RAG2‐suppressed lines compared to wild type. The protein content and amount of total lipids were increased and decreased, respectively, in the seeds of RAG2‐overexpressed and RAG2‐suppressed lines. Overexpression of RAG2 significantly increased grain size and improved grain quality and yield simultaneously. These results imply that RAG2 might play an important role in regulating grain weight and seed quality of rice. The functional characterization of rice RAG2 facilitates a further understanding of the mechanisms involved in grain size and seed quality and may be helpful in improving grain yield and quantity in cereal crops.
Keywords: Oryza sativa, RAG2, seed storage proteins, grain weight, grain quality
Introduction
Global food security is challenged by the convergence of multiple factors, including continuously growing population, reduced arable land, demand for biofuel production and global climate change (Zhang et al., 2008). Rice (Oryza sativa) is the most widely consumed staple food crop that feeds more than half of the world's human population. In addition, with the improved quality of life, people are increasingly concerned about the quality of food. Accordingly, increasing grain yield and improving grain quality of food crops are two important goals of basic and applied science research in plants (Sakamoto and Matsuoka, 2004; Zhang, 2007).
Grain yield of rice is determined by three major factors, including the number of tillers per plant, the number of filled grains per panicle and 1000‐grain weight (Sakamoto and Matsuoka, 2008). Grain weight is primarily determined by grain size (volume) and the degree of grain filling (plumpness) (Sakamoto and Matsuoka, 2008; Xing and Zhang, 2010). Grain size or shape is also an important quality trait of rice grains because of varied consumer preferences in different geographical areas (Zuo and Li, 2014). Grain size is specified by its three dimensions: grain length, grain width and grain thickness. As a quantitative trait, grain size is predominantly and tightly controlled by genetic factors. In recent years, several important genes that control the grain size in rice have been identified. GW2 encodes a previously unidentified RING‐type protein with E3 ubiquitin ligase activity and affects spikelet hull width by regulating cell numbers (Song et al., 2007). qGL3/qGL3.1 encodes a novel putative serine/threonine protein phosphatase. The reduced phosphatase activity may increase the cell number of the outer glume, resulting in longer grains (Zhang et al., 2012). GS3 is a major quantitative trait locus (QTL) that modulates grain length by controlling the number of the cell in the upper epidermis of the glume (Fan et al., 2006). GS5 encodes a putative serine carboxypeptidase and functions as a positive regulator of grain width (Li et al., 2011; Tan et al., 2000). GW5 encodes a novel nuclear protein that physically interacts with polyubiquitin. Within the ubiquitin–proteasome pathway, GW5 regulates cell division in the outer glumes during seed development (Wan et al., 2008). GW8 encodes a transcription factor OsSPL16 and positively regulates grain width and grain weight via the promotion of cell proliferation (Wang et al., 2012).
Grain weight is also affected by grain plumpness, which is mainly controlled by the accumulation of storage substances, such as starch, proteins and lipids. FLO2 plays a pivotal regulatory role in grain size and starch quality by affecting the accumulation of these substances in the rice endosperm (She et al., 2010). The mutation of rice starch regulator1 (RSR1) produces a larger seed and increases seed mass and yield (Fu and Xue, 2010). Mutation of OsSUT2, which encodes a tonoplast‐localized sucrose transporter, leads to a growth retardation phenotype with a notable reduction in grain weight (Eom et al., 2011). Maize (Zea mays) opaque2 (o2) encodes a basic Leu zipper transcription factor. Mutations of opaque2 result in a severe reduction in 22‐kDa α‐zein accumulation in seeds and grain filling (Prioul et al., 2008).
Seed storage proteins (SSPs) are one of the main factors determining the nutritional quality of rice. Based on their solubility properties, the SSPs of rice are classified into glutelin, prolamin, albumin and globulin (Yang et al., 2013). Glutelins are major SSPs of rice, accounting for 60%–80% by weight of the total seed protein content, and are encoded by 15 genes copies in the rice genome. Glutelins are classified into four subfamilies (GluA, GluB, GluC and GluD) based on amino acid sequence similarity (Kawakatsu et al., 2008). The prolamins make up 20%–30% of the seed protein and are encoded by a multigene family of 34 gene copies in three groups defined by their relative molecular weights (Saito et al., 2012; Xu and Messing, 2009). Rice seed albumin is classified as the water‐soluble fraction, which comprises about 5% of the total seed protein (Mawal et al., 1987). The major albumin exhibits heterogeneity in its molecular size (14–16 kDa) and isoelectric point (pI 6–8), and it shows immunological cross‐reactivity (Tsukasa Matsuda et al., 1991). The promoters of several GLUTELIN genes (GluA‐1, GluA‐2, GluA‐3 and GluB‐3) are detected in the peripheral region of the endosperm, whereas GluB‐5, GluC and GluD are active in various regions of the starch endosperm (Komatsu and Hirano, 1992; Lee et al., 2015). NRP33 encoding a 13‐kDa prolamin polypeptide has been cloned (Sha et al., 1996). Previous studies revealed that reducing the expression level of rice SSP‐related genes, such as GluA, GluB, RP10 (rice prolamins oryzein10) and RP16 (rice prolamins oryzein16), leads to changes in SSP content (Kawakatsu et al., 2010; Xu and Messing, 2009). Increasing the nutrient quality of rice through changing the SSP content is a critical goal in rice breeding. Expression of a soybean β‐globin gene in transgenic rice led to a 4% increase in total protein content in transgenic seeds (Zheng et al., 1995). Expression of a soybean glycinin gene in transgenic rice resulted in a significant improvement of glutelin storage in transgenic seed (Katsube et al., 1999). An interesting feature of the Lgc1 lines is that glutelin‐content mutants (Lgc1) are used to produce ‘super low‐protein rice’ for patients with kidney disease (Kusaba et al., 2003).
Fatty acid (FA) content is a quality‐related factor affecting rice appearance, eating quality and storage (Kim et al., 2015). FA deficiency is known to have a major impact on human health in developing countries. Down‐regulated expression of OsLTPL36 led to decreased FA content and reduced seed quality of rice (Wang et al., 2015). Overexpression of Arabidopsis thaliana SFD1/GLY1, a gene encoding plastid‐localized glycerol‐3‐phosphate dehydrogenase, increased seed lipid content in transgenic rice (Singh et al., 2016). These studies also contribute to rice molecular breeding focusing on improving grain quality.
The albumin gene family (RAGs) contains five genes, which are highly expressed at the seed maturation stage (Alvarez et al., 1995). Few studies of RAG genes have been reported. The expression of RAG1 is trans‐activated by RPBF (rice prolamin box binding factor) (Kawakatsu et al., 2009). The RAG2 gene was obtained by screening a rice genomic library with a probe clone RA17 (coding the 16‐kDa protein) (Adachi et al., 1993), and it is a member of the 14‐to‐16‐kDa α‐amylase/trypsin inhibitors of rice and contains 10 cysteine residues (Adachi et al., 1993; Alvarez et al., 1995). RAG2 has been reported to be a major allergen in rice (Kurokawa et al., 2014; Selgrade et al., 2009). RAG2 is specifically expressed in ripening seed (Kurokawa et al., 2014), but the biological functions of rice RAG2 remain elusive.
In this study, we examined the expression pattern of RAG2 using transcriptional profiling and in situ hybridization tests. The results show that RAG2 is specifically expressed in the developing seeds, with the highest expression level in seeds of 14–21 DAP. Overexpression and RNAi were carried out to increase and reduce the expression of RAG2, respectively. Grain size and 1000‐grain weight were obviously increased in RAG2‐OX lines compared to wild type (WT). Furthermore, three major storage substances of the seeds were changed in different ways in RAG2‐OX and RNAi lines. The content of proteins and lipids was increased significantly in RAG2‐OX lines. These results imply that RAG2, a seed‐specific expression protein, may play an important role in regulating storage substances and thereby affect seed plumpness and ultimately control the grain yield and quality of rice. The discovery of RAG2 may facilitate increased seed production and improvement of seed quality, and it can also be effectively applied to crop breeding programs.
Results
Structural and sequence analysis of RAG2
RAG2 is a member of 14‐to‐16‐kDa α‐amylase/trypsin inhibitors in rice, which are albumin proteins homologous to α‐amylase/trypsin inhibitor family proteins. The genomic DNA of RAG2 is 783 nucleotides with one exon (http://rice.plantbiology.msu.edu/cgi-bin/ORF_infopage.cgi?orf=LOC_Os07g11380). The cDNA contained a 498‐bp open reading frame (excluding the stop codon), which encoded a 166‐amino acid protein with a theoretical molecular mass of about 16 kDa and pI of 5.4 (AK107328) (Figure 1a). RAG2 displays high homology with another cDNA clone, RAG1 (Figure 1b). In addition, two 8‐bp direct repeat units (ATGCAAAA) existed in the RAG2 promoter (Figure 1a). This consensus sequence ATGCAAAA, which reminisces the heptamer sequence TGCAAAA, was identified in rice glutelin genes (Okita et al., 1989) and the ‐300‐bp element in cereal genes (Colot et al., 1987; Maier et al., 1987). CTTTCGTGTA has been identified as the recognition site of a DNA‐binding protein, particularly in the glutelin promoter. This site is similar to the sequence CTTTAGTCTT in the RAG2 promoter region (Figure 1a). The transcriptional initiation site of RAG2 gene might be the same as that of RAG1 gene because the nucleotide sequence around the comparable region is similar (Adachi et al., 1993). Previous studies found that RAG2 was localized mainly in protein bodies II (PB‐II) of the endosperm cells (Kurokawa et al., 2014). The protein annotation on the Pfam website indicated that RAG2 belonged to a protease inhibitor/seed storage/LTP family (CL0482).
Figure 1.
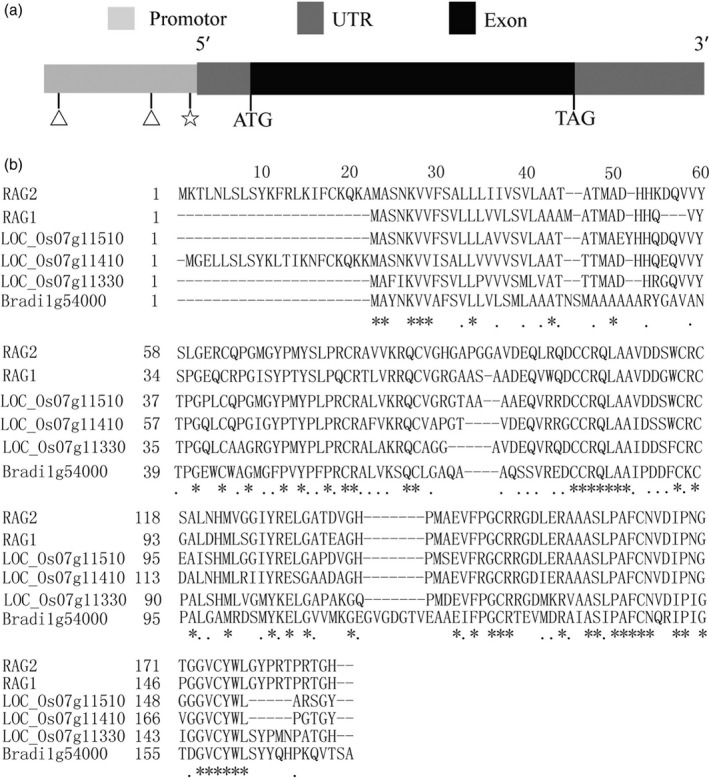
Structural and sequence analysis of RAG2. (a) A schematic representation of the exon and intron organization of RAG2. RAG2 consists of one exon (black box) with an 82‐bp 5′UTR (grey box) and a 200‐bp 3′UTR (grey box). Two ATGCAAAA (triangle, −1028 bp, −252 bp) and one CTTTAGTCTT (pentagon, −21 bp) cis‐element in RAG2 promoter region. (b) Protein sequence alignment of RAG2 with RAG1, LOC_Os07g11510, LOC_Os07g11410, LOC_Os07g11330 and Bradi1g54000. Residues marked with asterisks and dots are highly conserved and semiconserved, respectively. A dash ‘–’ denotes a gap in the alignment.
Specific high expression of RAG2 in developing rice seeds
The expression pattern of RAG2 was investigated in different tissues, including root, stem, leaf, panicle and seed of Zhonghua 11 (Oryza sativa ssp. japonica cv. Zhonghua 11). The analysis of qRT‐PCR indicated that RAG2 was specifically expressed in the developing seed with a higher expression level during 14–21 DAP; however, the expression levels in leaf, root and stem were much lower (Figure 2i). These results suggest that RAG2 may function in the developing seed of rice.
Figure 2.
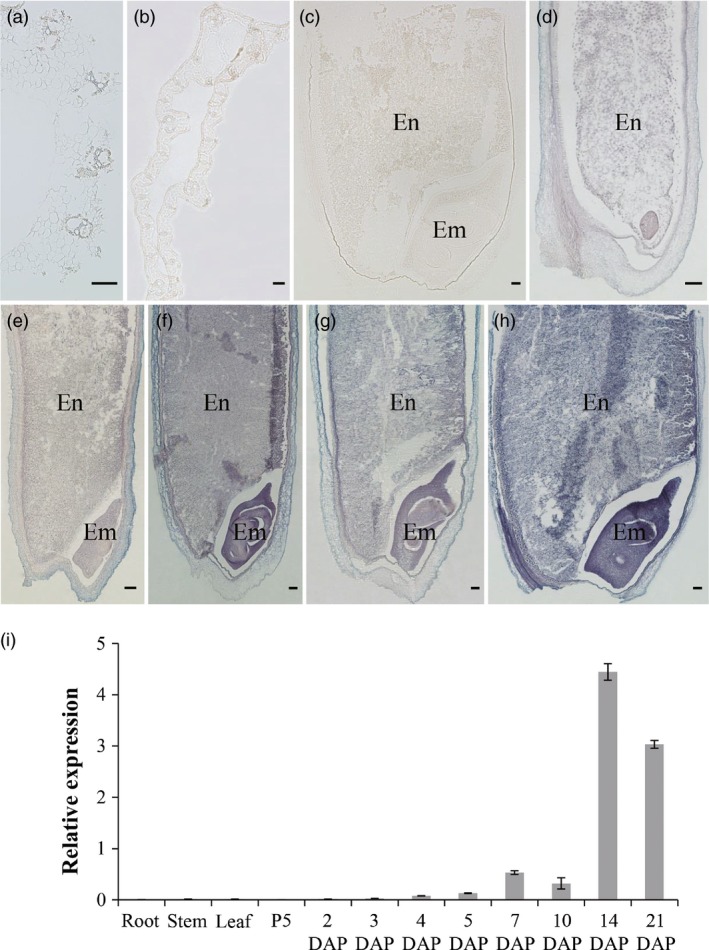
Spatial and temporal expression pattern of RAG2. (a–h) RNA in situ hybridization of RAG2. (a) Stem. (b) Leaf. (c) Sense. (d) 3‐d seed. (e) 5‐d seed. (f) 7‐d seed. (g) 10‐d seed. (h) 14‐d seed. En, Endosperm; Em, Embryo. Bar: 100 μm (a–h). (i) qRT‐PCR analysis of RAG2. Total RNAs were extracted from root (booting stages), stem (booting stages), leaf (1 day before heading), P5 is panicle at stage 5 (pollen mother cell formation stage), developing seeds (2, 3, 4, 5, 7, 10, 14 and 21 DAP). Data are mean ± SE for three replicates.
In addition, the temporal and spatial expression pattern of RAG2 was determined by in situ hybridization (Figure 2a–h). The strong expression signal of RAG2 was observed at 3, 5, 7, 10 and 14 DAP in seed, and gradually increased with seed development, which was consistent with the qRT‐PCR results (Figure 2d–h). In the 7‐, 10‐ and 14‐DAP endosperm, the expression signal was observed in aleurone layer (Figure 2f–h). In the 14‐DAP seed, the embryo and endosperm had the strongest hybridization signal of RAG2 (Figure 2h). Above all, the expression of RAG2 was specifically high in the developing seed, and it was strongly expressed in the developing embryo and endosperm. The temporal and spatial expression pattern of RAG2 further suggested that it might play a role in rice seed.
Production of RAG2‐overexpressed and RAG2‐suppressed lines
To investigate the function of RAG2 in rice seed development, we generated transgenic rice with RAG2 overexpression or suppression in the seed under the control of maize ubiquitin1 promoter. Successful transformants were confirmed by PCR. Twenty RAG2‐overexpressed lines (RAG2‐OX) and 45 RAG2‐suppressed lines (RAG2‐RNAi) were generated, respectively. Six lines (OX‐2, OX‐5, OX‐7, OX‐9, OX‐11 and OX‐15) with increased expression levels and six lines (Ri‐2, Ri‐7, Ri‐10, Ri‐17, Ri‐22 and Ri‐37) with reduced expression levels compared to WT were selected for the next generation. Additionally, three independent RAG2‐OX transgenic lines (OX‐2‐15, OX‐5‐5 and OX‐9‐22) with higher expression level, which originated from T0 generations (OX‐2, OX‐5 and OX‐9), were advanced to the T2 generation (Table S1) for functional analysis. Finally, three independent RAG2‐RNAi transgenic lines (Ri‐2‐21, Ri‐22‐12 and Ri‐37‐9) with lower expression level, which originated from T0 generations (Ri‐2, Ri‐22 and Ri‐37), were advanced to the T5 generation (Table S2) for functional analysis. To facilitate simplified drawing, three RAG2‐OX lines (OX‐2‐15, OX‐5‐5 and OX‐9‐22) with a higher expression level were renamed as OX‐1, OX‐2 and OX‐3, and three RAG2‐RNAi (Ri) lines (Ri‐22‐12, Ri‐2‐21 and Ri‐37‐9) with a lower expression level were renamed as Ri‐1, Ri‐2 and Ri‐3 (Figure 3a). DNA hybridization showed that pU2301‐Cflag (RAG2‐OX) and pDS1301‐RAG2 (RAG2‐RNAi) were integrated into the genomes of transgenic plants as only one transgene copy, whereas no cross‐hybridization was observed in WT (Figure 3b).
Figure 3.
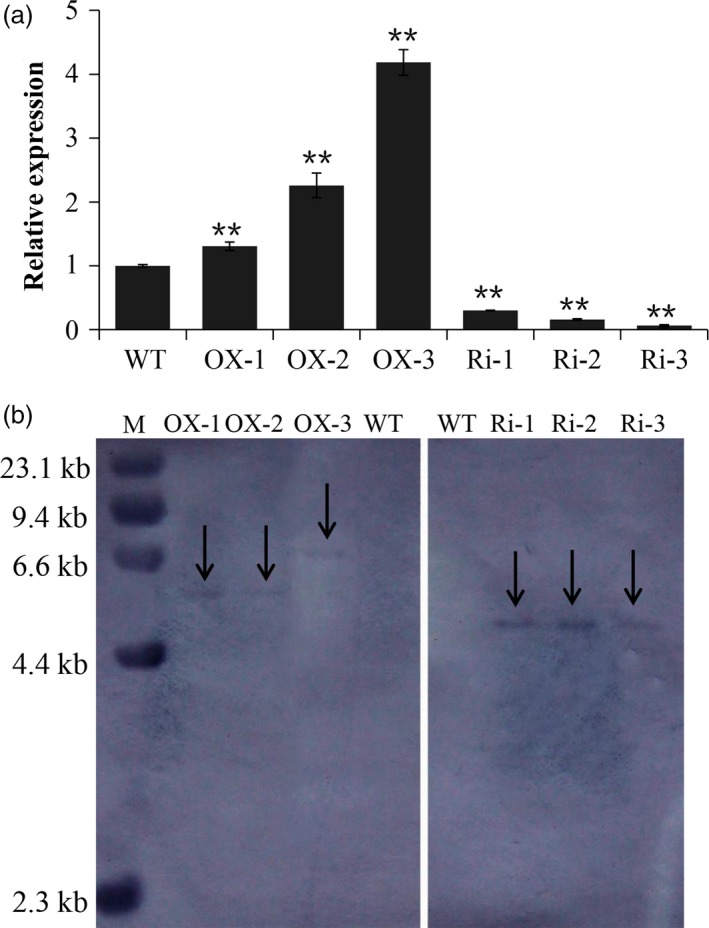
Analysis of WT and the transgenic lines. Real‐time quantitative RT‐PCR was used for analysis of RAG2 transcription in transgenic rice (a), and 10 μg of genomic DNA from each plant line was digested with HindIII for DNA blot hybridization (b). Data are mean ± SE for three replicates. *P < 0.05, **P < 0.01. P‐values produced by two‐tailed Student's t‐test.
Additionally, RAG1 is the paralog of RAG2 (Adachi et al., 1993), qRT‐PCR analysis showed that the expression level of RAG1 was not affected in the RAG2‐RNAi transgenic lines (Figure S1).
RAG2 regulates seed size and grain yield
To investigate the influence of altered expression of the RAG2 gene on yield‐related traits, we compared the seed setting rate and the 1000‐grain weight among WT, RAG2‐overexpressed (OX‐1, OX‐2 and OX‐3) and RAG2‐suppressed lines (Ri‐1, Ri‐2 and Ri‐3). Morphological/phenotypic differences were not observed between WT and the two types of transgenic lines at the vegetative stage. The RAG2‐OX lines and WT lines had the same seed setting rate and grain number per panicle. However, significant reductions were observed in the seed setting rate and grain number in the RAG2‐RNAi lines compared with the WT lines (Table 1). Notably, plants with overexpression of RAG2 produced larger seeds than the RNAi and WT plants (Figure 4a, Figure S2). We found that RAG2‐OX transgenic plants showed a substantial increase in grain length compared to the WT (+12.0%, +13.6%, +15.2%, Figure 4c), with smaller increases in grain width (+1.5%, +3.4%, +3.5%, Figure 4d) and thickness (+3.3%, +5.4%, +7.3%, Figure 4e). We also detected a significant increase in seed area (+12.6%, +16.3%, +18.0%, Figure 4f) and 1000‐grain weight (+10.4%, +11.2%, +14.0%, Figure 4b) in RAG2‐OX transgenic plants. Meanwhile, RAG2‐RNAi transgenic plants exhibited an almost opposite phenotype compared to the WT, including a slight reduction in grain length (−2.2%, −1.9%, −1.8%, Figure 4c), grain thickness (+1.7%, −0.4%, −1.5%, Figure 4e) and 1000‐grain weight (+0.7%, −4.1%, +0.1%, Figure 4b); however, the decreases in seed area (−5.7%, −6.1%, −6.4%, Figure 4f) and grain width (−4.4%, −4.7%, −5.1%, Figure 4d) were clear. Furthermore, qRT‐PCR analysis revealed that a consistency existed between the expression level of RAG2 and the phenotype of the seed, suggesting that regulation of the RAG2 resulted in pleiotropic traits (Figures 3a and 4). Increased RAG2 expression may possibly enlarge grain size by expanding the length, width and depth, hence, increasing grain weight.
Table 1.
Analysis of yield parameters of WT and the transgenic lines
| Line | No. of floret per panicles | No. of tillers per plant | Seed set rate (%) | 1000‐grain weight (g) | Chalkiness rate (%) | Chalkiness degree |
|---|---|---|---|---|---|---|
| WT | 156 ± 9 | 11.33 ± 1.18 | 82.38 ± 4.11 | 25.43 ± 1.04 | 10.49 ± 1.96 | 2.95 ± 0.08 |
| OX‐1 | 150 ± 15 | 11.25 ± 1.16 | 79.09 ± 2.52 | 28.08 ± 0.78** | 9.31 ± 3.18 | 3.84 ± 0.01** |
| OX‐2 | 151 ± 21 | 11.42 ± 0.95 | 84.71 ± 1.29 | 28.29 ± 1.03** | 12.82 ± 0.02* | 1.92 ± 0.03* |
| OX‐3 | 161 ± 18 | 11.33 ± 1.11 | 82.20 ± 5.90 | 29.00 ± 1.06** | 6.03 ± 0.28* | 2.66 ± 0.16 |
| Ri‐1 | 147 ± 11 | 11.33 ± 1.03 | 70.13 ± 3.83** | 25.617 ± 3.85 | 63.32 ± 4.14** | 28.60 ± 0.28** |
| Ri‐2 | 141 ± 19* | 11.17 ± 1.14 | 63.39 ± 7.17** | 25.451 ± 0.54 | 70.69 ± 0.05** | 31.14 ± 0.17** |
| Ri‐3 | 135 ± 16* | 11.41 ± 1.04 | 64.09 ± 12.15** | 24.391 ± 0.38* | 73.99 ± 4.56** | 32.64 ± 1.08** |
Grains of the fully filled seeds were removed and used for analysis of the 1000‐grain weight. At least 400 grains of each plant were used to measure the grain chalkiness rate. Data are presented as means ± SE of three biological replicates.
*P < 0.05, and **P < 0.01 by two‐tailed Student's t‐test.
Figure 4.
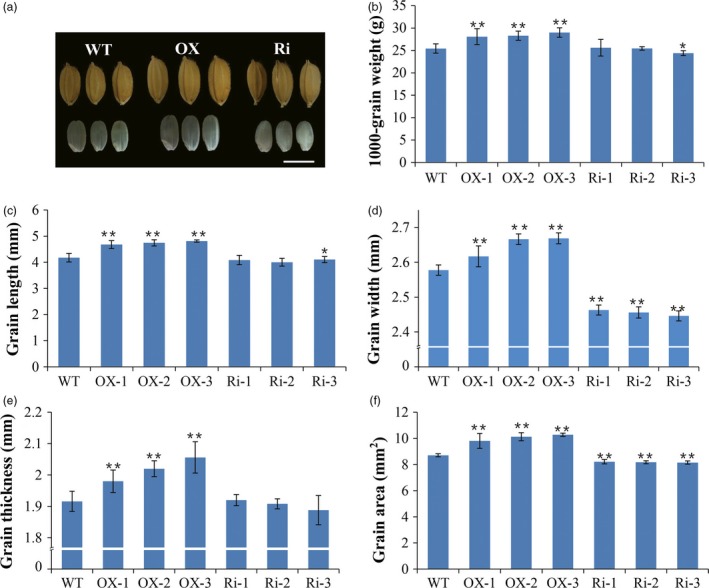
Phenotypes of grain yields of WT and the transgenic lines. (a) Grain size comparison among the RAG2‐OX,RAG2‐RNAi and WT seeds. (b) 1000‐grain weights of the RAG2‐OX,RAG2‐RNAi and WT seeds. (c) Grain lengths of the RAG2‐OX,RAG2‐RNAi and WT seeds. (d) Grain widths of the RAG2‐OX,RAG2‐RNAi and WT seeds. (e) Grain thicknesses of RAG2‐OX,RAG2‐RNAi and WT seeds. (f) Grain areas of the RAG2‐OX,RAG2‐RNAi and WT seeds. Scale bars (a) 5 cm. Values in (b–f) are means ± SD (n = 20). Two‐tailed Student's t‐tests were performed between the RAG2‐OX and WT, and RAG2‐RNAi and WT, respectively (*P < 0.05, **P < 0.01).
We also observed that the grains in RAG2‐RNAi lines displayed higher chalkiness compared with the WT (Table 1). The results of scanning electron microscopy (SEM) indicated that the starch granules in RAG2‐RNAi lines were loosely packed and irregularly polyhedron‐shaped compared with those in the WT (Figure 5), while in RAG2‐OX lines, starch granules had irregular polyhedron shapes similar to those of WT granules (Table 1, Figure S3, e1–e3, f1–f3). These results suggest that RAG2 might also be involved in chalky endosperm formation.
Figure 5.
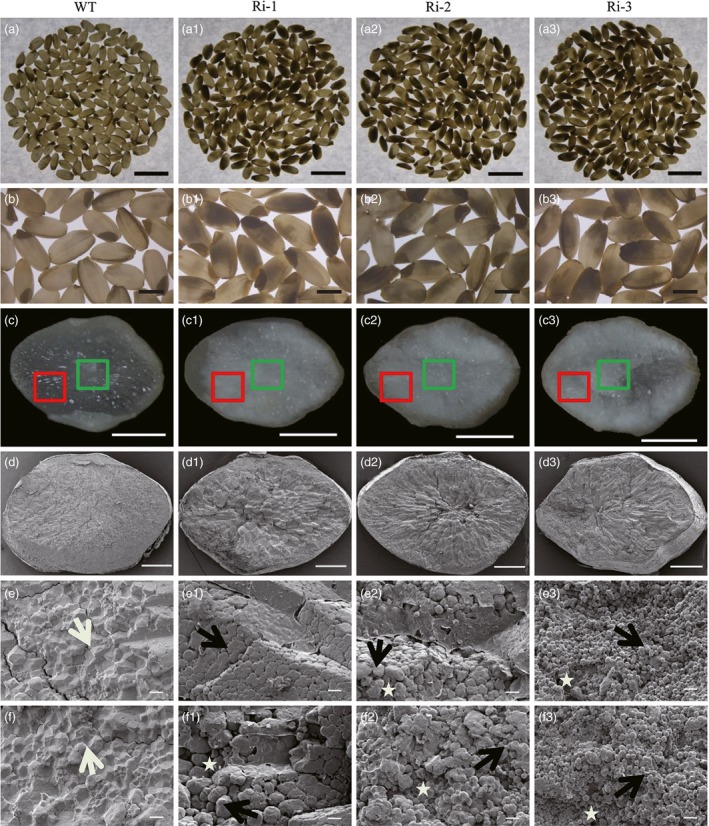
Phenotypes of seeds of WT and RAG2‐RNAi plant. WT: a, b, c, d, e, f. Ri‐1: a1, b1, c1, d1, e1, f1. Ri‐2: a2, b2, c2, d2, e2, f2. Ri‐3: a3, b3, c3, d3, e3, f3. (a‐a3) 150 grains of mature seeds in white light background. (b‐b3) Mature seeds. (c–c3) Cross sections of mature endosperm. RAG2‐RNAi line grains displayed higher chalkiness with less translucence compared with the WT. (d–d3) SEM of the central area of mature endosperm in (c–c3). (e–e3) SEM of the central area of mature endosperm, with the cross sections indicated by a green square in (c–c3). (f–f3) SEM of the central area of mature endosperm, with the cross sections indicated by a red square in (c–c3). White and black arrows in (e–e3, f‐f3) indicate polygonal and round starch granules, respectively. Areas in which starch granules are loosely packed are indicated by asterisks. The packaging of starch granules in the endosperm of RAG2‐RNAi lines (Ri‐1, Ri‐2 and Ri‐3) was loose, with round and smaller granules. Scale bars: 10 mm (a–a3), 3 mm (b–b3), 1 mm (c–c3), 500 μm (d–d3), 10 μm (e–e3, f–f3). Data are mean ± SE for three replicates. *P < 0.05, **P < 0.01. P‐values produced by two‐tailed Student's t‐test.
RAG2 affects the accumulation of seed storage substances
To analyse the significant increase in grain weight in RAG2‐OX lines and the effect of RAG2 on grain quality, we measured the contents of total protein, starch and total lipid.
Measurement by the chemical method revealed that the content of seed total protein was increased by 3, 4.8 and 4 mg/g in the OX‐1, OX‐2 and OX‐3 lines and reduced by 1.9, 2.4 and 1.6 mg/g in the Ri‐1, Ri‐2 and Ri‐3 lines, respectively, compared with the WT (Figure 7a, Table 2). The glutelin and prolamin contents were clearly increased in the RAG2‐OX lines (+28.5%, +31.0%, +55.5%) and reduced in the RAG2‐RNAi lines (+1.6%, −2.5%, −7.9%) (Table 2). Based on the SDS‐PAGE analysis, most of the SSPs, such as the glutelins and prolamins, were higher in the RAG2‐OX lines and lower in the RAG2‐RNAi lines (Figure 6). Previous research showed that RAG2 was an albumin. The 16‐kDa RAG2 was increased in the RAG2‐OX lines and almost disappeared in the RAG2‐RNAi lines (Figure 6a).
Figure 7.
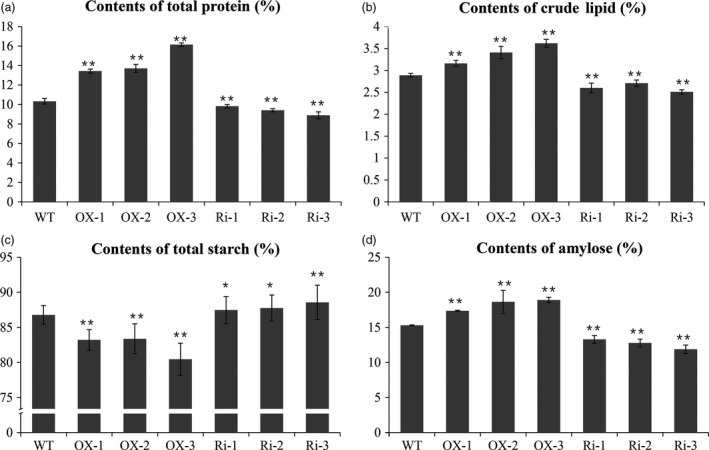
Comparisons of protein, lipid and starch contents in mature seeds of WT and the transgenic lines. Powder grains of fully filled seeds of the WT and the transgenic lines were used for analysis of the starch, protein and lipid content. (a) Contents of total protein. (b) Contents of total starch. (c) Contents of crude lipid. (d) Contents of amylose. Data are mean ± SE for three replicates. *P < 0.05, **P < 0.01. P‐values produced by two‐tailed Student's t‐test.
Table 2.
Contents of protein, starch and lipid in mature seeds
| Line | Contents of total protein (mg/g) | Contents of prolamin (%) | Contents of glutelin (%) | Contents of total starch (%) | Contents of total lipid (%) |
|---|---|---|---|---|---|
| WT | 102.14 ± 0.79 | 0.33 ± 0 | 10.00 ± 0.29 | 86.79 ± 1.33 | 2.89 ± 0.04 |
| OX‐1 | 105.16 ± 0.65** | 0.57 ± 0.01** | 12.85 ± 0.20** | 83.22 ± 1.46** | 3.16 ± 0.07** |
| OX‐2 | 106.15 ± 1.14** | 0.59 ± 0.02** | 13.10 ± 0.42** | 83.37 ± 2.13** | 3.41 ± 0.14** |
| OX‐3 | 106.94 ± 0.87** | 0.60 ± 0.02** | 15.55 ± 0.16** | 80.45 ± 2.30** | 3.62 ± 0.09** |
| Ri‐1 | 100.55 ± 0.37** | 0.30 ± 0.03** | 10.16 ± 0.20 | 87.47 ± 1.92* | 2.60 ± 0.11** |
| Ri‐2 | 99.79 ± 0.46** | 0.32 ± 0.02* | 9.75 ± 0.18* | 87.76 ± 1.86* | 2.71 ± 0.07* |
| Ri‐3 | 100.22 ± 0.31** | 0.31 ± 0.02* | 9.21 ± 0.38** | 88.57 ± 2.45** | 2.51 ± 0.05** |
Powder grains of the transgenic lines and WT fully filled seeds were used for analysis of the lipid, protein and starch contents. All data are given as means ± SE of three biological replicates. *P < 0.05. **P < 0.01. P‐values produced by two‐tailed Student's t‐test.
Figure 6.
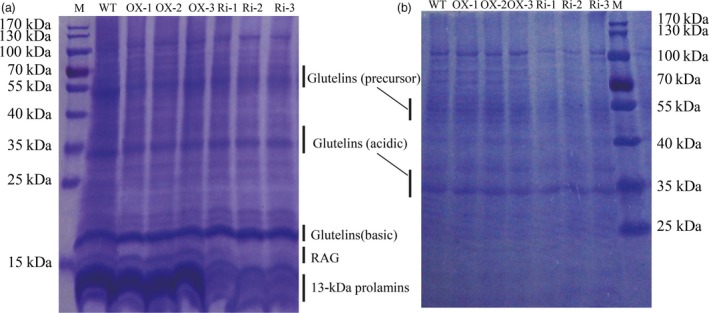
Seed storage proteins level in seeds of WT and the transgenic lines. SDS‐PAGE of seed proteins extracted from mature seeds. Glutelins (precursor, acidic and basic subunits), α‐globulin, 14‐to‐16‐kDa rice protein (RAG) and 13‐kDa prolamins are indicated. a: Add 10 μL sample; b: Add 5 μL sample.
Total starch content and apparent amylose content (AAC) were measured in transgenic rice seeds. Total starch content in Ri‐1 (88.57%), Ri‐2 (87.76%), Ri‐2 (87.47%), OX‐1 (83.22%), OX‐2 (83.37%) and OX‐3 (80.45%) was largely changed compared to the WT (86.79%; Figure 7c, Table 2). The AAC was lower in Ri‐1 (11.88%), Ri‐2 (12.78%) and Ri‐2 (13.29%), but higher in OX‐1 (17.37%), OX‐2 (18.63%) and OX‐3 (18.91%) compared with the WT (15.30%) (Figure 7d, Table 2). Thus, the AAC was higher, by 12.8%–24.3%, in the RAG2‐OX lines and lower, by 19.6%–26.4%, in the RAG2‐RNAi lines than in the WT.
The results demonstrate that the total lipid content of the seed in three RAG2‐OX lines (OX‐1, OX‐2 and OX‐3) was 27.70%, 28.33% and 31.52% higher than that in the WT. Conversely, the total lipid content of the seed was 10.03%, 6.22% and 13.15% lower in the three RAG2‐RNAi lines (Ri‐1, Ri‐2 and Ri‐3) (Figure 7b, Table 2). Crude FA components of rice include palmitic, oleic and linoleic acids, among others. The palmitic acid is a saturated FA with the highest content in rice seed (Ying et al., 2012), and the oleic acid is a predominant monounsaturated FA of rice seed oil. In view of this, we further tested the content of six main FAs composition using GC‐MS. The results showed that C16:0 (palmitic acid), C18:1 (oleic acid), C18:2 (linoleic acid) and C18:3 (linolenic acid) were significantly increased in the three RAG2‐OX lines, but decreased in the three RAG2‐RNAi lines (Table 3).
Table 3.
Contents of fatty acid in mature seeds
| Traits | Fatty acid composition (mg/g) | ||||||
|---|---|---|---|---|---|---|---|
| WT | OX‐1 | OX‐2 | OX‐3 | Ri‐1 | Ri‐2 | Ri‐3 | |
| C16:0 (Palmitic acid) | 4.84 ± 0.06 | 5.12 ± 0.05* | 5.38 ± 0.08** | 5.90 ± 0.02** | 4.53 ± 0.02** | 4.25 ± 0.01** | 4.04 ± 0.01** |
| C18:0 (Stearic acid) | 0.59 ± 0.03 | 0.60 ± 0.02 | 0.67 ± 0.06** | 0.76 ± 0.04** | 0.64 ± 0.03 | 0.64 ± 0.01 | 0.58 ± 0.03 |
| C18:1 (Oleic acid) | 12.52 ± 0.21 | 13.52 ± 0.16** | 14.36 ± 0.38** | 14.87 ± 0.15** | 10.34 ± 0.69** | 9.98 ± 0.31** | 9.85 ± 0.20** |
| C18:2 (Linoleic acid) | 10.23 ± 0.13 | 11.57 ± 0.08** | 12.84 ± 0.39** | 13.72 ± 0.14** | 10.33 ± 0.40 | 10.42 ± 0.26 | 10.01 ± 0.05* |
| C18:3 (Linolenic acid) | 0.36 ± 0.02 | 0.42 ± 0.02** | 0.46 ± 0. 01** | 0.48 ± 0.02** | 0.38 ± 0.01 | 0.37 ± 0.02 | 0.35 ± 0.01 |
| C20:0 (Eicosanoic acid) | 0.22 ± 0.01 | 0.24 ± 0.01 | 0.45 ± 0.07** | 0.27 ± 0.02** | 0.20 ± 0.03* | 0.20 ± 0.04* | 0.18 ± 0.02** |
Data are presented as means ± SE of three biological replicates.
*P < 0.05. **P < 0.01. P‐values produced by two‐tailed Student's t‐test.
Relative expression levels of the genes related to seed storage compounds
Storage compounds were altered in RAG2‐overexpressed and RAG2‐suppressed seeds, and we investigated the expression level of the genes related to these compounds. Total RNA for qRT‐PCR was extracted from the developing seeds at 14 DAF.
The expression levels of SSP genes were closely correlated with their protein level in each line (Figures 6 and 8). In the RAG2‐OX lines, the glutelin gene GluA was obviously increased, and the GluB and GluD were slightly enhanced (Figure 8). Conversely, in the RAG2‐RNAi lines, the levels of GluA and GluB were notably reduced, and those of GluD were slightly decreased compared to the WT (Figure 8). The prolamin genes, such as RM1 (Cys‐rich 13‐kDa prolamin), Prol14 (Cys‐poor 13‐kDa prolamin) and RP10 (10‐kDa prolamin), were slightly increased in the RAG2‐OX lines and decreased in the RAG2‐RNAi lines. These results further suggest that the changes in expression of RAG2 led to altered expression of rice SSP genes.
Figure 8.
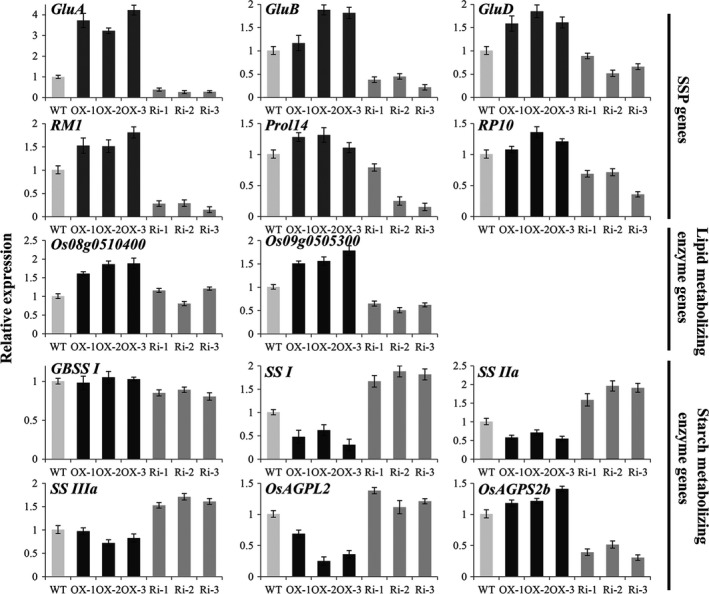
Expression analyses of SSP genes and starch and lipid‐metabolizing enzyme genes by quantitative RT‐PCR. RNA was extracted from developing seeds at 14 d after flowering (DAF) for SSP genes (GluA, GluB, GluD,RM1, Prol14 and RP10) and lipid (Os08g0510400 and Os09g0505300) and starch (GBSS I,SS I,SS IIa,SS IIIa, OsAGPL2 and OsAGPS2b)‐metabolizing enzyme genes. Light‐grey, black and dark‐grey bars represent the expression levels in WT,RAG2‐OX and RAG2‐RNAi line seeds, respectively. The relative expression levels were normalized to those of UBI.
We examined the expression level of starch‐related genes. The expression levels of the granule‐bound starch synthase gene GBSS I and the ADP glucose pyrophosphorylase gene OsAGPS2b were decreased in the RAG2‐RNAi line seeds and were comparable in the RAG2‐OX lines and the WT (Figure 8). The expression of genes encoding other starch‐metabolizing enzymes was increased in seeds in the RAG2‐RNAi lines and decreased in the RAG2‐OX lines. These findings are consistent with measurement of the previous components, suggesting that RAG2 influences starch biosynthesis.
We also examined the expression levels of the rice ketoacyI‐ACP reductase gene Os08g0510400 and the acyIACP thioesterase gene Os09g0505300, which are known to play crucial roles in lipid metabolism in Arabidopsis, and are rice homologues of Arabidopsis At1g62610 and At3g25110, respectively (Mu et al., 2008). The expression of Os08g0510400 and Os09g0505300 was markedly increased in RAG2‐OX line seeds and decreased slightly in RAG2‐RNAi line seeds (Figure 8). These results suggest that RAG2 influences the accumulation of fat.
A change in RAG2 expression leads to the altered expression of storage substance genes. These results are also consistent with the change in storage components.
Discussion
In the study, we demonstrated that RAG2, a 16‐kDa α‐amylase/trypsin inhibitor of rice, was expressed specifically in the developing seed and was possibly involved in the regulation of grain yield and grain quality of rice. The overexpression of RAG2 in transgenic rice was found to increase the content of storage proteins and lipids in seeds and to improve grain yield. Thus, our results may help future rice molecular breeding focused on improving grain yields and seed quality.
In this study, overexpression of RAG2 on the background of WT remarkably increased grain size (Figure 4, Figure S2) and 1000‐grain weight (Figure 4b, Table 1), while RNAi‐mediated knockdown of RAG2 resulted in decreased grain size (Figure 4, Table 1). GS3, GS5, GL3, GW2 and GW8 have been reported to control the grain weight by regulating grain size in rice (Fan et al., 2006; Li et al., 2011; Song et al., 2007; Tan et al., 2000; Wang et al., 2012; Zhang et al., 2012). However, the expression level of these genes was not significantly different between the WT and transgenic plants (Figure S4). Furthermore, histological analysis showed no obvious changes in the morphology phenotype of endosperm cells among WT, RAG2‐OX and RAG2‐RNAi lines (Figure S5). These results suggest that RAG2 regulates rice grain weight in a pathway distinct from those associated with previous cloned grain size regulators in rice. We speculated that RAG2 regulated grain weight by influencing the degree of grain filling (plumpness).
Grain plumpness is determined mainly by the accumulation of storage substances, such as starch, proteins and lipids (Zuo and Li, 2014). Previous studies showed that change in storage substance accumulation affected the grain size and grain weight. FLO2 as a transcription factor actives starch synthesis‐related genes for grain size and grain weight regulation in rice. Overexpression of FLO2 enlarged the size and weight of grains significantly (She et al., 2010). GIF1 encodes a cell‐wall invertase that is required for carbon partitioning during early grain filling. Overexpression of GIF1 increases grain size and grain weight (Wang et al., 2008). Loss function of the TGW6 allele increases grain weight through pleiotropic effects on source organs and results in significant yield increases (Ishimaru et al., 2013). Similarly, overexpression of the RAG2 gene was found to increase the content of storage proteins and lipids in transgenic rice. Therefore, overexpression of RAG2 may possibly improve the accumulation of seed storage substances and contribute to enlarging the size and weight of grains in rice. Further supports were from the expression pattern of RAG2. qRT‐PCR and in situ hybridization showed that RAG2 was specifically high expressed in seed at 14–21 DAP. This stage is an important period of accumulation of seed storage substances.
Rice is the main source of people's daily intake of protein, providing them with a total amount of protein that is three times that of beans (Mahmoud and El Anany, 2014). The content and composition of SSPs are important nutrient qualities of seeds (Kim et al., 2013). Research by the International Rice Research Institute (IRRI) showed that the average content of SSPs was 9.5% in rice (Gomez, 1979). Here, we found that overexpression of RAG2 increased total protein content by 3 mg/g to 4.8 mg/g. The content of glutelin and prolamin was significantly increased in the RAG2‐OX lines. The glutelin storage protein is an important digestible protein and is an index of nutrient quality of seeds (Kim et al., 2013). Moreover, qRT‐PCR analysis found that the expression level of most of the SSP‐related genes, including glutelin genes (GluA, GluB and GluD) and prolamin genes (RM1, Prol14 and RP10), was increased in the RAG2‐OX lines and decreased in the RAG2‐RNAi lines. These results suggest that RAG2 may involve in the accumulation of SSP in rice. Overexpression of RAG2 increased the protein content, thereby increasing the nutritional quality of seeds.
The FA content is another nutritional quality of seeds that determines the appearance and eating quality of rice (Boxer et al., 2010). The content of total lipids was increased (9%–25%) in the RAG2‐OX line seeds, but decreased in the RAG2‐RNAi line seeds (Figure 7a–c, Table 2). A recent report showed that OsLTPL36, a lipid‐transfer protein of rice, is essential for FA accumulation and seed quality (Wang et al., 2015). The analysis of the homologous sequence showed that the 19 amino acid residues of lipid‐transfer proteins are like those present in RAG2 of rice (Adachi et al., 1993), implying that RAG2 might be important in lipid metabolism for FA accumulation in rice seed.
The main seed storage substances of rice include starch, proteins and lipids (Fitzgerald et al., 2009). The total percentage of seed storage substances is constant, and a change in one component will certainly lead to a change in the percentage of the other contents (Fitzgerald et al., 2009; Zhu et al., 2003). OsPPDKB encodes a pyruvate orthophosphate dikinase that modulates carbon metabolism during grain filling in rice. The starch content is reduced, the total protein content is slightly higher, and the lipid is significantly increased in its mutant (flo4) (Kang et al., 2005). In a sweet wheat (SW) mutant lacking functional granule‐bound starch synthase I (GBSSI) and starch synthase IIa (SSIIa), the starch content is much lower, but the content of protein and lipid is higher compared to the WT (Shimbata et al., 2011). Similarly, our results showed that a change in the expression of RAG2 not only affected the accumulation of storage proteins, but also affected the lipid and starch contents.
A previous study showed that several rice proteins, such as 14‐to‐16‐, 26‐, 33‐ and 60‐kDa proteins, were recognized by serum IgE of patients showing hypersensitive reactions to rice ingestion (Tsukasa Matsuda et al., 1991). As the first reported allergen protein in rice (Ito et al., 2005), the protein content of RAG2 was obviously decreased in the RAG2‐RNAi lines (Figure 6), but the main nutritional protein glutelin was decreased less. In conclusion, although the content of the allergen protein RAG2 was increased in RAG2‐OX lines, both the grain yield and the nutritional quality were improved. This kind of transgenic rice, with high nutritional value and high yield, could be provided to the vast majority of nonallergic people. While in the RAG2‐RNAi lines, the content of RAG2 protein was obviously decreased, this type of transgenic rice, with lower allergen protein, could be a functional food to allergic patients.
Experimental procedures
Plasmid construction and transformation of rice
To construct the overexpression vector of RAG2, a 498‐bp cDNA fragment (accession number AK107328) without its stop codon encoding the full length of RAG2 was amplified, and was then linked to pU2301‐cFLAG, which carries a maize ubiquitin promoter.
To construct the RNA interference vector of RAG2, a 394‐bp cDNA fragment of RAG2 was amplified, and was then inserted into pDS1301. The recombinant with the first intron, splicing acceptor site and nos terminator gene was driven by maize ubiquitin promoter.
The recombinant constructs were introduced into Agrobacterium tumefaciens strain EHA105 that were further transferred into Zhonghua 11 (ZH11) (Oryza sativa ssp. japonica) as reported (Ma et al., 2015). Primers used in this study are shown in Table S3.
Plant materials and evaluation of the yield‐related traits
RAG2‐FLAG overexpression transgenic plants and RAG2‐RNAi transgenic plants were generated in Zhonghua11 (Oryza sativa ssp. japonica cv. Zhonghua11) background. All the requisite rice plants were grown in a field at the Huazhong Agricultural University, Wuhan and Hainan, China.
Fully filled grains were used for measuring grain size (length, width and thickness), grain quality and 1000‐grain weight. Grain size (length, width and area) of nearly 400 grains from each line was measured using a MRS‐9600TFU2L (MICROTEK) grain observation instrument. Twenty randomly chosen grains from each plant were lined up length‐wise and width‐wise along a Vernier caliper to measure grain length and grain width, respectively. Grain thickness of 20 randomly chosen grains from each plant was measured using Vernier caliper. Grain chalkiness was assessed by the MRS‐9600TFU2L (MICROTEK) grain observation instrument. Grain weight was calculated on the basis of 200 grains and then converted to 1000‐grain weight as previously described (Li et al., 2014). All of the experimental grains were dehulled grains, except the 1000‐grain weight grains. Phenotypic measurements of the positive transgenic plants were undertaken using three independent lines at least.
Sequence and structural analysis of RAG2
DNA and protein sequence of RAG2 were obtained from MSU database (http://rice.plantbiology.msu.edu/) and KOME database (http://cdna01.dna.affrc.go.jp/cDNA/). Blast similarity protein sequence according to the following criteria: query cover>30%, identity>40%, E value<1e‐5 in NCBI (http://www.ncbi.nlm.nih.gov/). Proteins are shown in the figure with the corresponding accession numbers. Alignments were performed in ClustalW.
RNA isolation and quantitative RT‐PCR analyses
Total RNA of various samples was extracted using the TRIzol reagent (Invitrogen, Waltham, MA). First‐strand cDNA was synthesized using PRIME Script Reverse Transcriptase (TaKaRa, Dalian, China). The expression levels were measured using an ABI StepOne™ Real‐time System (Applied Biosystems, Carlsbad, CA) with rice Ubiquitin as the internal control. Primers used in this study are shown in Table S3.
In situ hybridization
Paraffin sections were obtained according to the previous method (Ma et al., 2015). A 135‐bp specific fragment was obtained and cloned into pGM‐T vector. Hybridization of digoxigenin‐labelled antisense and sense probes and immunological detection were performed as described (Sang et al., 2012). The primers are listed in Table S3.
DNA blot hybridization
Southern blotting was performed as previously described (Chen et al., 2015). DNA probes specific to G418 (for pU2301‐RAG2‐cFLAG) and hph (for pDS1301‐RAG2) coding sequence were labelled using a PCR DIG probe synthesis kit (Roche Diagnostics). Detection of the signals was carried out using a Nikon camera (E60, Japan). The primers are listed in Table S3.
Scanning electron microscopy
The samples were transversely sectioned, natural dried, sputter‐coated with gold particles, observed and photographed using a scanning electron microscope (JSM‐6390LV; JEOL, Japan).
SDS‐PAGE
SDS‐PAGE was carried out using total proteins from seeds as per previous method (Yamamoto et al., 2006). After electrophoresis, SDS‐PAGE gels were stained using Coomassie Brilliant Blue G‐250 (Nacalai Tesque, http://www.nacalai.co.jp).
Statistical analysis
Data were subjected to a software package used for statistical analysis, and significant differences between individual means were established using a two‐tailed Student's t‐test in Microsoft Office Excel 2010. Significance was accepted at P < 0.05 and P < 0.01.
Analysis of the total protein, starch and total lipid contents
Hundred milligram, 500 mg and 3.4 g of grounded rice powder were used for the assays of total protein, starch and total lipid contents, respectively. All measurements were performed in three biological replicates.
Total protein content was detected using an XDS Near‐Infrared Rapid Content Analyzer (Foss® Analytical, Hilleroed, Denmark) (Li et al., 2014). The content of glutelin and prolamin was measured using the Bradford assay (Yang et al., 2013). Starch content was measured as previously described method (Li et al., 2014). Crude fat content was measured using the Soxtec method (Hijona et al., 2010). FA extraction was performed as previously described (Pan et al., 2011). Quantification of FA content was measured using GC‐MS (HP7890, California) (Ying et al., 2012).
Conflict of Interest
The authors declare no conflict of interests.
Supporting information
Figure S1 Expression analyses of RAG1 by quantitative RT‐PCR. RNA was extracted from developing seeds at 14 d after flowering (DAF). Light‐grey, black, and dark‐grey bars represent the expression levels in WT, RAG2‐OX, and RAG2‐RNAi line seeds, respectively. The relative expression levels were normalized to that of UBI.
Figure S2 Comparison of mature seeds of WT and the transgenic lines. (a) Comparison of 20 mature seeds length and width of WT, RAG2‐OX, and RAG2‐RNAi lines. Bar = 1 cm. (b) Statistics of 20 mature seeds length and width of WT, RAG2‐OX, and RAG2‐RNAi lines. Data are mean ± SE for three replicates. *P < 0.05, **P < 0.01. P‐values produced by two‐tailed Student's t‐test.
Figure S3 Phenotypes of seeds of WT and RAG2‐OX plant. WT: a, b, c, d, e, f. Ri‐1: a1, b1, c1, d1, e1, f1. Ri‐2: a2, b2, c2, d2, e2, f2. Ri‐3: a3, b3, c3, d3, e3, f3. (a‐a3) 150 grains of mature seeds in white light background. (b‐b3) Mature seeds. (c–c3) Cross sections of mature endosperm. RAG2‐OX lines grains displayed comparable chalkiness with WT. (d–d3) SEM of the central area of mature endosperm in (c–c3). (e–e3) SEM of the central area of mature endosperm, with the cross sections indicated by a green square in (c–c3). (f–f3) SEM of the central area of mature endosperm, with the cross sections indicated by a red square in (c–c3). Scale bars: 10 mm (a–a3), 3 mm (b–b3), 1 mm (c–c3), 500 μm (d–d3), 10 μm (e–e3, f–f3). (g) Grain chalkiness rate of WT and 3 RNAi lines. Data are mean ± SE for three replicates. *P < 0.05, **P < 0.01. P‐values produced by two‐tailed Student's t‐test.
Figure S4 Expression analyses of grain weight and grain size genes by quantitative RT‐PCR. RNA was extracted from developing seeds at 14 d after flowering (DAF) for these genes (GS3, GS5, GL3, GW2, and GW8). Light‐grey, black, and dark‐grey bars represent the expression levels in WT, RAG2‐OX, and RAG2‐RNAi line seeds, respectively. The relative expression levels were normalized to that of UBI.
Figure S5 Longitudinal section showing seed development of RAG2‐OX lines, RAG2‐RNAi lines and corresponding wild‐type plants. Longitudinal section seeds were stained with 0.05% haematoxylin to compare seed development in wild‐type Zhonghua11 (ZH11), RAG2‐OX and RAG2‐RNAi lines. Bars = 300 μm. Em, embryo; En, endosperm.
Table S2 Analysis of yield parameters of WT and RAG2‐RNAi T1 lines.
Table S3 Primers used for functional analysis of RAG2.
Table S1 Analysis of yield parameters of WT and RAG2‐OX T1 lines.
Acknowledgements
We gratefully acknowledge Prof. Jie Luo (Huazhong Agricultural University, Wuhan, China) for his helpful discussions. This work was supported by grant 31570321 from the National Natural Science Foundation of China.
References
- Adachi, T. , Izumi, H. , Yamada, T. , Tanaka, K. , Takeuchi, S. , Nakamura, R. and Matsuda, T. (1993) Gene structure and expression of rice seed allergenic proteins belonging to the α‐amylase/trypsin inhibitor family. Plant Mol. Biol. 21, 239–248. [DOI] [PubMed] [Google Scholar]
- Alvarez, A.M. , Adachi, T. , Nakase, M. , Aoki, N. , Nakamura, R. and Matsuda, T. (1995) Classification of rice allergenic protein cDNAs belonging to the α‐amylase/trypsin inhibitor gene family. Biochim. Biophys. Acta, 1251, 201–204. [DOI] [PubMed] [Google Scholar]
- Boxer, M.B. , Shen, M. , Zhang, Y. , Liu, L. , Auld, D.S. and Beller, M. (2010) Modulators of Lipid Storage In Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US) Available from: https://www.ncbi.nlm.nih.gov/books/NBK153217/ [Google Scholar]
- Chen, M. , Zhao, Y. , Zhuo, C. , Lu, S. and Guo, Z. (2015) Overexpression of a NF‐YC transcription factor from bermudagrass confers tolerance to drought and salinity in transgenic rice. Plant Biotechnol. J. 13, 482–491. [DOI] [PubMed] [Google Scholar]
- Colot, V. , Robert, L.S. , Kavanagh, T.A. , Bevan, M.W. and Thompson, R.D. (1987) Localization of sequences in wheat endosperm protein genes which confer tissue‐specific expression in tobacco. EMBO J. 6, 3559–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom, J.S. , Cho, J.I. , Reinders, A. , Lee, S.W. , Yoo, Y. , Tuan, P.Q. , Choi, S.B. et al (2011) Impaired function of the tonoplast‐localized sucrose transporter in rice, OsSUT2, limits the transport of vacuolar reserve sucrose and affects plant growth. Plant Physiol. 157, 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, C. , Xing, Y. , Mao, H. , Lu, T. , Han, B. , Xu, C. , Li, X. et al (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, M.A. , McCouch, S.R. and Hall, R.D. (2009) Not just a grain of rice: the quest for quality. Trends Plant Sci. 14, 133–139. [DOI] [PubMed] [Google Scholar]
- Fu, F.F. and Xue, H.W. (2010) Coexpression analysis identifies Rice Starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 154, 927–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, K.A. (1979) Effect of environment on protein and amylose content of rice In: Proc. Workshop on chemical aspects of rice grain quality. IRRI, 59–68. [Google Scholar]
- Hijona, E. , Hijona, L. , Larzabal, M. , Sarasqueta, C. , Aldazabal, P. , Arenas, J. and Bujanda, L. (2010) Biochemical determination of lipid content in hepatic steatosis by the Soxtec method. World J. Gastroenterol.: WJG, 16, 1495–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru, K. , Hirotsu, N. , Madoka, Y. , Murakami, N. , Hara, N. , Onodera, H. , Kashiwagi, T. et al (2013) Loss of function of the IAA‐glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 45, 707–711. [DOI] [PubMed] [Google Scholar]
- Ito, M. , Kato, T. and Matsuda, T. (2005) Rice allergenic proteins, 14–16 kDa albumin and alpha‐globulin, remain insoluble in rice grains recovered from rice miso (rice‐containing fermented soybean paste). Biosci. Biotechnol. Biochem. 69, 1137–1144. [DOI] [PubMed] [Google Scholar]
- Kang, H.G. , Park, S. , Matsuoka, M. and An, G. (2005) White‐core endosperm floury endosperm‐4 in rice is generated by knockout mutations in the C‐type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J.: Cell Molecul. Biol. 42, 901–911. [DOI] [PubMed] [Google Scholar]
- Katsube, T. , Kurisaka, N. , Ogawa, M. , Maruyama, N. , Ohtsuka, R. , Utsumi, S. and Takaiwa, F. (1999) Accumulation of soybean glycinin and its assembly with the glutelins in rice. Plant Physiol. 120, 1063–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu, T. , Yamamoto, M.P. , Hirose, S. , Yano, M. and Takaiwa, F. (2008) Characterization of a new rice glutelin gene GluD‐1 expressed in the starchy endosperm. J. Exp. Bot. 59, 4233–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu, T. , Yamamoto, M.P. , Touno, S.M. , Yasuda, H. and Takaiwa, F. (2009) Compensation and interaction between RISBZ1 and RPBF during grain filling in rice. Plant J. 59, 908–920. [DOI] [PubMed] [Google Scholar]
- Kawakatsu, T. , Hirose, A. , Yasuda, H. and Takaiwa, F. (2010) Reducing rice seed storage protein accumulation leads to changes in nutrient quality and storage organelle formation. Plant Physiol. 154, 1842–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.J. , Lee, J.Y. , Yoon, U.H. , Lim, S.H. and Kim, Y.M. (2013) Effects of reduced prolamin on seed storage protein composition and the nutritional quality of rice. Int. J. Mol. Sci. 14, 17073–17084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, N.H. , Kwak, J. , Baik, J.Y. , Yoon, M.R. , Lee, J.S. , Yoon, S.W. and Kim, I.H. (2015) Changes in lipid substances in rice during grain development. Phytochemistry, 116, 170–179. [DOI] [PubMed] [Google Scholar]
- Komatsu, S. and Hirano, H. (1992) Rice seed globulin: a protein similar to wheat seed glutenin. Phytochemistry, 31, 3455–3459. [DOI] [PubMed] [Google Scholar]
- Kurokawa, S. , Kuroda, M. , Mejima, M. , Nakamura, R. , Takahashi, Y. , Sagara, H. , Takeyama, N. et al (2014) RNAi‐mediated suppression of endogenous storage proteins leads to a change in localization of overexpressed cholera toxin B‐subunit and the allergen protein RAG2 in rice seeds. Plant Cell Rep. 33, 75–87. [DOI] [PubMed] [Google Scholar]
- Kusaba, M. , Miyahara, K. , Iida, S. , Fukuoka, H. , Takano, T. , Sassa, H. , Nishimura, M. et al (2003) Low glutelin content1: a dominant mutation that suppresses the glutelin multigene family via RNA silencing in rice. Plant Cell, 15, 1455–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.J. , Jo, Y.M. , Lee, J.Y. , Lim, S.H. and Kim, Y.M. (2015) Lack of globulin synthesis during seed development alters accumulation of seed storage proteins in rice. Int. J. Mol. Sci. 16, 14717–14736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Fan, C. , Xing, Y. , Jiang, Y. , Luo, L. , Sun, L. , Shao, D. et al (2011) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Fan, C. , Xing, Y. , Yun, P. , Luo, L. , Yan, B. , Peng, B. et al (2014) Chalk5 encodes a vacuolar H+‐translocating pyrophosphatase influencing grain chalkiness in rice. Nat. Genet. 46, 398–404. [DOI] [PubMed] [Google Scholar]
- Ma, K. , Zhang, D. , Liu, Y. , Ouyang, Y. , Li, J. , Hu, C. and Yao, J. (2015) Ectopic expression of EbFIE from apomictic Eulaliopsis binata in rice results in pleiotropic phenotypes likely due to interaction with OsCLF. Plant Sci. 234, 86–96. [DOI] [PubMed] [Google Scholar]
- Mahmoud, A.H. and El Anany, A.M. (2014) Nutritional and sensory evaluation of a complementary food formulated from rice, faba beans, sweet potato flour, and peanut oil. Food Nutrit. Bullet. 35, 403–413. [DOI] [PubMed] [Google Scholar]
- Maier, U.G. , Brown, J.W. , Tologcyzki, C. and Feix, G. (1987) Binding of a nuclear factor to a consensus sequence in the 5′ flanking region of zein genes from maize. EMBO J. 6, 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawal, Y.R. , Mawal, M.R. and Ranjekar, P.K. (1987) Biochemical and immunological characterization of rice albumin. Biosci. Rep. 7, 1–9. [DOI] [PubMed] [Google Scholar]
- Mu, J. , Tan, H. , Zheng, Q. , Fu, F. , Liang, Y. , Zhang, J. , Yang, X. et al (2008) LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis . Plant Physiol. 148, 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita, T.W. , Hwang, Y.S. , Hnilo, J. , Kim, W.T. , Aryan, A.P. , Larson, R. and Krishnan, H.B. (1989) Structure and expression of the rice glutelin multigene family. J. Biol. Chem. 264, 12573–12581. [PubMed] [Google Scholar]
- Pan, Y.Y. , Wang, S.T. , Chuang, L.T. , Chang, Y.W. and Chen, C.N. (2011) Isolation of thermo‐tolerant and high lipid content green microalgae: oil accumulation is predominantly controlled by photosystem efficiency during stress treatments in Desmodesmus . Bioresour. Technol. 102, 10510–10517. [DOI] [PubMed] [Google Scholar]
- Prioul, J.L. , Mechin, V. and Damerval, C. (2008) Molecular and biochemical mechanisms in maize endosperm development: the role of pyruvate‐Pi‐dikinase and Opaque‐2 in the control of C/N ratio. C.R. Biol. 331, 772–779. [DOI] [PubMed] [Google Scholar]
- Saito, Y. , Shigemitsu, T. , Yamasaki, R. , Sasou, A. , Goto, F. , Kishida, K. , Kuroda, M. et al (2012) Formation mechanism of the internal structure of type I protein bodies in rice endosperm: relationship between the localization of prolamin species and the expression of individual genes. Plant J.: Cell Molecul. Biol. 70, 1043–1055. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T. and Matsuoka, M. (2004) Generating high‐yielding varieties by genetic manipulation of plant architecture. Curr. Opin. Biotechnol. 15, 144–147. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T. and Matsuoka, M. (2008) Identifying and exploiting grain yield genes in rice. Curr. Opin. Plant Biol. 11, 209–214. [DOI] [PubMed] [Google Scholar]
- Sang, X. , Li, Y. , Luo, Z. , Ren, D. , Fang, L. , Wang, N. , Zhao, F. , Ling, Y. , Yang, Z. , Liu, Y. and He, G. (2012) CHIMERIC FLORAL ORGANS1, encoding a monocot‐specific MADS box protein, regulates floral organ identity in rice. Plant physiology. 160, 788–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selgrade, M.K. , Bowman, C.C. , Ladics, G.S. , Privalle, L. and Laessig, S.A. (2009) Safety assessment of biotechnology products for potential risk of food allergy: implications of new research. Toxicol. Sci. 110, 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha, S. , Sugiyama, Y. , Mitsukawa, N. , Masumura, T. and Tanaka, K. (1996) Cloning and sequencing of a rice gene encoding the 13‐kDa prolamin polypeptide. Biosci. Biotechnol. Biochem. 60, 335–337. [DOI] [PubMed] [Google Scholar]
- She, K.C. , Kusano, H. , Koizumi, K. , Yamakawa, H. , Hakata, M. , Imamura, T. , Fukuda, M. et al (2010) A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell, 22, 3280–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimbata, T. , Inokuma, T. , Sunohara, A. , Vrinten, P. , Saito, M. , Takiya, T. and Nakamura, T. (2011) High levels of sugars and fructan in mature seed of sweet wheat lacking GBSSI and SSIIa enzymes. J. Agric. Food Chem. 59, 4794–4800. [DOI] [PubMed] [Google Scholar]
- Singh, V. , Singh, P.K. , Siddiqui, A. , Singh, S. , Banday, Z.Z. and Nandi, A.K. (2016) Over‐expression of Arabidopsis thaliana SFD1/GLY1, the gene encoding plastid localized glycerol‐3‐phosphate dehydrogenase, increases plastidic lipid content in transgenic rice plants. J. Plant. Res. 129, 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X.J. , Huang, W. , Shi, M. , Zhu, M.Z. and Lin, H.X. (2007) A QTL for rice grain width and weight encodes a previously unknown RING‐type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. [DOI] [PubMed] [Google Scholar]
- Tan, Y.F. , Xing, Y.Z. , Li, J.X. , Yu, S.B. , Xu, C.G. and Zhang, Q. (2000) Genetic bases of appearance quality of rice grains in Shanyou 63, an elite rice hybrid. Theor. Appl. Genet. 101, 823–829. [DOI] [PubMed] [Google Scholar]
- Tsukasa Matsuda, R.N. , Sugiyama, Makoto and Nakamura, Ryo . (1991) Immunochemical studies on rice allergenic proteins. Agric. Biol. Chem. 55, 509–513. [Google Scholar]
- Wan, X. , Weng, J. , Zhai, H. , Wang, J. , Lei, C. , Liu, X. , Guo, T. et al (2008) Quantitative trait loci (QTL) analysis for rice grain width and fine mapping of an identified QTL allele gw‐5 in a recombination hotspot region on chromosome 5. Genetics, 179, 2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, E. , Wang, J. , Zhu, X.D. , Hao, W. , Wang, L.Y. , Li, Q. , Zhang, L.X. et al (2008) Control of rice grain‐filling and yield by a gene with a potential signature of domestication. Nat. Genet. 40, 1370–1374. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Wu, K. , Yuan, Q. , Liu, X. , Liu, Z. , Lin, X. , Zeng, R. et al (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Zhou, W. , Lu, Z. , Ouyang, Y. , O, C.S. and Yao, J. (2015) A lipid transfer protein, OsLTPL36, is essential for seed development and seed quality in rice. Plant Sci. 239, 200–208. [DOI] [PubMed] [Google Scholar]
- Xing, Y. and Zhang, Q. (2010) Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61, 421–442. [DOI] [PubMed] [Google Scholar]
- Xu, JH. and Messing, J. (2009) Amplification of prolamin storage protein genes in different subfamilies of the Poaceae. Theor. Appl. Genet. 119, 1397–1412. [DOI] [PubMed] [Google Scholar]
- Yamamoto, M.P. , Onodera, Y. , Touno, S.M. and Takaiwa, F. (2006) Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes. Plant Physiol. 141, 1694–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Dai, L. , Xia, H. , Zhu, K. , Liu, H. and Chen, K. (2013) Protein profile of rice (Oryza sativa) seeds. Genet. Molecul. Biol. 36, 87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying, J.Z. , Shan, J.X. , Gao, J.P. , Zhu, M.Z. , Shi, M. and Lin, H.X. (2012) Identification of quantitative trait Loci for lipid metabolism in rice seeds. Molecul. Plant, 5, 865–875. [DOI] [PubMed] [Google Scholar]
- Zhang, Q. (2007) Strategies for developing green super rice. Proc. Natl Acad. Sci. USA, 104, 16402–16409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Li, J. , Xue, Y. , Han, B. and Deng, X.W. (2008) Rice 2020: a call for an international coordinated effort in rice functional genomics. Mol. Plant, 1, 715–719. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Wang, J. , Huang, J. , Lan, H. , Wang, C. , Yin, C. , Wu, Y. et al (2012) Rare allele of OsPPKL1 associated with grain length causes extra‐large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA, 109, 21534–21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Z. , Sumi, K. , Tanaka, K. and Murai, N. (1995) The bean seed storage protein β‐phaseolin is synthesized, processed, and accumulated in the vacuolar Type‐II protein bodies of transgenic rice endosperm. Plant Physiol. 109, 777–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, T. , Budworth, P. , Chen, W. , Provart, N. , Chang, H.S. , Guimil, S. , Su, W. et al (2003) Transcriptional control of nutrient partitioning during rice grain filling. Plant Biotechnol. J. 1, 59–70. [DOI] [PubMed] [Google Scholar]
- Zuo, J. and Li, J. (2014) Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 48, 99–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Expression analyses of RAG1 by quantitative RT‐PCR. RNA was extracted from developing seeds at 14 d after flowering (DAF). Light‐grey, black, and dark‐grey bars represent the expression levels in WT, RAG2‐OX, and RAG2‐RNAi line seeds, respectively. The relative expression levels were normalized to that of UBI.
Figure S2 Comparison of mature seeds of WT and the transgenic lines. (a) Comparison of 20 mature seeds length and width of WT, RAG2‐OX, and RAG2‐RNAi lines. Bar = 1 cm. (b) Statistics of 20 mature seeds length and width of WT, RAG2‐OX, and RAG2‐RNAi lines. Data are mean ± SE for three replicates. *P < 0.05, **P < 0.01. P‐values produced by two‐tailed Student's t‐test.
Figure S3 Phenotypes of seeds of WT and RAG2‐OX plant. WT: a, b, c, d, e, f. Ri‐1: a1, b1, c1, d1, e1, f1. Ri‐2: a2, b2, c2, d2, e2, f2. Ri‐3: a3, b3, c3, d3, e3, f3. (a‐a3) 150 grains of mature seeds in white light background. (b‐b3) Mature seeds. (c–c3) Cross sections of mature endosperm. RAG2‐OX lines grains displayed comparable chalkiness with WT. (d–d3) SEM of the central area of mature endosperm in (c–c3). (e–e3) SEM of the central area of mature endosperm, with the cross sections indicated by a green square in (c–c3). (f–f3) SEM of the central area of mature endosperm, with the cross sections indicated by a red square in (c–c3). Scale bars: 10 mm (a–a3), 3 mm (b–b3), 1 mm (c–c3), 500 μm (d–d3), 10 μm (e–e3, f–f3). (g) Grain chalkiness rate of WT and 3 RNAi lines. Data are mean ± SE for three replicates. *P < 0.05, **P < 0.01. P‐values produced by two‐tailed Student's t‐test.
Figure S4 Expression analyses of grain weight and grain size genes by quantitative RT‐PCR. RNA was extracted from developing seeds at 14 d after flowering (DAF) for these genes (GS3, GS5, GL3, GW2, and GW8). Light‐grey, black, and dark‐grey bars represent the expression levels in WT, RAG2‐OX, and RAG2‐RNAi line seeds, respectively. The relative expression levels were normalized to that of UBI.
Figure S5 Longitudinal section showing seed development of RAG2‐OX lines, RAG2‐RNAi lines and corresponding wild‐type plants. Longitudinal section seeds were stained with 0.05% haematoxylin to compare seed development in wild‐type Zhonghua11 (ZH11), RAG2‐OX and RAG2‐RNAi lines. Bars = 300 μm. Em, embryo; En, endosperm.
Table S2 Analysis of yield parameters of WT and RAG2‐RNAi T1 lines.
Table S3 Primers used for functional analysis of RAG2.
Table S1 Analysis of yield parameters of WT and RAG2‐OX T1 lines.


