Abstract
Plant NADPH-dependent glyoxylate/succinic semialdehyde reductases 1 and 2 (GLYR1 and GLYR2) are considered to be involved in detoxifying harmful aldehydes, thereby preserving plant health during exposure to various abiotic stresses. Phylogenetic analysis revealed that the two GLYR isoforms appeared in the plant lineage prior to the divergence of the Chlorophyta and Streptophyta, which occurred approximately 750 million years ago. Green fluorescent protein fusions of apple (Malus x domestica Borkh.), rice (Oryza sativa L.) and Arabidopsis thaliana [L.] Heynh GLYRs were transiently expressed in tobacco (Nicotiana tabaccum L.) suspension cells or Arabidopsis protoplasts, as well in methoxyfenozide-induced, stably transformed Arabidopsis seedlings. The localization of apple GLYR1 confirmed that this isoform is cytosolic, whereas apple, rice and Arabidopsis GLYR2s were localized to both mitochondria and plastids. These findings highlight the potential involvement of GLYRs within distinct compartments of the plant cell.
Keywords: abiotic stress, γ-aminobutyrate, glyoxylate/succinic semialdehyde reductase, photorespiration, phylogenetic analysis, subcellular localization
Introduction
Plant NADPH-dependent glyoxylate/succinic semialdehyde (SSA) reductases (GLYR, EC 1.2.1.79) are hypothesized to detoxify harmful reactive aldehydes into their corresponding less toxic alcohols, thereby preserving plant health during various abiotic stresses (Allan et al., 2009). Glyoxylate is an intermediate in photorespiration and produced in peroxisomes via the oxidation of glycolate (Bauwe et al., 2010), whereas SSA is an intermediate in γ-aminobutyrate (GABA) metabolism and produced in mitochondria via the transamination of GABA (Shelp et al., 2012).
GLYRs were first isolated from spinach and pea leaves, and found in both cytosolic and purified plastid fractions, with 10–20% of the total leaf GLYR activity being present in isolated chloroplasts (Zelitch, 1953; Givan et al., 1988; also see review by Givan and Kleczkowski, 1992). To date, the best characterized plant GLYRs in terms of their subcellular localizations and biochemical properties are those from Arabidopsis. For instance, AtGLYR1 has been shown to be localized to cytosol, whereas AtGLYR2 is localized to plastids (Simpson et al., 2008; Ching et al., 2012). Both enzymes prefer NADPH over NADH as a cofactor and display a higher affinity for glyoxylate than for SSA (Breitkreuz et al., 2003; Hoover et al., 2007a; Simpson et al., 2008), and NADP+ competitively inhibits AtGLYR1, indicating that NADPH/NADP+ ratios may regulate GLYR activity in planta (Hoover et al., 2007b). In the current study, we studied evolutionary relationships between the two plant GLYRs and compared the subcellular localization of GLYRs from apple (Malus × domestica Borkh.), a dicotyledonous species, and rice (Oryza sativa L.), a monocotyledonous species, with those from Arabidopsis thaliana [L] Heynh. Our findings established that GLYR1s are exclusively cytosolic, whereas GLYR2s are localized to both mitochondria and plastids.
Materials and Methods
Phylogenetic Analysis
Arabidopsis GLYR1 and GLYR2 proteins were used as queries for a BLASTP search of the National Center for Biotechnology Information1, Phytozome2, and OneKP3 databases. To construct the phylogenetic tree, GLYR proteins were chosen from among chlorophytic and streptophytic species with identity above a 50% cutoff; their NCBI Reference Sequence IDs are given in Supplementary Tables S1, S2. The evolutionary history was inferred using the Maximum Likelihood method based on the JTT matrix-based model (Jones et al., 1992). The tree with the highest log likelihood (–8865.0032) is shown. All positions containing gaps and missing data were eliminated. Evolutionary analysis was conducted in MEGA7 (Kumar et al., 2016).
Plant Materials, RNA, and DNA Extraction, and Identification of Plant GLYRs
Arabidopsis thaliana (L.) Heynh ecotype Columbia (Col-0) was the genetic background of the wild type (WT) and the AtGLYR2- GREEN FLUORESCENT PROTEIN (GFP) transgenic line. Total RNA was extracted and used for synthesis of cDNA and quantitative PCR analysis as described previously (Zarei et al., 2011, 2014). The preparation of apple and rice RNA and cDNA has been described elsewhere (Brauer et al., 2011; Trobacher et al., 2013b). The primer sequences used to determine the abundance of GLYR2-GFP and the housekeeping transcript ELONGATION FACTOR-1 ALPHA (At5g60390; Czechowski et al., 2005) are listed in Supplementary Table S3. The extraction of Arabidopsis genomic DNA has been described (Zarei et al., 2011).
Identification and Cloning of cDNAs Encoding Apple and Rice GLYRs and Arabidopsis GLYR2
The Arabidopsis GLYR sequences were utilized as queries in the apple genome database4. Two GLYRs have been identified as MdGLYR1 (MDP0000149834) and MdGLYR2 (MDP0000158245). GFP-tagged versions of MdGLYR2, AtGLYR2, and OsGLYR2 were constructed for subcellular localization studies. The full-length open reading frame (ORF) of MdGLYR1 was amplified with CB-F1 and CB-R1 primers, whereas the MdGLYR2 ORF was amplified with CB-F2 and CB-R2 primers (Supplementary Table S3). The resulting PCR products were sub-cloned into the plant expression vector pUC18-GFP, resulting in both MdGLYRs being fused at their C-termini to a monomerized version of GFP. The full-length ORF of AtGLYR2 was amplified with NheI-AtGLYR2-F and NheI-AtGLYR2-R primers from Arabidopsis rosette leaf cDNA. The resulting PCR product was sub-cloned into pUC18/NheI-GFP, yielding AtGLYR2-GFP. The rice cv. Nipponbare (AK064876) cDNA was provided by the Rice Genome Resource Center, National Institute of Agrobiological Sciences (Tsukuba, Japan). The OsGLYR2 ORF was amplified using the primer sets NotI-OsGLYR2-F and XhoI-OsGLYR2-R (Supplementary Table S3). The resulting PCR product was sub-cloned into pTH-2 vector (Englert et al., 2007), yielding OsGLYR2-GFP.
Transient Expression and Subcellular Localization of Apple, Rice, and Arabidopsis and Rice GLYRs in Tobacco BY-2 Cells and Arabidopsis Protoplasts
A total of 5 μg of plasmid DNA encoding an individual GFP fusion protein with or without 1 μg of plasmid DNA encoding cytosolic Cherry (pRTL2/Cherry; Gidda et al., 2011) or the plastidial marker pSAT4/PDCpl-E2-Cherry (Park et al., 2012) was transiently expressed in tobacco BY-2 suspension cells via tungsten particle bombardment. Details on processing BY-2 cells for (immunofluorescence) microscopy, including cell fixation, as well as confocal laser-scanning microscopy (CLSM) are described in Trobacher et al. (2013a). Mitochondria were immunostained using rabbit anti-CoxII affinity-purified IgGs and goat anti-rabbit rhodamine red × secondary antibodies (Jackson Immunoresearch Laboratories), according to Frelin et al. (2012). GFP and chlorophyll [in Arabidopsis protoplasts or seedlings (see below)] were excited with a 488 nm argon ion laser at 25% power, and the emitted light was detected at 500–530 nm for GFP and 685–750 for chlorophyll. Cherry and Mitotracker were excited with a 543 nm Ar/HeNe laser at 85% power, and emitted light was detected at 590–650 nm for Cherry and 579–599 nm for Mitotracker.
Arabidopsis cell suspension protoplasts were prepared by enzyme digestion (Zarei et al., 2011). Ten microgram of plasmid DNA encoding an individual GFP fusion protein with or without 5 μg of pSAT4/PDCpl-E2-Cherry plasmid DNA was mixed with 125 μL of protoplast solution containing half a million cells. Protoplasts were transformed using polyethylene glycol as described elsewhere (Schirawski et al., 2000). Mitochondria were stained with Mitotracker Red CMXRos (Thermo Fisher Scientific) as described in the manufacturer’s manual.
All fluorescent images of BY-2 cells and protoplasts are representative of at least three independent transformations with a minimum of 10 transformed cells imaged per transformation. Fluorophore emissions were imaged sequentially, and no detectable bleed-through was observed with the same acquisition settings used in data collection.
Stable Expression and Subcellular Localization of AtGLYR2 in Arabidopsis Seedlings
A stable Arabidopsis line expressing methoxyfenozide-inducible AtGLYR2-GFP was generated as described by Dietrich et al. (2008). The AtGLYR2-GFP ORF was amplified from pUC18-AtGLYR2-GFP with F-PacI-LAtGR2-GFP and R-SpeI-LAtGR2-GFP primers and sub-cloned into the plasmid CD1660-1-5XG-M35S, resulting in the construct CD-1660-1-5XG-M35S::AtGLYR2-GFP. The 5XG-M35S-AtGLYR2-GFP cassette was digested with NotI and ApaI and subcloned into the plasmid CD1468-1 possessing a promoter-binding VGE element, resulting in the construct CD1468-1-VGE-5XG-M35S::AtGLYR2-GFP. This cassette was then digested with AscI and sub-cloned into the binary vector pEC291and transformed into EHA105 Agrobacterium cells. Arabidopsis plants were stably transformed with pEC291-AtGLYR2-GFP via the floral dip method (Clough and Bent, 1998) and PCR-positive transgenic plants were further tested for protein expression and phenotype. Highly expressing GFP lines of 14-days-old T2 seedlings were selected using an epifluorescent microscope (Leica DM-6000CS), then grown to maturity for collection of T3 seed.
Localization analysis of Arabidopsis stably expressing inducible-AtGLYR2-GFP was performed using 14-days-old T3 seedlings. Expression of AtGLYR2-GFP was induced by foliar application of 61 μM methoxyfenozide (Intrepid 2F insecticide, Dow AgroSciences). Seedlings were incubated for 48 h before imaging. Microscopy was performed using CLSM as described above.
Results and Discussion
Comparative Genomic Analysis of Plant GLYRs
The deduced amino acid sequences of GLYR1 and GLYR2 have 54–58% identity across 35 species of Viridiplantae, which is comprised of chlorophytic species, including both unicellular and multicellular algae, as well as charophytic and embryophytic plants (the latter two groups jointly known as the Streptophyta) (Becker, 2013). These sequences were used to construct a phylogenetic tree (Figure 1). The GLYR1 and GLYR2 proteins from the Embryophyta generated two distinct clusters with a relatively high degree of sequence identity within each cluster (78–97% for GLYR1s and 66–84% for GLYR2s). A small clade with a high degree of identity (85–91% for GLYR1 and 73–85% for GLYR2) is dedicated to monocotyledonous plants within each cluster. Most of the embryophytic genomes contain both GLYR1 and GLYR2 genes, the notable exceptions being Cucumis sativus and C. melo, which lack GLYR1, but apparently have two GLYR2s located in tandem on the same chromosome. The two GLYR2s in C. sativus and C. melo, which were designated as GLYR2A and GLYR2B, are 72–73% identical. Furthermore, in silico analysis of subcellular localization using TargetP (Emanuelsson et al., 2000) and WoLF PSORT (Horton et al., 2007) revealed that these cucurbit GLYR2As, like their Arabidopsis, apple and rice counterparts (Supplementary Figure S1A), possess a putative N-terminal mitochondrial/chloroplastidial targeting sequence, whereas the cucurbit GLYR2Bs do not. Physcomitrella patens, Klebsormidium flaccidum, Chlamydomonas reinhardtii, and Volvox carteri contain both GLYR1 and GLYR2 proteins. However, other members of the Chlorophyta (Chlorella variabilis, Coccomyxa subellipsoidea C-169, Micromonas pusila, Bathycoccus prasinos, and Ostreococcus lucimarinus) contain only a single GLYR, with 46–53% identity to AtGLYR1.
FIGURE 1.
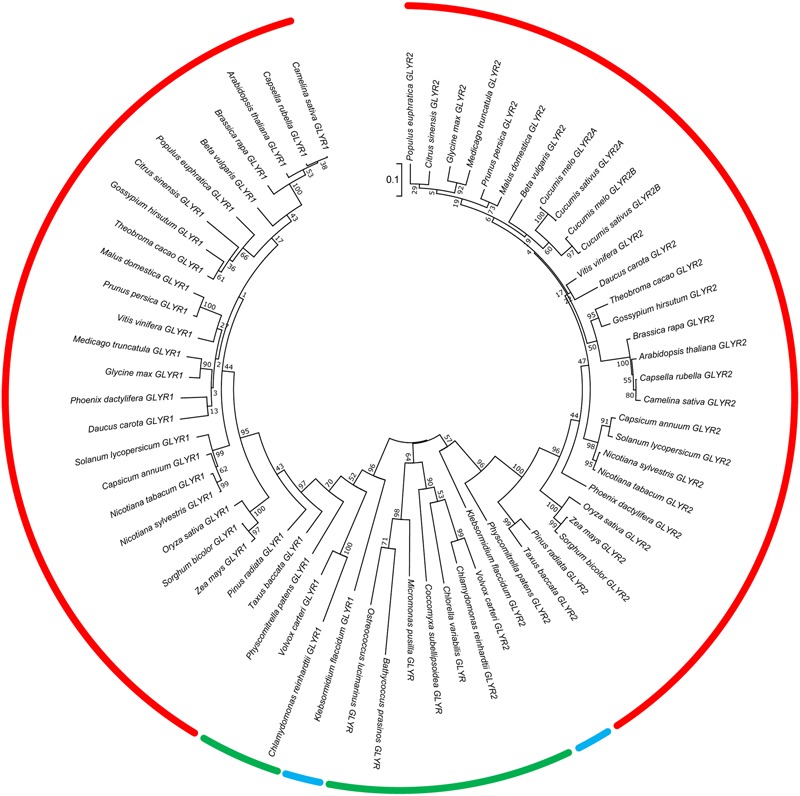
Phylogenetic analysis of plant GLYR amino acid sequences across plant species. The unrooted phylogenetic tree was depicted using the Maximum Likelihood method in MEGA7. The red, blue, and green bars represent species in the Embryophyta, Charophyta, and Chlorophyta, respectively. The numbers at each node represent support values obtained by a bootstrap test using 1000 replicates (Felsenstein, 1985). The tree is drawn to scale (0.1 representing a 10% change) with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The protein IDs for the GLYR1s and GLYR2s are given in Supplementary Tables S1, S2.
Overall, we found that both GLYR1 and GLYR2 appear in the plant lineage prior to the divergence of the Chlorophyta and Streptophyta, which occurred approximately 750 million years ago (Becker, 2013). Notably, only a single GLYR is found in the oldest members of the Chlorophyta examined and the number of GLYR proteins has not expanded during evolution of the Streptophyta. Thus, the GLYR1/2 orthologs probably arose from the duplication of a single GLYR-type protein in the Chlorophyta, which underwent functional diversification. Also, an extinction event, followed by duplication, probably occurred after the divergence of the Cucurbitaceae, resulting in subfunctionalization of the GLYR2 proteins (Brocker et al., 2013). Hoover et al. (2013) have identified six important amino acid residues for catalysis (Lys170 and Asn174 in AtGLYR1) and substrate binding (Phe231, Asp239, Ser121, Thr95 in AtGLYR1) and protein sequence alignment of the GLYRs studied here revealed a general conservation of these catalytic and substrate binding residues, with only two exceptions. C. reinhardtii GLYR1 lacks both catalytic residues (Lys170 and As174), whereas C. variabilis GLYR1 lacks Thr95, suggesting that the majority of the GLYRs investigated here are indeed functional. Interestingly, single GLYRs also appear in the bacterial lineage (e.g., Geobacter spp; Zhang et al., 2014), and these possess greater identity to the plant GLYR1s than the GLYR2s (approximately 50–52%). Based on conservation of all the plant catalytic and substrate binding residues, it seems likely that these GLYRs would also prefer glyoxylate over SSA as substrate, but further study is required to substantiate such a prediction.
Subcellular Localization of Apple, Rice, and Arabidopsis GLYRs in Transient and Stable Expression Plant (cell) Systems
Apple, rice, and Arabidopsis GLYR1s lack a predicted N-terminal organelle targeting signal (Supplementary Figure S1A) and are therefore considered to be cytosolic. Indeed, while AtGLYR1 was previously thought to contain a C-terminal tripeptide sequence (i.e., SRE) similar to the type 1 peroxisomal target signal (PTS1) motif (reviewed in Reumann et al., 2016), recent experimental evidence indicates that the protein is localized exclusively to the cytosol and not to peroxisomes (Ching et al., 2012). Both MdGLYR1 and OsGLYR1 lack a C-terminal PTS1-like peroxisomal targeting signal and are therefore presumed to be also exclusively cytosolic.
Compared to their GLYR1 counterparts, most higher plant GLYR2s examined to date (cucurbit GLYR2Bs being the exception known) possess N-terminal extensions of varying lengths and are generally predicted to be localized to plastids rather than to mitochondria (or the cytosol) by various subcellular localization prediction programs (Supplementary Figures S1A,B). A mitochondrial-specific prediction program such as MitoProt (Claros, 1995) predicts mitochondrial localization for all three GLYR2s, whereas another, MitoFates (Fukasawa et al., 2015) predicts mitochondrial localization for rice GLYR2 only. However, DualPred, a program specific for dual targeting to mitochondria and plastids (Saravanan and Lakshmi, 2014), predicts that MdGLYR2 and OsGLYR2, but not AtGLYR2, localize to both organelles. Moreover, AtGLYR2 is annotated at the Arabidopsis subcellular proteome database [SUBAcon (Hooper et al., 2014)] to be localized to plastids; however, this latter conclusion was drawn solely from mass spectrometry studies involving isolated plastids (i.e., chloroplast), but not mitochondrial organellar fractions.
In order to begin to assess the subcellular localization of GLYRs in vivo, MdGLYR-GFP fusion proteins were transiently expressed and visualized via CLSM in tobacco suspension-cultured BY-2 cells, serving as a well-established model system for protein subcellular localization studies (Brandizzi et al., 2003). Similar to the abovementioned previous results for AtGLYR1 (Ching et al., 2012), MdGLYR1-GFP displayed diffuse localization throughout the cell, similar to co-expressed Cherry, a red fluorescent protein that served as a cytosolic marker protein (Shaner et al., 2004) (Figures 2A–D). By contrast, MdGLYR2-GFP localized to distinct globular-shaped structures, which co-localized with the co-expressed plastid marker fusion protein, consisting of the E2 subunit of the pyruvate dehydrogenase complex fused to Cherry (PDCpl-E2-Cherry) (Park et al., 2012) (Figures 2E–H), indicating that, similar to previous results for AtGLYR2 (Simpson et al., 2008), MdGLYR2 is localized to plastids. However, MdGLYR2-GFP in the majority (>70%) of BY-2 cells examined localized also to numerous small puncta that did not co-localize with co-expressed PDCpl-E2-Cherry (Figure 3A), but did co-localize with the endogenous mitochondrial protein, cytochrome oxidase subunit II (CoxII) (Figure 3B). Taken together, these data suggest that MdGLYR2-GFP is localized to both plastids and mitochondria in BY-2 cells. Similar results were observed for OsGLYR2-GFP (Figures 3C,D). These findings also prompted us to revisit the subcellular localization of AtGLYR2, which had not been assessed for mitochondrial localization in our previous study (Simpson et al., 2008). As shown in Figures 3E,F, AtGLYR2-GFP localized to both co-expressed PDCpl-E2-Cherry-containing plastids, as well as to endogenous CoxII-containing mitochondria.
FIGURE 2.
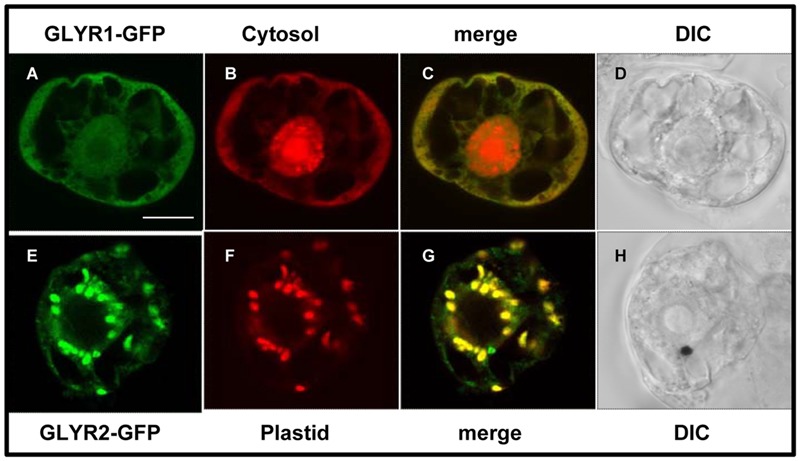
Subcellular localization of MdGLYRs in tobacco BY-2 cells. Representative CLSM images of BY-2 cells (co)expressing (as indicated by the panel labels) MdGLYR1-GFP (A) and the cytosolic marker Cherry (B) or MdGLYR2-GFP (E) and plastid marker PDCpl-E2-mCherry (F). Processing of cells and viewing using CLSM are described in the Section ‘Materials and Methods’. Shown also is the corresponding merged (C,G) and differential inference contrast (DIC) (D,H) images for each cell. The yellow color in the merged images indicates co-localization. Scale bar in A = 10 μm.
FIGURE 3.
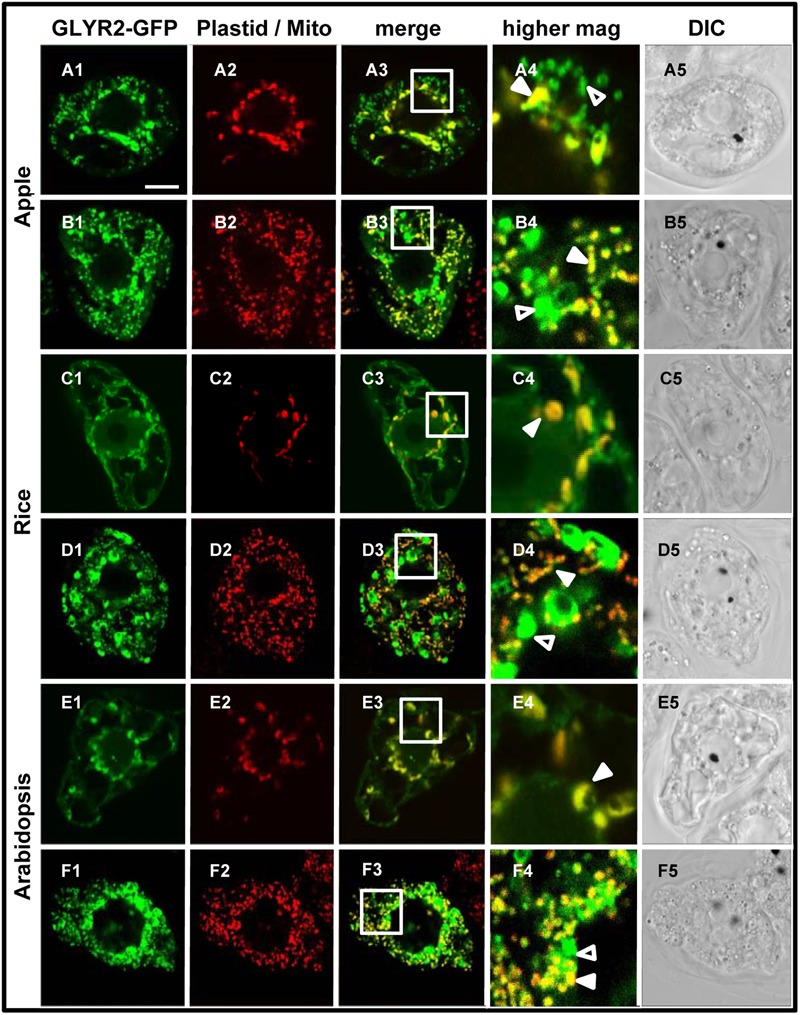
Plastid and mitochondrial localization of MdGLYR2, AtGLYR2, or OsGLYR2 in tobacco BY-2 cells. (A–F) Representative CLSM images of BY-2 cells (co)expressing (as indicated by the panel labels) MdGLYR2-GFP, OsGLYR2-GFP, or AtGLYR2-GFP and either the plastid marker PDCpl-E2-Cherry or immunostained for endogenous, mitochondrial CoxII. Processing of cells for immunofluorescence and viewing using CLSM are described in the ‘Materials and Methods’. Shown also is the corresponding merged and DIC images for each cell. Boxes in the merged images represent the portion of the cell shown at higher magnification in the panel to the right. The yellow color in the merged images indicates co-localization; solid arrowheads indicate obvious examples of co-localization, whereas open arrowheads indicate obvious examples of non-co-localization. Scale bar in A = 10 μm.
In order to assess the possibility that artifacts could be generated by heterologous protein expression in tobacco BY-2 cells (reviewed in Denecke et al., 2012 and Millar et al., 2009), the subcellular localization of MdGLYR2-GFP, OsGLYR2-GFP, and AtGLYR2-GFP was further investigated in transiently transformed Arabidopsis suspension cell-derived protoplasts. As shown in Figure 4, all three GLYR2-GFPs localized in protoplasts to both co-expressed PDCpl-E2-Cherry-containing plastids and Mitotracker-stained mitochondria.
FIGURE 4.
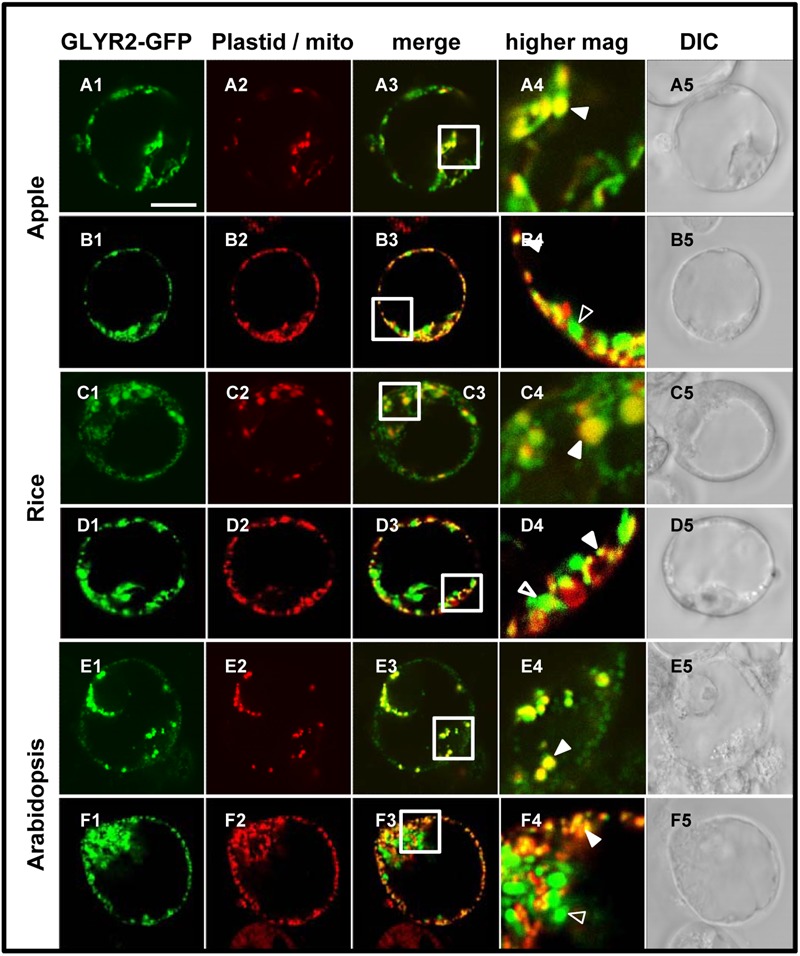
Plastid and mitochondrial localization of MdGLYR2, OsGLYR2, or AtGLYR2 in Arabidopsis protoplasts. (A–F) Representative CLSM images of Arabidopsis protoplasts (co)expressing (as indicated by the panel labels) MdGLYR2-GFP, OsGLYR2-GFP, or AtGLYR2-GFP and either the plastid marker PDCpl-E2-Cherry or stained with Mitotracker. Processing and viewing of protoplasts using CLSM are described in the Section ‘Materials and Methods’. Shown also is the corresponding merged and DIC images for each cell. Boxes in the merged images represent the portion of the cell shown at higher magnification in the panel to the right. The yellow color in the merged images indicates co-localization; solid arrowheads indicate obvious examples of co-localization, whereas open arrowheads indicate obvious examples of non-co-localization. Scale bar in A = 10 μm.
To further eliminate the possibility that the localization of GLYR2 to mitochondria was an artifact of expression in either a non-native system (i.e., tobacco BY-2 cells) or perhaps a distinct cell type (i.e., Arabidopsis suspension cell-derived protoplasts), we generated stably transformed Arabidopsis seedlings expressing methoxyfenozide-inducible AtGLYR2-GFP. The presence of the AtGLYR2-GFP transgene was confirmed via gene-specific PCR amplification, and the expression of AtGLYR2, as assessed by quantitative PCR, was observed to be approximately four times higher in transgenic seedlings induced with methoxyfenozide for 48 and 72 h, than in non-treated wild-type (WT) control seedlings or similarly treated WT seedlings, all of which displayed relatively low levels of AtGLYR2-GFP (Supplementary Figure S2). Furthermore, allied control experiments revealed that no fluorescence attributable to GFP was observed in WT seedlings in the presence or absence of methoxyfenozide, due to bleed-through from endogenous chlorophyll autofluorescence or Mitotracker staining, or in AtGLYR2-GFP-transformed seedlings in the absence of methoxyfenozide induction (Supplementary Figure S3). However, upon induction AtGLYR2-GFP localized consistently to both plastids and mitochondria, based on the co-localization of the fusion protein with chlorophyll autofluorescence and Mitotracker staining, respectively (Figure 5), corroborating results for the dual localization of AtGLYR2-GFP in tobacco BY-2 cells (Figure 3) and Arabidopsis protoplasts (Figure 4).
FIGURE 5.
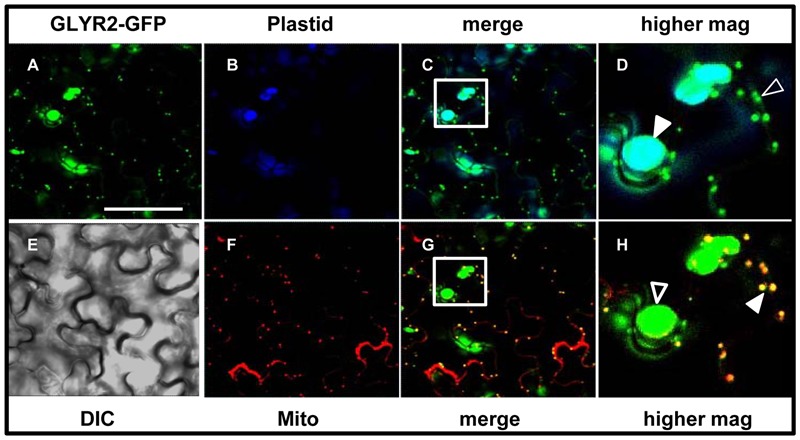
Dual localization of methoxyfenozide-inducible AtGLYR2-GFP in stably transformed Arabidopsis. Arabidopsis seedlings, stably expressing AtGLYR2-GFP were imaged (by CLSM) after induction with methoxyfenozide (A). (B,F) Represent the corresponding chlorophyll autofluorescence (plastid marker) and Mitotracker staining of the same cells, respectively. Co-localization of GFP and chlorophyll is observed based on the cyan color in the merged image in (C) and co-localization of GFP and Mitotracker is observed based on the yellow color in the merged image in (G). Boxes correspond to the regions of the cell shown at higher magnification in panels (D,H); solid arrowheads indicate obvious examples of co-localizations, whereas open arrowheads indicate obvious examples of non-co-localization. (E) The corresponding differential interference contrast image. Scale bar = 30 μm.
Dual targeting of proteins to mitochondria and chloroplasts could be a consequence of alternative gene splicing, transcription and/or translation initiation sites, as well as perhaps an ambiguous targeting signal that is recognized by the import machinery at both organelles (Carrie and Small, 2013). For example, the import of Thr-tRNA synthetase into both mitochondria and chloroplast is considered to involve a shared targeting signal domain and specific organelle receptors (Berglund et al., 2009), whereas the carrier protein Brittle 1 requires distinct targeting information that is recognized by different organelle receptors (Bahaji et al., 2011). In the case of GLYR2, the Arabidopsis Information Resource depicts only one version of the AtGLYR2 gene and no potential splice variants. Moreover, cloning of AtGLYR2, MdGLYR2, or OsGLYR2 from total cDNA resulted in only one clear sequence corresponding to GLYR2, indicating that GLYR2s in these three species are not alternatively spliced or transcribed, despite the presence of a conserved second methionine 16–18 amino acids downstream of the first N-terminal methionine of AtGLYR2 and MdGLYR2 (Supplemental Figure S1A). With respect to an ambiguous dual targeting signal peptide, while no consensus sequence(s) has been identified to date, N-terminal sequences enriched in positively charged residues (i.e., arginine, histidine and lysine) and deficient in glycine and negatively charged residues (i.e., aspartate and glutamate) (Carrie et al., 2009), or enriched in phenylalanine, leucine and serine and deficient in glycine (Berglund et al., 2009) have been reported. However, the N-terminal sequences of GLYR2s do not generally possess the same amino acid characteristics (Supplementary Figure S1A) and therefore may possess some other yet-to-be identified dual targeting signals, resulting in their localization to both plastids and mitochondria.
Compartmentation and Potential Function of GLYR Proteins
As depicted in the model in Figure 6, cytosolic GLYR1, as well as plastidial and mitochondrial GLYR2, could serve physiological roles in both glyoxylate and SSA metabolism. For example, abiotic stresses such as drought, heat and salinity cause plant stomata to close, leading to reduced intracellular CO2/O2 ratios and elevated rates of photorespiration, which might result in the accumulation of glycolate and glyoxylate (Allan et al., 2009). Arabidopsis plants exposed to these stresses could have elevated GLYR expression, as well as NAD(P)H/NAD(P)+ ratios (Allan et al., 2008), suggesting that NADPH-dependent GLYR activity would be stimulated (Allan et al., 2009). In contrast, chilling would be expected to decrease the absolute and relative rates of photorespiration (Foyer et al., 2009), and O2 deficiency stresses such as hypoxia and submergence would be expected to suppress the activity of glycolate oxidase (Narsai et al., 2009), so that glyoxylate would not be generated. However, the levels of GABA and GLYR expression could increase, together with the NAD(P)H/NAD(P)+ ratios, resulting in the diversion of SSA from succinate production to γ-hydroxybutyrate (Allan et al., 2008, 2012). Notably, the submergence-induced accumulation of GHB is decreased in both Arabidopsis glyr1 and glyr2 single knockout mutants (Allan et al., 2012). GABA and γ-hydroxybutyrate production has also been observed with many abiotic stresses (Allan et al., 2008). Thus, while the GLYRs are biochemically interchangeable, it can be hypothesized that they function in metabolically diverse cellular compartments.
FIGURE 6.
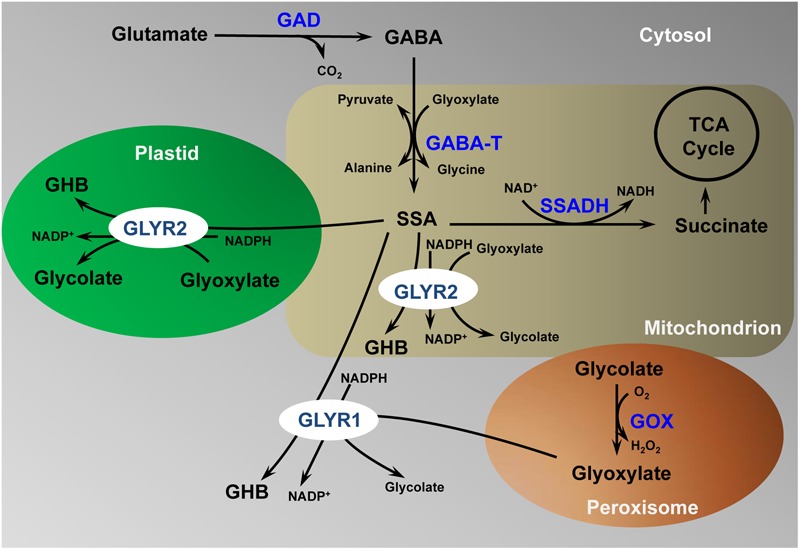
Proposed model for the detoxification of γ-aminobutyrate-derived succinic semialdehyde and photorespiration-derived glyoxylate in multiple subcellular compartments. γ-aminobutyrate, GABA; GABA transaminase, GABA-T; glutamate decarboxylase, GAD; glycolate oxidase, GOX; glyoxylate reductase, GLYR; γ-hydroxybutyrate, GHB; succinic semialdehyde, SSA; tri-carboxylic acid cycle, TCA. (Modified from Shelp et al. (2012); permission to reproduce not required)
Author Contributions
BS conceived the project; BS and GB supervised the project; CB and AZ conducted most of the experiments and data analysis; CT cloned the apple GLYR genes; JD, KA, and RM supplied reagents and/or discussed the project; CB, AZ, RM, GB, and BS wrote and/or edited the manuscript; and, all authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The research was funded in part by AgroFresh Inc., but they were not involved in experimental design or the decision to publish.
Acknowledgments
Thanks to Dr. Michael Shaw (Dow Agrosciences) for providing a sample of Intrepid 2F insecticide, Drs. William Plaxton and Joonho Park (Queen’s University) for providing the pSAT4-(PDCpl-E2)-Cherry plasmid, Dr. Edgar Cahoon (University of Nebraska-Lincoln) for providing plasmids CD1660-1-5XG-M35S, CD1468-1-VGE-5XG-M35S, and pEC291, Dr. Jaideep Mathur (University of Guelph) for the use of his epifluorescence microscope, Dr. Satinder Gidda (University of Guelph) for assistance with plasmid construction and fluorescence microscopy, and Dr. Katerina Jordan (University of Guelph) for use of equipment in her laboratory.
Funding. This research was supported by funding from the Natural Sciences and Engineering Research Council (NSERC) of Canada as a Strategic Project Grant (BS and GB) and Individual Discovery Grants (BS), and from the Ontario Ministry of Agriculture Food and Rural Affairs (BS and GB), AgroFresh Inc. (BS, GB, and JD), and MITACS Inc. (BS and GB).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00601/full#supplementary-material
References
- Allan W. L., Breitkreuz K. E., Waller J. C., Simpson J. P., Hoover G. J., Rochon A., et al. (2012). Detoxification of succinate semialdehyde in Arabidopsis glyoxylate reductase and NAD kinase mutants subjected to submergence stress. Botany 90 51–61. 10.1139/b11-083 [DOI] [Google Scholar]
- Allan W. L., Clark S. M., Hoover G. J., Shelp B. J. (2009). Role of plant glyoxylate reductases during stress: a hypothesis. Biochem. J. 423 15–22. 10.1042/BJ20090826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan W. L., Simpson J. P., Clark S. M., Shelp B. J. (2008). γ-Hydroxybutyrate accumulation in Arabidopsis and tobacco plants is a general response to abiotic stress: Putative regulation by redox balance and glyoxylate reductase isoforms. J. Exp. Bot. 59 2555–2564. 10.1093/jxb/ern122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahaji A., Ovecka M., Barany I., Risueno M. C., Munoz F. J., Baroja-Fernandez E., et al. (2011). Dual targeting to mitochondria and plastids of AtBT1 and ZmBT1, two members of the mitochondrial carrier family. Plant Cell Physiol. 52 597–609. 10.1093/pcp/pcr019 [DOI] [PubMed] [Google Scholar]
- Bauwe H., Hagemann M., Fernie A. R. (2010). Photorespiration: players, partners and origin. Trends Plant Sci. 15 330–336. 10.1016/j.tplants.2010.03.006 [DOI] [PubMed] [Google Scholar]
- Becker B. (2013). Snow ball earth and the split of Streptophyta and Chlorophyta. Trends Plant Sci. 18 180–183. 10.1016/j.tplants.2012.09.010 [DOI] [PubMed] [Google Scholar]
- Berglund A. K., Spånning E., Biverståhl H., Maddalo G., Tellgren-Roth C., Mäler L., et al. (2009). Dual targeting to mitochondria and chloroplasts: characterization of Thr-tRNA synthetase targeting peptide. Mol. Plant 2 1298–1309. 10.1093/mp/ssp048 [DOI] [PubMed] [Google Scholar]
- Brandizzi F., Irons S., Kearns A., Hawes C. (2003). BY-2 cells: culture and transformation for live cell imaging. Curr. Protoc. Cell Biol. 1 1–17. 10.1002/0471143030.cb0107s19 [DOI] [PubMed] [Google Scholar]
- Brauer E. K., Rochon A., Bi Y.-M., Bozzo G. B., Rothstein S. J., Shelp B. J. (2011). Reappraisal of nitrogen use efficiency in rice overexpressing glutamine synthetase 1. Physiol. Plant. 141 361–372. 10.1111/j.1399-3054.2011.01443.x [DOI] [PubMed] [Google Scholar]
- Breitkreuz K. E., Allan W. L., Van Cauwenberghe O. R., Jakobs C., Talibi D., André B., et al. (2003). A novel γ-hydroxybutyrate dehydrogenase: identification and expression of an Arabidopsis cDNA and potential role under oxygen deficiency. J. Biol. Chem. 278 41552–41556. 10.1074/jbc.M305717200 [DOI] [PubMed] [Google Scholar]
- Brocker C., Vasiliou M., Carpenter S., Carpenter C., Zhang Y., Wang X., et al. (2013). Aldehyde dehydrogenase (ALDH) superfamily in plants: gene nomenclature and comparative genomics. Planta 237 189–210. 10.1007/s00425-012-1749-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C., Giraud E., Whelan J. (2009). Protein transport in organelles: dual targeting of proteins to mitochondria and chloroplasts. FEBS J. 276 1187–1195. 10.1111/j.1742-4658.2009.06876.x [DOI] [PubMed] [Google Scholar]
- Carrie C., Small I. (2013). A reevaluation of dual-targeting of proteins to mitochondria and chloroplasts. Biochim. Biophys. Acta 1833 253–259. 10.1016/j.bbamcr.2012.05.029 [DOI] [PubMed] [Google Scholar]
- Ching S. L. K., Gidda S. K., Rochon A., Van Cauwenberghe O. R., Shelp B. J., Mullen R. T. (2012). Glyoxylate reductase isoform 1 is localized in the cytosol and not peroxisomes in plant cells. J. Integr. Plant Biol. 54 152–168. 10.1111/j.1744-7909.2012.01103.x [DOI] [PubMed] [Google Scholar]
- Claros M. G. (1995). MitoProt, a Macintosh application for studying mitochondrial proteins. Comput. Appl. Biosci. 11 441–447. 10.1093/bioinformatics/11.4.441 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M. K. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization. Plant Physiol. 139 5–17. 10.1104/pp.105.063743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecke J., Aniento F., Frigerio L., Hawes C., Hwang I., Mathur J., et al. (2012). Secretory pathway research: the more experimental systems the better. Plant Cell 24 1316–1326. 10.1105/tpc.112.096362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich C. R., Han G., Chen M., Berg R. H., Dunn T. M., Cahoon E. B. (2008). Loss-of-function mutations and inducible RNAi suppression of Arabidopsis LCB2 genes reveal the critical role of sphingolipids in gametophytic and sporophytic cell viability. Plant J. 54 284–298. 10.1111/j.1365-313X.2008.03420.x [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300 1005–1016. 10.1006/jmbi.2000.3903 [DOI] [PubMed] [Google Scholar]
- Englert M., Latz A., Becker D., Gimple O., Beier H., Akama K. (2007). Plant pre-tRNA splicing enzymes are targeted to multiple cellular compartments. Biochimie 89 1351–1365. 10.1016/j.biochi.2007.06.014 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39 783–791. 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Foyer C. H., Bloom A. J., Queval G., Noctor G. (2009). Photorespiratory metabolism genes, mutants, energetics, and redox signaling. Annu. Rev. Plant Biol. 60 455–484. 10.1146/annurev.arplant.043008.091948 [DOI] [PubMed] [Google Scholar]
- Frelin O., Agrimi G., Laera V. L., Castegna A., Richardson L. G. L., Mullen R. T., et al. (2012). Identification of mitochondrial thiamin diphosphate carriers from Arabidopsis and maize. Funct. Integr. Genomics 12 317–326. 10.1007/s10142-012-0273-4 [DOI] [PubMed] [Google Scholar]
- Fukasawa Y., Tsuji J., Fu S. C., Tomii K., Horton P., Imai K. (2015). MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteomics 14 1113–1126. 10.1074/mcp.M114.043083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidda S. K., Shockey J. M., Falcone M., Kim P. K., Rothstein S. J., Andrews D. W., et al. (2011). Hydrophobic-domain-dependent protein-protein interactions mediate the localization of GPAT enzymes to ER subdomains. Traffic 12 452–472. 10.1111/j.1600-0854.2011.01160.x [DOI] [PubMed] [Google Scholar]
- Givan C., Kleczkowski L. A. (1992). The enzymic reduction of glyoxylate and hydroxypyruvate in leaves of higher plants. Plant Physiol. 100 552–556. 10.1104/pp.100.2.552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givan C. V., Tsutakawa S., Hodgson J. M., David N., Randall D. D. (1988). Glyoxylate reductase activity in pea leaf protoplasts nucleotide specificity and subcellular location. J. Plant Physiol. 132 593–599. 10.1016/S0176-1617(88)80260-8 [DOI] [Google Scholar]
- Hooper C. M., Tanz S. K., Castleden I. R., Vacher M. A., Small I. D., Millar A. H. (2014). SUBAcon: a consensus algorithm for unifying the subcellular localization data of the Arabidopsis proteome. Bioinformatics 30 3356–3364. 10.1093/bioinformatics/btu550 [DOI] [PubMed] [Google Scholar]
- Hoover G. J., Jørgensen R., Rochon A., Bajwa V. S., Merrill A. R., Shelp B. J. (2013). Identification of catalytically important amino acid residues for enzymatic reduction of glyoxylate in plants. Biochim. Biophys. Acta 1834 2663–2671. 10.1016/j.bbapap.2013.09.013 [DOI] [PubMed] [Google Scholar]
- Hoover G. J., Prentice G. A., Merrill R. A., Shelp B. J. (2007a). Glyoxylate reductase: studies of initial velocity, dead-end inhibition and product inhibition. Can. J. Bot. 85 896–902. 10.1139/B07-082 [DOI] [Google Scholar]
- Hoover G. J., Van Cauwenberghe O. R., Breitkreuz K. E., Clark S. M., Merrill A. R., Shelp B. J. (2007b). Glyoxylate reductase: general biochemical properties and substrate specificity for the recombinant protein, and developmental expression and implications for glyoxylate and succinic semialdehyde metabolism in planta. Can. J. Bot. 85 883–895. 10.1139/B07-081 [DOI] [Google Scholar]
- Horton P., Park K.-J., Obayashi T., Fujita N., Harada H., Adams-Collier C. J., et al. (2007). Wolf PSORT: protein localization predictor. Nucleic Acids Res. 35 W585–W587. 10.1093/nar/gkm259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8 275–282. 10.1093/bioinformatics/8.3.275 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A. H., Carrie C., Pogson B., Whelan J. (2009). Exploring the function-location nexus: using multiple lines of evidence in defining the subcellular location of plant proteins. Plant Cell 21 1625–1631. 10.1105/tpc.109.066019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R., Howell K. A., Carroll A., Ivanova A., Millar A. H., Whelan J. (2009). Defining core metabolic and transcriptomic responses to oxygen availability in rice embryos and young seedlings. Plant Physiol. 151 306–322. 10.1104/pp.109.142026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Khuu N., Howard A. S., Mullen R. T., Plaxton W. C. (2012). Bacterial- and plant-type phosphoenolpyruvate carboxylase isozymes from developing castor oil seeds interact in vivo and associate with the surface of mitochondria. Plant J. 71 251–262. 10.1111/j.1365-313X.2012.04985.x [DOI] [PubMed] [Google Scholar]
- Reumann S., Chowdhary G., Lingner T. (2016). Characterization, prediction and evolution of plant targeting type 1 (PTS1s). Biochim. Biophys. Acta 1863 790–803. 10.1016/j.bbamcr.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Saravanan V., Lakshmi P. T. V. (2014). Dualpred: a webserver for predicting plant proteins dual-targeted to chloroplast and mitochondria using split protein-relatedness-measure feature. Curr. Bioinform. 10 323–331. 10.2174/1574893609666140226000041 [DOI] [Google Scholar]
- Schirawski J., Planchais S., Haenni A. L. (2000). An improved protocol for the preparation of protoplasts from an established Arabidopsis thaliana cell suspension culture and infection with RNA of turnip yellow mosaic tymovirus: a simple and reliable method. J. Virol. Methods 86 85–94. 10.1016/S0166-0934(99)00173-1 [DOI] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22 1567–1572. 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Shelp B. J., Mullen R. T., Waller J. C. (2012). Compartmentation of GABA metabolism raises intriguing questions. Trends Plant Sci. 17 57–59. 10.1016/j.tplants.2011.12.006 [DOI] [PubMed] [Google Scholar]
- Simpson J. P., Di Leo R., Dhanoa P. K., Allan W. L., Makhmoudova A., Clark S. M., et al. (2008). Identification and characterization of a plastid-localized Arabidopsis glyoxylate reductase isoform: comparison with a cytosolic isoform and implications for cellular redox homeostasis and aldehyde detoxification. J. Exp. Bot. 59 2545–2554. 10.1093/jxb/ern123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobacher C. P., Clark S. M., Bozzo G. G., Mullen R. T., DeEll J. R., Shelp B. J. (2013a). Catabolism of GABA in apple fruit: subcellular localization and biochemical characterization of two γ-aminobutyrate transaminases. Postharv. Biol. Technol. 75 106–113. 10.1016/j.postharvbio.2012.08.005 [DOI] [Google Scholar]
- Trobacher C. P., Zarei A., Liu J., Clark S. M., Bozzo G. G., Shelp B. J. (2013b). Calmodulin-dependent and calmodulin-independent glutamate decarboxylases in apple fruit. BMC Plant Biol. 13:144 10.1186/1471-2229-13-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarei A., Körbes A. P., Younessi P., Montiel G., Champion A., Memelink J. (2011). Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol. Biol. 75 321–331. 10.1007/s11103-010-9728-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarei A., Trobacher C. P., Cooke A. R., Meyers A. J., Hall J. C., Shelp B. J. (2014). Apple fruit copper amine oxidase isoforms: peroxisomal MdAO1 prefers diamines as substrates, whereas extracellular MdAO2 exclusively utilizes monoamines. Plant Cell Physiol. 56 137–147. 10.1093/pcp/pcu155 [DOI] [PubMed] [Google Scholar]
- Zelitch I. (1953). Oxidation and reduction of glycolic and glyoxylic acids in plants. II. Glyoxylic acid reductases. J. Biol. Chem. 201 719–726. [PubMed] [Google Scholar]
- Zhang Y., Zheng Y., Qin L., Wang S., Buchko G. W. (2014). Structural characterization of a β-hydroxyacid dehydrogenase from Geobacter sulfurreducens and Geobacter metallireducens with succinic semialdehyde activity. Biochimie 104 61–69. 10.1016/j.biochi.2014.05.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


