Abstract
Tumor necrosis factor-α (TNF) exerts its biological effect through two types of receptors, p55 TNF receptor (TNFR1) and p75 TNF receptor (TNFR2). An inflammatory response is known to be induced mainly by TNFR1, whereas an anti-inflammatory reaction is thought to be mediated by TNFR2 in some autoimmune diseases. We have been investigating the use of an antagonistic TNF mutant (TNFR1-selective antagonistic TNF mutant (R1antTNF)) to reveal the pharmacological effect of TNFR1-selective inhibition as a new therapeutic modality. Here, we aimed to further improve and optimize the activity and behavior of this mutant protein both in vitro and in vivo. Specifically, we examined a trimeric structural fusion of R1antTNF, formed via the introduction of short peptide linkers, as a strategy to enhance bioactivity and molecular stability. By comparative analysis with R1antTNF, the trimeric fusion, referred to as single-chain R1antTNF (scR1antTNF), was found to retain in vitro molecular properties of receptor selectivity and antagonistic activity but displayed a marked increase in thermal stability. The residence time of scR1antTNF in vivo was also significantly prolonged. Furthermore, molecular modification using polyethylene glycol (PEG) was easily controlled by limiting the number of reactive sites. Taken together, our findings show that scR1antTNF displays enhanced molecular stability while maintaining biological activity compared with R1antTNF.
Keywords: autoimmune disease, biomaterials, drug design, protein chemical modification, protein design, protein engineering, tumor necrosis factor (TNF), TNF, R1antTNF, single chain, molecular stability, thermal shift assay, PEGylation
Introduction
Recently, TNF inhibitors in various molecular formats such as a neutralizing antibody (infliximab or adalimumab), soluble receptor (etanercept), or a PEGylated antibody Fab fragment (certolizumab pegol), have been clinically used as anti-TNF drugs to treat autoimmune diseases such as rheumatoid arthritis, multiple sclerosis, and ulcerative colitis (1–3). These drugs elicit a highly beneficial therapeutic effect, but this treatment may lead to serious complications including bacterial or viral infection (4, 5), demyelination (6), and lupus-like syndrome (7). Therefore, biopharmaceutical development of this treatment with a new modality is sought.
TNF binds to two receptor subtypes to exert its biological functions: TNFR13 and TNFR2 (8). TNFR1 is expressed in diverse cell types and raises the inflammatory response, whereas TNFR2, which is limited to expression in immune cells, endothelial cells, and neuronal cells, induces cell survival and proliferation. For example, it has been reported that the incidence and severity of arthritis were lower and milder in TNFR1 knock-out mice than in wild-type mice (9). Moreover, TNFR1 knock-out mice or TNFR1/TNFR2 double knock-out mice display less severe symptoms of experimental autoimmune encephalomyelitis symptoms than wild-type mice (10). Previous studies demonstrated that transgenic mice overexpressing human TNF develop severe arthritis (11, 12) or demyelination (13). Thus, the involvement of TNFR1 in autoimmune diseases is strongly implicated as human TNF is unable to signal through the murine TNFR2. Furthermore, TNF/TNFR2 signaling was shown to be crucial for antigen-stimulated activation and proliferation of T cells (14) or suppression of bacterial infection (15, 16), indicating the importance of TNFR2 function in the homeostasis mechanism. However, more recently, TNFR2 was reported to be expressed at high levels in regulatory T cells, and the TNFR2 signal promoted expansion or activation of regulatory T cells (17–19). These findings highlight the importance of the TNFR2 signal for immune tolerance. Therefore, we generated the TNFR1-selective antagonistic TNF mutant (R1antTNF) derived from human TNF using a phage display technique to specifically inhibit the physiological activity of TNFR1 (20).
R1antTNF binds only to TNFR1 and inhibits multiple TNF functions mediated through this receptor both in vitro and in vivo, akin to TNF inhibitors such as anti-human TNF-neutralizing antibody or TNFR2-Fc fusion protein (21, 22). Unfortunately, R1antTNF, like TNF, has a very short half-life in the plasma of mice when administered intravenously. Indeed, frequent administration of R1antTNF is required to maintain its concentration in the blood. Therefore, to increase the half-life of the protein, we have applied site-specific PEGylation for R1antTNF, which is a modification technique using polyethylene glycol (PEG) developed in our previous work (23–25). PEGylation significantly improves the in vivo stability and therapeutic effect of R1antTNF without any associated loss of bioactivity (20, 21). However, given that R1antTNF exists as a trimeric structure, there are problems such as a reduction in biological activity caused by intermolecular dissociation. Moreover, there may be heterogeneous modification reactions associated with the PEGylation procedure.
Here, we have attempted to further improve and optimize R1antTNF activity and behavior both in vitro and in vivo by generating a trimeric structural fusion of R1antTNF using short peptide linker regions to enhance its molecular stability. The resultant trimeric form of the protein is termed single-chain R1antTNF (scR1antTNF). We then compared R1antTNF and scR1antTNF to examine the impact of this fusion on the molecular stability and PEGylation efficacy of the molecule.
Results
Generation and characterization of scR1antTNFs
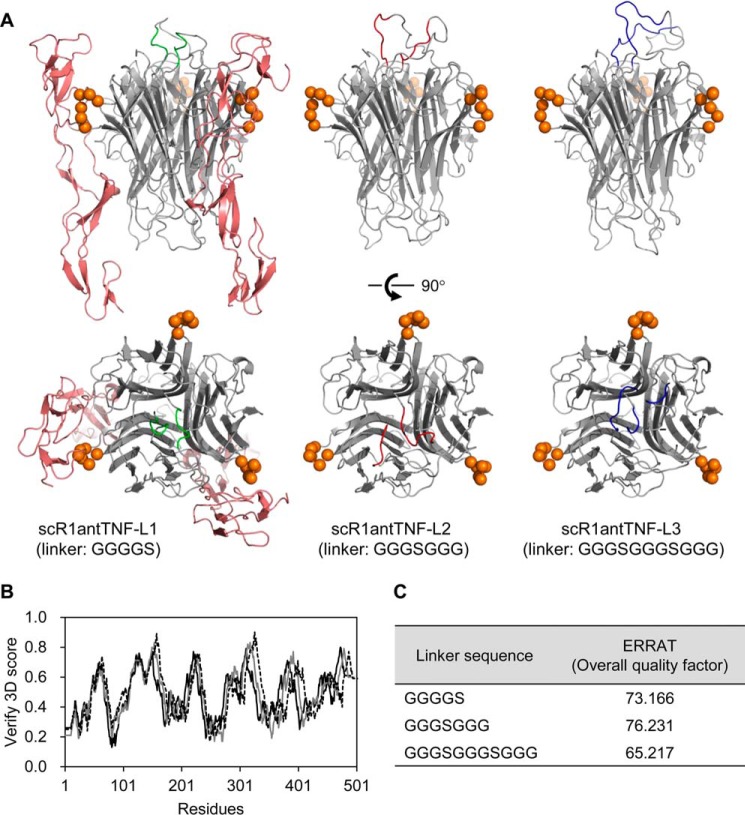
In this study, we designed an engineered version of R1antTNF, termed scR1antTNF, that cross-linked three R1antTNF monomers via short peptide linkers. Three linker sequences were initially chosen: GGGGS (scR1antTNF-L1), GGGSGGG (scR1antTNF-L2), and GGGSGGGSGGG (scR1antTNF-L3). Structural modeling of scR1antTNFs·TNFR1 complex using the Modeller program showed that the three R1antTNF monomers are conjugated by two peptide linkers present on the upper surface of the protein (Fig. 1A). Therefore, the fusion does not appear to have an effect on the TNF-TNFR1 interaction through the TNF side surface. Analysis of each structural model using Verify 3D found no conformational errors, i.e. no values less than 0.1 (Fig. 1B). Furthermore, the ERRAT score for all of the models was ∼70%, i.e. a typical value for a predicted structural model (Fig. 1C) (26). This modeling procedure indicated that scR1antTNF-L1, scR1antTNF-L2, and scR1antTNF-L3 display similar properties.
Figure 1.
Structural model of scR1antTNFs. A, a structural model of scR1antTNF-L1 (GGGGS linker) with a TNFR1 extracellular domain was constructed using Modeller. The R1antTNF was composed of three monomers (gray schematic). The N- and C-terminal peptide linkers are shown as a green schematic. Monomeric TNFR1, which interacts with the two TNF monomers, is shown as a red schematic. For clarity, only two parts of the three monomers are shown. Amino acids mutated in R1antTNF that are thought to interact with TNFR1 are shown as orange spheres. The peptide linkers of scR1antTNF-L2 (GGGSGGG) and scR1antTNF-L3 (GGGSGGGSGGG) are shown as red and blue schematics, respectively. Verify 3D (B) and ERRAT (C) were used to check the quality of the protein models. The structural models of scR1antTNF with three different linker sequences, GGGGS (gray line), GGGSGGG (dotted line), and GGGSGGGSGGG (black line), were compared using Verify 3D.
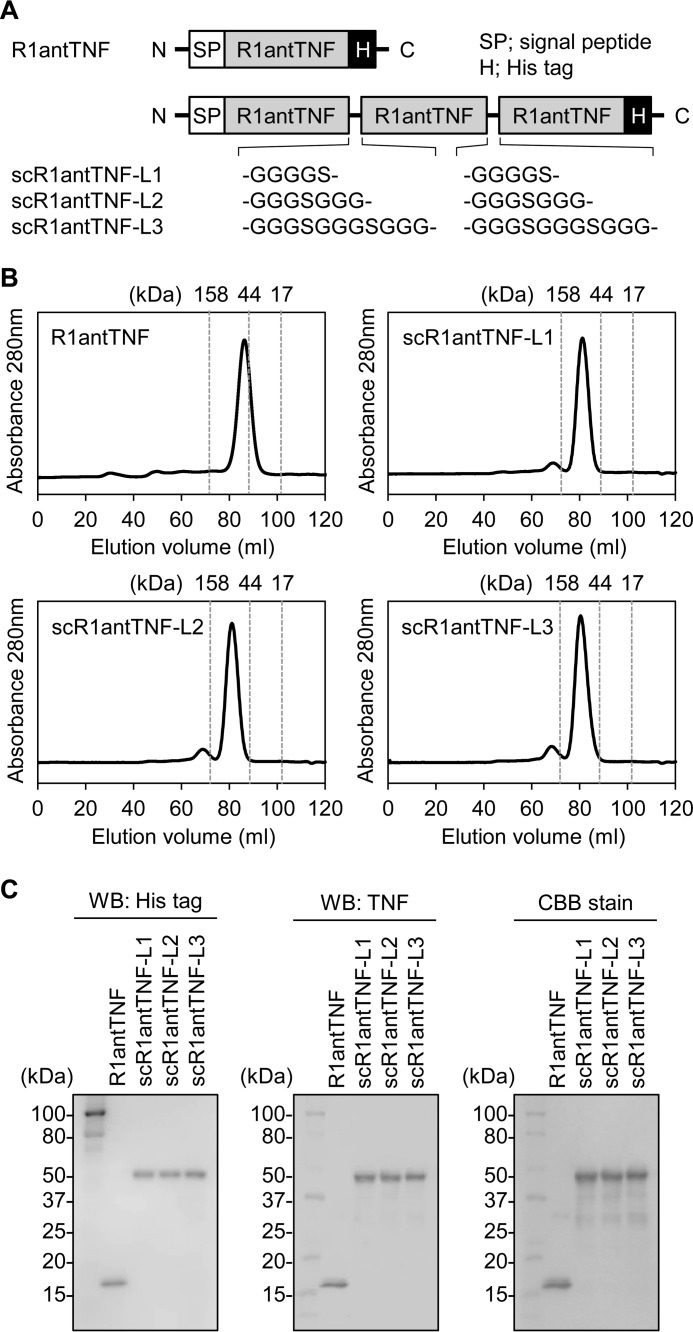
To confirm this conclusion experimentally, expression vectors for R1antTNF and the three types of scR1antTNF (Fig. 2A) were engineered and transfected into Expi293F cells. Each recombinant His-tagged fusion protein was then purified using immobilized metal ion chromatography (supplemental Fig. 1) and size-exclusion chromatography (SEC). In SEC analysis, R1antTNF and scR1antTNF-L1–L3 eluted at almost the same elution volume as might be expected from their anticipated molecular weight (Fig. 2B). Moreover, SDS-PAGE performed under reducing conditions followed by Western blotting showed that R1antTNF co-migrates with WT-TNF as a band with an apparent molecular mass of about 17 kDa, whereas scR1antTNF-L1–L3 runs with an apparent molecular mass of ∼52 kDa (Fig. 2C). Thus, it was confirmed that R1antTNF forms a trimeric structure made up of three 17-kDa monomers that associate in solution, whereas the three scR1antTNF proteins are composed of a single polypeptide comprising the three monomers in a fused arrangement generated by incorporation of short linker peptides.
Figure 2.
Generation and characterization of scR1antTNFs. A, schematic primary structures of R1antTNF, scR1antTNF-L1, -L2, and -L3. Each scR1antTNF comprises three R1antTNF domains fused together by short peptide linkers as indicated. Each mutant form of the protein was fused to a signal peptide derived from a mouse IgG VH and His tag in N- and C-terminal regions, respectively. SEC purification (B) and Western blotting (WB) with anti-His tag antibody and anti-TNF antibody followed by Coomassie Brilliant Blue (CBB) stain (C). These results confirmed the expression of the recombinant proteins.
In vitro receptor selectivity and bioactivity of scR1antTNFs
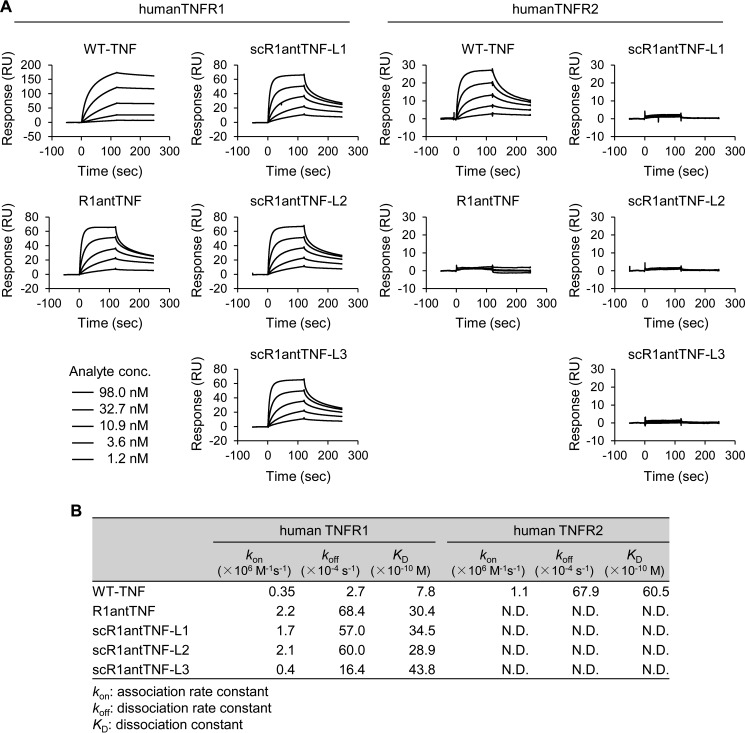
TNF can bind to both TNFR1 and TNFR2, but R1antTNF binds only TNFR1. Therefore, the selectivity of scR1antTNF-L1–L3 against TNF receptors was assessed using surface plasmon resonance (SPR) to confirm its binding specificity (Fig. 3A). At a 100 μg/ml concentration of each scR1antTNF, the maximum binding response (Rmax) values obtained from each sensorgram for TNFR1 or TNFR2 were 60 or <5 response units, respectively. Thus, these scR1antTNFs appear to selectively interact with TNFR1 in preference to TNFR2. The dissociation constant (KD) for the interaction of R1antTNF, scR1antTNF-L1, scR1antTNF-L2, and scR1antTNF-L3 with TNFR1 were similar (i.e. 3.0, 3.5, 2.9, and 4.4 nm, respectively) although greater than that for TNF (0.78 nm) (Fig. 3B). However, both these mutants have bigger kon and koff values than WT-TNF (0.35 × 106 m−1 s−1 and 0.3 ms−1, respectively). Therefore, TNF mutants compete with TNF for binding to the receptor by repeated rapid association and dissociation.
Figure 3.
In vitro receptor binding ability of scR1antTNFs. A, binding ability of WT-TNF, R1antTNF, scR1andTNF-L1, -L2, and -L3 to human TNFR1 and human TNFR2 was analyzed by SPR. Each sensorgram was measured with serially diluted samples (1.2, 3.6, 10.9, 32.7, and 98.0 nm). B, kinetic parameters of WT-TNF, R1antTNF, and three scR1antTNFs for human TNFR1/R2 were calculated using BIAcore T200 evaluation software. Avidity values of TNF as trimer are shown. RU, response units.
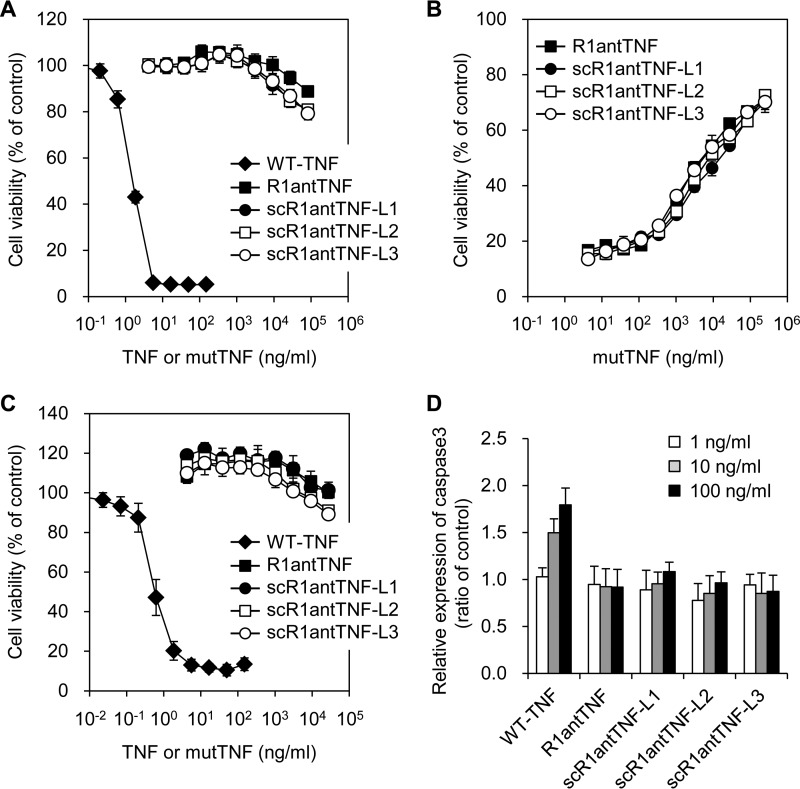
Furthermore, the cross-linking effect of the short peptide linkers was investigated in terms of the bioactivity of scR1antTNF. The R1antTNF and scR1antTNFs were found not to display agonistic activity through TNFR1 because these proteins showed only slight cytotoxicity for mouse fibroblasts (LM cells) (Fig. 4A). Moreover, R1antTNF and scR1antTNFs also suppressed TNF-induced cytotoxicity in a dose-dependent manner (Fig. 4B). These results indicated that the antagonistic activities of scR1antTNF-L1, -L2, and -L3 were almost equivalent to that of R1antTNF. In addition, a cytotoxic assay was performed using hTNFR2/mFas preadipocytes to confirm these bioactivities through TNFR2 (Fig. 4C). Cells overexpressing the fusion chimera receptor of human TNFR2 and mouse Fas can induce cell death through TNFR2 (27). As a result, TNF displayed concentration-dependent cytotoxicity, but R1antTNF and scR1antTNFs induced only a slight cytotoxic effect. Therefore, scR1antTNFs, like R1antTNF, do not transmit a signal via TNFR2.
Figure 4.
In vitro bioactivity of the scR1antTNFs through TNFR1 and TNFR2. Agonistic (A) and antagonistic (B) activities of R1antTNF (closed squares), scR1antTNF-L1 (closed circles), scR1antTNF-L2 (open squares), and scR1antTNF-L3 (open circles) through TNFR1 were confirmed by LM cell assay. The activity of WT-TNF (closed diamonds) was used as a control. C, agonistic activity through TNFR2 was evaluated by cell death of hTNFR2/mFas preadipocytes. D, caspase-3 activation induced by serially diluted WT-TNF, R1antTNF, or scR1antTNFs (1, 10, and 100 ng/ml) using A673 cells was measured with Ac-DEVD-p-nitroanilide. All error bars represent S.D.; n = 3.
Caspase-3 is an effector protease that increases apoptosis in the caspase cascade through TNFR1. Intracellular signaling caused by oligomerization between TNF and receptor on the cell surface was reported previously (28). Therefore, caspase-3 activation was tested to confirm whether receptor oligomerization is enhanced by the action of scR1antTNFs (Fig. 4D). The results clearly show that WT-TNF, but not scR1antTNFs, induces concentration-dependent activation of caspase-3. Taken together, these findings revealed that the receptor affinity and biological activity of R1antTNF were retained by scR1antTNFs despite the presence and the length of the peptide linkers within this trimeric structure.
In vitro thermal stability of scR1antTNFs
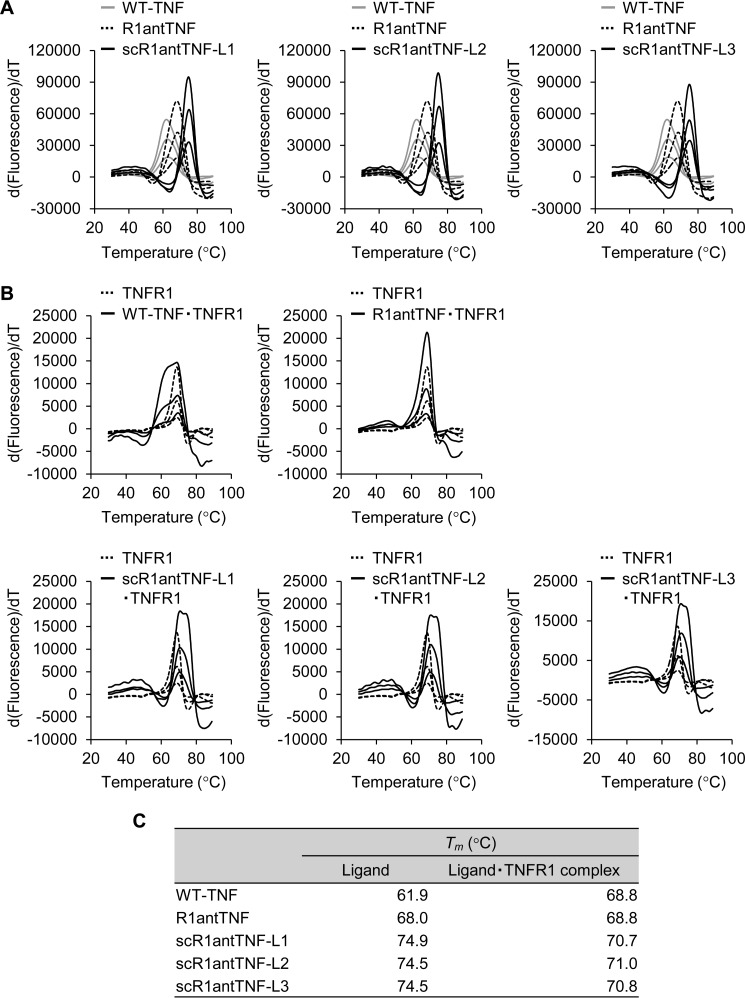
We also investigated the thermal stability of scR1antTNF in vitro by using a thermal shift assay (TSA) and differential scanning calorimetry (DSC). TSA can quickly screen for changes in the thermal denaturation temperature (Tm) of a protein either alone or as part of a complex (Fig. 5, A and B). Using this technique, the Tm values of WT-TNF and R1antTNF were determined to be 61.9 and 68.0 °C, respectively (Fig. 5C). By contrast, the Tm values of scR1antTNF-L1, -L2, and -L3 were found to be 74.9, 74.5, and 74.5 °C, respectively. Thus, the Tm values for scR1antTNF-L1, -L2, and -L3 are significantly higher by ∼12 and 6 °C than the values of WT-TNF and R1antTNF, respectively (Fig. 5B).
Figure 5.
In vitro thermal stability of scR1antTNFs measured by thermal shift assay. A, thermal stability of WT-TNF (gray line), R1antTNF (dotted line), and scR1antTNFs (black line) was evaluated by TSA. The protein was serially diluted from 700 μg/ml by 2-fold dilution. The inflection point of the peak indicates a protein denaturation temperature (Tm). B, thermal stability of the complex of WT-TNF, R1antTNF, or scR1antTNFs with human TNFR1-Fc was evaluated using TSA of human TNFR1-Fc fusion protein and its complex as shown by the dotted line and black line, respectively. C, Tm values of WT-TNF, R1antTNF, scR1antTNFs, and each complex with TNFR1 are shown.
In general, a ligand in complex with its receptor results in enhanced structural stability. Therefore, the stability of the ligand·receptor complex was analyzed (Fig. 5B). TNFR1-Fc chimera indicated a Tm value of 68.9 °C, and the TNFR1 complexes with TNF, R1antTNF, scR1antTNF-L1, scR1antTNF-L2, and scR1antTNF-L3 also indicated similar Tm values of 68.8, 68.8, 70.7, 71.0, and 70.8 °C, respectively (Fig. 5C). The melting curves of TNF and scR1antTNFs have some minor deviations from that of the receptor, and both curves form a single curve similar to each other. By contrast, because the curves of R1antTNF overlap closely with that of the receptor, a larger curve was formed. The Tm value for the scR1antTNFs·TNFR1 complex was greater than that of the other two proteins. However, the Tm of each protein complex did not vary significantly from the mean of the unbound ligand and solo receptor. Therefore, the stability of the TNFR1 complex might depend on the stability of the corresponding ligand, which in turn can affect the resulting complex.
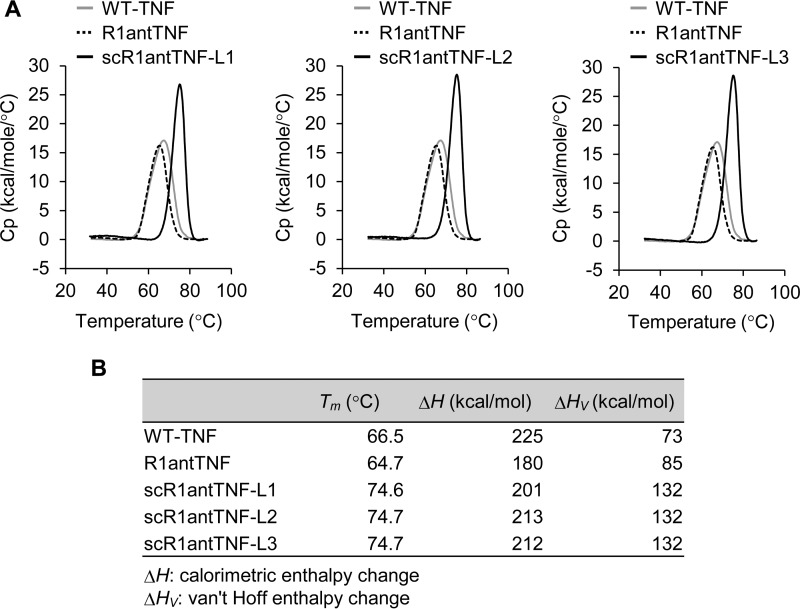
DSC was also performed to compare the thermal stability of the WT-TNF and TNF mutants. The peak of each scR1antTNF shifted to an elevated temperature, and the peak became sharper compared with that of WT-TNF (Fig. 6A). These observations suggested that scR1antTNFs display improved thermal stability because both the Tm increased and the temperature range of transition state narrowed. Indeed, the Tm values of scR1antTNF-L1 (74.6 °C), scR1antTNF-L2 (74.7 °C), and scR1antTNF-L3 (74.7 °C) were about 10 °C higher than those of TNF (66.5 °C) and R1antTNF (64.7 °C) (Fig. 6B). Furthermore, the difference between ΔH and ΔHV of scR1antTNFs was smaller than that of TNF and R1antTNF. In general, proteins with the largest difference between ΔH and ΔHV tend to undergo a denaturation process via intermediate states. It was assumed that scR1antTNFs behaved like a monomer structure because scR1antTNFs adopted a compact conformation and underwent highly cooperative denaturation by cross-linking of the trimer structure. However, there was no alteration in thermal stability due to the difference of linker length among scR1antTNF-L1, scR1antTNF-L2, and scR1antTNF-L3.
Figure 6.
In vitro thermal stability of scR1antTNFs by differential scanning calorimetry. A, thermal stability of WT-TNF (gray line), R1antTNF (dotted line), and scR1antTNF (black line) was evaluated by DSC. The WT-TNF, R1antTNF, and scR1antTNFs were diluted to 900, 900, and 700 μg/ml, respectively. The inflection point of the peak indicates a protein denaturation temperature (Tm). B, values of Tm, ΔH, and ΔHV of WT-TNF, R1antTNF, and scR1antTNFs are shown. Cp, heat capacity.
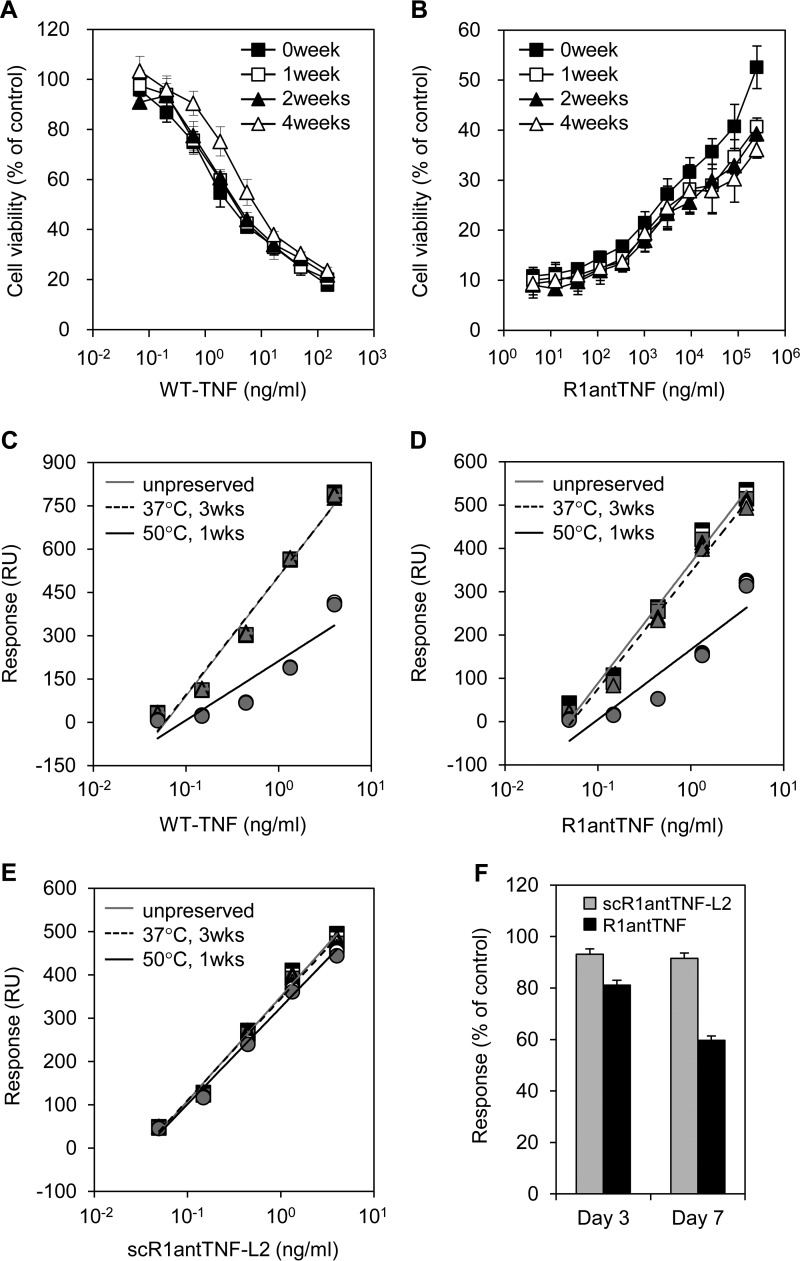
In our previous study, the agonistic activity of WT-TNF (Fig. 7A) and the antagonistic activity of R1antTNF (Fig. 7B) gradually decreased following prolonged storage at 37 °C for several weeks. Here, to evaluate the effect of a temperature shift on ligand function in vitro, we used scR1antTNF-L2 as a representative trimeric structural fusion of R1antTNF. Binding studies between TNFR1 and WT-TNF, R1antTNF, or scR1antTNF-L2 stored at 37 °C for 3 weeks or at 50 °C for 1 week were measured by SPR. The Rmax values, measured at 98 nm, were plotted as a concentration-response curve. The binding of WT-TNF and R1antTNF was largely unaffected by incubation at 37 °C for 3 weeks (Fig. 7, C and D). However, binding capacity of these ligands drastically decreased after incubation at 50 °C for 1 week. By contrast, full binding ability of scR1antTNF-L2 for TNFR1 was retained even after incubation of the ligand at 50 °C for 1 week (Fig. 7E). Indeed, the ability of R1antTNF to bind TNFR1 steadily diminished upon incubation at 50 °C in contrast to scR1antTNF-L2, which displayed no such decreased binding (Fig. 7F). These findings indicate that thermal stability is enhanced by fusion of the trimeric structure.
Figure 7.
Time-dependent alteration in bioactivity and binding ability of WT-TNF and TNF mutants. Agonistic activity of WT-TNF (A) and antagonistic activity of R1antTNF (B) was gradually reduced in a time-dependent manner by prolonged storage at 37 °C. Samples stored for 0, 1, 2, and 4 weeks are indicated by closed squares, open squares, closed triangles, and open triangles, respectively. WT-TNF (C), R1antTNF (D), and scR1antTNF-L2 (E) was incubated for 3 weeks at 37 °C or for 1 week at 50 °C. Each fitted line was calculated from the Rmax value of these proteins to human TNFR1 measured by SPR. Plots indicate serially diluted protein concentrations (0.05–5.00 ng/ml). Squares, triangles, and circles represent the concentration of protein at 4, 25, and 37 °C, respectively. Results were measured in triplicate. F, binding response alteration of TNF mutants stored at 50 °C for 3 and 7 days was evaluated. Error bars represent S.D.; n = 3. RU, response units.
In vivo molecular stability and inflammatory cytokine production
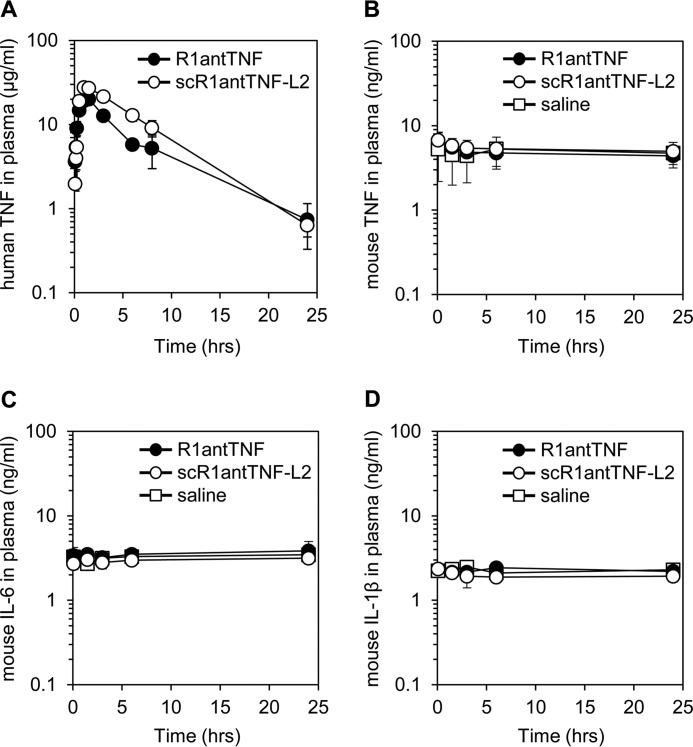
To evaluate the stability of R1antTNF and scR1antTNF-L2 in vivo, plasma clearances were measured after administration to a mouse (Fig. 8A). TNF mutants could be detected by ELISA using anti-human TNF antibody without being affected by mouse TNF (supplemental Fig. 2, A and B). Plasma concentrations of both TNF mutants peaked 90 min after i.p. injection. The plasma concentration of scR1antTNF-L2 after 20 h was maintained at a higher level than that of R1antTNF, indicating an increased blood half-life. Therefore, scR1antTNF-L2 appears to be tolerated by the protein elimination process.
Figure 8.
In vivo plasma clearance and cytokine induction of scR1antTNF. A, plasma concentrations of R1antTNF and scR1antTNF-L2 after i.p. injection were measured by ELISA. Error bars represent S.D.; n = 5. Mouse TNF (B), mouse IL-6 (C), and mouse IL-1β (D) in plasma were detected by ELISA. R1antTNF, scR1antTNF-L2, and saline are indicated by closed circles, open circles, and open squares, respectively. Error bars represent S.D.; n = 5.
Furthermore, administration of TNF mutants did not increase plasma concentrations of TNF, IL-6, and IL-1β (Fig. 8, B–D). Therefore, as observed for R1antTNF, scR1antTNF-L2 did not trigger acute induction of inflammatory cytokines by immune response activation. Taken together, these data suggest that scR1antTNF-L2 displays effective increased molecular stability compared with R1antTNF.
Molecular modification efficiency of scR1antTNF
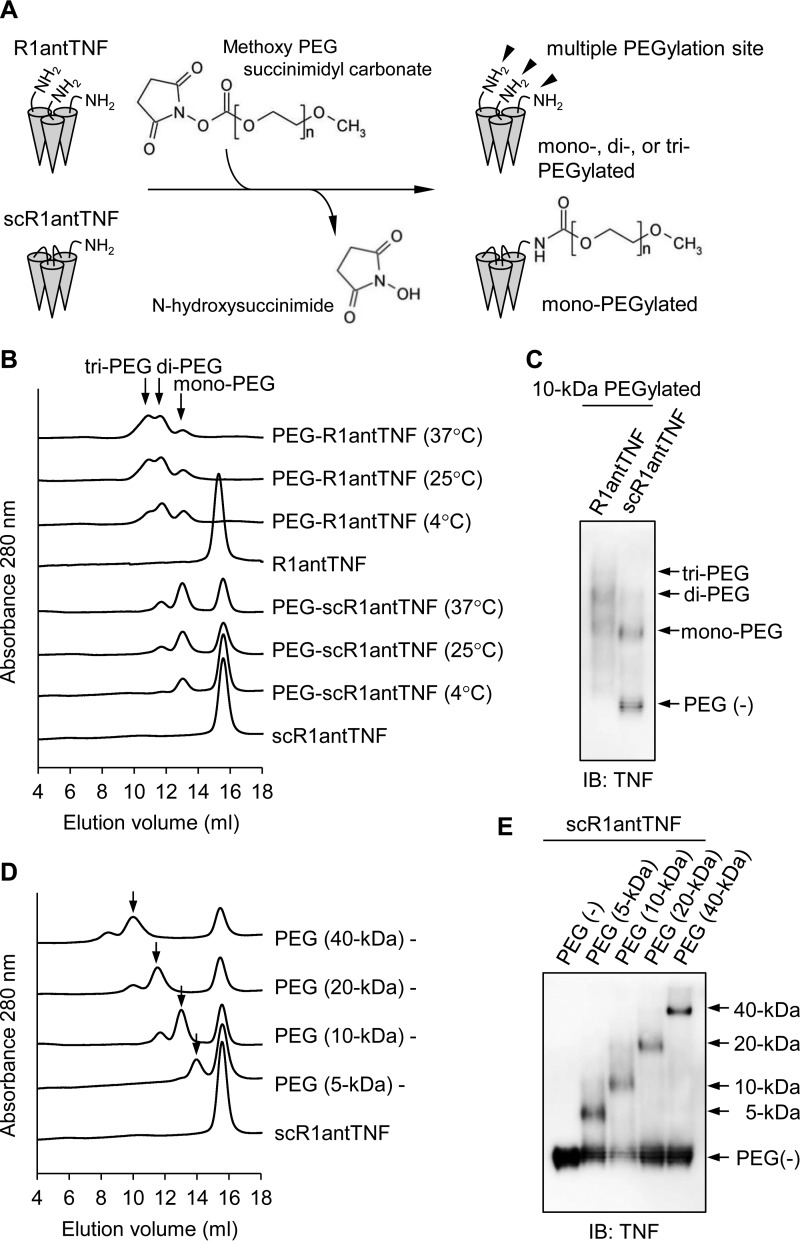
The superior efficiency of the molecular modification reaction is considered to be an advantage related to the single-chain structure. It is known that molecular modifications of proteins such as PEGylation are effective in suppressing immunogenicity, degradation by proteases, and excretion into the urine (24, 29). We previously reported that N terminus-specific mono-PEGylation of R1antTNF improved in vivo stability (20, 21). However, because R1antTNF adopts a homotrimeric structure composed of three monomers, there are three amino groups arising from each N-terminal residue that may be subject to modification by PEG (Fig. 9A). Consequently, the preparation of a mono-PEGylated product is technically challenging. By contrast, control of PEGylation for scR1antTNFs is relatively easy because only one N-terminal group is available for reaction (Fig. 9A). Thus, the PEGylation reaction conditions were examined.
Figure 9.
The difference of PEGylation efficiency between R1antTNF and scR1antTNF. A, the PEGylation reaction scheme of R1antTNF and scR1antTNF. R1antTNF, but not scR1antTNF, has multiple reactive sites as shown by arrowheads. Therefore, three types of PEGylated mutants were created. B, R1andTNF and scR1antTNF modified with 10-kDa PEG at different reaction temperatures (4, 25, and 37 °C) were analyzed by gel-filtration chromatography. The resultant chromatograms were overlaid. Peaks of mono-, di-, and tri-PEGylated protein are indicated by an arrow. C, each PEGylated mutant was also detected by native PAGE and Western blotting with an anti-human TNF antibody. Gel-filtration chromatography (D) and native PAGE followed by Western blotting (E) were performed to compare the PEGylated reaction efficacy due to differences of PEG size (5, 10, 20, and 40 kDa). Arrows indicate a peak of mono-PEGylated product. IB, immunoblotting.
R1antTNF and scR1antTNF-L2 were incubated with 10-kDa PEG-COO-NHS at 4, 25, and 37 °C. R1antTNF was found to react easily by PEGylation. However, SEC analysis identified significant amounts of di- or tri-PEGylated R1antTNF product even when the reaction was performed under mild conditions at 4 °C (Fig. 9B). Moreover, the amount of PEGylated products increased in a reaction temperature-dependent manner. By contrast, when using scR1antTNF-L2, the mono-PEGylated derivative was the dominant product, which increased in a temperature-dependent manner. Nonetheless, there was a significant proportion of unreacted scR1antTNF. At this stage, it is unclear whether the presence of a peptide linker is directly related to the buildup of unreacted scR1antTNF. However, the separation of unreacted scR1antTNF from PEGylated scR1antTNF is facile. The identity of these PEGylated TNF mutant peaks was subsequently confirmed by native PAGE (Fig. 9C). Furthermore, depending on the size of PEG, uniform reaction is difficult due to the associated steric hindrance. However, scR1antTNF preferentially produced a mono-PEGylated reaction product that was independent of the PEG size (Fig. 9, D and E). Therefore, the trimeric structural fusion of R1antTNF also displays a superiority in terms of molecular optimization by modification.
In vivo plasma clearance of 40-kDa branched PEGylated scR1antTNF
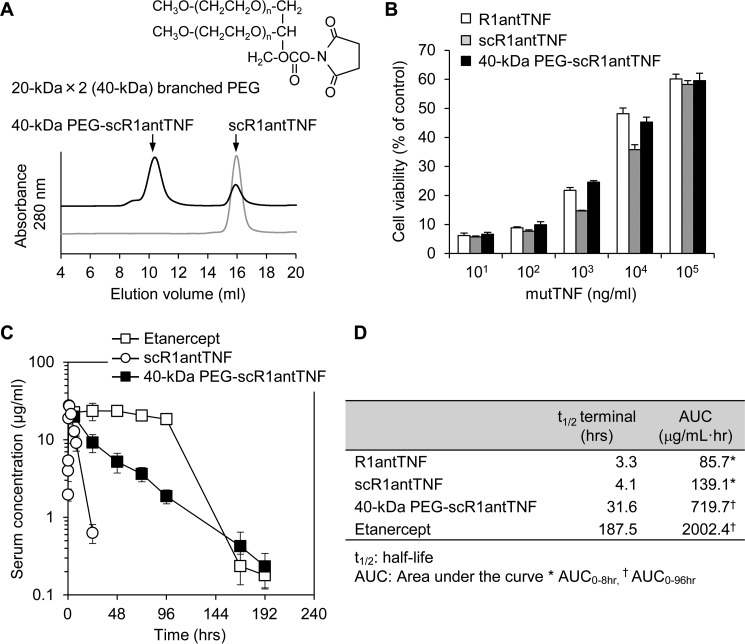
To prolong its stability in blood, scR1antTNF-L2 was modified with 20-kDa × 2 (40-kDa) branched PEG (Fig. 10A). The 40-kDa branched PEGylated scR1antTNF-L2 (40-kDa PEG-scR1antTNF) was detected as a single peak by SEC analysis. Purified 40-kDa PEG-scR1antTNF suppressed TNF-induced cytotoxicity in a dose-dependent manner as observed for R1antTNF and scR1antTNF (Fig. 10B). 40-kDa PEG-scR1antTNF and etanercept were also administered to mice. Blood samples were taken at various time points and then analyzed, which revealed a gradual decrease in the concentration of 40-kDa PEG-scR1antTNF compared with etanercept (Fig. 10C). Nonetheless, as expected, 40-kDa PEG-scR1antTNF had significantly improved stability over that of scR1antTNF. We also analyzed the difference in half-life among R1antTNF, scR1antTNF, and 40-kDa PEG-scR1antTNF by measuring the area under the curve (AUC) (Fig. 10D). Our analysis suggested that 40-kDa branched PEGylation increases the blood half-life by about 10-fold.
Figure 10.
Generation of 40-kDa branched PEGylated scR1antTNF and its in vitro activity and in vivo plasma clearance after a single dose. A, the scR1antTNF was incubated with 20-kDa × 2 (40-kDa) branched PEG to generate 40-kDa PEG-scR1antTNF. SEC analysis was performed to check the extent of reaction. B, the inhibition activity of TNF-induced cytotoxicity of 40-kDa PEG-scR1antTNF (black bars) was compared with R1antTNF (white bars) and scR1antTNF (gray bars) by LM cell assay. Error bars represent S.D.; n = 3. C, plasma concentrations of scR1antTNF (open circles) and 40-kDa PEG-scR1antTNF (closed squares) after a single i.p. injection were measured by ELISA. Etanercept (open squares), a human TNFR2-Fc fusion protein, was used as the positive control. Error bars represent S.D.; n = 5. D, the half-life and AUC of R1antTNF, scR1antTNF, and 40-kDa PEG-scR1antTNF were evaluated by moment analysis.
Discussion
We previously reported the generation of R1antTNF as a TNFR1-selective TNF inhibitor. This mutant form of TNF exerts superior pharmacological effects when used to treat a mouse model of rheumatoid arthritis or multiple sclerosis (21, 22). Unfortunately, however, R1antTNF, like TNF, displays a relatively short blood half-life, which is thought to arise from its trimeric structure.
In previous studies, several strategies were reported to increase the stability and activity of TNF. Fusion of trimeric TNF by the introduction of linker sequences increased its receptor affinity and antitumor activity (30, 31). Moreover, a disulfide-cross-linked TNF mutant enhanced its activity through TNFR2 (32).
Therefore, in this study, we have attempted to improve the molecular stability and PEGylation efficacy of R1antTNF. Our strategy involved generating scR1antTNF, which cross-links three monomers of R1antTNF via short amino acid linkers. We reasoned that the rigid structure of a trimeric TNF fusion might enhance its inhibitory effect by forming a stronger complex with TNFR1. However, there are no reports in the literature of applying this strategy to an antagonistic mutant.
TNF, which has a trimeric structure, binds to trimeric TNF receptors. Recently, it was suggested that clustering of the TNF·TNFR1 complex on the cell surface might be important for the incremental TNF signaling to stably transfer the intracellular signal through this receptor (28). As described earlier, we have been investigating the use of an antagonistic TNF mutant termed R1antTNF. R1antTNF has a homotrimeric structure and exerts an antagonistic activity by competing with endogenous TNF for the active binding site of TNFR1. However, dissociation of the trimeric structure of R1antTNF or resultant heterotrimerization of R1antTNF with endogenous TNF may decrease its antagonistic activity. Based on these considerations, we thought that scR1antTNF might inhibit TNFR1 function more strongly than R1antTNF by a receptor clustering effect and suppression of homotrimer dissociation or heterotrimerization with TNF.
Therefore, we designed three types of scR1antTNF with different linker sequences. Our analyses showed that (i) the three scR1antTNFs displayed similar molecular properties, and (ii) the suppression activity of scR1antTNFs was almost the same as that of R1antTNF (Fig. 4B). Interestingly, however, the trimeric protein fusion showed markedly increased stability over the non-fused protein (Figs. 5 and 6). To enhance the antagonistic activity, it may be necessary to add further mutations that can overcome the difference in binding properties between TNF and R1antTNF at the monomer level. Nonetheless, scR1antTNFs retains receptor selectivity and antagonistic activity of R1antTNF for TNFR1 despite the presence of linkers of different lengths while displaying enhanced molecular stability both in vitro and in vivo.
As a further strategy to enhance protein stability in vivo, PEGylation can contribute significantly to maintaining drug concentration in the blood (33). Several biopharmaceuticals that had been modified with water-soluble polymers such as PEG have been commercially produced (e.g. PEG-interferon α2a and certolizumab pegol) (34). We have also reported a bioconjugation technique for proteins that specifically modifies the N-terminal residue, dramatically prolonging the blood half-life (24). However, it remains technically difficult to generate a uniformly PEGylated product while retaining the bioactivity of a molecule such as TNF, which has a trimeric structure. In this regard, scR1antTNF has the distinct advantage that it is easy to control the mono-PEGylation reaction. Specifically, mono-PEGylation is achieved because scR1antTNF has a single amino group at its N terminus. Thus, even derivatization with a large molecule such as 40-kDa PEG becomes facile (Fig. 9D). The ability to uniformly PEGylate a protein drug is a significant advantage for pharmaceutical applications.
Indeed, two arm-branched PEG (20 kDa × 2) conjugated to scR1antTNF (40-kDa PEG-scR1antTNF) increased retention in the blood and enabled facile modification without loss of antagonistic activity (Fig. 10). Our previous report indicated that 5-kDa PEG-R1antTNF had a pharmacological effect in murine experimental autoimmune encephalomyelitis, which is utilized as an animal model for multiple sclerosis (21). Therefore, 40-kDa PEG-scR1antTNF may extend the administration interval substantially while maintaining efficacy. Furthermore, modification of high molecular weight PEG generally increases the viscosity of the protein solution. However, an increase in viscosity can be suppressed by using branched PEG of the same molecular weight. Thus, 40-kDa PEG-scR1antTNF was also optimized in terms of its pharmaceutical properties.
By generating a trimeric fusion of scR1antTNF with peptide linkers, we found that the thermal stability of R1antTNF was greatly improved without any associated loss of bioactivity. Furthermore, the presence of a single N terminus in the fused version of the trimeric protein is advantageous in terms of site-specific PEGylation. Thus, the pharmaceutical potential of R1antTNF is fully exploited by combining these two strategies.
Enhanced thermal stability improves the pharmaceutical applicability of a drug including prolonged half-life, preservation stability, ease of production, and even drug design. Furthermore, this methodology is appropriate for various cytokine-based drug seed proteins currently undergoing development. As such, the methods described here represent a new foundation technology for enhancing the molecular stability and biological activity of protein-based drugs.
Experimental procedures
Cell culture
LM cells (mouse fibroblast cell line) were maintained in Eagle's minimum essential medium (Wako Pure Chemical) supplemented with 1% fetal bovine serum (FBS) and 1% antibiotic mixture (10,000 units/ml penicillin, 10 mg/ml streptomycin, and 25 μg/ml amphotericin B) (Wako). hTNFR2/mFas preadipocytes (TNFR1−/−R2−/− mouse preadipocytes expressing a chimeric receptor of the extracellular and transmembrane domains of human TNFR2 and intracellular domain of mouse Fas), which were previously established (27), were cultured in RPMI 1640 medium (Wako) supplemented with 10% FBS, 1% antibiotic mixture, and 5 μg/ml blasticidin S HCl (Invitrogen). A673 cells (human Ewing's sarcoma cell line) were cultured in DMEM (Wako) supplemented with 10% FBS and 1% antibiotic mixture.
Modeling of R1antTNF and R1antTNF·human TNFR1 complex
The scR1antTNF structure was modeled on that of R1antTNF (Protein Data Bank code 2E7A) using UCSF Chimera and Modeller packages. Modeller is a widely used platform for homology modeling of three-dimensional structures of proteins. Chimera provides a graphical interface to running Modeller. The scR1antTNF structure was superimposed on that of lymphotoxin-α derived from the lymphotoxin-α·TNFR1 complex (Protein Data Bank code 1TNR).
Validation analysis of TNF mutants
The length of the linker sequence was determined by using Verify 3D and the ERRAT program. The Verify 3D analysis can evaluate the quality of 3D protein models and template structures. The ERRAT program can validate the probable existence of protein structures based on an “overall quality factor.” Increased overall quality factor scores reflect improved structures.
Cloning of TNF mutants
The gene sequence of R1antTNF is given in our previous report (20). scR1antTNF was alternatingly bounded in the N- and C-terminal regions of R1antTNF by three types of amino acid linkers (Figs. 1A and 2A). The gene mutants were fused to the signal peptide (MGWSLILLFLVAVATGVHS) at the N terminus and His tag sequences at the C terminus. Codon usage was optimized for eukaryotic expression (GenScript). The engineered sequences were then subcloned into the mammalian expression vector pcDNA3.1 (+) (Invitrogen) to give pcDNA3.1-R1antTNF-His6 and pcDNA3.1-scR1antTNF-L1 (-L2 or -L3)-His6.
Expression and purification of TNF mutants
TNF mutants were expressed using the Expi293 Expression System (Invitrogen) according to the manufacturer's protocol. In brief, the respective expression vector, pcDNA3.1-R1antTNF-His6 or pcDNA3.1-scR1antTNF-L1 (-L2 or -L3)-His6, was transfected into Expi293F cells by a lipofection method using ExpiFectamine 293 reagent. Seven days later, cultured medium was collected by centrifugation. The desired protein in the supernatant was then purified using Ni-Sepharose Excel (GE Healthcare). Bound TNF mutant protein was eluted with 20 mm sodium phosphate buffer (pH 7.4) containing 500 mm imidazole. Protein in the eluate was further purified by size-exclusion chromatography using a HiLoad 16/600 Superdex 200 prep grade column (GE Healthcare).
SDS-PAGE, native PAGE, and immunoblotting
SDS-PAGE and native PAGE were performed using Tris-glycine-SDS buffer (pH 8.3), and the resolved proteins were transferred onto a PVDF membrane (ClearTrans SP PVDF membrane, Wako). After blocking of nonspecific binding proteins, the mutant protein was probed with HRP-conjugated anti-penta-His antibodies (Qiagen) or biotinylated anti-human TNF antibodies (R&D Systems) in combination with streptavidin-HRP. Visualization was achieved by chemiluminescence with ECL Prime (GE Healthcare).
Binding kinetics analysis using SPR
The binding kinetics were analyzed by the SPR technique (BIAcore T200, GE Healthcare). Fc fusion proteins of human TNFR1 or TNFR2 (R&D Systems) were immobilized on sensor chip CM5 by an amine coupling reaction. The WT-TNF or TNF mutants diluted with HBS-EP buffer (HEPES-buffered saline with EDTA and polysorbate) (GE Healthcare) were injected for 2 min at a flow rate of 20 μl/min, and dissociation was monitored for a further 2 min. The data were subsequently analyzed using the 1:1 Langmuir binding model with BIAevaluation T200 software.
In vitro bioactivity of TNF mutants via TNFR1 or TNFR2
For the cytotoxicity assay, LM cells (3 × 104 cells/well) were cultured with serially diluted WT-TNF or TNF mutants at 37 °C in 96-well plates. For the inhibition assay, LM cells (1 × 104 cells/well) were cultured with a serial dilution of the TNF mutants in the presence of mouse WT-TNF (5 ng/ml) (Peprotech). Forty-eight hours later, cell viabilities were determined by a methylene blue assay (20). hTNFR2/mFas preadipocytes (1.5 × 104 cells/well) were cultured with serially diluted human WT-TNF or the TNF mutants in the presence of cycloheximide (1 μg/ml). After incubation for 48 h at 37 °C, cell survival was measured by a WST-8 colorimetric assay (Cell Counting Kit-8, Dojindo) according to the manufacturer's protocol.
A673 cells (5 × 104 cells/well) were cultured with human WT-TNF or TNF mutants containing actinomycin D (1 μg/ml) at 37 °C. Six hours later, caspase-3 activities were measured with Ac-DEVD-p-nitroanilide using the caspase-3 assay kit (Abcam) by following the manufacturer's protocol.
Thermal shift assay
Human TNF (700 μg/ml) and TNF mutants (700 μg/ml) were 2-fold serially diluted with PBS (pH 7.4). The diluted proteins were mixed with SYPRO Orange (Protein Thermal Shift Dye, Life Technologies) as samples. Samples were subsequently heat-denatured by raising the temperature from 25 to 90 °C at a rate of 0.16 °C/10 s, and fluorescence intensity was measured using a StepOnePlus Real Time PCR System (Applied Biosystems). To determine the Tm values, data were analyzed using Protein Thermal Shift Software v1.0 (Applied Biosystems).
Differential scanning calorimetry
Measurements were performed using a differential scanning calorimeter (MicroCal VP-Capillary DSC System, Malvern). Protein samples were dissolved to a measured concentration in phosphate-buffered saline (pH 7.4). The same PBS buffer was used as reference. Scans were recorded from 15 to 90 °C at a scan rate of 1 °C/min. The resultant DSC curves were normalized using the baseline as the reference, and the data were processed using the non-two-state model fitting with MicroCal Origin software.
Thermal stability test by protein preservation
Human TNF (1 mg/ml) and TNF mutants (1 mg/ml) were stored at 37 °C for 3 weeks or at 50 °C for 1 week. The binding ability of each protein to human TNFR1 was then measured by SPR (BIAcore). The Rmax values of serially diluted proteins (0.05, 0.14, 0.44, 1.33, and 5.00 ng/ml) were plotted and calculated by fitting a line. The degree of protein denaturation was determined from the slope of the line.
Measurement of TNF mutant and cytokine concentrations in plasma
Female BALB/c mice (8 weeks old) were purchased from Japan SLC. Mice were injected i.p. with 50 μg of each R1antTNF, scR1antTNF, or 40-kDa branched PEGylated scR1antTNF. Saline and etanercept (a commercially available TNF inhibitor) were injected as negative and positive controls, respectively. To examine plasma clearance, blood was collected from the tail vein at 5, 10, 15, 30, 60, 90, 180, 360, 480, and 1,440 min after injection. Furthermore, clearance of 40-kDa PEGylated scR1antTNF was assessed in blood samples collected at 6, 24, 48, 72, 96, 168, and 192 h. The concentration of each TNF mutant or etanercept in plasma samples was measured by ELISA with a human TNF ELISA kit (BD Biosciences) or a human IgG ELISA kit (Bethyl Laboratories), respectively. Half-life (t½) and AUC were calculated by moment analysis. Acute induction of inflammatory cytokines was evaluated by TNF, IL-6, and IL-1β concentrations on ELISA for each mouse cytokine (OptEIA, BD Biosciences) at selected time points (5, 90, 180, 360, and 1,440 min).
PEGylation analysis of TNF mutants by size-exclusion chromatography
For PEGylation, TNF mutants were incubated with a 10-fold molar excess of methoxy polyethylene glycol succinimidyl carbonate (SUNBRIGHT TS series, NOF Corp.) at 4, 25, or 37 °C for 10 min. Reactions were quenched by addition of 6-aminohexanoic acid. The PEGylated products were confirmed by size-exclusion chromatography using a Superdex 200 10/300 GL column (GE Healthcare). Proteins were eluted with a PBS buffer (pH 7.4) at a flow rate of 0.5 ml/min, and the eluate was monitored by absorption at 280 nm.
Author contributions
M. I. and H. K designed the experiments and wrote the manuscript. M. I., D. A., S. T., H. K., and S.-i. T. performed in vitro and in vivo experiments. T. T. and K. M. measured the stability of proteins using DSC and provided informative advice. M. N., Y. M., T. N., Y. K., and T. I. contributed to the homology modeling of the protein three-dimensional structure. Y. M. and Y. T. gave valuable advice on recombinant protein preparation. S.-i. T. directed the study and assisted in writing and editing the manuscript. All authors provided comments on the manuscript.
Supplementary Material
Acknowledgment
We thank Dr. Keizo Fukushima (Kobe Gakuin University) for valuable advice and for providing the analysis tool of moment analysis.
This work was supported by the Japan Society for the Promotion of Science KAKENHI, Hoansya Foundation, Takeda Science Foundation, Project for Cancer Research and Therapeutic Evolution (P-CREATE), and the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from the Japan Agency for Medical Research and Development (AMED). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. 1 and 2.
- TNFR
- TNF receptor
- R1antTNF
- TNFR1-selective antagonistic TNF mutant
- scR1antTNF
- single-chain R1antTNF
- SEC
- size-exclusion chromatography
- SPR
- surface plasmon resonance
- Rmax
- maximum binding response
- hTNFR2/mFas
- human TNFR2/mouse Fas
- TSA
- thermal shift assay
- DSC
- differential scanning calorimetry
- Tm
- thermal denaturation temperature
- AUC
- area under the curve
- ΔH
- calorimetric enthalpy change
- ΔHV
- van't Hoff enthalpy change.
References
- 1. Thalayasingam N., and Isaacs J. D. (2011) Anti-TNF therapy. Best Pract. Res. Clin. Rheumatol. 25, 549–567 [DOI] [PubMed] [Google Scholar]
- 2. Bradley J. R. (2008) TNF-mediated inflammatory disease. J. Pathol. 214, 149–160 [DOI] [PubMed] [Google Scholar]
- 3. Liang S., Dai J., Hou S., Su L., Zhang D., Guo H., Hu S., Wang H., Rao Z., Guo Y., and Lou Z. (2013) Structural basis for treating tumor necrosis factor α (TNFα)-associated diseases with the therapeutic antibody infliximab. J. Biol. Chem. 288, 13799–13807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gómez-Reino J. J., Carmona L., Valverde V. R., Mola E. M., Montero M. D., and BIOBADA SER Group (2003) Treatment of rheumatoid arthritis with tumor necrosis factor inhibitors may predispose to significant increase in tuberculosis risk: a multicenter active-surveillance report. Arthritis Rheum. 48, 2122–2127 [DOI] [PubMed] [Google Scholar]
- 5. Lubel J. S., Testro A. G., and Angus P. W. (2007) Hepatitis B virus reactivation following immunosuppressive therapy: guidelines for prevention and management. Intern. Med. J. 37, 705–712 [DOI] [PubMed] [Google Scholar]
- 6. Mercieca C., Vella N., and Borg A. A. (2012) Demyelination during anti-TNFα therapy for ankylosing spondylitis. Mod. Rheumatol. 22, 303–307 [DOI] [PubMed] [Google Scholar]
- 7. Shakoor N., Michalska M., Harris C. A., and Block J. A. (2002) Drug-induced systemic lupus erythematosus associated with etanercept therapy. Lancet 359, 579–580 [DOI] [PubMed] [Google Scholar]
- 8. Cabal-Hierro L., and Lazo P. S. (2012) Signal transduction by tumor necrosis factor receptors. Cell. Signal. 24, 1297–1305 [DOI] [PubMed] [Google Scholar]
- 9. Mori L., Iselin S., De Libero G., and Lesslauer W. (1996) Attenuation of collagen-induced arthritis in 55-kDa TNF receptor type 1 (TNFR1)-IgG1-treated and TNFR1-deficient mice. J. Immunol. 157, 3178–3182 [PubMed] [Google Scholar]
- 10. Eugster H. P., Frei K., Bachmann R., Bluethmann H., Lassmann H., and Fontana A. (1999) Severity of symptoms and demyelination in MOG-induced EAE depends on TNFR1. Eur. J. Immunol. 29, 626–632 [DOI] [PubMed] [Google Scholar]
- 11. Butler D. M., Malfait A. M., Mason L. J., Warden P. J., Kollias G., Maini R. N., Feldmann M., and Brennan F. M. (1997) DBA/1 mice expressing the human TNF-α transgene develop a severe, erosive arthritis: characterization of the cytokine cascade and cellular composition. J. Immunol. 159, 2867–2876 [PubMed] [Google Scholar]
- 12. Keffer J., Probert L., Cazlaris H., Georgopoulos S., Kaslaris E., Kioussis D., and Kollias G. (1991) Transgenic mice expressing human tumour necrosis factor: a predictive genetic model of arthritis. EMBO J. 10, 4025–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Akassoglou K., Bauer J., Kassiotis G., Pasparakis M., Lassmann H., Kollias G., and Probert L. (1998) Oligodendrocyte apoptosis and primary demyelination induced by local TNF/p55TNF receptor signaling in the central nervous system of transgenic mice: models for multiple sclerosis with primary oligodendrogliopathy. Am. J. Pathol. 153, 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim E. Y., Priatel J. J., Teh S. J., and Teh H. S. (2006) TNF receptor type 2 (p75) functions as a costimulator for antigen-driven T cell responses in vivo. J. Immunol. 176, 1026–1035 [DOI] [PubMed] [Google Scholar]
- 15. Olleros M. L., Guler R., Corazza N., Vesin D., Eugster H. P., Marchal G., Chavarot P., Mueller C., and Garcia I. (2002) Transmembrane TNF induces an efficient cell-mediated immunity and resistance to Mycobacterium bovis bacillus Calmette-Guerin infection in the absence of secreted TNF and lymphotoxin-α. J. Immunol. 168, 3394–3401 [DOI] [PubMed] [Google Scholar]
- 16. Saunders B. M., Tran S., Ruuls S., Sedgwick J. D., Briscoe H., and Britton W. J. (2005) Transmembrane TNF is sufficient to initiate cell migration and granuloma formation and provide acute, but not long-term, control of Mycobacterium tuberculosis infection. J. Immunol. 174, 4852–4859 [DOI] [PubMed] [Google Scholar]
- 17. Chen X., Bäumel M., Männel D. N., Howard O. M., and Oppenheim J. J. (2007) Interaction of TNF with TNF receptor type 2 promotes expansion and function of mouse CD4+CD25+ T regulatory cells. J. Immunol. 179, 154–161 [DOI] [PubMed] [Google Scholar]
- 18. Chen X., and Oppenheim J. J. (2011) The phenotypic and functional consequences of tumour necrosis factor receptor type 2 expression on CD4+ FoxP3+ regulatory T cells. Immunology 133, 426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Okubo Y., Mera T., Wang L., and Faustman D. L. (2013) Homogeneous expansion of human T-regulatory cells via tumor necrosis factor receptor 2. Sci. Rep. 3, 3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shibata H., Yoshioka Y., Ohkawa A., Minowa K., Mukai Y., Abe Y., Taniai M., Nomura T., Kayamuro H., Nabeshi H., Sugita T., Imai S., Nagano K., Yoshikawa T., Fujita T., et al. (2008) Creation and x-ray structure analysis of the tumor necrosis factor receptor-1-selective mutant of a tumor necrosis factor-α antagonist. J. Biol. Chem. 283, 998–1007 [DOI] [PubMed] [Google Scholar]
- 21. Nomura T., Abe Y., Kamada H., Shibata H., Kayamuro H., Inoue M., Kawara T., Arita S., Furuya T., Yamashita T., Nagano K., Yoshikawa T., Yoshioka Y., Mukai Y., Nakagawa S., et al. (2011) Therapeutic effect of PEGylated TNFR1-selective antagonistic mutant TNF in experimental autoimmune encephalomyelitis mice. J. Control. Release 149, 8–14 [DOI] [PubMed] [Google Scholar]
- 22. Shibata H., Yoshioka Y., Abe Y., Ohkawa A., Nomura T., Minowa K., Mukai Y., Nakagawa S., Taniai M., Ohta T., Kamada H., Tsunoda S., and Tsutsumi Y. (2009) The treatment of established murine collagen-induced arthritis with a TNFR1-selective antagonistic mutant TNF. Biomaterials 30, 6638–6647 [DOI] [PubMed] [Google Scholar]
- 23. Morishige T., Yoshioka Y., Inakura H., Tanabe A., Yao X., Tsunoda S., Tsutsumi Y., Mukai Y., Okada N., and Nakagawa S. (2010) Creation of a lysine-deficient LIGHT mutant with the capacity for site-specific PEGylation and low affinity for a decoy receptor. Biochem. Biophys. Res. Commun. 393, 888–893 [DOI] [PubMed] [Google Scholar]
- 24. Yamamoto Y., Tsutsumi Y., Yoshioka Y., Nishibata T., Kobayashi K., Okamoto T., Mukai Y., Shimizu T., Nakagawa S., Nagata S., and Mayumi T. (2003) Site-specific PEGylation of a lysine-deficient TNF-α with full bioactivity. Nat. Biotechnol. 21, 546–552 [DOI] [PubMed] [Google Scholar]
- 25. Yoshioka Y., Tsutsumi Y., Ikemizu S., Yamamoto Y., Shibata H., Nishibata T., Mukai Y., Okamoto T., Taniai M., Kawamura M., Abe Y., Nakagawa S., Nagata S., Yamagata Y., and Mayumi T. (2004) Optimal site-specific PEGylation of mutant TNF-α improves its antitumor potency. Biochem. Biophys. Res. Commun. 315, 808–814 [DOI] [PubMed] [Google Scholar]
- 26. Rouached H., Berthomieu P., El Kassis E., Cathala N., Catherinot V., Labesse G., Davidian J. C., and Fourcroy P. (2005) Structural and functional analysis of the C-terminal STAS (sulfate transporter and anti-σ antagonist) domain of the Arabidopsis thaliana sulfate transporter SULTR1.2. J. Biol. Chem. 280, 15976–15983 [DOI] [PubMed] [Google Scholar]
- 27. Abe Y., Yoshikawa T., Kamada H., Shibata H., Nomura T., Minowa K., Kayamuro H., Katayama K., Miyoshi H., Mukai Y., Yoshioka Y., Nakagawa S., Tsunoda S., and Tsutsumi Y. (2008) Simple and highly sensitive assay system for TNFR2-mediated soluble- and transmembrane-TNF activity. J. Immunol. Methods 335, 71–78 [DOI] [PubMed] [Google Scholar]
- 28. Schütze S., Tchikov V., and Schneider-Brachert W. (2008) Regulation of TNFR1 and CD95 signalling by receptor compartmentalization. Nat. Rev. Mol. Cell Biol. 9, 655–662 [DOI] [PubMed] [Google Scholar]
- 29. Kamada H., Tsutsumi Y., Tsunoda S., Kihira T., Kaneda Y., Yamamoto Y., Nakagawa S., Horisawa Y., and Mayumi T. (1999) Molecular design of conjugated tumor necrosis factor-α: synthesis and characteristics of polyvinyl pyrrolidone modified tumor necrosis factor-α. Biochem. Biophys. Res. Commun. 257, 448–453 [DOI] [PubMed] [Google Scholar]
- 30. Boschert V., Krippner-Heidenreich A., Branschädel M., Tepperink J., Aird A., and Scheurich P. (2010) Single chain TNF derivatives with individually mutated receptor binding sites reveal differential stoichiometry of ligand receptor complex formation for TNFR1 and TNFR2. Cell. Signal. 22, 1088–1096 [DOI] [PubMed] [Google Scholar]
- 31. Krippner-Heidenreich A., Grunwald I., Zimmermann G., Kühnle M., Gerspach J., Sterns T., Shnyder S. D., Gill J. H., Männel D. N., Pfizenmaier K., and Scheurich P. (2008) Single-chain TNF, a TNF derivative with enhanced stability and antitumoral activity. J. Immunol. 180, 8176–8183 [DOI] [PubMed] [Google Scholar]
- 32. Ban L., Kuhtreiber W., Butterworth J., Okubo Y., Vanamee É. S., and Faustman D. L. (2015) Strategic internal covalent cross-linking of TNF produces a stable TNF trimer with improved TNFR2 signaling. Mol. Cell. Ther. 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shibata H., Yoshioka Y., Ikemizu S., Kobayashi K., Yamamoto Y., Mukai Y., Okamoto T., Taniai M., Kawamura M., Abe Y., Nakagawa S., Hayakawa T., Nagata S., Yamagata Y., Mayumi T., et al. (2004) Functionalization of tumor necrosis factor-α using phage display technique and PEGylation improves its antitumor therapeutic window. Clin. Cancer Res. 10, 8293–8300 [DOI] [PubMed] [Google Scholar]
- 34. Talpaz M., O'Brien S., Rose E., Gupta S., Shan J., Cortes J., Giles F. J., Faderl S., and Kantarjian H. M. (2001) Phase 1 study of polyethylene glycol formulation of interferon α-2B (Schering 54031) in Philadelphia chromosome-positive chronic myelogenous leukemia. Blood 98, 1708–1713 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.