Abstract
The autoimmune regulator (AIRE) protein is the key factor in thymic negative selection of autoreactive T cells by promoting the ectopic expression of tissue-specific genes in the thymic medullary epithelium. Mutations in AIRE cause a monogenic autoimmune disease called autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. AIRE has been shown to promote DNA breaks via its interaction with topoisomerase 2 (TOP2). In this study, we investigated topoisomerase-induced DNA breaks and chromatin structural alterations in conjunction with AIRE-dependent gene expression. Using RNA sequencing, we found that inhibition of TOP2 religation activity by etoposide in AIRE-expressing cells had a synergistic effect on genes with low expression levels. AIRE-mediated transcription was not only enhanced by TOP2 inhibition but also by the TOP1 inhibitor camptothecin. The transcriptional activation was associated with structural rearrangements in chromatin, notably the accumulation of γH2AX and the exchange of histone H1 with HMGB1 at AIRE target gene promoters. In addition, we found the transcriptional up-regulation to co-occur with the chromatin structural changes within the genomic cluster of carcinoembryonic antigen-like cellular adhesion molecule genes. Overall, our results suggest that the presence of AIRE can trigger molecular events leading to an altered chromatin landscape and the enhanced transcription of low-expressed genes.
Keywords: autoimmune disease, chromatin structure, DNA sequencing, DNA topoisomerase, immunology, RNA splicing, tolerance, transcription
Introduction
Immunological tolerance protects against harmful immune responses to endogenous body constituents (1). The initial stages of T cell tolerance occur in the thymus, where thymic stromal cells present self-peptides to thymocytes (2). The repertoire of self-peptides and the potential to detect and eliminate autoreactive thymocytes is critically dependent on the autoimmune regulator (AIRE)3 protein. In thymic medullary epithelial cells, AIRE enhances the expression of genes encoding the tissue-specific antigens (TSAs) that are restricted to peripheral tissues (3). In humans, mutations in the AIRE gene cause an autosomal recessive disease known as autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED), which is associated with destructive autoimmune reactions against multiple (primarily endocrine) organs (4, 5). The monogenic nature of the disease has rendered AIRE deficiency an important model for studying the mechanisms of autoimmunity and central tolerance. The mechanism by which AIRE targets and regulates the transcription of a broad repertoire of TSA genes remains largely unknown. AIRE enhances transcription and interacts with multiple proteins implicated in the modification of chromatin structure, activation of transcription, and mRNA splicing (6, 7). At the epigenetic level, it acts via binding to hypomethylated H3K4 residues (8, 9), although a later study demonstrated a broader AIRE protein presence on genomic sites, linking RNA polymerase II enrichment at transcriptional start sites with AIRE binding (10).
According to the proteomic screens, a top-ranked AIRE interaction partner is topoisomerase IIα (TOP2A), which, together with its paralog TOP2B, plays a significant role in relaxing torsional tensions of the DNA double helix (6). Eukaryotic TOP proteins are large homodimeric enzymes that generate transient DNA breaks that result in the removal of positive and negative DNA supercoils (11). The TOP2 proteins can be blocked at different functional stages by various small-molecule inhibitors such as etoposide and merbarone (12). Studies in various model systems have demonstrated a role for TOP2 in regulating transcription (13–15), and Abramson et al. (6) linked its catalytic activity to the changes in the expression of AIRE target genes.
In this study, we applied the RNA-seq approach to systematically analyze the effect of etoposide, which specifically blocks the religation activity of the TOP2 enzymes, in the doxycycline-inducible AIRE-Tet cell line (16). We found that low-dose treatment of AIRE-expressing cells with etoposide results in a robust and synergistic up-regulation of AIRE target genes. Furthermore, we observed the up-regulation of an additional large set of genes with low expression levels, the characteristic feature of genes activated by AIRE. Etoposide treatment did not enhance AIRE-dependent gene expression after TOP2A depletion by RNAi, nor could it enhance transcription when the AIRE CARD domain contained a patient mutation. Furthermore, the CARD domain mutation disrupted the protein interaction between AIRE and TOP2A, as assayed by co-IP. Additionally, we observed AIRE-related structural rearrangements in the chromatin structure, such as the enrichment of γH2AX and HMGB1, and the reduction of histone H1 at AIRE target gene promoters. In light of these findings, we performed FAIRE-seq to assess AIRE-related genome-wide changes of chromatin accessibility and found that a large fraction of differentially enriched FAIRE peaks overlap with CTCF sites. Finally, we tested whether this overlap implies that the expression of AIRE could entail changes in CTCF-mediated chromatin interactions using the 3C method, and we were able to determine altered chromatin looping in the CEACAM gene cluster.
Results
The TOP2 inhibitor etoposide enhances the expression of AIRE target genes
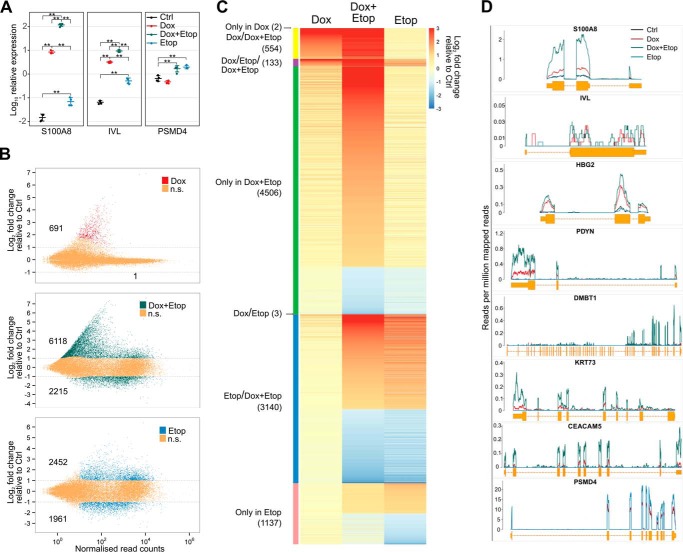
In an earlier study, etoposide, a drug that specifically stabilizes the TOP2-DNA complex and promotes DNA breaks, was demonstrated to induce the expression of AIRE target genes in HEK293 cells. However, the study did not find an additive or synergistic effect of AIRE and etoposide (6). In contrast to the previous study, we detected a strong synergistic increase in target gene S100A8 and IVL transcription with low-dose etoposide (2 μm) treatment of doxycycline-induced, AIRE-positive AIRE-Tet cells after 2 h of treatment (supplemental Fig. 1A). After 24 h of 2 μm etoposide treatment, this up-regulation reached an even higher increase in expression: over 8000-fold and 150-fold for S100A8 and IVL genes, respectively (Fig. 1A). Treatment with etoposide did not cause apoptosis in AIRE-Tet cells (supplemental Fig. 1B), and the low concentration of etoposide (2 μm) induced the largest change in expression, as we could not establish further increase with higher etoposide concentrations (10 and 50 μm) (supplemental Fig. 1A). We noticed, in agreement with Abramson et al. (6), that etoposide alone (Etop) was able to increase, albeit to a lesser extent (4-, 7-, and 3-fold, respectively), the expression of AIRE-dependent S100A8 and IVL and AIRE-independent PSMD4 genes (Fig. 1A). These results suggest a strong synergistic effect of AIRE and low-dose etoposide on target gene expression, which is much higher than etoposide alone. We performed subsequent experiments by treating cells with 2 μm etoposide for 24 h, i.e. under conditions where we observed the strongest synergistic effect.
Figure 1.
Etoposide treatment expands the repertoire of AIRE target genes and enhances their expression in AIRE-Tet cells. A, expression of the AIRE target genes S100A8 and IVL and the AIRE-independent gene PSMD4 after AIRE induction (Dox), etoposide treatment (Etop), or their combination (Dox+Etop) relative to the untreated control sample (Ctrl) in the AIRE-Tet cell line. Log10-transformed data points together with their mean ± S.D. are from four independent experiments. Statistical significance was assessed by two-sample t test (**, p < 0.01). B, assessment of genome-wide gene expression changes with RNA-seq after Dox (top), Etop (bottom), or Dox+Etop treatment (center) in the AIRE-Tet cell line relative to Ctrl. The colors and numbers depict DE genes with an adjusted p < 0.1 and absolute log2 -fold change >1 (gray lines). n.s., not significant. C, heatmap representation of log2 -fold changes of DE genes across all conditions relative to Ctrl. The text and color bar on the left show the division of genes between one or multiple samples (indicated by slashes). D, expression of select AIRE-dependent genes (S100A8, IVL, HBG2, PDYN, DMBT1, KRT73, and CEACAM5) and the AIRE-independent PSMD4 gene based on read counts from the RNA-seq experiment in AIRE-Tet cells after Dox, Etop, and Dox+Etop treatment or in Ctrl.
Genes that are synergistically activated by AIRE and the TOP2 inhibitor have low expression levels
The observation that AIRE induction and etoposide strongly enhanced the transcription of select genes posed the question of whether immobilizing TOP2 enzymes in the DNA-cleavable complex could induce AIRE-dependent gene transcription on a genome-wide scale. To assess the impact of etoposide treatment on the global level of gene expression, we performed RNA-seq with AIRE-Tet cells. Consistent with earlier results (16, 17), doxycycline (Dox)-induced AIRE was responsible for the up-regulation of otherwise low or moderately expressed genes in AIRE-Tet cells (Fig. 1B, top panel). AIRE up-regulated 691 of 692 differentially expressed (DE) genes and thus acted almost exclusively as an activator. Strikingly, AIRE activation together with 2 μm etoposide led to substantial genome-wide transcriptional activation, as altogether 6118 genes were activated in the Dox+Etop-treated cells (Fig. 1, B, center panel, and C). AIRE together with etoposide still predominantly acted as an activator, as 73% of DE genes were up-regulated and 27% of genes (2215) were down-regulated in Dox+Etop cells. The finding that AIRE had a mostly up-regulating effect is in agreement with previous RNA-seq with sorted primary thymic epithelial cells from Aire-deficient and wild-type mice (18, 19). We found that a large proportion of the genes activated by AIRE and etoposide were also normally expressed at low levels (81% of the 4506 genes in the Dox+Etop only group, Fig. 1C). In contrast, etoposide treatment alone influenced the expression of large number of genes with various levels of expression, and the numbers of up- and down-regulated genes were approximately similar (56% and 44% of the 4413 genes, respectively) (Fig. 1B, bottom panel).
As expected, AIRE primarily activated protein-coding genes (93%), in agreement with its role to activate self-antigen proteins to be presented by thymic epithelial cells, whereas etoposide treatment increased the fraction of pseudogenes and RNA genes (supplemental Fig. 2A). We did not identify non-canonical transcription start sites among AIRE target genes (data not shown), in contrast to an earlier report (20). In agreement with our initial qPCR results, the RNA-seq data analysis confirmed that the S100A8 and IVL genes were up-regulated under Dox and Dox+Etop conditions (Fig. 1D). In subsequent experiments, we included an additional subset of AIRE-activated genes (HBG2, PDYN, DMBT1, KRT73, and CEACAM5) for analysis. The AIRE-mediated expression of these genes was enhanced by etoposide, as demonstrated by the RNA-seq experiment (Fig. 1D) and validated by qPCR (data not shown).
AIRE with etoposide enhanced the activation of tissue-specific genes and gene clusters
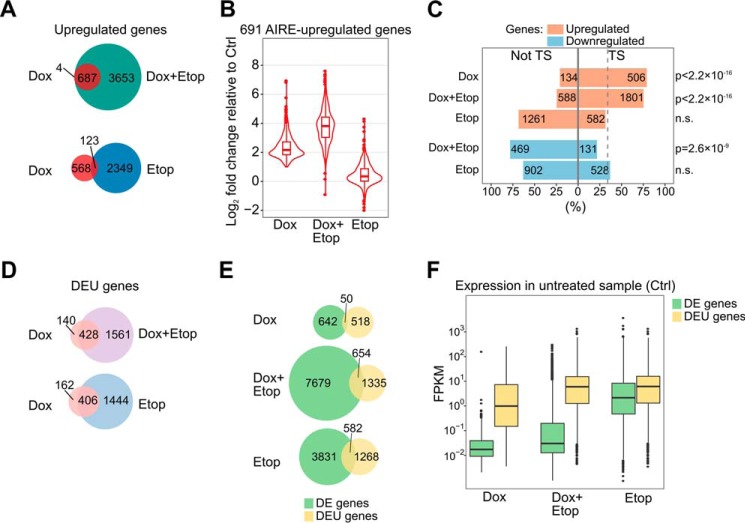
In further analyses of AIRE-regulated genes, we found that 99.4% of genes up-regulated by Dox were also up-regulated in Dox+Etop samples (687 of 691) (Fig. 2A). This almost complete inclusion of Dox up-regulated genes within the Dox+Etop group was remarkable, in contrast to the overlap between the Dox and Etop gene lists that shared 123 genes (Fig. 2A). Thus, although AIRE and etoposide independently regulate distinct subsets of genes, the addition of etoposide to AIRE-expressing cells strongly enhances AIRE transcriptional activity. The genes up-regulated by AIRE alone showed a further, ∼4-fold increase in the presence of AIRE and etoposide (Fig. 2B, Dox+Etop). In contrast, overall there was no increase in the expression of AIRE target genes after treatment with etoposide only (Fig. 2B), although few AIRE target genes (including S100A8 and IVL) showed an elevation in expression, as seen in Fig. 1A.
Figure 2.
Features of genes affected by the induction of AIRE, treatment with etoposide, or the AIRE/etoposide combination in AIRE-Tet cells. A, the number of unique and overlapping up-regulated genes in Dox-, Etop-, and Dox+Etop-treated AIRE-Tet cells. The proportion of AIRE-up-regulated genes overlapping up-regulated genes in Dox+Etop (687 of 691) was significantly greater than the overlap with up-regulated genes in Etop (123 of 691) (p < 2.2 × 10−16, χ2 test). B, the distribution of the effect sizes (log2 -fold changes) of the 691 AIRE-up-regulated genes after Dox, Dox+Etop, or Etop treatment. C, percentage and number of tissue-specific (TS) and broadly expressed (not TS) genes among the DE genes in each condition. The dashed line marks the percentage of tissue-specific genes in the genome (34%), which was compared with the proportions of tissue-specific genes in each condition (χ2 test). D, the number of genes featuring DEU in Dox AIRE-Tet cells that are unique or in common with DEU genes in Dox+Etop and Etop cells. The proportions of overlapping Dox genes (428 of 568 with Dox+Etop and 406 of 568 with Etop) were not significantly different (p = 0.16, χ2 test). E, the overlap between DE and DEU genes under Dox, Dox+Etop, and Etop conditions. The proportion of DEU genes overlapping DE genes in Dox (50 of 568) was significantly smaller than the overlap between DE and DEU genes in both Dox+Etop (654 of 1989) and Etop (582 of 1850) (both comparisons resulted in p < 2.2 × 10−16, χ2 test). F, comparison of the basal expression of DE and DEU genes by using the expression levels measured in Ctrl AIRE-Tet cells. The x axis shows the experimental conditions under which the genes were detected as differentially expressed or spliced. The y axis shows the normalized expression level in fragments per kilobase of exon per million mapped reads (FPKM).
Consistent with previous studies, most of the AIRE-induced genes were associated with tissue-specific expression patterns (79%), and this phenomenon was largely preserved after etoposide co-treatment (75%, Fig. 2C). However, only 32% of etoposide alone-activated genes were associated with a tissue-specific expression pattern (Fig. 2C), which is comparable with the overall proportion of tissue-specific genes (34%) determined by the Human Protein Atlas Project. A similarly low percentage of tissue-specific genes was observed in the down-regulated gene groups (22% and 37% in Dox+Etop and Etop, respectively) (Fig. 2C).
The genes activated by AIRE in Dox and Dox+Etop cells were enriched for known APECED autoantigens (6 of 692 and 25 of 8333 genes, respectively, of 31 autoantigens reported in Ref. 4) compared with four autoantigens among the 4413 DE genes under Etop conditions (p = 4 × 10−5 and 0.03, respectively). We observed highly significant chromosomal clustering among 21% of AIRE-regulated genes, with an average of 4.5 genes/cluster. In comparison, 27% of DE genes that were only in Dox+Etop cells were in clusters (on average 4.2 genes/cluster). Only 14.2% of DE genes in Etop cells were in clusters, which was significantly less than in Dox or Dox+Etop (p = 5.3 × 10−6 and p < 2.2 × 10−16, respectively), and these clusters tended to be smaller in gene numbers (on average 3.5 genes/cluster).
AIRE regulates alternative splicing in a different subset of genes
AIRE interacts with the proteins involved in splicing (6, 7), and recent studies demonstrated AIRE-dependent differential inclusion of exons in transcripts whose overall levels were only slightly affected or unaffected by AIRE (18, 21). In our RNA-seq data, we found that the number of genes featuring differential exon usage (DEU) following AIRE induction was in approximately the same range (568 genes) with DE genes. Etoposide treatment increased the number of DEU genes both in Dox+Etop and Etop cells, but the increase was very similar (1989 and 1850 DEU genes, respectively) (Fig. 2D). Furthermore, a large fraction of the DEU genes found in Dox cells overlapped with DEU genes both in Dox+Etop and Etop cells (428 of 568 and 406 of 568, respectively) (Fig. 2D), which suggests that the DEU genes are not synergistically affected by AIRE and etoposide. Additionally, there was very little overlap between the DE and DEU genes in Dox samples (Fig. 2E). The overlap was larger in Dox+Etop cells (Fig. 2E), but this was mostly due to a greater number of DEU genes the Dox+Etop cells share with Etop cells. In general, the alternatively spliced genes tended to be highly expressed (Fig. 2F) and were less likely to represent tissue-specific genes compared with AIRE target genes (supplemental Fig. 2B). However, it should be noted that the initial low expression of AIRE target genes might hinder the analysis of alternative splicing.
Gene activation requires the interaction of TOP2A with intact AIRE
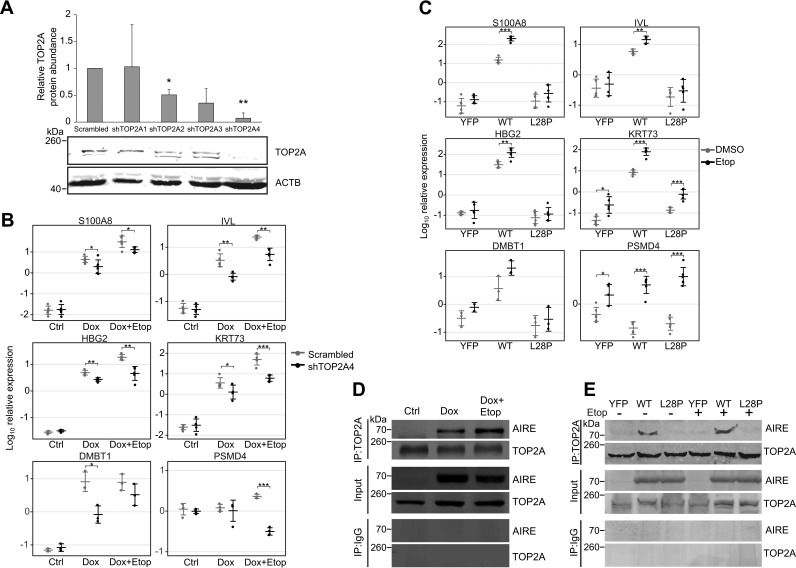
To confirm that TOP2A is directly involved in enhancement of AIRE target gene activation, we used shRNA to knock down TOP2A expression in AIRE-Tet cells (Fig. 3A). The effect of gene transcription was significantly reduced in cells transfected with a TOP2A-specific shRNA (shTOP2A4) compared with the scrambled control shRNA (Scrambled) that retained the ability to enhance AIRE target gene expression (Fig. 3B).
Figure 3.
Knockdown of TOP2A reduces the activation of AIRE target genes, and AIRE CARD domain mutation abolishes the AIRE-TOP2A interaction. A, top panel, the effect of four TOP2A-specific shRNAs (shTOP2A1–4) on the expression of TOP2A. The TOP2A protein signal was normalized to the loading control protein β-actin (ACTB) and set relative to the negative control shRNA (Scrambled) treatment. Data show the mean ± S.D. of three independent experiments. Statistical significance was assessed by one-sample t test comparing the values to the reference value of 1 (*, p < 0.05; **, p < 0.01). Bottom panel, a representative Western blot of the TOP2A knockdown experiment using anti-TOPa and anti-β-actin antibodies as described under “Experimental Procedures.” B, expression analysis of AIRE-dependent genes (S100A8, IVL, HBG2, KRT73, and DMBT1) and an AIRE-independent PSMD4 gene in AIRE-Tet cells using qPCR after shRNA-mediated knockdown of TOP2A. Log10-transformed data points together with their mean ± S.D. are from three to six independent experiments. Statistical significance was assessed by two-sample t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001). C, expression analysis of AIRE-dependent genes (S100A8, IVL, HBG2, KRT73, and DMBT1) and an AIRE-independent gene (PSMD4) in HEK293 cells stably expressing YFP, WT AIRE, or mutant AIRE (L28P). Log10-transformed data points together with their mean ± S.D. are from three to six independent experiments. Statistical significance was assessed by two-sample t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001). D, analysis of the AIRE and TOP2A interaction in Ctrl, Dox, and Dox+Etop AIRE-Tet cells by co-IP. E, co-IP analysis of the AIRE and TOP2A interaction in HEK293 cells stably expressing YFP, WT AIRE, or mutant AIRE (L28P) with or without etoposide treatment. In both co-IP experiments, anti-TOP2A antibody was used for immunoprecipitation, and anti-TOP2A and anti-AIRE antibodies were subsequently used in the Western blot. Input was used as a protein loading control, and immunoprecipitation with anti-IgG antibody was used to determine the background binding of AIRE.
We also wanted to demonstrate that functional AIRE is required for the enhancement of gene activation by etoposide. For this, we investigated the effect of etoposide on the transcription-enhancing activity of AIRE carrying the L28P APECED mutation in the CARD domain and compared it with the intact AIRE protein. We used stably transfected HEK293 cells constitutively expressing wild-type AIRE, the AIRE-L28P mutant protein, or YFP as a negative control. In a previous study, we demonstrated that, although AIRE-L28P localizes to the nucleus, the mutated protein lacks the speckled pattern observed with wild-type AIRE and is unable to activate AIRE target genes (22). Following etoposide treatment of AIRE-L28P cells, we did not observe changes in expression of most of the genes studied, whereas cells expressing wild-type AIRE reacted to etoposide treatment by enhancing the transcription of AIRE target genes similarly to the doxycycline-induced AIRE in AIRE-Tet cells (Fig. 3C). Although the intact AIRE protein in doxycycline-induced AIRE-Tet or in stably transfected HEK293 cells was able to interact with TOP2A with or without etoposide addition (Fig. 3, D, second and third lanes, and E, second and fifth lanes), the mutant AIRE-L28P was unable to interact with TOP2A (Fig. 3E, third and sixth lanes).
AIRE-mediated gene activation utilizes the DNA cleavage activity of both type 1 and 2 topoisomerases
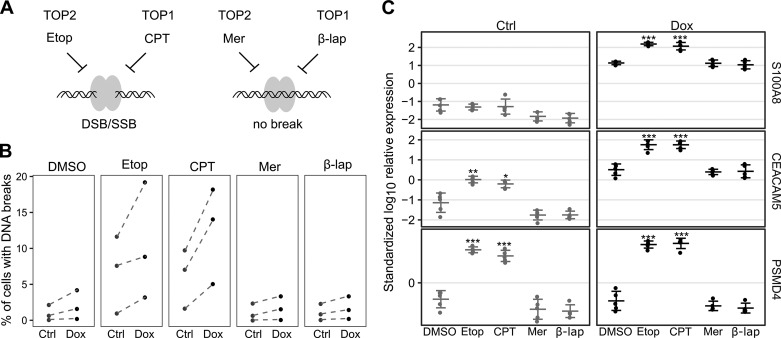
Our results prompted us to confirm that 2 μm etoposide is sufficient to significantly affect the chromosomal DNA integrity as a determinant of AIRE-mediated transcriptional activity. For this, we treated AIRE-Tet cells with doxycycline and etoposide and analyzed the Br-dUTP-labeled DNA breaks by flow cytometry. Additionally, we treated the cells with 1 μm camptothecin, a topoisomerase I (TOP1)-specific inhibitor that, similarly to etoposide, freezes the TOP-DNA cleavable complex; 1 μm merbarone, a TOP2 inhibitor that blocks enzymatic activity before the DNA cleavage reaction; and 1 μm β-lapachone, a TOP1 inhibitor that blocks enzymatic activity before the DNA cleavage reaction (Fig. 4A). In this way, two of the inhibitors studied, etoposide and camptothecin, cause TOP2- and TOP1-related DNA breaks, respectively, and two other inhibitors, merbarone and β-lapachone, inhibit TOP2 and TOP1, respectively, without causing DNA breaks. We observed an increase, although not significant, in the percentage of cells containing DNA breaks in doxycycline-induced cells after treatment with etoposide and camptothecin (Fig. 4B). As expected, and in agreement with the inhibitory mechanism of topoisomerases before DNA cleavage, neither merbarone nor β-lapachone had an effect on the occurrence of DNA lesions (Fig. 4B). Based on these findings, we tested the effect of TOP1 inhibitors on AIRE-dependent gene expression and found that camptothecin treatment essentially mimicked that of etoposide by strongly and significantly up-regulating AIRE target genes (Fig. 4C), suggesting that both types of topoisomerases can influence AIRE-related transcriptional processes via DNA breaks. Although the role of TOP1 with respect to the function of AIRE remains to be elucidated, TOP1 is known to support the expression of genes with low transcription levels (23).
Figure 4.
AIRE-dependent gene expression relies on the DNA cleavage activity of both TOP2 and TOP1. A, schematic of the mechanisms of TOP1/2 inhibition by Etop, camptothecin (CPT), merbarone (Mer), and β-lapachone (β-lap). The gray ellipses and the helix represent the topoisomerase homodimer and the DNA molecule, respectively. SSB, single-stranded break; DSB, double-stranded break. B, percentage of uninduced (Ctrl) and induced (Dox) AIRE-Tet cells with DNA breaks after mock treatment with DMSO, TOP2-specific (Etop and Mer), and TOP1-specific inhibitors (CPT and β-lap) using Br-dUTP-labeled nicked DNA measured by FACS. The data are derived from three independent experiments. Dashed lines connect experimental groups from the same experiment. C, expression of the AIRE target genes S100A8 and CEACAM5 and the AIRE-independent gene PSMD4 after DMSO, Etop, CPT, Mer, and β-lap treatment in Ctrl and Dox AIRE-Tet cells. Log10-transformed data points together with their mean ± S.D. are from five independent experiments. Statistical significance was assessed by two-sample t test comparing the inhibitor treatments to the mock treatment under Ctrl and Dox conditions separately (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Induction of AIRE causes structural changes in chromatin at AIRE target gene promoters
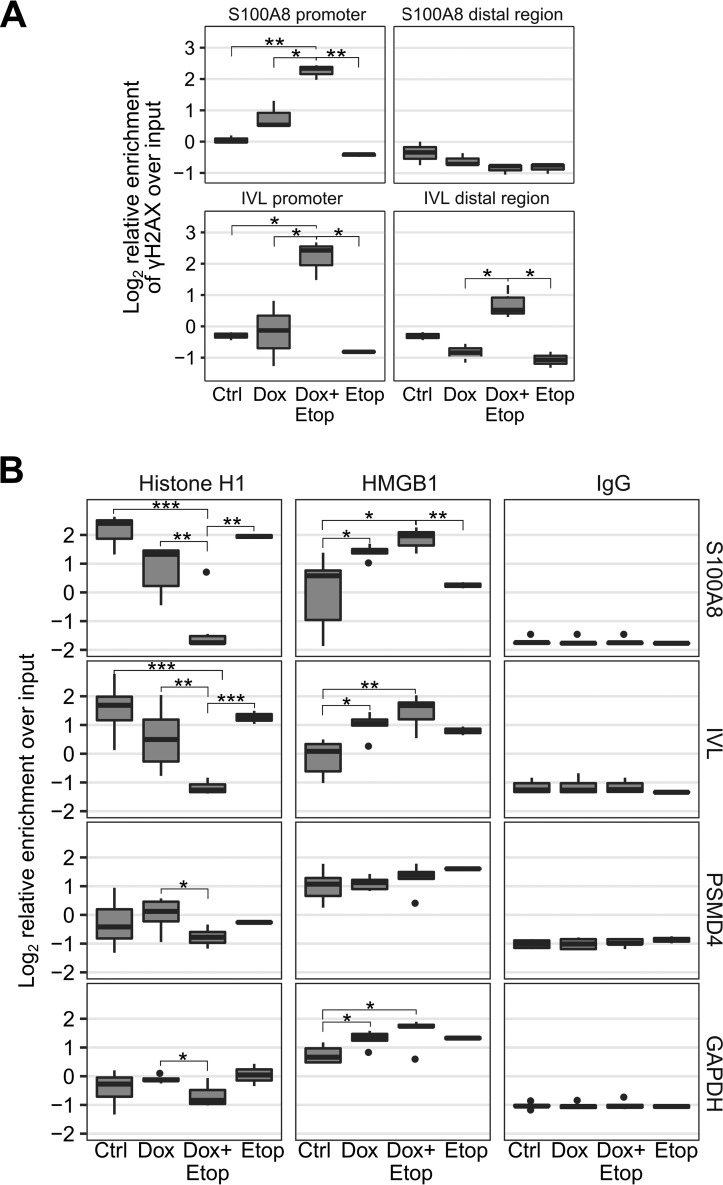
Consistent with the increased proportion of cells with DNA breaks, we observed a significant enrichment of γH2AX, a known marker of DNA double-stranded breaks (24), at the promoter regions of the AIRE-dependent S100A8 and IVL genes in Dox+Etop cells compared with Ctrl and Etop cells (Fig. 5A). The γH2AX signal was weaker in control regions located ∼10 kb away from IVL and S100A8 promoters (Fig. 5A).
Figure 5.
Inhibition of TOP2 with etoposide increases the occurrence of double-stranded DNA breaks and facilitates H1/HMGB1 exchange in AIRE target gene promoters. A, assessment of DNA breaks as measured by the enrichment of the Ser139-phosphorylated histone H2AX (γH2AX) in AIRE-dependent S100A8 and IVL gene promoters and distal control regions in Ctrl, Dox, Dox+Etop, and Etop AIRE-Tet cells using qPCR. The box and whiskers plots show the median and interquartile range of log2-transformed data from three independent experiments. Whiskers cover data points within a 1.5 × interquartile range. Statistical significance was assessed by two-sample t test (*, p < 0.05; **, p < 0.01). B, qPCR analysis of H1 and HMGB1 enrichment at AIRE target gene (S100A8, IVL, and DMBT1) and AIRE-independent gene (PSMD4 and GAPDH) promoters in Ctrl, Dox, Dox+Etop, and Etop AIRE-Tet cells. IgG was used as a negative control for ChIP experiments. The box and whiskers plots show the median and interquartile range of log2-transformed data from five independent experiments. Whiskers cover data points within a 1.5 × interquartile range. Statistical significance was assessed by two-sample t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
The TOP2A-induced DNA breaks generally attract and activate the poly(ADP-ribose) polymerase 1 (PARP1) enzyme (13). Activated PARP1 can use histone H1 as a substrate for poly(ADP-ribosyl)ation, thereby excluding it from a subset of PARP1-containing promoters (25). Histone H1 is widely viewed as a transcriptional repressor (26), and removal of histone H1 or exchange of H1 with the high-mobility group protein B1 (HMGB1) facilitates gene expression (27, 28). We investigated whether the transcriptional activation observed with AIRE, especially in the presence of etoposide, is accompanied by changes in the abundance of histone H1 and HMGB1 by performing ChIP analyses using histone H1 and HMGB1-specific antibodies at S100A8 and IVL promoters. Our results indicated that, in the presence of AIRE, the linker histone H1 signal was significantly reduced at AIRE target gene promoters compared with Ctrl, and it further declined after treatment with etoposide (Dox+Etop) (Fig. 5B). Additionally, we found that more HMGB1 was recruited to AIRE target gene promoters in Dox and Dox+Etop cells (Fig. 5B). In contrast, the HMGB1 signal at the promoters of AIRE-independent PSMD4 and GAPDH genes was consistently higher than histone H1 under all experimental conditions, in agreement with their high expression in AIRE-Tet cells (Fig. 5B).
AIRE induces changes in CTCF-mediated chromatin looping in the CEACAM gene cluster
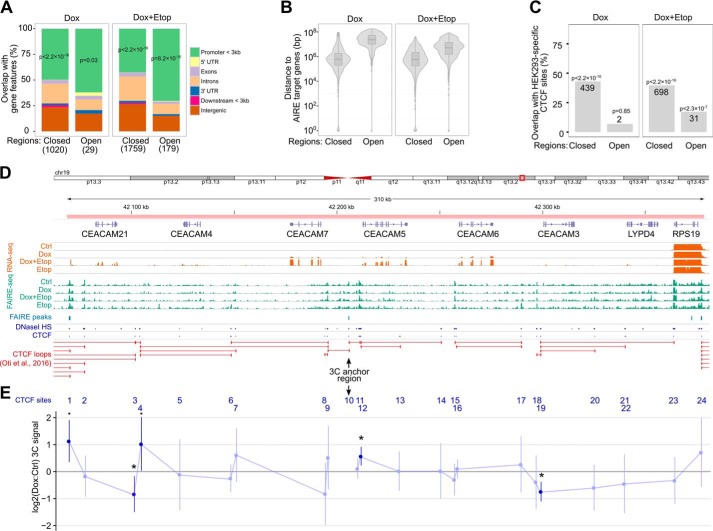
Topoisomerase and RNA polymerase activities remodel DNA supercoiling, influencing the folding of large-scale chromatin structures and creating domains that are flanked by CTCF insulator protein binding sites (28). To address the genome-wide changes in chromatin structure induced by AIRE and etoposide, we adopted the FAIRE-seq approach, which allowed us to analyze chromatin accessibility in AIRE-Tet cells and compare this in AIRE-induced and etoposide-treated cells. We found changes in ∼1000 and 1900 genomic regions in Dox and Dox+Etop cells, respectively, whereas, in Etop cells, we could not detect any FAIRE peaks that were differentially accessible compared with Ctrl. Interestingly, >90% of the FAIRE peaks in Dox- and Dox+Etop-treated cells became less accessible, and 42–70% of the detected changes occurred in gene promoters, as assessed by the overlapping of FAIRE peaks with genic and intergenic features (Fig. 6A). However, very few of the differentially accessible regions in Dox and Dox+Etop cells were in the vicinity of AIRE-activated genes (Fig. 6B). A significant fraction of the changed regions in Dox and Dox+Etop cells overlapped with HEK293-specific chromatin architectural protein CTCF binding sites (Fig. 6C).
Figure 6.
FAIRE-seq and 3C reveal AIRE-dependent changes in chromatin structure. A, percentages of FAIRE peaks that overlap with genic and intergenic regions in Dox and Dox+Etop AIRE-Tet cells relative to Ctrl. The x axis labels refer to the FAIRE peaks that show either loss or gain of chromatin accessibility (Closed and Open, respectively). The numbers of the peaks are shown in brackets. The overlaps between the peaks and promoters up to 3 kb from the transcription start site were assessed with the relative distance test implemented in the GenometriCorr R package. B, distance between DE genes and the closest FAIRE peaks in Dox and Dox+Etop AIRE-Tet cells. The statistical analysis did not determine a significant overlap between the genomic regions. C, differentially enriched FAIRE peaks show a strong enrichment of CTCF sites in AIRE-Tet cells both with Dox and Dox+Etop treatment. The numbers of FAIRE peaks in the overlap are shown on the columns. The statistical significance of the overlaps between the FAIRE-seq peaks and HEK293-specific CTCF sites was assessed with the relative distance test implemented in the GenometriCorr R package. D, annotated overview of the genomic region that comprises the CEACAM cluster at chr19:42068524-42378450. The tracks below the gene annotations show the AIRE-Tet-specific gene expression (RNA-seq), chromatin accessibility (FAIRE-seq), differentially enriched FAIRE peaks, HEK293-specific DNase sequencing, and CTCF ChIP sequencing peaks (ENCODE) and computationally predicted CTCF loops (32). E, the chromatin interactions of the 24 CTCF sites in the CEACAM gene cluster depicted in D were analyzed by 3C. The interaction frequencies were quantified by qPCR and are shown as log2 -fold changes between the signals from Dox and Ctrl AIRE-Tet cells. CTCF site 10 was used as an anchor region in the qPCR assays. The data show the mean ± S.D. from five independent experiments. Statistical significance was assessed by one-sample t test comparing the log2 -fold changes with the reference value of 0 (■, p < 0.1; *, p < 0.05).
As the induction of AIRE changed chromatin accessibility in CTCF binding sites, we next focused on a 310-kb genomic region on chr19 containing CEACAM genes to investigate the potential impact of AIRE on CTCF binding and CTCF-mediated chromatin interactions (Fig. 6D). The genomic region contains CEACAM3, CEACAM4, CEACAM5, CEACAM6, CEACAM7, and CEACAM21 genes, which are co-expressed with AIRE in human medullary thymic epithelial cells (29, 30). The cluster structure together with RNA-seq, FAIRE-seq, and CTCF binding data is depicted in Fig. 6D. Our RNA-seq data showed CEACAM genes as AIRE-activated genes in AIRE-Tet cells, prompting the analysis of genomic changes in 24 CTCF binding sites within this locus by chromosome conformation capture (3C) assay (31). The 3C assay combines protein cross-linking and proximity ligation of DNA to detect long-range chromatin interactions between genomic loci. We set a FAIRE peak (site 10 between CEACAM5 and CEACAM7 genes, Fig. 6E) that overlapped with the CTCF binding site and became less accessible after AIRE induction as the anchor region for the 3C experiments. The chromatin interactions between the anchor region and the remaining 23 CTCF sites were analyzed by qPCR using primers that corresponded to Sau3AI-specific restriction fragments that either overlapped or flanked the CTCF sites. We compared the 3C signal from Dox cells with Ctrl AIRE-Tet cells and found that, in AIRE-expressing cells, the anchor CTCF site formed stronger chromatin contacts with sites 1 and 4 flanking CEACAM21 and with site 12 near CEACAM5. In addition, contacts with sites 3 and 19 close to the CEACAM21 and CEACAM3 genes, respectively, became significantly weaker in AIRE-expressing cells. This suggests that AIRE affects long-range restructuring of chromatin and determines the partitioning of the CEACAM cluster into chromatin loops.
Discussion
Topological and structural changes of chromatin play a critical role in the regulation of gene expression and in promoting the accessibility of genomic DNA. DNA topoisomerases, the key enzymes that affect chromatin architecture and influence transcription, control the topological state of genomic DNA (11). Among the various topoisomerases, TOP2A has been shown to interact with AIRE (6). Here we report an analysis of chromatin-related molecular events in the context of AIRE-dependent gene activation that involve the activities of the topoisomerases.
In this study, we describe the genome-wide effect of etoposide treatment in the doxycycline-inducible AIRE-Tet cell line. In contrast to a previous study (6), we found a strong synergistic effect on transcription when AIRE-expressing cells were treated with low-dose etoposide. We observed a remarkable increase in the number of genes expressed and in the fold difference of individual genes. The genes that were synergistically activated displayed features of AIRE target genes, including low expression, tissue-specific expression under physiological conditions, and chromosomal clustering (16, 18, 32–34). Subsequent analyses of the AIRE-expressing AIRE-Tet cells revealed another distinct set of genes that was prone to alternative splicing in AIRE-induced cells. Earlier studies with primary thymic cells have established that alternative exon usage affects central tolerance (35) and that AIRE knock-out mouse thymic epithelial cells have a different gene splicing pattern than wild-type cells (18, 21). However, although treatment of AIRE-expressing cells with etoposide increased the number of alternatively spliced genes, these genes did not display features of typical AIRE-regulated genes, and they largely overlapped with genes that became alternatively spliced in the presence of etoposide alone.
Transcription is associated with severe topological perturbations of DNA, and locally targeted TOP2-associated DNA breaks generate a permissive chromatin setting that facilitates gene transcription (11). The interaction between AIRE and TOP2A, which has been determined by co-immunoprecipitation in Ref. 6 and in our study, appears to be one key event in AIRE target gene expression, as TOP2A knockdown significantly reduced the expression of AIRE target genes. Furthermore, the APECED L28P mutation in the AIRE CARD domain abolished the multi-fold induction of AIRE target genes, which might be partially explained by the disruption of the AIRE-TOP2A interaction, although the direct proof for this remains to be shown. These results suggest that DNA breaks induced in the regions of AIRE target genes are needed to activate the transcription of these genes. As etoposide enhances the AIRE and TOP2A interaction and stabilizes TOP2A-DNA complexes, it is conceivable that an increased presence of DNA breaks in AIRE gene promoters would facilitate positive transcription elongation factor b (P-TEFb) recruitment to these loci and activate their transcription, as reported earlier (36, 37). In this respect, the recruitment of the bromodomain protein BRD4 to target genes was shown to connect AIRE and P-TEFb to release paused promoter-proximal RNA polymerase II and to induce AIRE-dependent transcripts (38). Interestingly, AIRE-dependent transcription appears to be equally well stimulated by low concentrations of camptothecin, which, analogously to etoposide, freezes the TOP1-DNA cleavable complex covalently to the chromatin. How TOP1 enhances AIRE-mediated gene regulation will have to be elucidated in subsequent experiments. Nevertheless, the idea that single-stranded DNA breaks may play a role at least in the early steps of AIRE-dependent gene expression in etoposide-treated cells would be feasible, given that the low concentration of etoposide predominantly results in single-stranded DNA lesions (39). Importantly, our results highlight the functional difference of the inhibitors (regardless of their targeting of TOP1 or TOP2) between those that generate DNA breaks and bind the protein covalently to DNA (etoposide and camptothecin) and compounds that inhibit TOP catalytic activity but do not generate the breaks (merbarone and β-lapachone). It should be noted that, in addition to TOP2, the TOP1 activity in association with transcription is well known (11), and a recent study demonstrated that the initiation of transcription requires RNA polymerase II-mediated activation of the TOP1 activity at transcription start sites by BRD4 (40). Thus, it is very likely that both TOP2 and TOP1 inhibitors that induce DNA breaks enhance AIRE target gene expression by similar mechanisms and transcriptional complexes.
Our ChIP experiments demonstrated that γH2AX, a marker of double-stranded DNA breaks (24, 41), binds the promoter regions of AIRE-activated genes. AIRE co-immunoprecipitates with γH2AX and DNA-PK, and DNA-PK was found to be essential for the recruitment of AIRE to gene promoters (42, 43). We detected the exchange of histone H1 with the HMGB1 protein at target gene promoters. The enrichment of HMGB1 is an indicator of a more relaxed chromatin environment and associated with actively transcribed genes, whereas histone H1 is crucial for the proper compaction of the chromatin structure (13, 28). This exchange requires the enzymatic activity of PARP1, which is part of the DNA repair complex, including the aforementioned DNA-PK, and is stimulated by the TOP2-generated DNA breaks. These interrelated associations were further strengthened by the observation that etoposide treatment of AIRE-expressing cells caused almost complete replacement of histone H1 with HMGB1. Altogether, these results suggest that the effect of etoposide is associated with DNA breaks at AIRE target genes. Although we were able to show the presence of relatively high levels of DNA breaks and break-associated proteins, it has been difficult to demonstrate a substantial increase of AIRE at the target genes. Then how do we explain the relatively low binding of AIRE at target genes? One explanation would be that AIRE interaction with chromatin could occur only at early stages of transcriptional activation or via other proteins such as histone H3, as shown earlier, making the detection of AIRE difficult by the ChIP method. Alternatively, the modest increase in AIRE at target genes versus the higher abundance of DNA break-associated proteins might be explained by the recent findings that TOP-induced breaks can be long-lasting, in contrast to AIRE interaction with chromatin. Earlier studies have identified genomic regions that are targeted by using TOP poisons such as etoposide and camptothecin, although their chromatin features are still not understood (12). In addition, the enrichment of γH2AX at activated genes after etoposide treatment in AIRE-expressing cells could be attributed to the action of the passing RNA polymerase II that denatures the topoisomerase-DNA cleavable complexes (44). For more in vivo settings, ChIP sequencing studies in cultured neuronal cells found increased γH2AX signals largely confined to actively transcribed genes but not to Polycomb-repressed regions or heterochromatin (45). Consistent with this hypothesis, more DNA breaks were present in MHCIIhi-expressing medullary thymic epithelial cells (mTECs) from AIRE-positive wild-type mice compared with AIRE knockout littermates (6). It remains to be studied whether DNA breaks are critical for the expression of a distinct set of thymic TSAs regulated by the transcription factor Fezf2 (46).
We tested the putative chromatin relaxation implied by the ChIP experiments and expression analyses by genome-wide FAIRE-seq approach to investigate chromatin accessibility. To our surprise, we detected relatively few changes within or near the genes whose expression was enhanced by AIRE. Instead, we found a large proportion of AIRE-related changes in chromatin structure occurring in intergenic regions at CTCF sites that are known as chromatin insulator regions, which set boundaries between nearby genomic regions. These intergenic locus-specific structural changes at CTCF sites are involved in the arrangement of chromosomal architecture and can affect interactions between promoter and enhancer sequences (47). We therefore analyzed the changes in CTCF-mediated chromatin interactions that could explain the enhanced transcription of AIRE-dependent genes. We found that, in AIRE-expressing cells, the select anchor region in the AIRE-regulated CEACAM gene cluster forms several chromatin loops over a large distance that are distinct from those in uninduced AIRE-Tet cells and that partially overlap with the CTCF loops that were predicted earlier by computational analysis (48). Thus, the long-distance interactions between CTCF sites within the gene clusters containing AIRE target genes may modulate their transcriptional permissiveness.
Experimental procedures
Inducible and stable AIRE cell lines
Doxycycline-inducible AIRE expression in the HEK293 cell line (AIRE-Tet) has been described previously (16). Cells were cultured in DMEM supplemented with 10% tetracycline-negative fetal calf serum, 100 units/ml penicillin/streptomycin, and 0.15 mg/ml G418 for at least 24 h before any additional treatments. Untreated cells served as a negative control material, and AIRE expression was induced with 1.5 μm doxycycline (631311, Clontech). After 24 h, the control and induced cells were either mock-treated with DMSO or with 2 μm etoposide (E1383, Sigma) for another 24 h. In some experiments, the cells were additionally incubated with 1 μm merbarone (M2070, Sigma), 1 μm camptothecin (C9911, Sigma), and 1 μm β-lapachone (L2307, Sigma) for 24 h. Stable HEK293 cells expressing WT AIRE, mutant AIRE (L28P), or YFP were cultured as described previously in Ref. 49.
RNA isolation and qPCR
Total RNA was isolated from AIRE-Tet cells at different time points during etoposide using TRIzol reagent according to the protocol of the manufacturer (Life Technologies). The yield and purity of RNA were determined by NanoDrop 1000. Subsequently, 5 μg of total RNA from each sample was used as a template for cDNA synthesis using the SuperScriptTM III First-Strand Synthesis kit according to the protocol of the manufacturer (Life Technologies). The gene expression levels of AIRE target genes were detected by quantitative real-time PCR (qPCR) using Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific) and the ViiA 7 real-time PCR system (Life Technologies). The expression of AIRE target genes was normalized to the housekeeping gene HPRT1 and analyzed using the comparative Ct method (50). For statistical analysis, the -fold change values were log-transformed, standardized, and scaled according to the procedure described in Ref. 51. Experimental groups were compared with a two-sample t test implemented in R. The primers are listed in supplemental Table 1.
RNA-seq sample preparation and data analysis
RNA was isolated with the miRNAeasy Mini Kit (Qiagen) using on-column DNase digestion from 106 AIRE-Tet cells induced with 1.5 μm doxycycline for 48 h, 2 μm etoposide for 24 h, or a combination of both. Uninduced DMSO-treated cells were used as a negative control. Two independent biological replicates from each treatment were prepared from 500 ng of RNA with RNA integrity number (RIN) > 9. Sequencing libraries were generated with the TruSeq Stranded Total RNA LT (with Ribo Zero Gold) Sample Prep Kit (Illumina) according to the instructions of the manufacturer, with the exception that 12 PCR cycles were used for the DNA enrichment step. Paired-end sequencing (2 × 100 bp) was performed with Illumina HiSeq 2000, which generated 80–100 × 106 sequenced fragments/sample. Prior to alignment, the reads were trimmed to remove adapter sequences and bases with a Phred score lower than 30 using Trim Galore!. Reads were mapped to the human reference genome GRCh37 (Ensembl release 75) using STAR aligner version 2.4.2a with the two-pass mode (52). Read count tables were generated with the STAR aligner option -quantMode geneCounts. Differential gene expression between the experimental samples was analyzed with DESeq2 (53). Read count tables and group comparisons for differential exon usage were performed with DEXSeq (54). Determination of tissue specificity was based on data from the Human Protein Atlas (55). Chromosomal clusters of genes were detected with the command line tool CROC (56).
Co-IP
Uninduced and doxycycline-induced AIRE-Tet cells, either untreated or treated with etoposide, were cultured on 10-cm plates. Cells were lysed with hypotonic lysis buffer (0.05% Nonidet P-40, 10 mm HEPES (pH 7.4), 1.5 mm MgCl2, 10 mm KCl, 5 mm EDTA, and 1:100 EDTA-free complete protease inhibitor mixture, Thermo Scientific) at a concentration of 30 × 106 cells/ml and incubated on ice for 15 min. Cell lysates were centrifuged for 10 min at 1000 × g at 4 °C. The supernatant was discarded, and the pelleted nuclei were resuspended in nuclear extraction buffer (20 mm HEPES (pH 7.4), 300 mm NaCl, 20 mm KCl, 4 mm CaCl2, 1:100 EDTA-free complete protease inhibitor mixture, and 1 unit MNase) at a concentration of 30 × 106 cells/ml. The lysates were incubated on ice for 1 h and centrifuged for 10 min at 13,000 × g at 4 °C. Lysates (1 mg) were kept overnight on a rotary shaker at 4 °C in Pierce spin columns with 10 μg of rabbit anti-TOP2A (Abcam, ab2987) or IgG control (Diagenode, AIP-103-110) antibodies. The following morning, 150 μl of protein G-Sepharose 4B beads (Invitrogen, 50% suspension in IP lysis buffer) was added, and the lysates were further incubated for 5 h on a rotary shaker at 4 °C. Beads were then washed five times with IP lysis buffer, and the immunoprecipitates were eluted in Laemmli buffer at 99 °C for 10 min.
TOP2A knockdown
Uninduced and doxycycline-induced AIRE-Tet cells, either untreated or treated with etoposide, were grown on 6-well plates and transfected with four SureSilencing shRNA vectors along with one scrambled negative control shRNA vector (KH01520N, SABiosciences) using Turbofect reagent (Thermo Scientific). The efficiency of knockdown was checked by Western blotting with anti-TOPA antibody 48 h after transfection. Anti-β-actin antibody (A5441, Sigma) was used to determine protein loading.
SDS-PAGE and Western blotting
Uninduced and doxycycline-induced AIRE-Tet cells, either untreated or treated with etoposide, were lysed in radioimmunoprecipitation assay buffer containing protease inhibitor mixture (Thermo Scientific). Equal amounts of proteins were resolved on 10% polyacrylamide gels by SDS-PAGE. Resolved proteins were transferred to Hybond-P PVDF membranes (Millipore) in 0.1 m Tris base (pH 8.3), 0.192 m glycine, and 10% (v/v) methanol using an electrophoretic transfer system with cold block. The membranes were blocked with Odyssey blocking buffer (LICOR Bioscience, 927-40000) at room temperature for 1 h. After blocking, the membranes were sequentially incubated overnight with the following primary antibodies: mouse polyclonal anti-AIRE (1:10,000, sc-17985, Santa Cruz Biotechnology), rabbit polyclonal anti-TOP2a (1:1000, ab2987, Abcam), and mouse monoclonal anti-β-actin (1:10,000, A5441, Sigma). Immunoreactive bands were detected using enhanced chemiluminescence and the Odyssey infrared imaging system (Li-Cor Bioscience).
TUNEL assay
Uninduced and doxycycline-induced AIRE-Tet cells, either untreated or treated with etoposide, merbarone, camptothecin, or β-lapachone, were fixed with 4% formaldehyde. The occurrence of DNA breaks was quantitatively assessed with the APO-BrdU TUNEL assay (Pharmingen). Briefly, the cells were labeled with Br-dUTP using TdT enzyme for 60 min at 37 °C and then incubated with FITC-labeled anti-BrdU antibody for 30 min at room temperature. After washing with PBS, the cells were analyzed by LSR Fortessa using FACSDiva software (both from BD Biosciences). The efficacy of the assay was determined with the positive and negative control samples included in the kit.
Apoptosis assay
Uninduced and doxycycline-induced AIRE-Tet cells, either untreated or treated with etoposide, were labeled with the Annexin V-FITC kit (130-092-052, Miltenyi Biotec) according to the protocol of the manufacturer, but propidium iodide was replaced with 7-aminoactinomycin D. The percentages of dead, viable, and early apoptotic cells were determined by FACS with LSR Fortessa using FACSDiva software (both from BD Biosciences).
ChIP
ChIP experiments were performed using 107 cells/experiment according to a protocol described previously with slight modifications (57). Briefly, uninduced and doxycycline-induced AIRE-Tet cells, either untreated or treated with etoposide, were cross-linked with 1% formaldehyde for 10 min at room temperature, and the formaldehyde was quenched by addition of glycine to a final concentration of 0.125 m. Cells were lysed in 200 μl of lysis buffer (50 mm Tris-HCl (pH 8), 10 mm EDTA, 1% SDS, and 1× Halt protease inhibitor mixture; Thermo Scientific). The samples were diluted 10× in radioimmune precipitation assay buffer containing 1× Halt protease inhibitor mixture. Sonication was performed with a Diagenode Bioruptor for 15 min using 30-s on/off cycles at a high voltage setting. After chromatin preparation, ChIP was performed with IP-Star (Diagenode) using the Chip Direct method (Diagenode), in which the antibodies are first coated onto the surface of magnetic beads, and then the bound antibodies are added to the sheared chromatin. ChIP-grade antibodies against HMGB1 (ab18256) and γH2AX (ab2893) were obtained from Abcam and against CTCF from Millipore (07-729). The antibody against histone H1 (sc-34464) was obtained from Santa Cruz Biotechnology. The ChIP signals were measured by qPCR, and the values were log-transformed, standardized, and scaled according to the procedure described in Ref. 51. Experimental groups were compared with a two-sample t test implemented in R. Primers for the ChIP DNA analysis are listed in supplemental Table 1.
Formaldehyde-assisted isolation of regulatory elements coupled with high-throughput sequencing (FAIRE-seq)
Uninduced and doxycycline-induced AIRE-Tet cells, either untreated or treated with etoposide, were cultured and processed as in the ChIP experiments up to the preparation of sonicated lysates. Lysates were then spun at 20,000 × g for 10 min at 4 °C to remove cellular debris, and 10% of the sample volume was collected for input sample preparation. The input samples were treated with 10 μg of DNase-free RNase A for 30 min at 37 °C, followed by 10 μg of proteinase K incubation overnight at 65 °C. The DNA from the remaining lysates and the proteinase K-treated input samples was prepared with standard phenol-chloroform extraction and additionally purified with a Qiagen PCR purification kit. The FAIRE-seq libraries were prepared following the Illumina Truseq ChIP sample preparation guide and subjected to 100-bp paired-end sequencing on an Illumina HiSeq 2000. The quality of the raw reads was checked with FastQC software. Adapter sequences and low-quality base calls (Phred score < 30) were removed with Trim Galore!. The remaining paired-end reads were mapped to the human genome (hg19) with bowtie2 in “local” mode (58). Properly paired reads with a mapping quality of >30 were used for detecting differentially accessible chromatin regions with the csaw R package (59). The differentially accessible regions were annotated with the ChIPseeker R package (60). Statistical significance of overlap between different sets of genomic intervals was assessed with the GenometriCorr R package (61).
3C
Uninduced and doxycycline-induced AIRE-Tet cells were processed according to the detailed protocol in Ref. 31, but the Sau3AI (Thermo) restriction enzyme was used in the DNA digestion step. The chromatin interactions were calculated as the ratio of the qPCR signal in the doxycycline-induced sample relative to the signal in the uninduced control sample. The anchor region was selected based on the FAIRE-seq results. The 3C signals were measured by qPCR, and the values were log-transformed, standardized, and scaled according to the procedure described in Ref. 51. Statistical significance of the log2 -fold change was assessed with a one-sample t test implemented in R. Primers for the 3C qPCR analysis were designed according to the guidelines in Ref. 31 and are listed in supplemental Table 1.
Author contributions
M. G. conducted the ChIP, co-IP, apoptosis, and siRNA assays and most of the gene expression analyses with qPCR. M. S, conducted 3C and some of the gene expression experiments. J. M. performed the RNA-seq experiment. K. K. and U. H. conducted the FACS analyses, and I. L. provided technical assistance and contributed to the preparation of figures. M. S. and T. T. analyzed the RNA-seq and FAIRE-seq data. M. G., J. M., and M. S. wrote the paper. A. M., L. M., and P. P. conceived and supervised the project.
Supplementary Material
Acknowledgments
We thank Drs. Ana Rebane, Martti Laan, Hedi Peterson, and Tauno Metsalu for help and thoughtful discussions. We also thank Maire Pihlap for excellent technical support and Dr. Vinay Choubey for help with the Odyssey detection system.
This work was supported by the Estonian Research Council funding IUT2-2, by the European Union through the European Regional Development Fund (Project No. 2014–2020.4.01.15-0012), by Estonian Research Council Mobilitas Grant MJD164, and ERA-Net.Rus project EGIDA. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. 1 and 2 and Table 1.
- AIRE
- autoimmune regulator
- TSA
- tissue-specific antigen
- APECED
- autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy
- TOP2A
- topoisomerase IIα
- RNA-seq
- RNA sequencing
- IP
- immunoprecipitation
- FAIRE-seq
- formaldehyde-assisted isolation of regulatory elements coupled with high-throughput sequencing
- 3C
- chromosome conformation capture
- CEACAM
- carcinoembryonic antigen-like cellular adhesion molecule
- Etop
- etoposide
- Dox
- doxycycline
- DE
- differentially expressed
- qPCR
- quantitative real-time PCR
- DEU
- differential exon usage
- Ctrl
- control
- PARP
- poly(ADP-ribose) polymerase
- CPT
- camptothecin
- Mer
- merbarone
- β-lap
- β-lapachone
- CARD
- caspase recruitment domain
- CTCF
- CCCTC-binding factor
- Br-dUTP
- bromodeoxyuridine triphosphate
- DNA-PK
- DNA-dependent protein kinase
- MHCII
- major histocompatibility complex class I.
References
- 1. Goodnow C. C. (2007) Multistep pathogenesis of autoimmune disease. Cell 130, 25–35 [DOI] [PubMed] [Google Scholar]
- 2. Klein L., Kyewski B., Allen P. M., and Hogquist K. A. (2014) Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see). Nat. Rev. Immunol. 14, 377–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson M. S., and Su M. A. (2016) AIRE expands: new roles in immune tolerance and beyond. Nat. Rev. Immunol. 16, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kisand K., and Peterson P. (2015) Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy. J. Clin. Immunol. 35, 463–478 [DOI] [PubMed] [Google Scholar]
- 5. Abramson J., and Husebye E. S. (2016) Autoimmune regulator and self-tolerance: molecular and clinical aspects. Immunol. Rev. 271, 127–140 [DOI] [PubMed] [Google Scholar]
- 6. Abramson J., Giraud M., Benoist C., and Mathis D. (2010) Aire's partners in the molecular control of immunological tolerance. Cell 140, 123–135 [DOI] [PubMed] [Google Scholar]
- 7. Gaetani M., Matafora V., Saare M., Spiliotopoulos D., Mollica L., Quilici G., Chignola F., Mannella V., Zucchelli C., Peterson P., Bachi A., and Musco G. (2012) AIRE-PHD fingers are structural hubs to maintain the integrity of chromatin-associated interactome. Nucleic Acids Res. 40, 11756–11768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Org T., Chignola F., Hetényi C., Gaetani M., Rebane A., Liiv I., Maran U., Mollica L., Bottomley M. J., Musco G., and Peterson P. (2008) The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 9, 370–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koh A. S., Kuo A. J., Park S. Y., Cheung P., Abramson J., Bua D., Carney D., Shoelson S. E., Gozani O., Kingston R. E., Benoist C., and Mathis D. (2008) Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc. Natl. Acad. Sci. U.S.A. 105, 15878–15883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Giraud M., Yoshida H., Abramson J., Rahl P. B., Young R. A., Mathis D., and Benoist C. (2012) Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 109, 535–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pommier Y., Sun Y., Huang S. N., and Nitiss J. L. (2016) Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 17, 703–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nitiss J. L. (2009) Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 9, 338–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ju B. G., Lunyak V. V., Perissi V., Garcia-Bassets I., Rose D. W., Glass C. K., and Rosenfeld M. G. (2006) A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 312, 1798–1802 [DOI] [PubMed] [Google Scholar]
- 14. Mondal N., Zhang Y., Jonsson Z., Dhar S. K., Kannapiran M., and Parvin J. D. (2003) Elongation by RNA polymerase II on chromatin templates requires topoisomerase activity. Nucleic Acids Res. 31, 5016–5024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sperling A. S., Jeong K. S., Kitada T., and Grunstein M. (2011) Topoisomerase II binds nucleosome-free DNA and acts redundantly with topoisomerase I to enhance recruitment of RNA Pol II in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 108, 12693–12698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Org T., Rebane A., Kisand K., Laan M., Haljasorg U., Andreson R., and Peterson P. (2009) AIRE activated tissue specific genes have histone modifications associated with inactive chromatin. Hum. Mol. Genet. 18, 4699–4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liiv I., Haljasorg U., Kisand K., Maslovskaja J., Laan M., and Peterson P. (2012) AIRE-induced apoptosis is associated with nuclear translocation of stress sensor protein GAPDH. Biochem. Biophys. Res. Commun. 423, 32–37 [DOI] [PubMed] [Google Scholar]
- 18. Meredith M., Zemmour D., Mathis D., and Benoist C. (2015) Aire controls gene expression in the thymic epithelium with ordered stochasticity. Nat. Immunol. 16, 942–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sansom S. N., Shikama-Dorn N., Zhanybekova S., Nusspaumer G., Macaulay I. C., Deadman M. E., Heger A., Ponting C. P., and Holländer G. A. (2014) Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res. 24, 1918–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Villaseñor J., Besse W., Benoist C., and Mathis D. (2008) Ectopic expression of peripheral-tissue antigens in the thymic epithelium: probabilistic, monoallelic, misinitiated. Proc. Natl. Acad. Sci. U.S.A. 105, 15854–15859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. St-Pierre C., Trofimov A., Brochu S., Lemieux S., and Perreault C. (2015) Differential features of AIRE-induced and AIRE-independent promiscuous gene expression in thymic epithelial cells. J. Immunol. 195, 498–506 [DOI] [PubMed] [Google Scholar]
- 22. Ferguson B. J., Alexander C., Rossi S. W., Liiv I., Rebane A., Worth C. L., Wong J., Laan M., Peterson P., Jenkinson E. J., Anderson G., Scott H. S., Cooke A., and Rich T. (2008) AIRE's CARD revealed, a new structure for central tolerance provokes transcriptional plasticity. J. Biol. Chem. 283, 1723–1731 [DOI] [PubMed] [Google Scholar]
- 23. Kouzine F., Gupta A., Baranello L., Wojtowicz D., Ben-Aissa K., Liu J., Przytycka T. M., and Levens D. (2013) Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat. Struct. Mol. Biol. 20, 396–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lowndes N. F., and Toh G. W. (2005) DNA repair: the importance of phosphorylating histone H2AX. Curr. Biol. 15, R99-R102 [DOI] [PubMed] [Google Scholar]
- 25. Krishnakumar R., Gamble M. J., Frizzell K. M., Berrocal J. G., Kininis M., and Kraus W. L. (2008) Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 319, 819–821 [DOI] [PubMed] [Google Scholar]
- 26. Harshman S. W., Young N. L., Parthun M. R., and Freitas M. A. (2013) H1 histones: current perspectives and challenges. Nucleic Acids Res. 41, 9593–9609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ragab A., and Travers A. (2003) HMG-D and histone H1 alter the local accessibility of nucleosomal DNA. Nucleic Acids Res. 31, 7083–7089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cato L., Stott K., Watson M., and Thomas J. O. (2008) The interaction of HMGB1 and linker histones occurs through their acidic and basic tails. J. Mol. Biol. 384, 1262–1272 [DOI] [PubMed] [Google Scholar]
- 29. Cloosen S., Arnold J., Thio M., Bos G. M., Kyewski B., and Germeraad W. T. (2007) Expression of tumor-associated differentiation antigens, MUC1 glycoforms and CEA, in human thymic epithelial cells: implications for self-tolerance and tumor therapy. Cancer Res. 67, 3919–3926 [DOI] [PubMed] [Google Scholar]
- 30. Pinto S., Michel C., Schmidt-Glenewinkel H., Harder N., Rohr K., Wild S., Brors B., and Kyewski B. (2013) Overlapping gene coexpression patterns in human medullary thymic epithelial cells generate self-antigen diversity. Proc. Natl. Acad. Sci. U.S.A. 110, E3497–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hagège H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., and Forné T. (2007) Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2, 1722–1733 [DOI] [PubMed] [Google Scholar]
- 32. Anderson M. S., Venanzi E. S., Klein L., Chen Z., Berzins S. P., Turley S. J., von Boehmer H., Bronson R., Dierich A., Benoist C., and Mathis D. (2002) Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401 [DOI] [PubMed] [Google Scholar]
- 33. Derbinski J., Pinto S., Rösch S., Hexel K., and Kyewski B. (2008) Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc. Natl. Acad. Sci. U.S.A. 105, 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johnnidis J. B., Venanzi E. S., Taxman D. J., Ting J. P., Benoist C. O., and Mathis D. J. (2005) Chromosomal clustering of genes controlled by the aire transcription factor. Proc. Natl. Acad. Sci. U.S.A. 102, 7233–7238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Klein L., Klugmann M., Nave K. A., Tuohy V. K., and Kyewski B. (2000) Shaping of the autoreactive T-cell repertoire by a splice variant of self protein expressed in thymic epithelial cells. Nat. Med. 6, 56–61 [DOI] [PubMed] [Google Scholar]
- 36. Oven I., Brdicková N., Kohoutek J., Vaupotic T., Narat M., and Peterlin B. M. (2007) AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol. Cell. Biol. 27, 8815–8823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Žumer K., Plemenitaš A., Saksela K., and Peterlin B. M. (2011) Patient mutation in AIRE disrupts P-TEFb binding and target gene transcription. Nucleic Acids Res. 39, 7908–7919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yoshida H., Bansal K., Schaefer U., Chapman T., Rioja I., Proekt I., Anderson M. S., Prinjha R. K., Tarakhovsky A., Benoist C., and Mathis D. (2015) Brd4 bridges the transcriptional regulators, Aire and P-TEFb, to promote elongation of peripheral-tissue antigen transcripts in thymic stromal cells. Proc. Natl. Acad. Sci. U.S.A. 112, E4448–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Muslimović A., Nyström S., Gao Y., and Hammarsten O. (2009) Numerical analysis of etoposide induced DNA breaks. PLoS ONE 4, e5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Baranello L., Wojtowicz D., Cui K., Devaiah B. N., Chung H. J., Chan-Salis K. Y., Guha R., Wilson K., Zhang X., Zhang H., Piotrowski J., Thomas C. J., Singer D. S., Pugh B. F., Pommier Y., et al. (2016) RNA Polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell 165, 357–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pinto D. M., and Flaus A. (2010) Structure and function of histone H2AX. Subcell. Biochem. 50, 55–78 [DOI] [PubMed] [Google Scholar]
- 42. Žumer K., Low A. K., Jiang H., Saksela K., and Peterlin B. M. (2012) Unmodified histone H3K4 and DNA-dependent protein kinase recruit autoimmune regulator to target genes. Mol. Cell. Biol. 32, 1354–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liiv I., Rebane A., Org T., Saare M., Maslovskaja J., Kisand K., Juronen E., Valmu L., Bottomley M. J., Kalkkinen N., and Peterson P. (2008) DNA-PK contributes to the phosphorylation of AIRE: importance in transcriptional activity. Biochim. Biophys. Acta 1783, 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang A., Lyu Y. L., Lin C. P., Zhou N., Azarova A. M., Wood L. M., and Liu L. F. (2006) A protease pathway for the repair of topoisomerase II-DNA covalent complexes. J. Biol. Chem. 281, 35997–36003 [DOI] [PubMed] [Google Scholar]
- 45. Madabhushi R., Gao F., Pfenning A. R., Pan L., Yamakawa S., Seo J., Rueda R., Phan T. X., Yamakawa H., Pao P. C., Stott R. T., Gjoneska E., Nott A., Cho S., Kellis M., and Tsai L. H. (2015) Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell 161, 1592–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takaba H., Morishita Y., Tomofuji Y., Danks L., Nitta T., Komatsu N., Kodama T., and Takayanagi H. (2015) Fezf2 orchestrates a thymic program of self-antigen expression for immune tolerance. Cell 163, 975–987 [DOI] [PubMed] [Google Scholar]
- 47. Ong C. T., and Corces V. G. (2014) CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 15, 234–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Oti M., Falck J., Huynen M. A., and Zhou H. (2016) CTCF-mediated chromatin loops enclose inducible gene regulatory domains. BMC Genomics 17, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saare M., Rebane A., Rajashekar B., Vilo J., and Peterson P. (2012) Autoimmune regulator is acetylated by transcription coactivator CBP/p300. Exp. Cell Res. 318, 1767–1778 [DOI] [PubMed] [Google Scholar]
- 50. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔ C(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 51. Willems E., Leyns L., and Vandesompele J. (2008) Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 379, 127–129 [DOI] [PubMed] [Google Scholar]
- 52. Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T. R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Love M. I., Anders S., Kim V., and Huber W. (2015) RNA-seq workflow: gene-level exploratory analysis and differential expression. F1000Res 4, 1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Anders S., Reyes A., and Huber W. (2012) Detecting differential usage of exons from RNA-seq data. Genome Res. 22, 2008–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Uhlén M., Fagerberg L., Hallström B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C. A., et al. (2015) Proteomics: tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- 56. Pignatelli M., Serras F., Moya A., Guigó R., and Corominas M. (2009) CROC: finding chromosomal clusters in eukaryotic genomes. Bioinformatics 25, 1552–1553 [DOI] [PubMed] [Google Scholar]
- 57. Dahl J. A., and Collas P. (2007) A quick and quantitative chromatin immunoprecipitation assay for small cell samples. Front. Biosci. 12, 4925–4931 [DOI] [PubMed] [Google Scholar]
- 58. Langmead B., and Salzberg S. L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lun A. T., and Smyth G. K. (2016) csaw: a Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res. 44, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yu G., Wang L. G., and He Q. Y. (2015) ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 [DOI] [PubMed] [Google Scholar]
- 61. Favorov A., Mularoni L., Cope L. M., Medvedeva Y., Mironov A. A., Makeev V. J., and Wheelan S. J. (2012) Exploring massive, genome scale datasets with the GenometriCorr package. PLoS Comput. Biol. 8, e1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.