Abstract
We examined whether the scaffolding protein sodium-hydrogen exchanger regulatory factor 1 (NHERF1) interacts with the calcium pump PMCA2 and the tyrosine kinase receptor ErbB2/HER2 in normal mammary epithelial cells and breast cancer cells. NHERF1 interacts with the PDZ-binding motif in PMCA2 in both normal and malignant breast cells. NHERF1 expression is increased in HER2-positive breast cancers and correlates with HER2-positive status in human ductal carcinoma in situ (DCIS) lesions and invasive breast cancers as well as with increased mortality in patients. NHERF1 is part of a multiprotein complex that includes PMCA2, HSP90, and HER2 within specific actin-rich and lipid raft-rich membrane signaling domains. Knocking down NHERF1 reduces PMCA2 and HER2 expression, inhibits HER2 signaling, dissociates HER2 from HSP90, and causes the internalization, ubiquitination, and degradation of HER2. These results demonstrate that NHERF1 acts with PMCA2 to regulate HER2 signaling and membrane retention in breast cancers.
Keywords: breast cancer, calcium transport, heat shock protein 90 (Hsp90), PDZ domain, receptor internalization
Introduction
Sodium-hydrogen exchanger regulatory factor 1 (NHERF1) is one of four related scaffolding proteins (NHERF1-NHERF4) that contain tandem PSD-95/Drosophila discs large/ZO-1 (PDZ) domains and a C-terminal ezrin/radixin/moesin/merlin (ERM) binding domain (1–4). NHERF1 interacts with a variety of membrane proteins through interactions with a canonical PDZ-binding motif (1, 2, 5, 6) and facilitates the formation of multiprotein complexes that are tethered to the actin cytoskeleton (2).
NHERF1 has been reported to have variable functions in breast cancer cells (7–16), and different NHERF1 mutations have been shown to either inhibit or to promote breast cancer (9, 17–20). In several studies, tumor NHERF1 levels have been demonstrated to correlate with HER2 expression (7, 12, 13). It has also been shown to influence signaling pathways involving β-catenin, platelet-derived growth factor, and RhoA-p38 MAP kinase in breast cancer cells (8, 10, 11, 14, 20, 21). The mechanisms governing the diverse actions of NHERF1 in breast cancers are poorly understood.
ErbB2/HER2 is overexpressed in 25–30% of human breast cancers, and transgenic expression of HER2 in the mouse mammary gland is sufficient to cause invasive mammary carcinomas (22, 23). HER2 has no recognized ligands and acts as an obligate heterodimer with other ErbB family receptors, especially with EGFR2 (ErbB1/HER1) and ErbB3/HER3 in breast cancer cells (24, 25). In contrast to other ErbB family members, HER2 is resistant to internalization and degradation and signals at the cell surface for prolonged periods after it is activated (26–29). Although the mechanisms underlying the retention of HER2 at the cell surface are not fully understood, it must interact with the chaperone HSP90 and the plasma membrane calcium ATPase2 (PMCA2) to avoid internalization and continue to signal at the plasma membrane (27, 30, 31).
PMCA2 pumps calcium across the plasma membrane into the extracellular fluid (32–34). It is highly expressed at the apical surface of lactating breast cells and transports calcium into milk (35–37). The splice variant of PMCA2 expressed by the mammary gland (PMCA2wb) contains an extended C-terminal domain ending in a canonical PDZ recognition sequence (ETSL) (38, 39). In this study, we demonstrate that NHERF1 interacts with PMCA2 in breast cancer cells and maintains interactions between PMCA2, HSP90, and HER2 within specific actin- and lipid raft-rich membrane domains. NHERF1 is required for the localization and retention of HER2 within these membrane domains; loss of NHERF1 expression alters the membrane structure, promotes HER2 internalization and degradation, and inhibits HER2 signaling.
Results
NHERF1 expression correlates with HER2 and PMCA2 expression in breast cancers
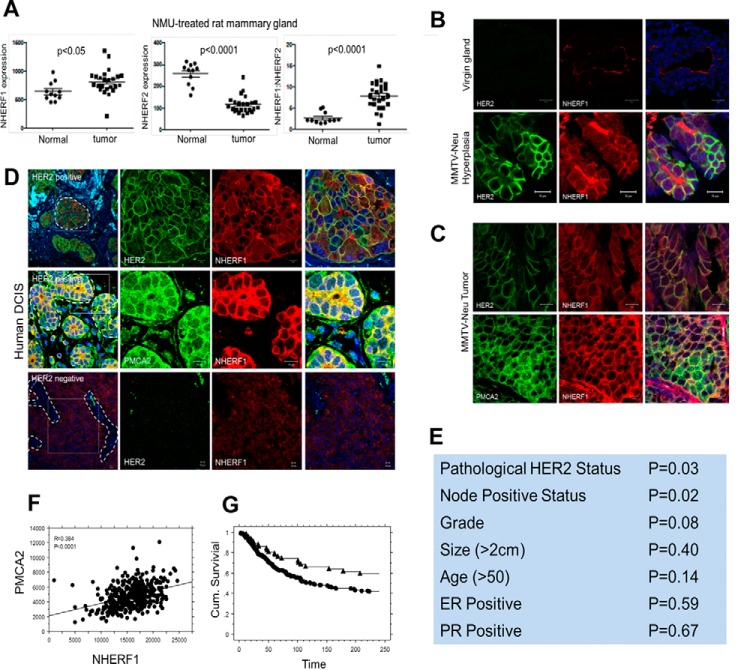
PMCA2 is prominently expressed on the apical surface of mammary epithelial cells. Prior studies showed that PMCA2 interacted with NHERF1 and NHERF2 in renal cells and that interactions with NHERF2 contributed to the apical retention of PMCA2 (38, 39). Therefore, we reasoned that similar interactions with NHERFs might anchor PMCA2 and HER2 at the cell surface in breast cancer cells. To explore this hypothesis, we first examined the expression of NHERF1 and NHERF2 mRNA in mammary tumors in rats (40). As shown in Fig. 1A, NHERF1 expression increased in N-methyl-N-nitrosourea-induced mammary tumors compared with normal mammary glands, whereas NHERF2 expression decreased in tumors relative to normal glands. As a result, the ratio of NHERF1 to NHERF2 expression increased in tumors. Therefore, we concentrated further studies on potential interactions between PMCA2, HER2, and NHERF1.
Figure 1.
A, NHERF1 mRNA levels, NHERF2 mRNA levels, and NHERF1/NHERF2 mRNA ratios from normal mammary glands versus mammary tumors harvested from N-methyl-N-nitrosourea-treated rats. B, immunofluorescence staining for HER2 (green) and NHERF1 (red) in mammary ducts from normal mice or hyperplastic regions in the mammary glands from MMTV-Neu mice. The right panels show co-staining with DAPI. Scale bars = 10 μm. C, immunofluorescence staining for HER2 (green) and NHERF1 (red) (top row) and for PMCA2 (green) and NHERF1 (red) (bottom row) in tumors from MMTV-Neu mice. Panels on the right show co-staining with DAPI. Scale bars = 10 μm. D, top row, immunofluorescence staining for HER2 (green) or NHERF1 (red) or merge in human HER2-positive DCIS. The right panels show a magnified view of the boxed area in the left panel. Center row, staining for PMCA2 (green) or NHERF1 (red) or merge in human HER2-positive DCIS. The right panels show a magnified view of the boxed area in the left panel. Bottom row, immunofluorescence staining for HER2 (green) or NHERF1 (red) or merge in human HER2-negative DCIS. The right panels show a magnified view of the boxed area in the left panel. Scale bars = 10 μm. E, correlations between NHERF1 expression and clinical pathological tumor characteristics. F, correlation between PMCA2 and NHERF1 AQUA scores in the YTMA49 tissue microarray. G, survival curves for tumors with NHERF1 AQUA scores above (circles) versus below (triangles) the optimal cut point as defined by the X-tile bioinformatics tool.
We saw a clear up-regulation of NHERF1 immunofluorescence in hyperplastic regions and in tumors in MMTV-Neu mice (Fig. 1, B and C) compared with the relatively weak NHERF1 immunofluorescence at the apical surface of normal luminal epithelial cells. Furthermore, NHERF1 was not localized at the apical surface of the cells but was expressed uniformly within the plasma membrane, where it co-localized with both HER2 and PMCA2 in hyperplastic regions and/or tumors (Fig. 1, B and C). We also examined immunofluorescence staining for NHERF1 in a series of 16 HER2-positive and four HER2-negative human DCIS samples. NHERF1 levels were clearly increased in all HER2-positive DCIS samples compared with HER2-negative DCIS samples (Fig. 1D). Furthermore, NHERF1 co-localized with HER2 and PMCA2 at the membrane of the malignant cells. As with mouse tumors, NHERF1 was uniformly expressed along the plasma membrane in DCIS lesions and was not limited to the apical surfaces of the cells.
We assessed NHERF1 immunofluorescence in invasive breast cancers by utilizing a previously described tumor microarray (YTMA-49) consisting of 652 invasive breast cancers annotated to clinical outcome data covering a mean of 8.9 years (30, 41, 42). NHERF1 fluorescence staining intensity was measured as a continuous variable using the semiautomated AQUA system (41). We found that NHERF1 expression correlated with clinical pathology HER2-positive status (p = 0.03) and positive nodal status (p = 0.02) (Fig. 1E). Compared with the previous analysis for PMCA2, we found a significant positive correlation between NHERF1 and PMCA2 AQUA scores (p < 0.001, Fig. 1F). When NHERF1 expression was considered as a continuous variable, there was a trend toward higher NHERF1 levels predicting shorter survival, but the association was not statistically significant (Cox hazard, p = 0.094). However, when the X-tile bioinformatics tool (43) was used to define an optimal cut point between high and low NHERF1 levels, NHERF1 AQUA levels above this threshold were associated with a statistically significant decreased length of survival (Fig. 1G, Cox hazard, p = 0.015). We examined the interactions between NHERF1, PMCA2, and HER2 and found that the relationships between NHERF1 AQUA scores and survival were lost when either PMCA2 or HER2 was included in a multivariate analysis. These results suggest that the ability of NHERF1 to predict mortality in this cohort is related to its associations with HER2 status and/or PMCA2 levels.
NHERF1 interacts with PMCA2 and HER2 in breast cancer cells
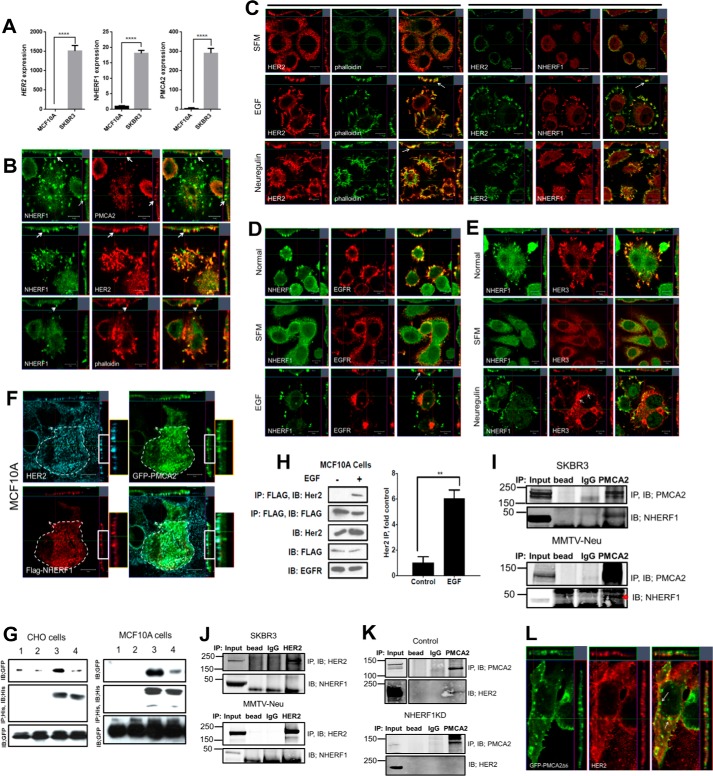
Next we examined whether NHERF1 interacted directly with PMCA2 and/or HER2. First, we examined NHERF1 mRNA levels and immunofluorescence staining in immortalized MCF10A human mammary epithelial cells and the HER2-positive human breast cancer cell line SKBR3. As expected, HER2 mRNA expression was much higher in SKBR3 cells than in MCF10A cells, as were both NHERF1 and PMCA2 mRNA levels (Fig. 2A). We then performed confocal immunofluorescence staining for NHERF1, HER2, and PMCA2 in SKBR3 cells. As shown in Fig. 2B, NHERF1 co-localized with PMCA2 and HER2 within punctate areas of the plasma membrane that protruded from the apical aspect of the cells. NHERF1 also co-localized with actin (Fig. 2, B and C) as well as with the HER2 signaling partners EGFR and HER3 in similarly appearing membrane domains (Fig. 2, D and E). Interestingly, in serum-starved cells, HER2 co-localized with NHERF1 and actin more broadly at the apical membrane of the cells, but acute pharmacologic activation of HER2 signaling with either EGF or Nrg1 treatment caused both NHERF1 and HER2 to coalesce within discrete actin-enriched membrane protrusions, whereas EGFR and HER3 became internalized, as described previously (26, 28, 30) (Fig. 2, C–E). To determine directly whether NHERF1, PMCA2, and HER2 co-localized within the same membrane domains, we transiently expressed GFP-tagged PMCA2 and FLAG-tagged NHERF1 in MCF10A cells that constitutively overexpressed HER2. As with SKBR3 cells, HER2, PMCA2 (GFP), and NHERF1 (FLAG tag) co-localized within membrane protrusions at the apical surface of MCF10A cells (Fig. 2F).
Figure 2.
A, HER2, NHERF1, and PMCA2 mRNA levels in MCF10A and SKBR3 cells as assessed by QPCR. B, confocal images of immunofluorescence co-staining of SKBR3 cells for NHERF1 (green) with PMCA2, HER2, or actin (red) as indicated. The images on the right show merged staining. The top and right panels of each image demonstrate optical sections in different orientations. Arrows point to apical membrane protrusions. Scale bars = 10 μm. C, confocal images of co-immunofluorescence for HER2 and actin (phalloidin) or NHERF1 in SKBR3 cells in serum-free medium (SFM, top row) or after treatment with EGF (center row) or NRG1 (bottom row). The top and right panels of each image represent optical sections through the cells in two different orientations. The left three columns show staining for HER2 (red), phalloidin (green), and merged staining. The right three columns show staining for HER2 (green), NHERF1 (red), and merged staining. Arrows point to the more prominent apical membrane protrusions that are actin-rich and contain HER2 and NHERF1 that form after treatment with growth factors. D, confocal images of co-immunofluorescence for NHERF1 (green) and EGFR in SKBR3 cells in growth medium (top row), in serum-free medium (center row), and following acute treatment with EGF. E, confocal images of co-immunofluorescence for NHERF1 (green) and HER3 (red) in SKBR3 cells in growth medium (top row), in serum-free medium (center row), and following acute treatment with NRG1. The top and right panels of each image depict optical sections through the cells in two different orientations. Scale Bars = 10 μm. F, confocal images of immunofluorescence for HER2 (aqua), PMCA2 (green), or NHERF1 (red) in MCF10A cells constitutively overexpressing HER2 and transfected with GFP-labeled, WT PMCA2, and FLAG-tagged NHERF1. The bottom right image shows merged images for all three stains. The top and right panels of each image demonstrate optical sections in different orientations. Boxed areas are magnified at the right. G, co-immunoprecipitation experiments in CHO or MCF10A cells transiently transfected with GFP-tagged PMCA2 and His-tagged NHERF1. All cells were transfected with either WT GFP-PMCA2 (lanes 1 and 3) or Δ6-mutant GFP-PMCA2 lacking the C-terminal six amino acids that include the PDZ interaction motif (lanes 2 and 4). Lanes 1 and 2 represent cells without NHERF1, whereas lanes 3 and 4 represent cells transfected with his-NHERF1. IB, immunoblot. H, co-immunoprecipitation of HER2 and FLAG-tagged NHERF1 in MCF10A cells with or without treatment with EGF. IP of FLAG-tagged NHERF1 from MCF10A cells pulled down HER2 only after acute treatment with EGF. The left panel represents the mean ± S.E. of the relative increase in immunoprecipitated HER2 after EGF in four different experiments. **, p < 0.005. I, co-immunoprecipitation of PMCA2 and NHERF1. IP of PMCA2 pulled down NHERF1 from lysates of SKBR3 cells (top panel) and MMTV-Neu mammary tumors (bottom panel). J, co-immunoprecipitation of HER2 and NHERF1. IP of HER2 failed to pull down NHERF1 from lysates of SKBR3 cells (top panel) or MMTV-Neu mammary tumors (bottom panel). K, co-immunoprecipitation of PMCA2 and HER2. In control cells (top panel), IP of PMCA2 pulled down HER2, but in NHERF1KD-SKBR3 cells (bottom panel), IP of PMCA2 failed to pull down HER2. L, confocal images of immunofluorescence for PMCA2 (green) and HER2 (red) in MCF10A cells constitutively overexpressing HER2 and transfected with GFP-labeled mutant Δ6PMCA2. The top and right panels of each image demonstrate optical sections in different orientations. The arrow points to Δ6PMCA2, not associated with HER2 and internalized away from the cell surface.
Next, we transiently expressed His-tagged NHERF1 with GFP-tagged PMCA2 in CHO cells and MCF10A cells. As shown in Fig. 2G, when we immunoprecipitated NHERF1 using anti-His tag antibody, we also pulled down PMCA2. Expression of a GFP-tagged, mutant PMCA2 lacking the six C-terminal amino acids encompassing the PDZ recognition sequence (Δ6PMCA2) disrupted the ability to co-IP PMCA2 (GFP) with NHERF1 (His) in both cell lines, demonstrating that interactions between PMCA2 and NHERF1 depend on the PDZ recognition domain of PMCA2. We also introduced the same constructs into MCF10A cells overexpressing HER2. At baseline, immunoprecipitation of NHERF1 from MCF10A cells did not pull down HER2 (Fig. 2H). However, given that activation of HER2 appeared to increase immunofluorescence co-localization of NHERF1 with HER2 (Fig. 2C), we also treated the cells with EGF, which led to the ability to co-IP HER2 with NHERF1 (Fig. 2H).
To determine whether endogenous NHERF1, PMCA2, and HER2 were contained within the same protein complex, we performed co-immunoprecipitation experiments in SKBR3 cells and in tumors harvested from MMTV-Neu mice. As shown in Fig. 2I, immunoprecipitation of PMCA2 pulled down NHERF1 from SKBR3 cells and from MMTV-Neu tumors. Despite their co-localization by immunofluorescence, immunoprecipitation of HER2 did not pull down NHERF1 from either SKBR3 cells at baseline or from MMTV-Neu tumors (Fig. 2J). Nevertheless, NHERF1 appears to be necessary for stable interactions between PMCA2 and HER2 in SKBR3 cells. As reported previously, in control cells, immunoprecipitation of PMCA2 pulled down HER2 (Fig. 2K) (30), but, in SKBR3 cells in which we stably knocked down expression of NHERF1, immunoprecipitation of PMCA2 no longer pulled down appreciable amounts of HER2 (Fig. 2K). Furthermore, although GFP-labeled wild-type PMCA2 co-localized with HER2 at the apical cell membrane (Fig. 2F), membrane co-localization was diminished in MCF10A-HER2 cells transfected with GFP-labeled Δ6PMCA2, and staining for GFP and HER2 was found in membrane invaginations and in intracellular vesicles (Fig. 2L). These results suggest that PDZ-mediated interactions between PMCA2 and NHERF1 are necessary for PMCA2 to stably interact with HER2 within the protruding membrane domains. Taken together, these data suggest that, in SKBR3 and MCF10A cells, NHERF1 is contained with a complex that also contains PMCA2 and HER2. Furthermore, activation of the EGFR and/or HER2 appears to increase the amount of NHERF1 within this complex, where it is necessary to stabilize interactions between HER2 and PMCA2.
NHERF1 is necessary for HER2 signaling in breast cancer cells
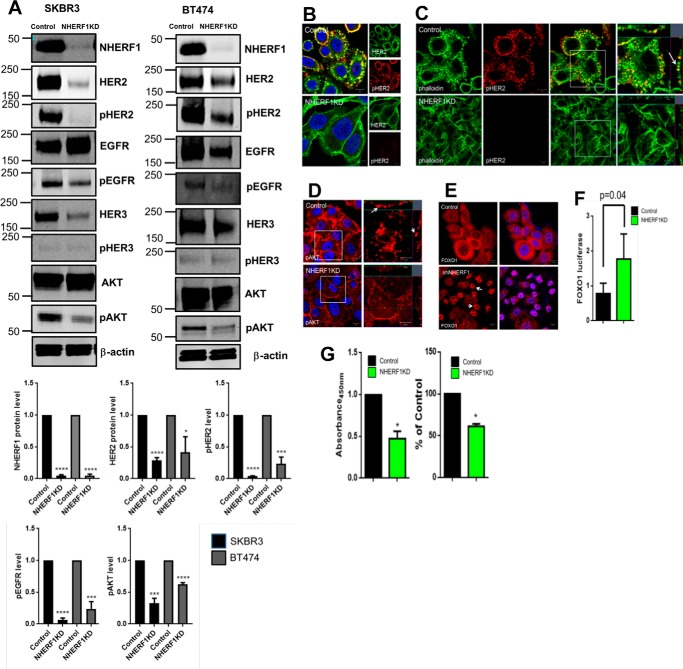
We next examined the effects of knocking down NHERF1 on HER2 signaling in SKBR3 and BT474 cells, two HER2-positive breast cancer cell lines. As shown in Fig. 3A, we achieved over 90% stable reductions in NHERF1 levels in NHERF1 knockdown (NHERF1KD) SKBR3 cells and NHERF1KD BT474 cells. Control cells expressed nonspecific shRNAs. In both cell lines, loss of NHERF1 significantly reduced total HER2 and pHER2 levels (Fig. 3A). There was a reduction in total EGFR expression in BT474 cells but not in SKBR3 cells. However, in both cell lines, knocking down NHERF1 dramatically reduced phospho-EGFR levels. NHERF1KD cells also showed reductions in total HER3 and pHER3 expression. Knocking down NHERF1 expression reduced AKT phosphorylation, although total AKT levels were unchanged. We also used immunofluorescence staining to assess HER2 activation in SKBR3 NHERF1KD cells. As shown in Fig. 3, B and C, in control cells, activated HER2 (pHER2 staining) was most prominent in actin-rich membrane protrusions. However, pHER2 staining was practically absent in NHERF1KD cells. Similarly, pAkt staining was located in prominent membrane protrusions in control cells and reduced in the NHERF1KD cells (Fig. 3D). To assess AKT bioactivity, we examined the localization of FOXO1 by immunofluorescence (Fig. 3E) and measured the expression of a FOXO1-luciferase reporter gene (Fig. 3F) (30). In control cells, FOXO1 was expressed primarily in the cytoplasm, whereas NHERF1KD-SKBR3 cells showed prominent nuclear FOXO1 staining. Knockdown cells also showed increased FOXO1-luciferase gene expression. Given that AKT activation excludes FOXO1 from the nucleus and inhibits reporter gene expression, these results document reduced AKT bioactivity in response to reduced NHERF1 expression (44). Taken together, these findings show that suppressing NHERF1 expression reduces HER2 levels, impairs HER2 activation, and decreases downstream AKT signaling. As might be expected from diminished HER2/AKT signaling, knocking down NHERF1 also led to a reduction in cell growth, as assessed by cell accumulation and BrdU incorporation (Fig. 3G).
Figure 3.
A, immunoblots showing levels of NHERF1 and various HER2 signaling components in whole-cell lysates of control and NHERF1KD SKBR3 and BT474 cells. Bar graphs below demonstrate the quantification of selected signaling components in 3 different experiments with NHERF1KD cells expressed relative to baseline in control cells. B, immunofluorescence staining for pHER2 (red) and total HER2 (green) in control versus NHERF1KD SKBR3 cells. Scale bars = 10 μm. C, immunofluorescence staining for pHER2 (red) and actin (phalloidin, green) in control versus NHERF1KD SKBR3 cells. The right panels show magnifications of the boxed areas in the merged images. The arrow points to co-staining in membrane protrusions. Scale bars = 10 μm. D, pAKT staining in control versus NHERF1KD SKBR3 cells. Arrows point to pAKT in membrane protrusions. Scale bars = 10 μm. E, FOXO1 in control (top row) or NHERF1KD SKBR3 cells (bottom row). Scale bars = 10 μm. F, activity of the FOXO1-luciferase construct transfected into either control or NHERF1KD SKBR3 cells. Error bars show the mean ± S.E. for n = 5. G, cell proliferation measured by cell accumulation assayed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (left) or BrdU incorporation (right) for control versus NHERF1KD SKBR3 cells. Data are expressed as percentage of control cells, and error bars represent the mean ± S.E. for three experiments. *, p < 0.05, **, p < 0.005, ***, p < 0.0005, ****, p < 0.00005.
Loss of NHERF1 and HER2 result in similar changes in global gene expression
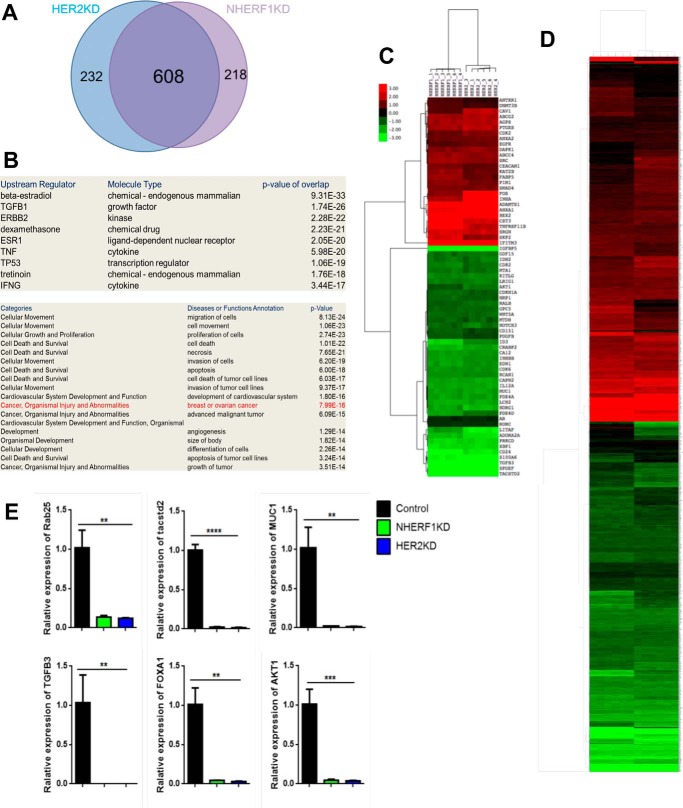
To test whether NHERF1 regulated HER2 function more systematically, we compared changes in global gene expression caused by knocking down NHERF1 with those caused by knocking down HER2. We performed oligonucleotide microarray analyses to define changes in baseline gene expression between control SKBR3 cells and SKBR3 cells with stable knockdown of either NHERF1 or HER2 expression (NHERF1KD and HER2KD). We identified 826 transcripts that were significantly altered more than 2-fold in either direction in NHERF1KD cells and 840 transcripts in HER2KD cells. The changes in gene expression in each knockdown cell line were remarkably similar; 82% of the transcripts altered in HER2KD cells also changed in NHERF1KD cells (Fig. 4A), and a heat map of the changes in 1058 total genes altered in either cell line or both (Fig. 4D) underscores the remarkable concordance in global gene expression caused by knocking down NHERF1 versus HER2. We validated changes in the expression of six separate genes altered in the microarray by QPCR. As shown in Fig. 4E, the expression of each of these genes was reduced in both NHERF1KD and HER2KD cells, as predicted by the microarray results. Functional annotation of the changes in gene expression demonstrated a strong correlation with HER2/ERBB2 signaling, and the altered genes were enriched for cancer-associated transcripts (Fig. 4B). For example, when we compared changes in the expression of the 74 genes in the “advanced malignant tumor” category, we found them to be virtually identical between NHERF1KD and HER2KD cells (Fig. 4C). These results demonstrate that knocking down either NHERF1 or HER2 expression results in fundamentally similar changes in global gene expression, supporting the notion that NHERF1 functions within a HER2-regulated biologic pathway.
Figure 4.
A, Venn diagram demonstrating overlap in changes in gene expression caused by knocking down HER2 versus NHERF1 in SKBR3 cells. B, signaling pathways and cellular processes predicted to be changed by functional annotation of gene expression changes common to HER2KD and NHERF1KD SKBR3 cells. C, heat map of changes in the expression of 74 genes within the advanced malignant tumor list caused by knocking down either NHERF1 or HER2 in SKBR3 cells. D, heat map of changes in mRNA levels for all 1058 transcripts showing a significant change in either HER2KD or NHERF1KD cells compared with control SKBR3 cells. E, validation of changes in gene expression by QPCR for six genes noted to be altered in gene array data for HER2KD and NHERF1KD cells compared with control SKBR3 cells. **, p < 0.005; ***, p < 0.0005; ****, p < 0.00005.
Loss of NHERF1 Increases intracellular calcium levels and alters HER2-containing membrane protrusions
Given that knocking down NHERF1 or PMCA2 produces similar alterations in HER2 signaling (30), we next asked whether loss of NHERF1 alters PMCA2 levels. As shown in Fig. 5A, knocking down NHERF1 reduced total cellular PMCA2 levels, increased intracellular calcium concentrations approximately 4-fold, and increased the sensitivity of the cells to calcium-induced apoptosis (Fig. 5, B and C). Reductions in NHERF1 also caused a profound change in membrane structure. At baseline, control SKBR3 cells displayed actin-rich protrusions from their apical aspect that contained HER2, EGFR, and HER3 (Fig. 5, D–F). Knockdown of NHERF1 resulted in the effacement of these structures, as shown by confocal immunofluorescence staining for actin (phalloidin) as well as by scanning and transmission electron microscopy (Fig. 5, D and G). At baseline, HER2 and EGFR as well as HER2 and HER3 co-localized within punctate membrane protrusions (Fig. 5, E and F). However, knocking down NHERF1 led to an overall reduction and more uniform distribution of staining for EGFR and HER2 as well as an increase in their intracellular localization (Fig. 5E). Knocking down NHERF1 extensively reduced membrane staining for HER3, which, instead, accumulated in an intracellular compartment where it no longer co-localized with HER2 (Fig. 5F). HER2 localizes to lipid raft regions within the plasma membrane (45–47), and the HER2-containing punctate membrane domains also stained for cholera toxin B, a histologic marker of lipid rafts (Fig. 5H). As before, co-localization between cholera toxin B and HER2 was particularly distinct after acute activation of HER2 signaling with either EGF or NRG1 (Fig. 5I). Strikingly, knocking down NHERF1 expression caused a large reduction in staining intensity for cholera toxin B and loss of co-localization with HER2 both at baseline and after treatment with EGF and NRG (Fig. 5, H and I). We confirmed these findings by examining flotillin 1 and HER2 expression in Triton X-100-insoluble membrane fractions from control and NHERF1KD SKBR3 cells. As shown in Fig. 5J, HER2, PMCA2, and NHERF1 were all contained within flotillin 1-positive, Triton X-100-insoluble fractions from control cells. By contrast, in NHERF1KD cells, there were reductions in the expression of all three proteins in Triton X-100-insoluble fractions and also a reduction in flotillin 1 expression itself within the Triton X-100-insoluble fractions as well as in total cellular lysates (Fig. 5, J and K). These data demonstrate that NHERF1 is important for establishing and/or maintaining actin- and lipid raft rich membrane protrusions that contain critical components of the HER2 signaling pathway.
Figure 5.
A, typical immunoblot showing PMCA2 levels in control versus NHERF1KD SKBR3 cells. β-actin served as a loading control. Bar graph shows quantification of 3 experiments. Bar for NHERF1KD represents mean ± S.E. of values relative to baseline in control cells. ****, p < 0.00005. B, relative intracellular calcium levels in NHERF1KD compared with control SKBR3 cells. Columns represent the mean ± S.E. for three separate measurements. The mean absolute calcium concentrations are included above the columns. *, p < 0.05. C, change in TUNEL-positive cells in control versus NHERF1KD SKBR3 cells at either 2 mm or 10 mm extracellular calcium with or without ionomycin. Each column represents the mean ± S.E. relative to control cells at 2 mm calcium without ionomycin. ‡, p < 0.0005; #, p < 0.00005. D, left panel, confocal images of immunofluorescence staining for HER2 (red) and actin (green, phalloidin) in control (top row) versus NHERF1KD (bottom row) SKBR3 cells. The images on the right show merged staining. The top and side panels represent optical sections through cells in two different orientations. Scale bars = 10 μm. Right panel, the percentage of individual cells showing HER2-positive membrane protrusions. The columns represent the mean ± S.E. for three experiments counting 150 individual cells. ****, p < 0.00005. E, confocal images for immunofluorescence staining for HER2 (green) and EGFR (red) in control and NHERF1KD cells. The right panels show merged staining. The arrow points to an area of HER2/EGFR co-localization in intracellular vesicles in knockdown cells. Scale bars = 10 μm. F, confocal images for immunofluorescence staining for HER2 (green) and HER3 (red) in control and NHERF1KD cells. The right panels show merged staining. The arrows point to an area of isolated HER3 staining in intracellular vesicles in knockdown cells. Scale bars = 10 μm. G, scanning (top row) and transmission (bottom row) electron microscopy of control versus NHERF1KD cells. White dotted lines demonstrate cell borders. Scale bars = 10 μm. H, confocal images for immunofluorescence staining for HER2 (green) and lipid raft regions of the membrane (cholera toxin B, red) in control and NHERF1KD cells. The right panels show merged staining. I, confocal images for immunofluorescence staining for HER2 (green) and lipid rafts (cholera toxin B, red) in control and NHERF1KD cells after treatment with EGF or NRG1. J, immunoblots showing Triton X-100-insoluble and -soluble fractions of plasma membrane from control (top panel) or NHERF1KD (bottom panel) SKBR3 cells. In control cells, HER2, PMCA2, and NHERF1 are found in flotillin-positive Triton X-100 insoluble membrane fractions. In NHERF1KD cells, all markers are significantly less abundant in the Triton X-100-insoluble fractions. K, immunoblot showing total cellular flotillin 1 expression in control versus NHERF1KD cells. Bar graph shows quantification of 3 experiments. Bar for NHERF1KD represents mean ± S.E. of values relative to baseline in control cells. **, p < 0.005.
NHERF1 is required for the retention of HER2 at the cell surface
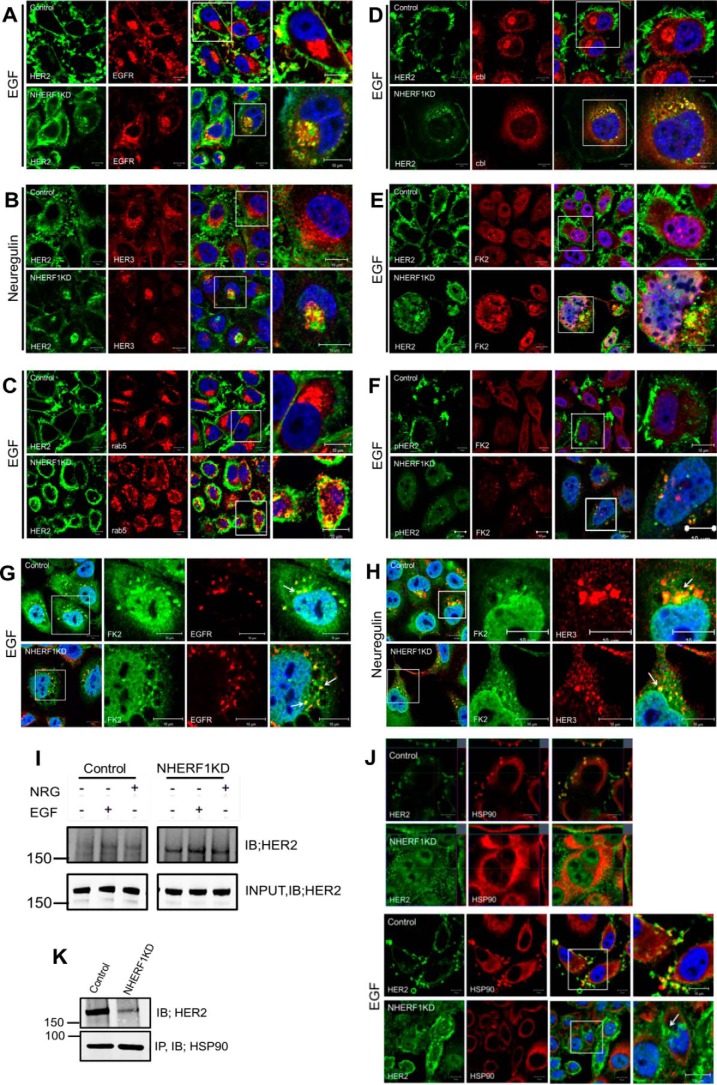
As shown in Fig. 6A, treating control cells with EGF led to the internalization of EGFR but not HER2. In contrast, treating NHERF1KD cells with EGF caused internalization of HER2 with EGFR, and both continued to co-localize within the cytoplasm. Likewise, treatment of control cells with Nrg1 led to the internalization of HER3, but HER2 remained on the cell surface within membrane protrusions (Fig. 6B). However, in NHERF1KD cells, NRG1 caused internalization of HER2 with HER3, and both co-localized in an intracellular compartment. The internalized HER2 in NHERF1KD cells was localized within an endocytic compartment, as demonstrated by co-localization between HER2 and rab5 in intracellular vesicles (Fig. 6C). By contrast, rab5 never co-localized with HER2 in control cells. Knocking down NHERF1 led to the recruitment of the ubiquitin ligase c-cbl and polyubiquitination and degradation of HER2. As can be appreciated, HER2 did not co-localize with c-cbl in control cells but did so in NHERF1KD cells after treatment with EGF (Fig. 6D). In addition, we co-stained control and NHERF1KD cells for HER2 and with the antibody FK2, which recognizes mono- and polyubiquitinated proteins. FK2 staining co-localized with HER2 and phosphorylated HER2 after EGF activation (Fig. 6, E and F) only in knockdown cells. In contrast, FK2 staining co-localized with EGFR or HER3 in control and NHERF1KD cells after treatment with either EGF or NRG1, respectively (Fig. 6, G and H). Co-immunoprecipitation of polyubiquitin and HER2 demonstrated an increase in HER2 ubiquitination in NHERF1KD cells both at baseline and after treatment with EGF or NRG1 (Fig. 6I). Finally, HSP90 has been shown to be important for stabilizing HER2 at the cell surface, and, as expected, we observed co-localization of HER2 and HSP90 in membrane protrusions in SKBR3 cells at baseline or after treatment with EGF (Fig. 6J) (27, 30, 31). However, in NHERF1KD cells, HER2 and HSP90 no longer co-localized, and HER2 was internalized. Likewise, immunoprecipitation of HSP90 pulled down HER2 in control cells, but the ability to co-IP HER2 with HSP90 was much reduced in NHERF1 KD cells (Fig. 5K). Together, these data suggest that loss of NHERF1 disrupts interactions between HER2 and HSP90 at the plasma membrane and leads to the ubiquitination and internalization of HER2 upon its activation (Fig. 7).
Figure 6.
A, immunofluorescence for HER2 (green) and EGFR (red) in control (top row) and NHERF1KD (bottom row) SKBR3 cells after treatment with EGF. B, immunofluorescence for HER2 (green) and HER3 (red) in control (top row) and NHERF1KD (bottom row) cells after treatment with NRG1. C, immunofluorescence for HER2 (green) and rab5 (red) in control (top row) and NHERF1KD cells (bottom row) after treatment with EGF. D, immunofluorescence for HER2 (green) and cbl (red) in control (top row) and NHERF1KD (bottom row) cells after treatment with EGF. E, immunofluorescence for HER2 (green) and polyubiquitin (FK2, red) in control (top row) and NHERF1KD (bottom row) cells after treatment with EGF. F, immunofluorescence for pHER2 (green) and polyubiquitin (FK2, red) in control (top row) and NHERF1KD (bottom row) after treatment with EGF. In A–F, the third panel in each row shows merged staining with DAPI (blue), and the fourth panel in each row represents a magnified view of the boxed region of the third panels. G, immunofluorescence for polyubiquitin (FK2, green) and EGFR (red) after acute treatment of control or NHERF1KD cells with EGF. The images on the right show merged staining for both antibodies and DAPI (blue) to mark nuclei. H, immunofluorescence for polyubiquitin (FK2, green) and HER3 (red) after acute treatment of control or NHERF1KD cells with NRG1. The images on the right show merged staining for both antibodies and DAPI (blue) to mark nuclei. Scale bars = 10 μm. I, co-IP for polyubiquitin complexes and HER2. IP for ubiquitinated proteins using FK2 antibody pulled down more HER2 from NHERF1KD cells than from control cells with or without treatment with EGF or NRG1. J, immunofluorescence for HER2 (green) and HSP90 (red) in control (first and third rows) and NHERF1KD (second and fourth rows) cells without (first and second rows) or with (third and fourth rows) treatment with EGF. The arrow points to internalized HER2, dissociated from HSP90, after EGF treatment of NHERF1KD cells. Scale bars = 10 μm. K, co-immunoprecipitation of HER2 and HSP90 from control and NHERF1KD cells. Less HER2 is pulled down with HSP90 in NHERF1 KD cells. IB, immunoblot.
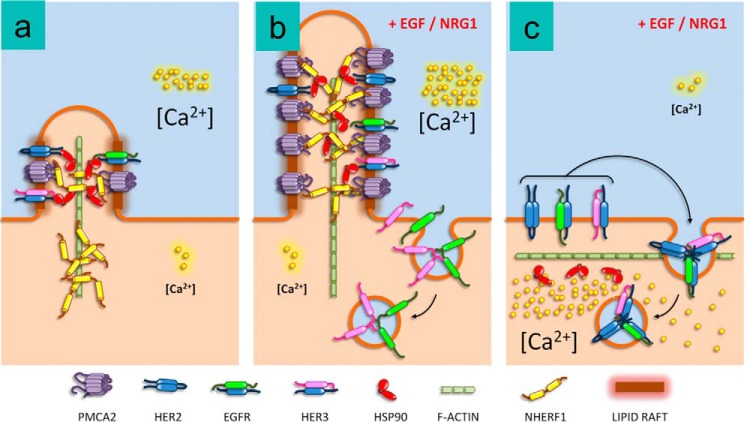
Figure 7.
Model for the interaction of NHERF1 with PMCA2 and HER2 in breast cancer cells. a, at baseline, NHERF1 helps maintain PMCA2, HSP90, and HER2 in small, lipid raft-rich protrusions from the membrane. b, acute activation of HER2 leads to enlargement of the membrane protrusions with recruitment of NHERF1 to support interactions between PMCA2, HSP90, and HER2 that allow HER2 to remain within the protrusions, whereas EGFR and HER3 are internalized into the cell. c, in the absence of NHERF1, there is a reduction in PMCA2 expression and the lipid raft content of the membranes as well as an effacement of the membrane protrusions. In addition, upon stimulation, HER2 no longer remains at the cell surface but is internalized with EGFR and HER3.
Discussion
Breast cells express the PMCA2w/b splice variant, which contains a C-terminal PDZ interaction motif and traffics to the apical membrane (34, 36, 39). During lactation, PMCA2 transports calcium into milk and prevents calcium-induced involution of secretory epithelial cells (35–37, 42, 48). In breast cancer cells, PMCA2 interacts with HER2 within specific membrane domains to maintain a low intracellular calcium concentration that supports active HER2 signaling and prevents HER2 internalization (30). Therefore, the mechanisms that regulate the membrane localization of PMCA2 and allow it to interact with HER2 are of considerable importance to normal lactation and breast cancer. The presence of a C-terminal PDZ-binding motif within PMCA2 suggests that a member of the NHERF scaffolding family might interact with PMCA2 and influence its membrane localization. We found that NHERF1 is prominently expressed in HER2-positive breast cancers, and, in CHO cells and MCF10A cells, we found that PMCA2 interacts with NHERF1 through its PDZ-binding motif. In breast cancer cells, we demonstrate that NHERF1 interacts with a complex that includes PMCA2, HSP90, and HER2 within lipid raft- and actin-rich membrane domains (Fig. 7). NHERF1 stabilizes interactions between PMCA2 and HER2 and between HER2 and HSP90 and maintains the structure and lipid raft content of membrane signaling domains in which activated HER2 resides after treatment with either EGF or NRG1. These data suggest that NHERF1 has a dominant role in determining the membrane localization of both PMCA2 and HER2.
We found that PMCA2 co-immunoprecipitates with both HER2 and NHERF1 and that knocking down NHERF1 disrupts the ability to co-IP PMCA2 and HER2. However, we could only detect co-immunoprecipitation of NHERF1 and HER2 after acute stimulation of HER2 signaling in MCF10A cells. Likewise, immunofluorescence imaging of PMCA2, HER2, NHERF1, and actin demonstrated increased localization to more prominent membrane protrusions after treatment with EGF or NRG1. These findings suggest that activation of HER2 may increase the recruitment of NHERF1 to a complex that includes PMCA2 and HER2 and that the addition of NHERF1 then helps to stabilize and/or augment interactions between PMCA2 and HER2 as well as the structure of the lipid raft-rich membrane protrusions (Fig. 7). Our prior work showed that PMCA2 regulates HER2 cell surface localization and activation as well as the structure of the membrane protrusions in part by maintaining low intracellular calcium concentrations (30). Given that knocking down NHERF1 decreases PMCA2 levels, increases intracellular calcium levels, and mimics the effects of loss of PMCA2 on HER2 signaling, we think it most likely that the ability of NHERF1 to regulate HER2 signaling is mediated directly by its capacity to bind to the C terminus of PMCA2 and retain both PMCA2 and HER2 within the overall signaling complex. However, NHERF1 has also been reported to regulate the assembly of microvilli and rho-mediated cytoskeletal remodeling (8, 49), and it is possible that NHERF1 stabilizes interactions between PMCA2 and HER2 indirectly by regulating the cytoskeletal structure of the membrane signaling domains rather than directly by scaffolding PMCA2 and/or HER2. More work will be needed to completely decipher the complex interactions between PMCA2, HER2, NHERF1, lipid rafts, and the actin cytoskeleton.
Knocking down NHERF1 expression in breast cancer cells inhibits HER2/Akt activation, inhibits cell proliferation, sensitizes cells to apoptosis, and results in changes in global gene expression that closely mimic those caused by knocking down HER2 itself. These data suggest that NHERF1 may contribute to the genesis or progression of HER2-positive breast cancers. Consistent with this idea, we found that NHERF1 expression correlated with HER2 status and PMCA2 expression and with metastatic spread to regional lymph nodes in a cohort of 652 breast cancers. In addition, high levels of NHERF1 predicted poorer survival in this same cohort, although, based on multivariate analysis, this may be a function of its correlation with PMCA2 and HER2 expression. These findings agree with several previous studies that documented positive correlations between HER2 and NHERF1 levels in human breast cancers and at least one report showing that NHERF1 correlates with a worse outcome in HER2-positive luminal B breast cancers (7, 12, 13, 50). However, other studies suggested that NHERF1 inhibits breast cancer growth by scaffolding the phosphate and tensin homolog (PTEN) protein and inhibiting PI3K signaling downstream of the PDGF receptor (11, 20). By contrast, we found that NHERF1 is required to support AKT activation downstream of HER2. Given that NHERFs can interact with a variety of different signaling molecules, it may be that NHERF1 exerts diverse effects on breast cancers in different signaling contexts. Larger clinical studies with the power to examine associations between NHERF1 and outcome in different molecular subtypes of breast cancer will be needed to more fully understand these contrasting results.
Receptor internalization followed by recycling or degradation is important for regulating signaling from receptor tyrosine kinases (26, 28). For reasons that remain incompletely understood, HER2 normally resists internalization and degradation upon activation and tends to remain at the cell surface, where it can continue to signal for prolonged periods, a property thought to be important for its transforming activity (26, 29). Interactions between HER2 and HSP90 as well as the localization of HER2 within membrane protrusions or microvillus-like structures have been reported previously to contribute to its resistance to internalization (27, 30, 31, 51–53). Our previous studies established that interactions between PMCA2 and HER2 are critical for HER2 to both interact with HSP90 and remain associated with membrane protrusions, and to avoid internalization, ubiquitination and degradation after its activation (30). We now demonstrate that NHERF1 is a key component of the same complex that stabilizes the membrane protrusions and HER2 surface localization and activity. These results suggest that it may be possible to devise new therapies for breast cancer by targeting NHERF1 to promote the internalization of HER2 and terminate its signaling.
Experimental procedures
Cell culture
SKBR3 cells and BT474 cells were obtained from the ATCC and maintained in culture in DMEM plus GlutaMAX-1 (Gibco/Life Technologies) containing 10% FBS and penicillin/streptomycin (Gibco/Life Technologies) at 37 °C in 5% CO2 (30). MCF10A and MCF10A-HER2 cells (a gift from the Stern laboratory, Yale University) were cultured in DMEM/F12 (Gibco/Life Technologies) containing 5% horse serum, EGF (100 μg/ml), hydrocortisone (1 mg/ml), cholera toxin (1 mg/ml), insulin (10 mg/ml), and penicillin/streptomycin (Gibco/Life Technologies) at 37 °C in 5% CO2 (30). In some experiments, cells were cultured as above but in medium without FBS or growth factors for 16 h and then treated with 100 ng/ml EGF (Cell Signaling Technology) or 50 ng/ml NRG1 (Cell Signaling Technology) for 1 or 2 h.
Knockdown cell lines
Stable cell lines expressing shRNA directed against NHERF1 and ErbB2 (HER2) were generated by transducing cells with commercially prepared lentiviruses encoding shRNAs targeting NHERF1 (sc-63330, Santa Cruz Biotechnology) or HER2 (318–328, Amsbio). Cells were infected by adding the various shRNA lentiviral particles to the culture for 48 h. Clones expressing the specific shRNAs were selected using 5 μg/ml of puromycin (Gibco/Life Technologies, NHERF1) or 5 μg/ml blasticidin S HCl (Gibco/Life Technologies, HER2).
Immunofluorescence
Cells were grown on coverslips, fixed in 4% paraformaldehyde for 20 min, permeabilized with 0.2% Triton X-100 for 10 min, washed three times with PBS, and incubated with primary antibody overnight at 4 °C. The cells were washed three times with PBS and incubated with secondary antibody for 1 h at room temperature. After washing, coverslips were mounted using Prolong Gold antifade reagent with DAPI (Invitrogen). Paraffin-embedded tissue sections were cleared with Histoclear (National Diagnostics) and graded alcohol using standard techniques. Antigen retrieval was performed using 7 mm citrate buffer (pH 6.0) under pressure. Sections were incubated with primary antibody overnight at 4 °C and with secondary antibody for 1 h at room temperature. Coverslips were mounted using Prolong Gold antifade reagent with DAPI (Invitrogen). All images were obtained using a Zeiss 780 confocal microscope. Primary antibodies were against the following: HER2 (sc-284), NHERF1 (sc-134485), flotillin 1 (sc-25506), phosphor-EGFR (sc-12351), FOXO1 (sc-11350), cbl (sc-170), ubiquitin (sc-8017), PMCA2 (PA1–915), and HER2 (MA1–35720) from Thermo Scientific (Waltham, MA); phospho-HER2 (2243S), phospho-AKT (4060S), AKT (4691S), EGFR (4267S), HER3 (12708P), phospho-HER3 (4791S), and FK2 (2325026) from Millipore (Temecula, CA); and P62 (610832) from BD Transduction Laboratories. We also stained for lipid rafts using cholera toxin subunit B-Alexa Fluor 555 conjugate (1423066) from Invitrogen and for actin using phalloidin-Atto 488 (49409) from Sigma.
Immunoblotting
Protein extracts were prepared using standard methods (30), subjected to SDS-PAGE, and transferred to a nitrocellulose membrane by wet Western-blotting transfer (Bio-Rad). The membrane was blocked in TBST buffer (TBS and 1% Tween) containing 5% milk for 1 h at room temperature. The blocked membranes were incubated overnight at 4 °C with primary antibodies in Odyssey blocking buffer (927-40000), washed three times with TBST buffer, and then incubated with secondary antibodies provided by LI-COR for 2 h at room temperature. After three washes with TBST buffer, the membranes were analyzed using the Odyssey infrared imaging system (LI-COR). Triton X-100-insoluble fractions were isolated using the caveolae/rafts isolation kit (CS0750) from Sigma. Samples were then analyzed for HER2 and flotillin1 by Western-blotting analysis as above. All immunoblot experiments were performed at least three times, and representative blots are shown in the figures.
RNA extraction and real-time RT-PCR
RNA was isolated using TRIzol (Invitrogen). Quantitative RT-PCR was performed with the SuperScript III Platinum One-Step quantitative RT-PCR kit (Invitrogen) using a Step One Plus real-time PCR system (Applied Biosystems) and the following TaqMan primer sets: ERBB2, Hs01001580_m1; PMCA2, Hs01090453_m1; NHERF1, Hs00188594_m1; Rab25, Hs01040784_m1; Tacstd2, Hs01922976_s1; Muc1, Hs00159357_m1; TGFB3, Hs01086000_m1; FOXA1, Hs04187555_m1; and AKT1, Hs00178289_m1. Human HPRT1 (4326321E) was used as reference genes (Invitrogen). Relative mRNA expression was determined using Step One software v2.2.2 (Applied Biosystems).
Co-immunoprecipitation
Cells were lysed with radioimmune precipitation assay buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 20 mm Tris-HCl, and 150 mm NaCl), and cell extracts were incubated overnight at 4 °C with protein A/G beads (sc-2003, Santa Cruz Biotechnology) and the specific antibody. After centrifugation, the immunoprecipitated proteins were eluted with LDS sample buffer (Invitrogen) containing 10% β-mercaptoethanol. The resulting samples were then analyzed by Western blotting (30).
Cell proliferation and apoptosis
Cell proliferation was assessed by measuring BrdU incorporation using the cell proliferation ELISA kit (11647229001) from Roche. Apoptosis was measured by TUNEL assay using the cell death detection ELISA kit (11544675001) from Roche (Genentech Inc.). Cell viability and cell numbers were quantified using the XTT cell viability assay (9095) from Cell Signaling Technology (Danvers, MA).
Tissue microarray
The breast carcinoma tissue microarray (YTMA49) consisted of 652 primary breast cancer specimens retrospectively obtained from 1953 to 1983. Cases were evenly divided between lymph node-positive and -negative, with a median follow-up of 8.9 years. Clinicopathologic data were extracted from Yale and Connecticut Tumor Registries, including disease-specific survival. Tissue microarray slides were stained as described previously (42). The primary antibodies were anti-NHERF1 (Affinity BioReagents) and a mouse anti-cytokeratin (Dako, Carpinteria, CA) to distinguish the tumor from stroma. The slides were counterstained with DAPI. Details of the automated image acquisition and analysis using AQUA have been described previously (41).
Gene array
Total RNA was prepared from control SKBR3 cells as well as NHERF1KD and HER2KD cells using TRIzol reagent (Invitrogen), and oligonucleotide microarray analysis was performed by the Yale Center for Genomic Analysis. The isolated RNA was purified using the RNeasy cleanup kit (Qiagen, Valencia, CA). RNA was reverse-transcribed and hybridized to the HumanHT-12 v3 Expression BeadChip (Illumina, San Diego, CA) and then scanned using the Illumina BeadArray reader. The images were analyzed by Beadstudio software. The instructions for quality control and data analysis were provided by Illumina.
Intracellular calcium measurements
Ratiometric intracellular calcium imaging was performed using 5 μm Fura-2/AM (Life Technologies) as described previously (30, 42). Cells were loaded with 5 μm Fura-2/AM (Life Technologies) for 30 min at 37 °C and then imaged at a frequency of 1 Hz on a Zeiss Axiovert 100 microscope. Intracellular calcium concentrations were calculated from the background-subtracted fluorescent ratio (R) at 340 and 380 nm of Fura-2/AM-loaded cells using the formula Kd × (R − Rmin)/(Rmax − R) × (Ff380/Fb380), where Kd is the dissociation constant of Fura 2 for calcium (225 nm), Rmin and Rmax are the empirically determined minimum and maximum fluorescent ratios, and Ff380/Fb380 is the fluorescence intensity at 380 nm in calcium-free conditions divided by the fluorescence intensity at 380 nm in saturating calcium concentrations (bound).
Statistics
Analyses were performed with Prism 6.0 (GraphPad Software, La Jolla, CA). Error bars represent standard error. Significance for all comparisons between two conditions was calculated using paired t tests, and significance for multiple comparisons was determined using one-way analysis of variance with Turkey post-test corrections. Significance for Kaplan-Meier analyses were calculated using the log-rank test, and correlations between PMCA2 and NHERF1 were calculated using Pearson's correlation coefficient.
Author contributions
J. J., J. N. V., W. K., and P. D. participated in the design, collection, and analysis of vital experiments. J. J. was responsible for the day-to-day planning and execution of the majority of the experiments. J. C. contributed to the analysis and interpretation of the microarray data. W. B. S. and P. A. F. contributed to the design and analysis of experiments examining the interactions between HER2 and NHERF1. J. J. W. was responsible for the oversight and analysis of all experiments and initiated the hypotheses that led to these experiments. J. J., P. A. F., and J. J. W. contributed to the writing of the manuscript.
Acknowledgments
We thank Dr. Emmanuel Strehler for the kind gift of PMCA2 expression vectors, Dr. David Stern for MCF10A-Her2 cells, Dr. David Rimm for assistance with TMA and AQUA analyses, Dr. Sihem Khelif for providing access to human DCIS samples, and Dr. Barbara Ehrlich for help with calcium imaging.
This work was supported by National Institutes of Health Grants CA153702 and HD076248 (to J. J. W.) and DK105811 (to P. A. F.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- EGFR
- EGF receptor
- MMTV
- murine mammary tumor virus
- IP
- immunoprecipitation
- AQUA
- automated quantitative analysis
- QPCR
- quantitative PCR
- DCIS
- ductal carcinoma in situ.
References
- 1. Weinman E. J., Hall R. A., Friedman P. A., Liu-Chen L. Y., and Shenolikar S. (2006) The association of NHERF adaptor proteins with G protein-coupled receptors and receptor tyrosine kinases. Annu. Rev. Physiol. 68, 491–505 [DOI] [PubMed] [Google Scholar]
- 2. Ardura J. A., and Friedman P. A. (2011) Regulation of G protein-coupled receptor function by Na+/H+ exchange regulatory factors. Pharmacol. Rev. 63, 882–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Voltz J. W., Weinman E. J., and Shenolikar S. (2001) Expanding the role of NHERF, a PDZ-domain containing protein adapter, to growth regulation. Oncogene 20, 6309–6314 [DOI] [PubMed] [Google Scholar]
- 4. Bretscher A., Chambers D., Nguyen R., and Reczek D. (2000) ERM-Merlin and EBP50 protein families in plasma membrane organization and function. Annu. Rev. Cell Dev. Biol. 16, 113–143 [DOI] [PubMed] [Google Scholar]
- 5. Brône B., and Eggermont J. (2005) PDZ proteins retain and regulate membrane transporters in polarized epithelial cell membranes. Am. J. Physiol. Cell Physiol. 288, C20–29 [DOI] [PubMed] [Google Scholar]
- 6. Nourry C., Grant S. G., and Borg J. P. (2003) PDZ domain proteins: plug and play! Sci. STKE 2003, RE7. [DOI] [PubMed] [Google Scholar]
- 7. Bellizzi A., Malfettone A., Cardone R. A., and Mangia A. (2010) NHERF1/EBP50 in breast cancer: clinical perspectives. Breast Care 5, 86–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cardone R. A., Bellizzi A., Busco G., Weinman E. J., Dell'Aquila M. E., Casavola V., Azzariti A., Mangia A., Paradiso A., and Reshkin S. J. (2007) The NHERF1 PDZ2 domain regulates PKA-RhoA-p38-mediated NHE1 activation and invasion in breast tumor cells. Mol. Biol. Cell 18, 1768–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dai J. L., Wang L., Sahin A. A., Broemeling L. D., Schutte M., and Pan Y. (2004) NHERF (Na+/H+ exchanger regulatory factor) gene mutations in human breast cancer. Oncogene 23, 8681–8687 [DOI] [PubMed] [Google Scholar]
- 10. Georgescu M. M., Morales F. C., Molina J. R., and Hayashi Y. (2008) Roles of NHERF1/EBP50 in cancer. Curr. Mol. Med. 8, 459–468 [DOI] [PubMed] [Google Scholar]
- 11. Georgescu M. M. (2008) NHERF1: molecular brake on the PI3K pathway in breast cancer. Breast Cancer Res. 10, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karn T., Ruckhäberle E., Hanker L., Müller V., Schmidt M., Solbach C., Gätje R., Gehrmann M., Holtrich U., Kaufmann M., and Rody A. (2011) Gene expression profiling of luminal B breast cancers reveals NHERF1 as a new marker of endocrine resistance. Breast Cancer Res. Treat. 130, 409–420 [DOI] [PubMed] [Google Scholar]
- 13. Mangia A., Chiriatti A., Bellizzi A., Malfettone A., Stea B., Zito F. A., Reshkin S. J., Simone G., and Paradiso A. (2009) Biological role of NHERF1 protein expression in breast cancer. Histopathology 55, 600–608 [DOI] [PubMed] [Google Scholar]
- 14. Maudsley S., Zamah A. M., Rahman N., Blitzer J. T., Luttrell L. M., Lefkowitz R. J., and Hall R. A. (2000) Platelet-derived growth factor receptor association with Na+/H+ exchanger regulatory factor potentiates receptor activity. Mol. Cell. Biol. 20, 8352–8363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pan Y., Weinman E. J., and Dai J. L. (2008) Na+/H+ exchanger regulatory factor 1 inhibits platelet-derived growth factor signaling in breast cancer cells. Breast Cancer Res. 10, R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Song J., Bai J., Yang W., Gabrielson E. W., Chan D. W., and Zhang Z. (2007) Expression and clinicopathological significance of oestrogen-responsive ezrin-radixin-moesin-binding phosphoprotein 50 in breast cancer. Histopathology 51, 40–53 [DOI] [PubMed] [Google Scholar]
- 17. Cheng S., Li Y., Yang Y., Feng D., Yang L., Ma Q., Zheng S., Meng R., Wang S., Wang S., Jiang W. G., and He J. (2013) Breast cancer-derived K172N, D301V mutations abolish Na+/H+ exchanger regulatory factor 1 inhibition of platelet-derived growth factor receptor signaling. FEBS Lett. 587, 3289–3295 [DOI] [PubMed] [Google Scholar]
- 18. Du G., Gu Y., Hao C., Yuan Z., He J., Jiang W. G., and Cheng S. (2016) The cellular distribution of Na+/H+ exchanger regulatory factor 1 is determined by the PDZ-I domain and regulates the malignant progression of breast cancer. Oncotarget 7, 29440–29453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Du G., Hao C., Gu Y., Wang Z., Jiang W. G., He J., and Cheng S. (2016) A novel NHERF1 mutation in human breast cancer inactivates inhibition by NHERF1 protein in EGFR signaling. Anticancer Res. 36, 1165–1173 [PubMed] [Google Scholar]
- 20. Pan Y., Weinman E. J., and Dai J. L. (2008) Na+/H+ exchanger regulatory factor 1 inhibits platelet-derived growth factor signaling in breast cancer cells. Breast Cancer Res. 10, R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lazar C. S., Cresson C. M., Lauffenburger D. A., and Gill G. N. (2004) The Na+/H+ exchanger regulatory factor stabilizes epidermal growth factor receptors at the cell surface. Mol. Biol. Cell 15, 5470–5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arteaga C. L., Sliwkowski M. X., Osborne C. K., Perez E. A., Puglisi F., and Gianni L. (2011) Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev. Clin. Oncol. 9, 16–32 [DOI] [PubMed] [Google Scholar]
- 23. Ursini-Siegel J., Schade B., Cardiff R. D., and Muller W. J. (2007) Insights from transgenic mouse models of ERBB2-induced breast cancer. Nat. Rev. Cancer 7, 389–397 [DOI] [PubMed] [Google Scholar]
- 24. Arteaga C. L., and Engelman J. A. (2014) ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell 25, 282–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hynes N. E., and MacDonald G. (2009) ErbB receptors and signaling pathways in cancer. Curr. Opin Cell Biol. 21, 177–184 [DOI] [PubMed] [Google Scholar]
- 26. Bertelsen V., and Stang E. (2014) The mysterious ways of ErbB2/HER2 trafficking. Membranes 4, 424–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hommelgaard A. M., Lerdrup M., and van Deurs B. (2004) Association with membrane protrusions makes ErbB2 an internalization-resistant receptor. Mol. Biol. Cell 15, 1557–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sorkin A., and Goh L. K. (2009) Endocytosis and intracellular trafficking of ErbBs. Exp. Cell Res. 315, 683–696 [DOI] [PubMed] [Google Scholar]
- 29. Stern D. F., Heffernan P. A., and Weinberg R. A. (1986) p185, a product of the neu proto-oncogene, is a receptorlike protein associated with tyrosine kinase activity. Mol. Cell. Biol. 6, 1729–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jeong J., VanHouten J. N., Dann P., Kim W., Sullivan C., Yu H., Liotta L., Espina V., Stern D. F., Friedman P. A., and Wysolmerski J. J. (2016) PMCA2 regulates HER2 protein kinase localization and signaling and promotes HER2-mediated breast cancer. Proc. Natl. Acad. Sci. U.S.A. 113, E282–E290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zagouri F., Sergentanis T. N., Chrysikos D., Papadimitriou C. A., Dimopoulos M. A., and Psaltopoulou T. (2013) Hsp90 inhibitors in breast cancer: a systematic review. Breast 22, 569–578 [DOI] [PubMed] [Google Scholar]
- 32. Brini M., and Carafoli E. (2009) Calcium pumps in health and disease. Physiol. Rev. 89, 1341–1378 [DOI] [PubMed] [Google Scholar]
- 33. Strehler E. E. (2013) Plasma membrane calcium ATPases as novel candidates for therapeutic agent development. J. Pharm. Pharm. Sci. 16, 190–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Strehler E. E., and Zacharias D. A. (2001) Role of alternative splicing in generating isoform diversity among plasma membrane calcium pumps. Physiol. Rev. 81, 21–50 [DOI] [PubMed] [Google Scholar]
- 35. Reinhardt T. A., Lippolis J. D., Shull G. E., and Horst R. L. (2004) Null mutation in the gene encoding plasma membrane Ca2+-ATPase isoform 2 impairs calcium transport into milk. J. Biol. Chem. 279, 42369–42373 [DOI] [PubMed] [Google Scholar]
- 36. VanHouten J. N., Neville M. C., and Wysolmerski J. J. (2007) The calcium-sensing receptor regulates plasma membrane calcium adenosine triphosphatase isoform 2 activity in mammary epithelial cells: a mechanism for calcium-regulated calcium transport into milk. Endocrinology 148, 5943–5954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. VanHouten J. N., and Wysolmerski J. J. (2007) Transcellular calcium transport in mammary epithelial cells. J. Mammary Gland Biol. Neoplasia 12, 223–235 [DOI] [PubMed] [Google Scholar]
- 38. Antalffy G., Caride A. J., Pászty K., Hegedus L., Padanyi R., Strehler E. E., and Enyedi A. (2011) Apical localization of PMCA2w/b is enhanced in terminally polarized MDCK cells. Biochem. Biophys. Res. Commun. 410, 322–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Padányi R., Xiong Y., Antalffy G., Lór K., Pászty K., Strehler E. E., and Enyedi A. (2010) Apical scaffolding protein NHERF2 modulates the localization of alternatively spliced plasma membrane Ca2+ pump 2B variants in polarized epithelial cells. J. Biol. Chem. 285, 31704–31712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chan M. M., Lu X., Merchant F. M., Iglehart J. D., and Miron P. L. (2005) Gene expression profiling of NMU-induced rat mammary tumors: cross species comparison with human breast cancer. Carcinogenesis 26, 1343–1353 [DOI] [PubMed] [Google Scholar]
- 41. Camp R. L., Chung G. G., and Rimm D. L. (2002) Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat. Med. 8, 1323–1327 [DOI] [PubMed] [Google Scholar]
- 42. VanHouten J., Sullivan C., Bazinet C., Ryoo T., Camp R., Rimm D. L., Chung G., and Wysolmerski J. (2010) PMCA2 regulates apoptosis during mammary gland involution and predicts outcome in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 107, 11405–11410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Camp R. L., Dolled-Filhart M., and Rimm D. L. (2004) X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin. Cancer Res. 10, 7252–7259 [DOI] [PubMed] [Google Scholar]
- 44. Yang J. Y., and Hung M. C. (2009) A new fork for clinical application: targeting forkhead transcription factors in cancer. Clin. Cancer Res. 15, 752–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Simons K., and Toomre D. (2000) Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1, 31–39 [DOI] [PubMed] [Google Scholar]
- 46. Alonso M. A., and Millán J. (2001) The role of lipid rafts in signalling and membrane trafficking in T lymphocytes. J. Cell Sci. 114, 3957–3965 [DOI] [PubMed] [Google Scholar]
- 47. Patra S. K. (2008) Dissecting lipid raft facilitated cell signaling pathways in cancer. Biochim. Biophys. Acta 1785, 182–206 [DOI] [PubMed] [Google Scholar]
- 48. Reinhardt T. A., and Lippolis J. D. (2009) Mammary gland involution is associated with rapid down regulation of major mammary Ca2+-ATPases. Biochem. Biophys. Res. Commun. 378, 99–102 [DOI] [PubMed] [Google Scholar]
- 49. Garbett D., LaLonde D. P., and Bretscher A. (2010) The scaffolding protein EBP50 regulates microvillar assembly in a phosphorylation-dependent manner. J. Cell Biol. 191, 397–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Paradiso A., Scarpi E., Malfettone A., Addati T., Giotta F., Simone G., Amadori D., and Mangia A. (2013) Nuclear NHERF1 expression as a prognostic marker in breast cancer. Cell Death Dis. 4, e904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Carraway C. A., Carvajal M. E., Li Y., and Carraway K. L. (1993) Association of p185neu with microfilaments via a large glycoprotein complex in mammary carcinoma microvilli: evidence for a microfilament-associated signal transduction particle. J. Biol. Chem. 268, 5582–5587 [PubMed] [Google Scholar]
- 52. Ivanova J. L., Edelweiss E. F., Leonova O. G., Balandin T. G., Popenko V. I., and Deyev S. M. (2012) Application of fusion protein 4D5 scFv-dibarnase:barstar-gold complex for studying P185HER2 receptor distribution in human cancer cells. Biochimie 94, 1833–1836 [DOI] [PubMed] [Google Scholar]
- 53. Mori S., Akiyama T., Morishita Y., Shimizu S., Sakai K., Sudoh K., Toyoshima K., and Yamamoto T. (1987) Light and electron microscopical demonstration of c-erB-2 gene product-like immunoreactivity in human malignant tumors. Virchows Arch. B. Cell Pathol. Incl. Mol. Pathol 54, 8–15 [DOI] [PubMed] [Google Scholar]