Abstract
Drosophila gp93 was identified as the ortholog of the mammalian endoplasmic reticulum-resident chaperone gp96. gp93 was found capable of rescuing gp96 client proteins, such as Toll-like receptors (TLRs) and integrins, in a gp96-deficient murine cell line. Mammalian gp96 was further found to require the cochaperone canopy 3 (CNPY3) for proper folding and expression of TLRs, but not integrins. In Drosophila, two possible CNPY family members have been identified but have not yet been characterized. Therefore, we sought to determine the role of Drosophila CNPYa and CNPYb in gp93 biology. Because of higher similarities between CNPY3 and CNPYb, we postulated that CNPYb would be a TLR-specific cochaperone of gp93. Indeed, CNPYb addition in gp93-expressing cells improved TLR expression. CNPYb and gp93 were further found to physically interact. Mutational analysis of cysteine residues in CNPYb identified differential dependence of these cysteines in chaperone function. Our study is the first to characterize Drosophila CNPY molecules. We further uncover more gp93 biology by identifying CNPYb as a cochaperone. A better understanding of this simpler Drosophila system will enable application to the mammalian system, such as has been done with Escherichia coli, yeast, and mammalian HSP90.
Keywords: chaperone, Drosophila, innate immunity, integrin, protein folding, toll receptor, toll-like receptor (TLR), gp93, gp96
Introduction
There exists a small family of canopy (CNPY)2 molecules that are highly conserved across animal kingdoms. There are four mammalian members (CNPY1, CNPY2, CNPY3, and CNPY4) but only two Drosophila members (CNPYa and CNPYb), which have yet to be characterized. Mammalian CNPY1 and CNPY2 have short C-terminal sequences as compared with CNPY3 and CNPY4 and are thus divided into two subgroups. CNPY molecules all share a similar pattern of six cysteine residues, which is characteristic of the saposin-like proteins. All CNPY molecules are further targeted to the endoplasmic reticulum (ER) via a signal peptide and ER retention sequence. Additionally, CNPY3 has a highly basic C-terminal region rich in lysine residues. CNPYs are assumed to form saposin-like dimers because of their cysteine residues and because CNPY1 harbors self-binding properties (1).
CNPY3 was found to be a Toll-like receptor (TLR)-specific cochaperone of mammalian gp96 (glycoprotein of 96 kilodaltons) (2). Also known as PRAT4A (protein associated with TLR4 A), CNPY3 can regulate TLR4 signaling and cell surface expression (3). Its role in regulating TLR1, TLR2, TLR5, TLR7, and TLR9 was also later uncovered (4, 5). Curiously, similar to gp96, CNPY3 does not appear to play a role in TLR3 regulation (4). The characterization of three single-nucleotide polymorphisms (R95L, S231I, and M145K) in CNPY3 led to the discovery that M145K could alter function (6). As expected of a cochaperone, CNPY3 and gp96 have been determined to physically interact, although the complex is dissociated with ATP. CNPY3 presumably promotes client loading, because it binds the ATP-sensitive form of gp96 (2). Additionally, IFNγ appears to induce CNPY3 expression in human monocytes (7).
Although CNPY3 is the most studied CNPY family member, some information has been uncovered about the other CNPY members. Similar to CNPY3 for instance, CNPY4 can also regulate TLR4 cell surface expression and has thus been called PRAT4B (protein associated with TLR4 B). It is ubiquitously expressed in various leukocytes and tissues, and it is conserved among different vertebrate species (8). Contrary to the complimentary roles of CNPY3 and CNPY4 in TLR4 regulation, these CNPY molecules have contradictory roles in TLR1 biology. Although CNPY3 aids cell surface trafficking of TLR1, CNPY4 negatively impacts it, demonstrating both positive and negative regulatory roles of CNPYs in TLR biology (7).
CNPY1 appears to be the most unique family member in terms of expression. Instead of being ubiquitously expressed, this molecule appears to be restricted to the nervous system. CNPY1 has been found to interact with FGFR1 to regulate FGF signaling in zebrafish along the midbrain-hindbrain boundary (1). This positive feedback was found to control dorsal forerunner cell clustering, which generates the Kupffer's vesicle organ, responsible for left-right body plan asymmetry in zebrafish development (9). Additionally, FGF21 was found to enhance CNPY2 expression. CPNY2 can interact with Idol/Mylip, an E3 ubiquitin ligase that degrades LDL receptors, and overexpression of CNPY2 can increase LDL receptors, which are critical in regulating blood cholesterol levels (10).
We previously identified Drosophila gp93 as the ortholog of mammalian gp96. Utilizing a mutant gp96 cell line, we determined that gp93 was capable of interacting with and rescuing various gp96 client proteins (integrins and TLRs) (11). Later, Maynard et al. (12) found that gp93 knock-out in Drosophila is late-larval lethal. Specifically, defects were seen in the midgut with abnormal copper cell and septate junction structures, decreased motility, reduced acidification, deficient nutrient uptake, depressed insulin signaling, and suppressed triglyceride and amino acid mobilization. Collectively, mutant flies exhibited a starvation-like phenotype (12).
Because gp96 chaperones mammalian TLRs and integrins, we thought that gp93 might chaperone the various Tolls and integrins in Drosophila (13–15). We further postulate that gp93 requires a CNPY cochaperone; for example, gp96 requires CNPY3 for TLR expression (2). We therefore sought to uncover the role of Drosophila CNPYa and CNPYb in gp93 biology. Biochemical analysis reveals that gp93 can interact with CNPYb and CNPY3, and that CNPYb improves TLR expression in gp93-expressing cells. Furthermore, cysteine residues in CNPYb were proven to be important for chaperoning TLR4, whereas Cys-39 and Cys-183 were also required for TLR2 expression. Moving to a Drosophila system using RNAi, we were unable to determine whether or not gp93 plays a role in Toll signaling, although this warrants further examination. This study is the first to characterize the Drosophila CNPY molecules and uncovers a possible cochaperone for gp93.
Results
Murine gp96 and Drosophila gp93 can dimerize
Mammalian gp96 chaperone function is highly conserved, such that Drosophila gp93 is able to rescue gp96 client proteins (TLRs and integrins) in a gp96 mutant murine pre-B cell line (11). Upon a closer look, the gp96 dimerization domain, amino acids 697–740, is highly conserved in gp93 with 24 of 44 residues the same (55%; Fig. 1A). Because of this high level of conservation coupled with the fact that gp96 normally functions as a homodimer, we hypothesized that gp96 and gp93 might dimerize. We therefore utilized wild-type murine pre-B cells (14.GFP) already containing endogenous gp96 and further transduced these cells with Flag-tagged Drosophila gp93 or Flag-tagged murine gp96 as a control. Transgene expression was verified by GFP and Flag intracellular staining. The cells were also analyzed for gp96 expression, which remained similar between the two cell lines (Fig. 1B). Gp96 and Flag expression were further verified by Western blotting analysis, where β-actin expression was used as an endogenous loading control (Fig. 1C). We then performed immunoprecipitation experiments on these cells, either pulling down gp96 or using an isotype control antibody, and then performing Western blotting analysis for both gp96 and Flag expression. Expectedly, dimerization can be seen in WT cells with gp96-Flag. Furthermore, we found that gp93 can indeed interact with gp96 under cross-linking conditions (Fig. 1D). To verify this data, we then performed the reverse immunoprecipitation, pulling down Flag and blotting for gp96. In this scenario, gp93 was found to interact with gp96 in both cross-linking and non-cross-linking conditions (Fig. 1E). To note, the gp96 antibody does not recognize gp93, and the gp96 antibody used for Western blotting does not recognize gp96-Flag because of disruption of the epitope by Flag insertion (data not shown). By demonstrating that gp96 and gp93 can dimerize, we unveil further conserved characteristics between the two molecules.
Figure 1.
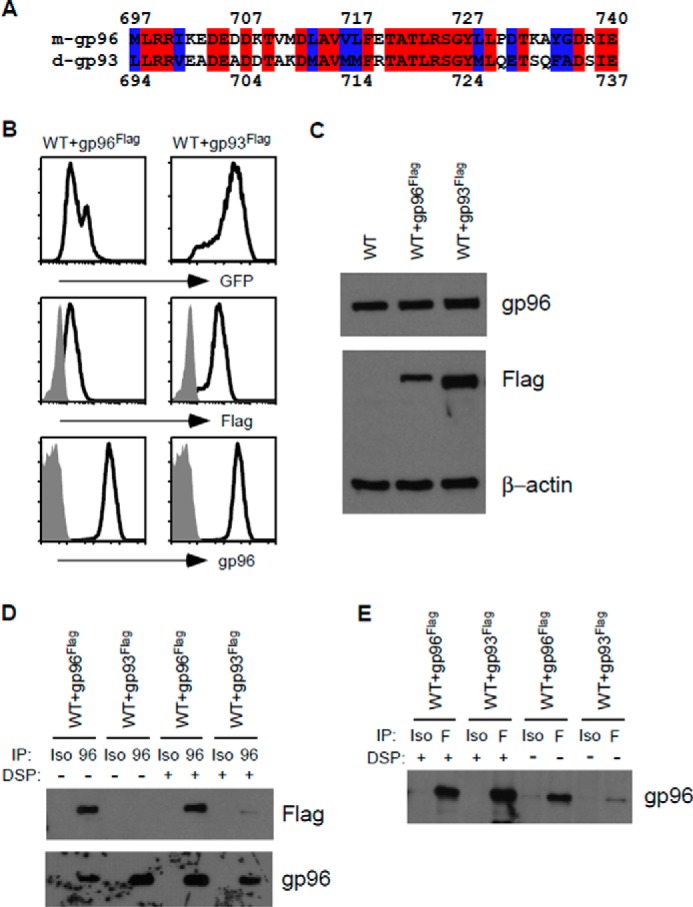
Drosophila gp93 can dimerize with murine gp96. A, amino acid sequence alignment of murine gp96 697–740 and Drosophila gp93 694–737, corresponding to the known gp96 dimerization domain. B, gp96-sufficient WT pre-B cells were retrovirally transduced with either gp96Flag or gp93Flag. Transgene expression was analyzed via flow cytometry for GFP and intracellular staining for Flag expression. gp96 expression was further probed. Gray histograms correspond to isotype controls. C, Western blotting analysis of the cells in B alongside parent WT pre-B cells. The membranes were probed for gp96, Flag, and β-actin. D, lysates from WT+gp96Flag or WT+gp93Flag cells were immunoprecipitated with either the gp96 or isotype (Iso) control antibody in the presence or absence of DSP and then resolved on a SDS-PAGE. Membranes were then blotted for Flag and gp96 expression. E, the reverse of D. Lysates were immunoprecipitated with Flag or isotype control antibody and then probed for gp96 expression.
CNPY conservation
Mammalian gp96 was found to require the cochaperone CNPY3 for proper folding of various TLRs (2). Although mammals have four CNPY family members (CNPY1–4), two such family members have been identified in Drosophila: CNPYa and CNPYb (1). Similar to their mammalian counterparts, both Drosophila CNPYs contain a signal peptide, an ER-retention signal, and six characteristic cysteine residues similar to the saposin family. CNPYb further contains a small basic region of amino acids at its C terminus, similar to that found in mammalian CNPY3 (Fig. 2A). Structurally, these CNPYs are proposed to fold upon themselves in a clam shape, where corresponding cysteine residues are able to form disulfide bonds (Fig. 2B). However, it is also possible that these residues form intermolecular bonds of either CNPY homo- or heterodimers. Although not as highly conserved as gp96-gp93, we still observe a high degree of amino acid conservation between CNPYb and CNPY3 or CNPY4 (50.2 and 49.3%, respectively). The amino acid sequence alignment demonstrates that the most conserved residues are located in the middle region (Fig. 2C and supplemental Fig. S1). Because of this high degree of conservation, and in particular the presence of the basic region, we hypothesize that CNPYb may serve as an ortholog of CNPY3 and hence as a cochaperone for gp93. In comparison, CNPYb is only 12.2% conserved compared with CNPY1, and 21.7% to CNPY2. Additionally, conservation numbers for CNPYa are 9% to CNPY1, 25.4% to CNPY2, 24.3% to CNPY3, and 26.5% to CNPY4 (supplemental Fig. S1). A phylogenetic tree further indicates similarities among canopy 3, 4, and b, and 1, 2, and a (Fig. 2D).
Figure 2.
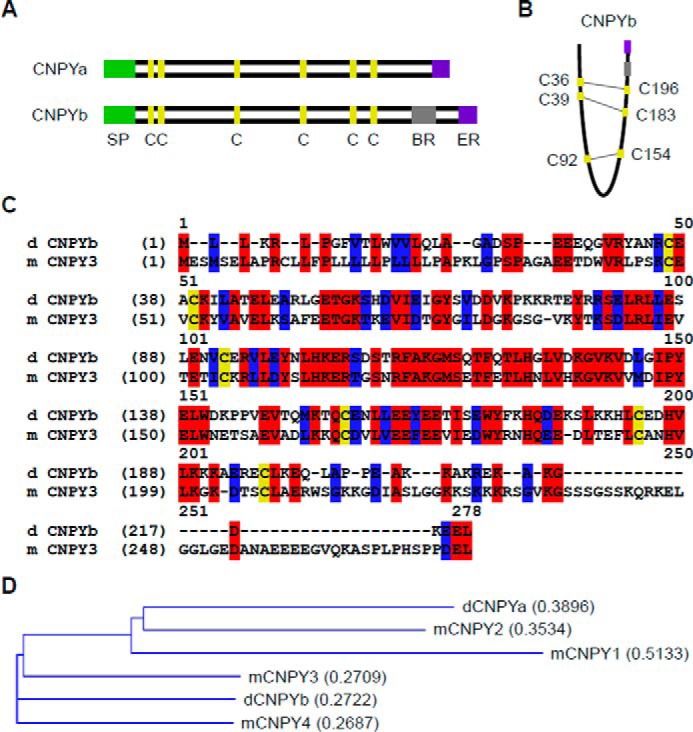
Drosophila CNPYb resembles murine CNPY3. A, schematic representation of Drosophila CNPYa and CNPYb. SP, signal peptide; C, cysteine residue. BR, basic region; ER, ER retention signal. B, schematic of the postulated folding structure of CNPYb and interacting cysteine residues. Numbers denote amino acid positions. C, amino acid sequence alignment of Drosophila CNPYb and murine CNPY3. Red residues are conserved, whereas blue residues are similar. Cysteine residues are highlighted in yellow. Numbers denote amino acid positions. D, phylogenetic tree comparing the indicated CNPY members.
CNPYb improves TLR expression
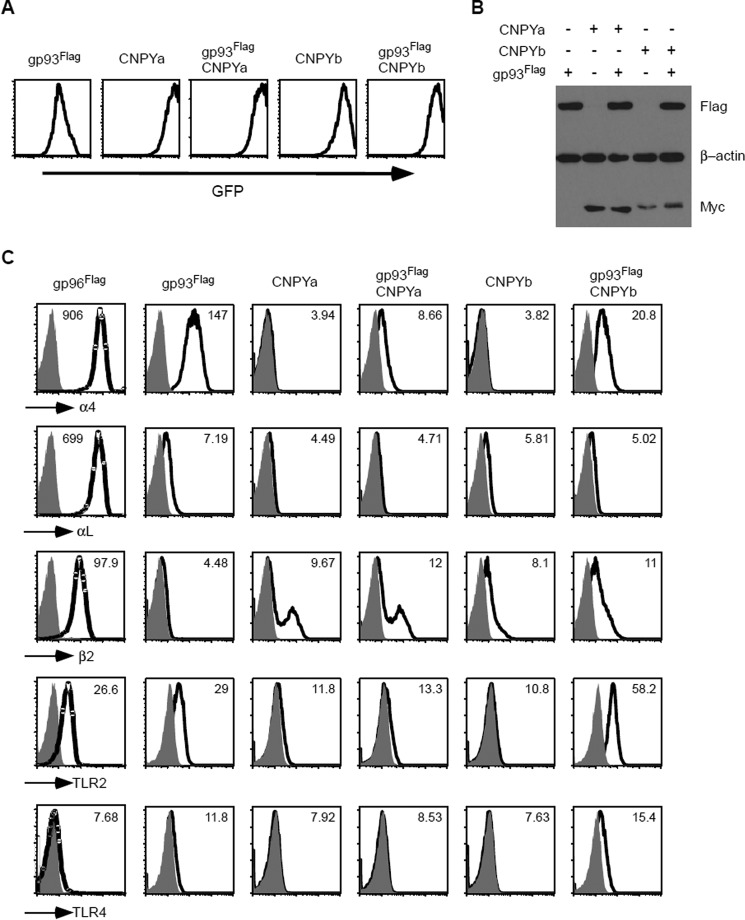
To study the role of CNPYa and CNPYb in gp93 biology, we again used a gp96 mutant murine pre-B cell line. CNPYa or CNPYb, both Myc-tagged, were retrovirally transduced into cells with or without gp93-Flag. Transgene expression was demonstrated by analysis of GFP reporter expression by Flow cytometry (Fig. 3A). Transgene expression was further verified by Western blotting for Flag and Myc expression, using β-actin as a loading control (Fig. 3B). Each cell line was then assayed for gp96-client expression. First, we show gp96 for comparison with gp93-expressing cells, the results of which have been previously discussed (11). Interestingly, CNPYa cells without gp93 had partial rescue of β2 integrin, yet the addition of gp93 had no effect. CNPYa did not improve any other client examined (Fig. 3C). Similarly, CNPYb alone increased expression of β2 integrin. Additionally, CNPYa decreased α4, αL, TLR2, and TLR4 expression in gp93-expressing cells. Further, cells expressing gp93 and CNPYb exhibited decreased α4 and αL yet increased β2, TLR2, and TLR4 expression (Fig. 3C). Therefore, CNPYb appears to be similar to CNPY3 in improving TLR expression, whereas CNPYa negatively impacts TLR expression. However, we see additional gp93-independent effects of CNPYa/b on integrin expression.
Figure 3.
Increased TLR expression with CNPYb. A, gp96-deficient pre-B cells were retrovirally transduced with gp93Flag, CNPYaMyc, CNPYbMyc, or combinations as indicated. Transgene expression was analyzed via flow cytometry for GFP expression. B, lysates from the cells in A were resolved on a SDS-PAGE and probed for Flag, Myc, and β-actin expression. C, cells from A were analyzed for α4, αL, β2, TLR2, and TLR4 expression via flow cytometry. The cells with gp96Flag were shown for comparison. Shaded histograms indicate isotype controls. Numbers denote mean fluorescence intensity.
CNPY3 and CNPYb bind both gp96 and gp93
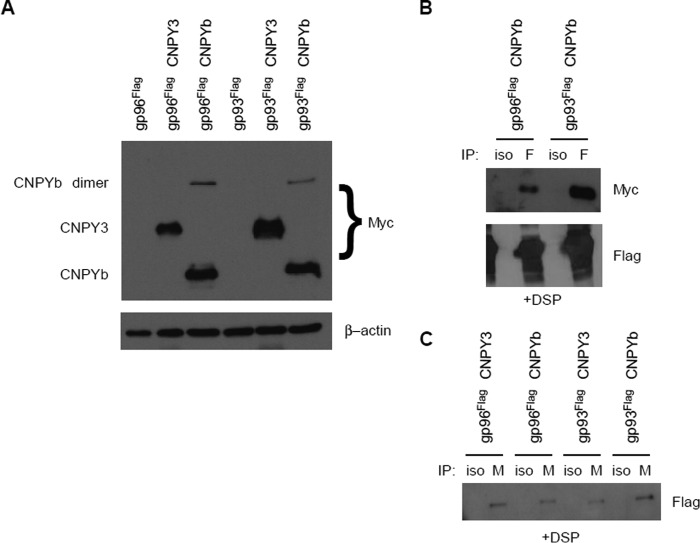
By definition, a cochaperone should physically bind to a chaperone to perform its function. Therefore, we sought to determine whether CNPYb interacts with gp93. We took gp96 mutant pre-B cells already transduced with either gp96-Flag or gp93-Flag and then further transduced them with either CNPY3-Myc or CNPYb-Myc. Transgene expression was verified by Western blotting analysis for myc. CNPY3 is ∼37 kDa, but CNPYb is ∼25 kDa in size. In cells with CNPYb, there appeared a second band at 50 kDa, which we presumed to be homodimers (Fig. 4A). We next performed Flag immunoprecipitation of gp96-Flag and gp93-Flag cells with CNPYb, followed by Western blotting for Myc. Under cross-linking conditions, not only did we see gp93 interact with CNPYb, but we even saw gp96 interact with CNPYb. We then probed the blots for Flag, demonstrating high pulldown efficiency (Fig. 4B). We next performed the reverse immunoprecipitation with all cells by pulling down Myc and blotting for Flag. Not only did we confirm that both gp96 and gp93 interact with CNPYb, but we further demonstrate that gp93 also interacts with CNPY3 (Fig. 4C). These differential interactions further demonstrate the high level of conservation in this system.
Figure 4.
gp93 and CNPYb dimerize. A, gp96-deficient pre-B cells were retrovirally transduced with gp96Flag or gp93Flag alone or in combination with CNPY3Myc or CNPYbMyc. Lysate was resolved on a SDS-PAGE and probed for Myc and β-actin expression. B, lysate from cells expressing CNPYb and either gp96 or gp93 were immunoprecipitated (IP) with Flag (F) or isotype (iso) control antibody in the presence of DSP, resolved on a SDS-PAGE, and blotted for Myc followed by Flag. C, lysate from cells coexpressing either gp96 or gp93 with CNPY3 or CNPYb were immunoprecipitated with Myc (M) or isotype control antibody in the presence of DSP, resolved on a SDS-PAGE, and probed for Flag expression.
Conserved cysteine residues in CNPYb are important for cochaperone function
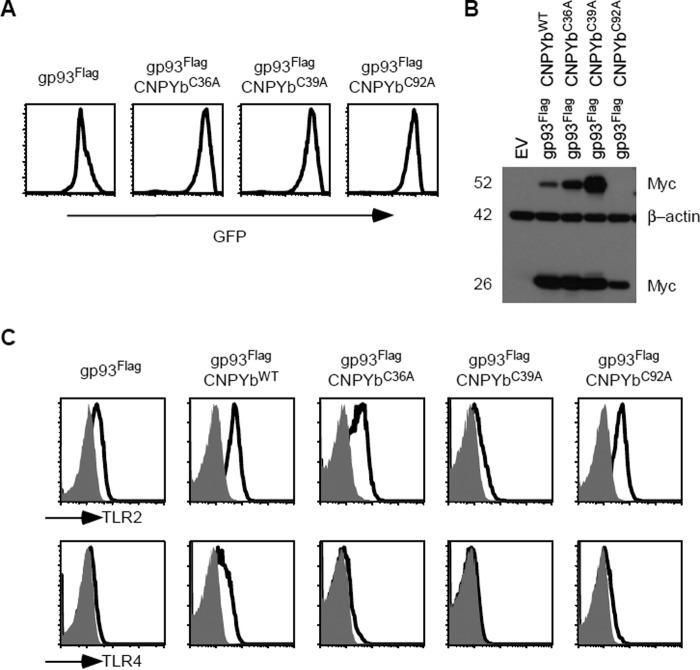
Although CNPYb is only ∼50% homologous to CNPY3, there must be some important conserved residues that maintain cochaperone function. Because the six cysteine residues are conserved, we decided to determine whether these residues were in fact important for function. Furthermore, if the proposed clam shape structure is correct, then mutating the first cysteine should phenocopy a mutation in the last cysteine (Fig. 2B). Therefore, we first performed mutational analysis on only the first three cysteine residues in CNPYb: at positions 36, 39, and 92. Using site-directed mutagenesis, these cysteines were mutated to alanine residues, and the Myc-tagged constructs were then transduced into gp96 mutant pre-B cells containing gp93-Flag. GFP reporter expression was first analyzed via flow cytometry (Fig. 5A). Transgene expression was then verified by Western blotting for Myc expression. Here, not only do we see dimers with WT CNPYb, but we also see dimers with the CNPYb mutants C36A and C39A. Although no dimers were seen with CNPYb C92A, the expression level in these cells was substantially lower and therefore easier to reduce (Fig. 5B). Next, we sought to determine whether or not these mutations would affect TLR expression. Via flow cytometry, TLR4 expression was reduced in cells containing gp93 and any of the CNPYb mutants. However, only CNPYb C39A had decreased TLR2 expression (Fig. 5C). These data imply the usage of different CNPYb cysteine residues for cochaperoning different client proteins.
Figure 5.
Differential cysteine dependence of CNPYb for TLR expression. A, gp96-deficient pre-B cells expressing gp93Flag were further transduced with various CNPYb mutants: C36A, C39A, or C92A. CNPY transgene expression was analyzed via increased GFP expression by flow cytometry. B, the indicated cell lysates were resolved on SDS-PAGE and probed for Myc and β-actin expression. Numbers denote kilodaltons. C, flow cytometry of the indicated cells was preformed to analyze TLR2 and TLR4 expression. Shaded histograms indicate isotype control staining.
Because only CNPYb C39A had the most severe phenotype of decreased TLR2 and TLR4 expression and because this residue is postulated to interact with Cys-183, we therefore decided to make the new construct CNPYb C183A. Again, we transduced the mutant CNPYb into gp96 mutant pre-B cells containing gp93-Flag. Increased GFP reporter expression was verified by flow cytometry (Fig. 6A), and transgene expression was demonstrated by Western blotting analysis of myc. This mutant also displayed a dimer at 50 kDa (Fig. 6B). Flow cytometry then confirmed a phenocopy of gp93-Flag + CNPYb C39A cells, with reduced TLR2 and TLR4 expression as compared with cells with gp93-Flag alone (Fig. 6C). This implies that Cys-39 and Cys-183 of CNPYb may indeed form a disulfide bridge important for cochaperone function.
Figure 6.
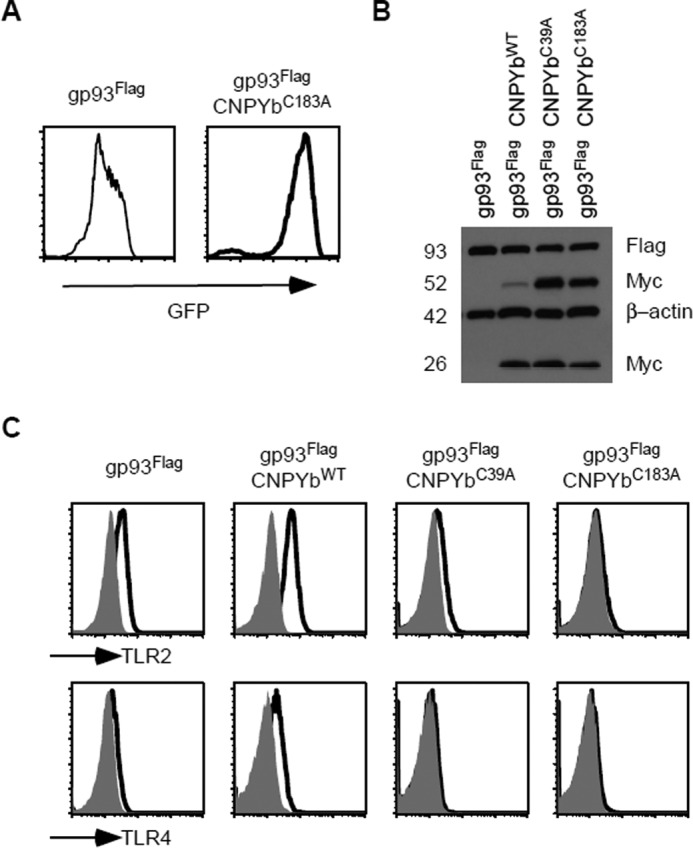
CNPYbC183A phenocopies the C39A mutant. A, gp93Flag expressing gp96-deficient pre-B cells were further transduced with CNPYbC183A. CNPYb transgene expression was verified by improved GFP expression analyzed via flow cytometry. B, lysates from the indicated cells were resolved on a SDS-PAGE and blotted for Flag, Myc, and β-actin. Numbers denote kilodaltons. C, TLR2 and TLR4 expression was analyzed via flow cytometry of the indicated cells. Shaded histograms indicate isotype control staining.
Do gp93 and CNPYb chaperone Drosophila Toll?
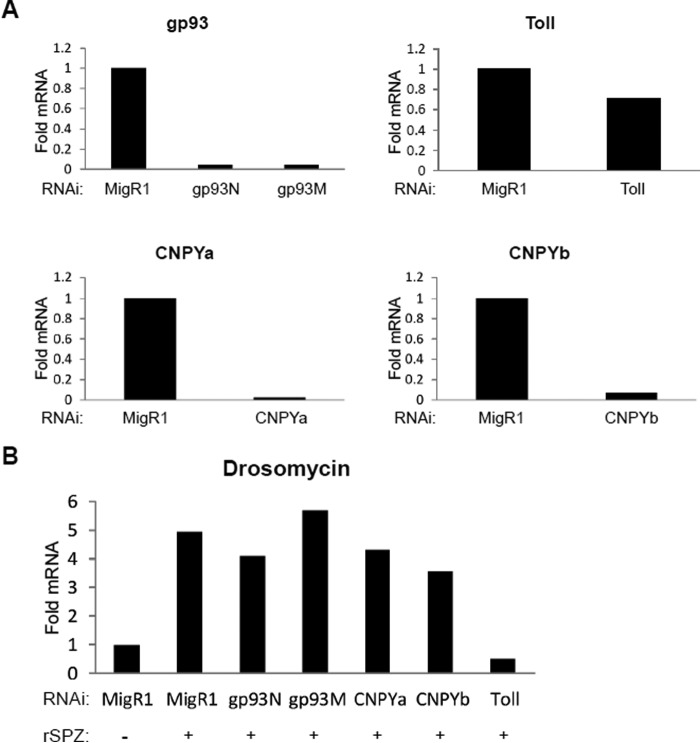
Because gp93 and CNPYb have the ability to cooperatively chaperone murine TLR2 and TLR4, we hypothesized that gp93 and CNPYb would work together endogenously in Drosophila to chaperone Toll. To this end, we switched to a Drosophila system utilizing the Drosophila hemacyte cell line S2*. We then used two different RNAi constructs to knockdown gp93 expression: gp93N, N terminus, and gp93M, middle domain. We also used RNAi constructs specific for Toll, CNPYa, and CNPYb. As a negative control, we utilized an RNAi construct specific to a retroviral vector, with no sequence homology to the Drosophila genome. To verify efficient knockdown of the different molecules, we performed qRT-PCR analysis of the cells. Moderate mRNA knockdown of Toll was observed, whereas knockdown of gp93, CNPYa, and CNPYb were near complete (Fig. 7A). To determine whether loss of gp93, CNPYa, or CNPYb affected Toll expression, we probed for Toll function. RNAi-treated cells were treated with recombinant spatzle (rSPZ), the endogenous Toll ligand. The cells were then analyzed for the downstream production of the antimicrobial peptide drosomycin by qRT-PCR. In cells treated with the negative control MigR1 RNAi construct, drosomycin production was induced with rSPZ addition. Furthermore, cells treated with Toll RNAi did in fact lose drosomycin induction. However, drosomycin induction was not altered in cells receiving gp93, CNPYa, or CNPYb RNAi (Fig. 7B). However, because no specific antibodies exist for gp93, CNPYa, or CNPYb, we are unable to determine whether or not protein levels are in fact knocked down. Because gp96 has a long half-life, it is therefore plausible that gp93 will also have a long half-life. Also, it is possible that CNPYa or CNPYb could compensate for loss of the other. Therefore, further experimentation is warranted to definitively prove the role of gp93, CNPYa, and CNPYb in Drosophila Toll biology.
Figure 7.
Knockdown of neither gp93, CNPYa, nor CNPYb alone decreases Toll signaling. A, Drosophila S2* cells were treated with RNAi constructs targeting the N terminus of gp93 (gp93N), the middle region of gp93 (gp93M), Toll, CNPYa, or CNPYb. A MigR1 negative control RNAi construct was also included. The cells were then analyzed for gp93, Toll, CNPYa, and CNPYb RNA expression via qRT-PCR. B, cells from A were treated with rSPZ, and drosomycin induction was analyzed via qRT-PCR. Samples were run in duplicate. The data are representative of two independent experiments.
Discussion
Further supporting our report of gp93 being the ortholog of gp96 and highlighting the high level of sequence conservation, we find that gp93 and gp96 can dimerize (11). Turning our attention to the CNPY family, we discover ∼50% sequence homology between CNPY3 and CNPYb, with a similar pattern of 6 cysteines and a C-terminal basic region. Utilizing our previously published model, we find that the addition of CNPYb in gp93-expressing cells, but not CNPYa, improves TLR expression. Unexpectedly, the presence of either CNPYa or CNPYb diminishes α integrin expression yet improves β integrin expression, even in the absence of gp93. Biochemical analysis finds that both gp96 and gp93 can bind both CNPY3 and CNPYb, highlighting further similarities between the mammalian and Drosophila systems.
Because gp93 and CNPYb can interact and because CNPYb improves TLR expression, we believe that CNPYb is acting as a cochaperone. Furthermore, because both CNPYa and CNPYb can improve β2 integrin expression in the absence of gp93, they appear to positively regulate its expression in a more dominant manner than gp93 itself. Conversely, both Drosophila CNPYs seem to negatively regulate αL and α4 expression. It is possible that these CNPYs are chaperones for β integrins and negative regulators for α integrins, perhaps via inhibiting ATPase activity or the client-binding domain of gp93. This would be comparable with the various cochaperones for HSP90, which can either inhibit or activate ATP hydrolysis to differentially affect chaperone function of specific client proteins (16).
Cysteine analysis determines that Cys-36, Cys-39, Cys-92, and Cys-183 in CNPYb are all important for TLR4 expression; yet only Cys-39 and Cys-183 play a role in TLR2 expression. These data suggest that Cys-39 and Cys-183 may interact via a disulfide bridge and that this interaction may be important for general cochaperone function. Thus, ablation of either residue disrupts the interaction and hence phenocopies the other. This further supports the idea of a folded clam structure and even mirrors the investigation of single nucleotide polymorphisms in CNPY3 where differential dependence of a residue was seen for various TLRs (6). This may indeed be the same case with CNPYb.
Moving to a Drosophila system utilizing S2* cells, RNAi administration was effective at knocking down RNA expression of gp93, CNPYa, and CNPYb. However, with the addition of the Toll ligand SPZ to the cells, we did not see diminished Toll signaling as evidenced by drosomycin expression. Although gp93 and CNPYb can chaperone mammalian TLRs, it is possible that they do not chaperone Drosophila Toll. However, we are unable to make a conclusion because we technically cannot verify protein knockdown. Because gp96 has a long half-life, it is quite possible that gp93 also has a long half-life. Therefore, although RNA levels are affected, protein levels may not be affected. Furthermore, it is possible that CNPYa or CNPYb can compensate for loss of the other or that gp93 and CNPYb may chaperone the other Drosophila Toll members. Although we looked specifically at Toll-1, there is also Toll-2 to Toll-9. However, TLR4 is the most similar to Toll-1 and only Toll-5 and Toll-9 may also be involved in immunity (17). Therefore, it seems unlikely to us that gp93 would not chaperone Toll-1. Unfortunately, this delineation requires further investigation.
In addition to Drosophila Toll molecules, it is also possible that gp93 may chaperone any or all of the seven Drosophila integrins, because supporting evidence exists in the literature. Specifically, several Drosophila integrins are expressed in the midgut (αPS1, αPS3, βPS, and βν) (18–21). Furthermore, migration of the primordial midgut cells is defected in βPS KO flies. This phenotype is then exacerbated in βPS/βν double KO flies where migration of the midgut cells is completely blocked (19). Similarly, gp93 KO flies have severe defects in larval midgut development (12). Because gp93 KO and βPS/βν double KO flies seem to phenocopy each other, we believe that gp93 must chaperone some or all of the Drosophila integrins similar to how gp96 chaperones various mammalian integrins (13, 15).
From our data contained herein and previously published, we have started to uncover gp93 biology. We find that gp93 alone is required for β and α integrin expression; yet gp93 and CNPYb together are required for optimal TLR expression. We also find that CNPYa and CNPYb alone can rescue β2 expression (Fig. 8). In Drosophila, there exists only five α integrins (αPS1, αPS2, αPS3, αPS4, and αPS5) and two β integrins (βPS and βν) (22). Future studies should focus on a Drosophila system to identify the endogenous role of gp93, CNPYa, and CNPYb in relation to the nine Tolls and seven integrin subunits. We believe that gp93 will be found to chaperone one or all of the Drosophila Tolls along with CNPYb and that CNPYa and CNPYb will further play a role in α and β integrin regulation. Uncovering the relationship among these molecules will shed light on the function of the mammalian gp96, a molecule implicated in various diseases.
Figure 8.
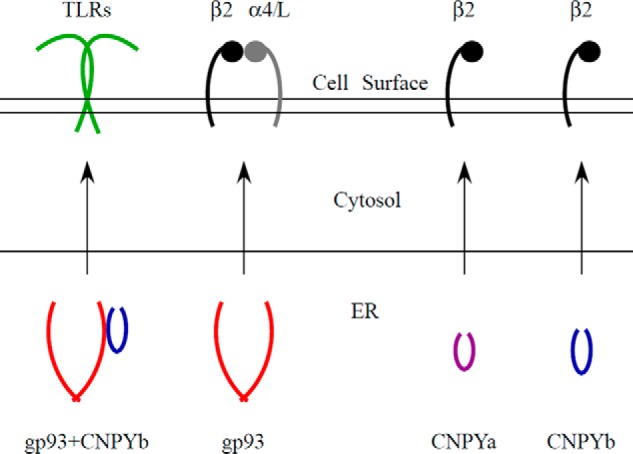
Model of Drosophila chaperone function. Both CNPYa and CNPYb alone can rescue β2 expression, whereas gp93 alone can rescue β2, α4, and αL expression. However, gp93 together with CNPYb is required for optimal TLR2 and TLR4 expression.
Experimental procedures
Sequence alignment
The sequences for murine gp96 (NP_035761), CNPY1 (NM_175651), CNPY2 (NM_019953), CNPY3 (NM_028065), and CNPY4 (NM_178612) and Drosophila gp93 (NP_651601), CNPYa (NM_136703), and CNPYb (NM_140832) were obtained from the National Center for Biotechnology Information database. Vector NTI (Invitrogen) software was used to align gene sequences and generate a phylogenetic tree.
Cloning constructs
Murine gp96 and Drosophila gp93 were cloned into the MigR1 retrovector as previously described (11). The vector contains a GFP reporter, which is expressed in a bicistronic fashion, and both constructs were engineered with C-terminal Flag tags (DYKDDDDK). Murine CNPY3 was amplified from murine genomic cDNA using the following primers: CNPY3 Not1 forward, catgcggccgcgccaccatggagtccatgtctgag; and CNPY3 myc Not1 reverse, tagcggccgcctacagttcatctttcagatcctcttctgagatgagtttttgttcggggctgtgtgggaggggcga. Drosophila CNPYa and CNPYb were amplified from S2 cDNA using the following primers: CNPYa Not1 forward, tagcggccgcgccaccatgctgacgaaggcgcttatc; CNPYa myc Not1 reverse, tagcggccgcctacagttcatctttcagatcctcttctgagatgagtttttgttcgccatcaaagtcatactcctc; CNPYb Not1 forward, tagcggccgcgccaccatgctcctcaaaagactgcct; and CNPYb myc Not1 reverse, tagcggccgctcacagctcatccttcagatcctcttctgagatgagtttttgttcgtcgcccttggctttttccct. All CNPY constructs were designed with C-terminal Myc tags (EQKLISEEDL) and directly cloned into the MigR1 vector.
Site-directed mutagenesis
A Stratagene QuikChange II XL site-directed mutagenesis kit was used to create all CNPYb mutants, following the manufacturer's protocol. Primers were designed with the Stratagene online primer design program, and mutants were PCR-amplified from wild-type CNPYb myc-MigR1. The following are the primer sequences: CNPYb C36A forward, tcgctacgcaaaccgcgccgaagcctgcaaaatc; CNPYb C36A reverse, gattttgcaggcttcggcgcggtttgcgtagcga; CNPYb C39A forward, aaccgctgcgaagccgccaaaatcctggccac; CNPYb C39A reverse, gtggccaggattttggcggcttcgcagcggtt; CNPYb C92A forward, ccctggagaacgtggccgagcgagtgttgg; CNPYb C92A reverse, ccaacactcgctcggccacgttctccaggg; CNPYb C183A forward, cactgaagaagcatctcgccgaggaccatgtgctca; and CNPYb C183A reverse, tgagcacatggtcctcggcgagatgcttcttcagtg. All retrovectors were transfected into Phoenix-Eco cells for virus production, and virus was then spin-transduced into the cell line of interest as previously described (11).
Cell culture
Phoenix Eco cells were grown in 5% CO2 at 37 °C in a humidified incubator. The cells were cultured in DMEM (Sigma) supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% FBS (Atlas Biologicals). Wild-type 14.GFP and gp96 mutant (KO) E4.126 murine pre-B cells harboring an inducible NF-κB-controlled GFP reporter were a gift from Randow and Seed (Harvard University) (13). They created the KO cells via mutagenesis with ICR191. The cells were grown in 5% CO2 at 37 °C in a humidified incubator. The cells were cultured in RPMI 1640 medium (Sigma) supplemented with 0.055 mm 2-mercaptoethanol (Life Technologies), 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% FBS (Atlas Biologicals). The Drosophila hemocyte cell line S2* was a gift from Dr. Neal Silverman (University of Massachusetts). The cells were grown at 27 °C without shaking. The cells were cultured in Schneider's Drosophila medium (PromoCell) supplemented with 12.33 mm l-glutamine, 1% GlutaMAX (Gibco), 20 units/ml penicillin, 20 μg/ml streptomycin, and 10% FBS. Sterile technique was used for all cells.
Flow cytometry
Flow cytometry of cells was performed as previously described (11). Briefly, the cells were blocked and then stained sequentially with primary and secondary antibodies. Propidium iodine was added for dead cell exclusion. For intracellular staining, the cells were fixed with 4% formaldehyde and permeabilized with methanol. The cells were then blocked with serum alone, followed by antibody staining. Stained cells were then ran on a FACSCalibur (BD Biosciences) with Cell Quest and analyzed by FlowJo software.
Western blotting and immunoprecipitation
Western blotting analysis of cells was performed as previously described (11). Cell lysates were obtained, quantified, resolved on 10% SDS-PAGE, and then transferred to a polyvinylidene difluoride membrane. The membranes were then blocked with 5% milk, followed by primary and secondary antibody application. Chemiluminescent substrate (Pierce) was added, and the membrane was exposed to film for visualization. For immunoprecipitation, the cells were treated with or without DSP cross-linker, as previously described (11), and then lysed. Lysates were then precleared with protein G beads and incubated with antibody. Fresh protein G beads were then added, and the mixture was washed after incubation. Protein was eluted from the beads and then resolved on 10% SDS-PAGE as above.
RNAi and Toll signaling
RNAi primers were designed to span 600–800 bp of the gene of interest and all primers contain a T7 promoter site. As a negative control, one primer set was designed to amplify a region of the MigR1 vector that, upon BLAST verification, would not recognize anything within the Drosophila genome. The following primers were used: CNPYa T7 forward, taatacgactcactataggaacgctgacccagacccactga; CNPYa T7 reverse, taatacgactcactataggaccggcgattcgtcgcagtag; CNPYb T7 forward, taatacgactcactataggaataccggcgcagcgaactgcv; CNPYb T7 reverse, taatacgactcactataggtaagccaatggccccaggcgaa; gp93N T7 forward, taatacgactcactatagggaggccgccacaacggagac; gp93N T7 reverse, taatacgactcactataggggcacctcctcctcgacggt; gp93M T7 forward, taatacgactcactataggaagtactcgcagttcatcaactt; gp93M T7 reverse, taatacgactcactataggatgtagtagatgtgctcctgctt; MigR1 T7 forward, taatacgactcactataggacctaaaacagttcccgcct; MigR1 T7 reverse, taatacgactcactataggaggaactgcttccttcacga; Toll T7 forward, taatacgactcactataggatatattcagcaacttgggcaac; and Toll T7 reverse, taatacgactcactataggccaagtgaagttccattaattgc. CNPYa, CNPYb, and Toll RNAi constructs were PCR-amplified from S2 cDNA and then cloned into the pGEM-T easy vector (Promega). These vectors, along with an empty vector MigR1 and gp93-MigR1, were then used as templates for RNAi production. After PCR amplification, products were purified and then in vitro transcribed into RNA using an Epicenter kit and T7 polymerase. RNA was incubated at 75 °C for 15 min to anneal sense and antisense strands and then allowed to cool to room temperature. To digest the remaining DNA, DNase was added to RNA and incubated at 37 °C for 15 min. Final RNAi preparations were then purified with Centri Spin-20 columns (Princeton Separations). RNAi was either used immediately or frozen at −80 °C.
At day −1, S2* cells were split to a density of 1 million cells/ml. At day 0, the cells were seeded in a 6-well plate at a density of 2 million cells in 3 ml of medium. Then at day 1, 6 μg of annealed RNAi in PBS with 125 mm CaCl2 was vortexed, incubated for 15 min, and then added dropwise to cells. The wells were mixed by gently swirling the plates and left overnight. At day 2, the cells were again split to 1 million cells/ml to dilute out the CaCl2. On day 4, an aliquot of cells was taken for determination of RNAi efficiency by qRT-PCR.
On day 6 after RNAi addition, 1 μm 20-hydroxyecdysone (Sigma) was added to cells and incubated for 24 h to amplify Toll signaling as previously described (23). On day 7, 2.4 nm of recombinant Spaetzle C106 was added to induce the Toll pathway. After 6 h, mRNA was isolated from cells, and qRT-PCR was performed to analyze drosomycin induction.
qRT-PCR
qRT-PCR was performed as previously described (11). Briefly, mRNA was isolated from cells using TRIzol reagent and then reverse transcribed to cDNA. SYBR Green supermix and a Bio-Rad iCyler were used for qRT-PCRs. The data were analyzed on Microsoft Excel using the dCT method, and Drosophila Rp49 was used as an internal control. The following primers were utilized: CNPYa forward, tgcagagcatcaagaaccac; CNPYa reverse, gtgaagctgtaaccttgggc; CNPYb forward, ggaggtcacccaaatgaaga; CNPYb reverse, gctcacagctcctccttgtc; drosomycin forward, gcagatcaagtacttgttcgccc; drosomycin reverse, cttcgcaccagcacttcagactgg; Gp93 forward, actacctgagcttcattcgtgg; Gp93 reverse, atcctccatcacgcccaatttg; Rp49 forward, agatcgtgaagaagcgcaccaag; Rp49 reverse, caccaggaacttcttgaatccgg; Toll forward, gccggtttcacagcggcact; and Toll reverse, gtgctccagttgcggcacca.
Author contributions
C. M. performed all the experiments. Z. L. and C. M. conceived the idea. Z. L. supervised the work. Both C. M. and Z. L. contributed to the writing of the manuscript and interpretation of the data.
Supplementary Material
Acknowledgments
We thank Dr. Anthony Vella, Dr. Linda Cauley, and members of the Li lab for insightful input, as well as Ephraim Ansa-Addo for help preparing the revised manuscript. We also thank Dr. Neil Silverman (University of Massachusetts) for providing cells and reagents and training of C. M. in Drosophila techniques.
This work was supported by National Institutes of Health Grant R01AI070603. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Fig. S1.
- CNPY
- canopy
- TLR
- Toll-like receptor
- ER
- endoplasmic reticulum
- qRT-PCR
- quantitative RT-PCR.
References
- 1. Hirate Y., and Okamoto H. (2006) Canopy1, a novel regulator of FGF signaling around the midbrain-hindbrain boundary in zebrafish. Curr. Biol. 16, 421–427 [DOI] [PubMed] [Google Scholar]
- 2. Liu B., Yang Y., Qiu Z., Staron M., Hong F., Li Y., Wu S., Li Y., Hao B., Bona R., Han D., and Li Z. (2010) Folding of Toll-like receptors by the HSP90 paralogue gp96 requires a substrate-specific cochaperone. Nat. Commun. 1, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wakabayashi Y., Kobayashi M., Akashi-Takamura S., Tanimura N., Konno K., Takahashi K., Ishii T., Mizutani T., Iba H., Kouro T., Takaki S., Takatsu K., Oda Y., Ishihama Y., Saitoh S., et al. (2006) A protein associated with toll-like receptor 4 (PRAT4A) regulates cell surface expression of TLR4. J. Immunol. 177, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 4. Takahashi K., Shibata T., Akashi-Takamura S., Kiyokawa T., Wakabayashi Y., Tanimura N., Kobayashi T., Matsumoto F., Fukui R., Kouro T., Nagai Y., Takatsu K., Saitoh S., and Miyake K. (2007) A protein associated with Toll-like receptor (TLR) 4 (PRAT4A) is required for TLR-dependent immune responses. J. Exp. Med. 204, 2963–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shibata T., Takemura N., Motoi Y., Goto Y., Karuppuchamy T., Izawa K., Li X., Akashi-Takamura S., Tanimura N., Kunisawa J., Kiyono H., Akira S., Kitamura T., Kitaura J., Uematsu S., et al. (2012) PRAT4A-dependent expression of cell surface TLR5 on neutrophils, classical monocytes and dendritic cells. Int. Immunol. 24, 613–623 [DOI] [PubMed] [Google Scholar]
- 6. Kiyokawa T., Akashi-Takamura S., Shibata T., Matsumoto F., Nishitani C., Kuroki Y., Seto Y., and Miyake K. (2008) A single base mutation in the PRAT4A gene reveals differential interaction of PRAT4A with Toll-like receptors. Int. Immunol. 20, 1407–1415 [DOI] [PubMed] [Google Scholar]
- 7. Hart B. E., and Tapping R. I. (2012) Cell surface trafficking of TLR1 is differentially regulated by the chaperones PRAT4A and PRAT4B. J. Biol. Chem. 287, 16550–16562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Konno K., Wakabayashi Y., Akashi-Takamura S., Ishii T., Kobayashi M., Takahashi K., Kusumoto Y., Saitoh S., Yoshizawa Y., and Miyake K. (2006) A molecule that is associated with Toll-like receptor 4 and regulates its cell surface expression. Biochem. Biophys. Res. Commun. 339, 1076–1082 [DOI] [PubMed] [Google Scholar]
- 9. Matsui T., Thitamadee S., Murata T., Kakinuma H., Nabetani T., Hirabayashi Y., Hirate Y., Okamoto H., and Bessho Y. (2011) Canopy1, a positive feedback regulator of FGF signaling, controls progenitor cell clustering during Kupffer's vesicle organogenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 9881–9886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Do H. T., Tselykh T. V., Mäkelä J., Ho T. H., Olkkonen V. M., Bornhauser B. C., Korhonen L., Zelcer N., and Lindholm D. (2012) Fibroblast growth factor-21 (FGF21) regulates low-density lipoprotein receptor (LDLR) levels in cells via the E3-ubiquitin ligase Mylip/Idol and the Canopy2 (Cnpy2)/Mylip-interacting saposin-like protein (Msap). J. Biol. Chem. 287, 12602–12611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morales C., Wu S., Yang Y., Hao B., and Li Z. (2009) Drosophila glycoprotein 93 Is an ortholog of mammalian heat shock protein gp96 (grp94, HSP90b1, HSPC4) and retains disulfide bond-independent chaperone function for TLRs and integrins. J. Immunol. 183, 5121–5128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maynard J. C., Pham T., Zheng T., Jockheck-Clark A., Rankin H. B., Newgard C. B., Spana E. P., and Nicchitta C. V. (2010) Gp93, the Drosophila GRP94 ortholog, is required for gut epithelial homeostasis and nutrient assimilation-coupled growth control. Dev. Biol. 339, 295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Randow F., and Seed B. (2001) Endoplasmic reticulum chaperone gp96 is required for innate immunity but not cell viability. Nat. Cell Biol. 3, 891–896 [DOI] [PubMed] [Google Scholar]
- 14. Yang Y., Liu B., Dai J., Srivastava P. K., Zammit D. J., Lefrançois L., and Li Z. (2007) Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity 26, 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Staron M., Yang Y., Liu B., Li J., Shen Y., Zúñiga-Pflücker J. C., Aguila H. L., Goldschneider I., and Li Z. (2010) gp96, an endoplasmic reticulum master chaperone for integrins and Toll-like receptors, selectively regulates early T and B lymphopoiesis. Blood 115, 2380–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pearl L. H., and Prodromou C. (2006) Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 75, 271–294 [DOI] [PubMed] [Google Scholar]
- 17. Valanne S., Wang J.-H., and Rämet M. (2011) The Drosophila Toll signaling pathway. J. Immunol. 186, 649–656 [DOI] [PubMed] [Google Scholar]
- 18. Yee G. H., and Hynes R. O. (1993) A novel, tissue-specific integrin subunit, beta nu, expressed in the midgut of Drosophila melanogaster. Development 118, 845–858 [DOI] [PubMed] [Google Scholar]
- 19. Devenport D., and Brown N. H. (2004) Morphogenesis in the absence of integrins: mutation of both Drosophila β subunits prevents midgut migration. Development 131, 5405–5415 [DOI] [PubMed] [Google Scholar]
- 20. Wehrli M., DiAntonio A., Fearnley I. M., Smith R. J., and Wilcox M. (1993) Cloning and characterization of α PS1, a novel Drosophila melanogaster integrin. Mech. Dev. 43, 21–36 [DOI] [PubMed] [Google Scholar]
- 21. Stark K. A., Yee G. H., Roote C. E., Williams E. L., Zusman S., and Hynes R. O. (1997) A novel α integrin subunit associates with βPS and functions in tissue morphogenesis and movement during Drosophila development. Development 124, 4583–4594 [DOI] [PubMed] [Google Scholar]
- 22. Takada Y., Ye X., and Simon S. (2007) The Integrins. Genome Biol. 8, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tanji T., Hu X., Weber A. N., and Ip Y. T. (2007) Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol. Cell Biol. 27, 4578–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.