Abstract
Reactive oxygen species (ROS) can damage DNA, proteins, and lipids, so cells have antioxidant systems that regulate ROS. In many bacteria, a dedicated peroxiredoxin reductase, alkyl hydroperoxide reductase subunit F (AhpF), catalyzes the rapid reduction of the redox-active disulfide center of the antioxidant protein peroxiredoxin (AhpC) to detoxify ROS such as hydrogen peroxide, organic hydroperoxide, and peroxynitrite. AhpF is a flexible multidomain protein that enables a series of electron transfers among the redox centers by accepting reducing equivalents from NADH. A flexible linker connecting the N-terminal domain (NTD) and C-terminal domain (CTD) of AhpF suggests that the enzyme adopts a large-scale domain motion that alternates between the closed and open states to shuttle electrons from the CTD via the NTD to AhpC. Here, we conducted comprehensive mutational, biochemical, and biophysical analyses to gain insights into the role of the flexible linker and the residues critical for the domain motions of Escherichia coli AhpF (EcAhpF) during electron transfer. Small-angle X-ray scattering studies of linker mutants revealed that a group of charged residues, 200EKR202, is crucial for the swiveling motion of the NTD. Moreover, NADH binding significantly affected EcAhpF flexibility and the movement of the NTD relative to the CTD. The mutants also exhibited a decrease in H2O2 reduction by the AhpF-AhpC ensemble. We propose that a concerted movement involving the NTD, C-terminal NADH, and FAD domains, and the flexible linker between them is essential for optimal intra-domain cross-talk and for efficient electron transfer to the redox partner AhpC required for peroxidation.
Keywords: antioxidant, bioenergetics, electron transfer, enzyme mechanism, flavoprotein, protein structure, small-angle X-ray scattering (SAXS)
Introduction
Reactive oxygen species (ROS)3 are inevitable byproducts of normal aerobic metabolism, which at high levels can inflict damage on DNA, lipids, and proteins. Biological cells have developed antioxidant systems to regulate the endogenous level of ROS (1, 2). Peroxiredoxins (Prxs), a class of thiol-specific antioxidant proteins, together with peroxiredoxin reductases (PrxR) are the predominant cellular defenses against oxidative stress and are involved in cellular signaling pathways (3, 4). Many bacteria possess an alkyl hydroperoxide reductase (AhpR) system, composed of two enzymes called AhpC (Prx) and AhpF (PrxR) (5, 6). AhpC catalyzes the reduction of H2O2, organic hydroperoxide, and peroxinitrite (6–8), wherein the active site cysteine, named as peroxidatic cysteine (CP), becomes selectively oxidized by peroxide to the CP–SOH intermediate. This further reacts with the so-called resolving cysteine (CR) of an adjacent subunit of the basic functional dimer, forming an intermolecular disulfide (9, 10). Regeneration of the oxidized AhpC is ensured by AhpF, a dedicated AhpC reductase in most bacteria, which enables the continuous peroxidase catalytic cycle (11). The coordinated interplay between AhpC and AhpF is essential to alleviate the oxidative stress (12, 13).
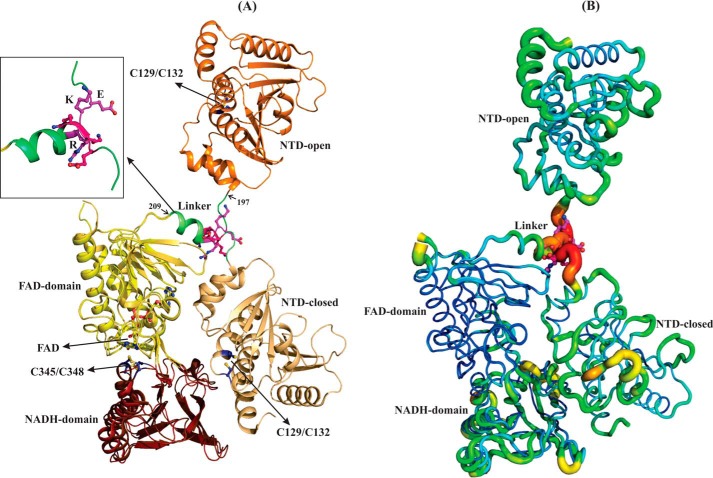
AhpF is a homodimeric protein encompassed by an N-terminal domain (NTD) and a C-terminal domain (CTD), which includes an FAD- and NADH-binding site as well as the redox-active center (Cys345/Cys348 in Escherichia coli AhpF (EcAhpF) (14) (Fig. 1A). The NTD contains a redox-active center (Cys129/Cys132 in EcAhpF), which is responsible for reducing the oxidized disulfide center of AhpC (Cys47/Cys166 in E. coli) (14). Both NTD and CTD are connected via a linker consisting of amino acids 197–209 in EcAhpF. The recent NADH-bound crystal structure of EcAhpF reveals that the CTD, analogues to that of thioredoxin reductase (TrxR), alternates between the flavin-oxidized and flavin-reducing states (15). In addition, the two AhpF structures available from Salmonella typhimurium (StAhpF) and EcAhpF show a compact (“closed state”) and extended form (“open state”), respectively (Fig. 1A) (14, 16). In the closed state, the NTD redox-active center is in proximity to the CTD and the open state allows the NTD redox-active center to be exposed for AhpC reduction (14). Furthermore, we verified the anticipated large-scale structural flexibility of EcAhpF in solution using small angle X-ray scattering (SAXS) studies (15). Based on these studies we proposed that the cross-talk of the NTD and CTD requires multiple substeps of movements between the closed and open state to shuttle electrons from the CTD to the AhpC redox center (15, 17).
Figure 1.
Structural comparison of EcAhpF and StAhpF. A, superimposition of the extended and compact conformations of monomeric EcAhpF (14) and StAhpF (16), respectively, reveals the most significant structural difference in the position of NTD. The linker region connects the NTD with the CTD, which contains the FAD- and NADH-binding sites. The linker adopts a loop-helix structure (inset) and the group of charged residues, 200EKR202, in the interface between the loop and helix region, might be involved in the large-scale domain movements of the NTD in solution. B, temperature factor (B-factor) putty representation of the crystal structures of EcAhpF and StAhpF colored by B-factor from low (blue) to high (red), shows the high B-factor for the linker region in both structures. For clarity, AhpF is shown as a monomer.
In general, linkers interconnecting different modules of multidomain proteins often provide sufficient flexibility to allow domain motions, which is essential to carry out a wide range of functions, including catalysis and protein-protein interactions (18, 19). Previous studies have shown that AhpF is a flexible modular protein (15) and the separate expression of the NTD and CTD modules of intact AhpF affects AhpC-mediated peroxide reduction (20). In AhpF crystal structures, the linker residues 197–209 adopt a loop-helix structure, with residues Lys201-Lys209 and Arg202-Lys209 of EcAhpF (14) and StAhpF (16), respectively, forming an α-helix, respectively, which might have a structural role in modulating the domain motion of the NTD (Fig. 1A) (14). So far, detailed insight into (i) the precise role of the linker and its residues facilitating the flexibility for domain motion of the NTD, (ii) the mode of domain arrangement of NTD with respect to specific functional states, and (iii) the essential role of the linker flexibility on the peroxidase catalytic cycle are missing. To address these questions and to gain deeper insight into the role of the linker on the structural flexibility, we carried out mutational, biochemical, and biophysical analyses on the EcAhpF linker region, in particular the charged amino acids 200EKR202, which might come in proximity to the CTD and provide interactions. By generating and analyzing the single, double, and triple mutations EcAhpF(E200A), EcAhpF(K201A), EcAhpF(R202A), EcAhpF(K201A/R202A), and EcAhpF(E200A/K201A/R202A), we identified that residues Glu200, Lys201, and Arg202 are essential for function, and that these residues may act as a hinge for NTD movements. Moreover, the significant hampering in structural flexibility is deduced due not merely to the linker mutations but also due to the presence of NADH, indicating a substrate-induced cross-talk, which might favor EcAhpF to adopt a closed state. Overall, the flexible linker is crucial to maintain the rapid equilibrium of NTD motion, including the upwards movements of the NTD for an efficient electron-transfer reaction to finally regenerate AhpC for peroxide reduction.
Results
Rational design of the linker region mutants
The closed (StAhpF, Protein Data Bank (PDB) code 1HYU (16)) and open states (EcAhpF, PDB code 4O5Q (14)) of AhpF differ by a rotation and translation of the NTD about 178° and 1.25 Å, respectively (Fig. 1A) (14). A closer look at the linker regions highlights conformational differences for the residues 197TGAEKR202, which are poorly ordered and have high B-factors in both the structures (Fig. 1B). By contrast, the B-factors are low for the helix region 203AAEELNK209 of the linker. A group of charged residues, 200EKR202, lies at the interface of the flexible loop that contains the small side chain residues 197TGA199 and helix 203AAEELNK209. Therefore, we hypothesized that 200EKR202 may play a role as a mechanical swivel allowing the NTD to move up and down. To verify our proposal, we have generated the mutants, EcAhpF(E200A), EcAhpF(K201A), EcAhpF(R202A), EcAhpF(K201A/R202A), and EcAhpF(E200A/K201A/R202A), wherein the 200EKR202 residues are mutated to alanine.
Solution X-ray scattering experiments of EcAhpF linker mutants
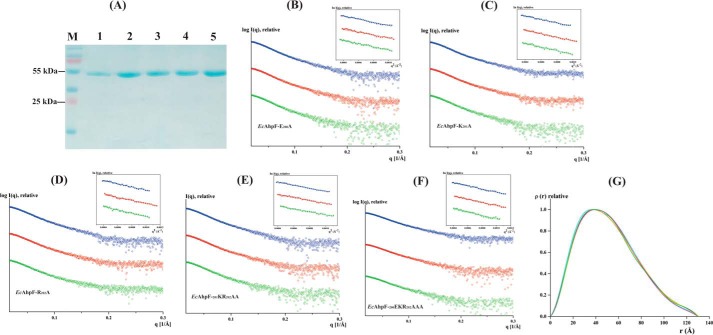
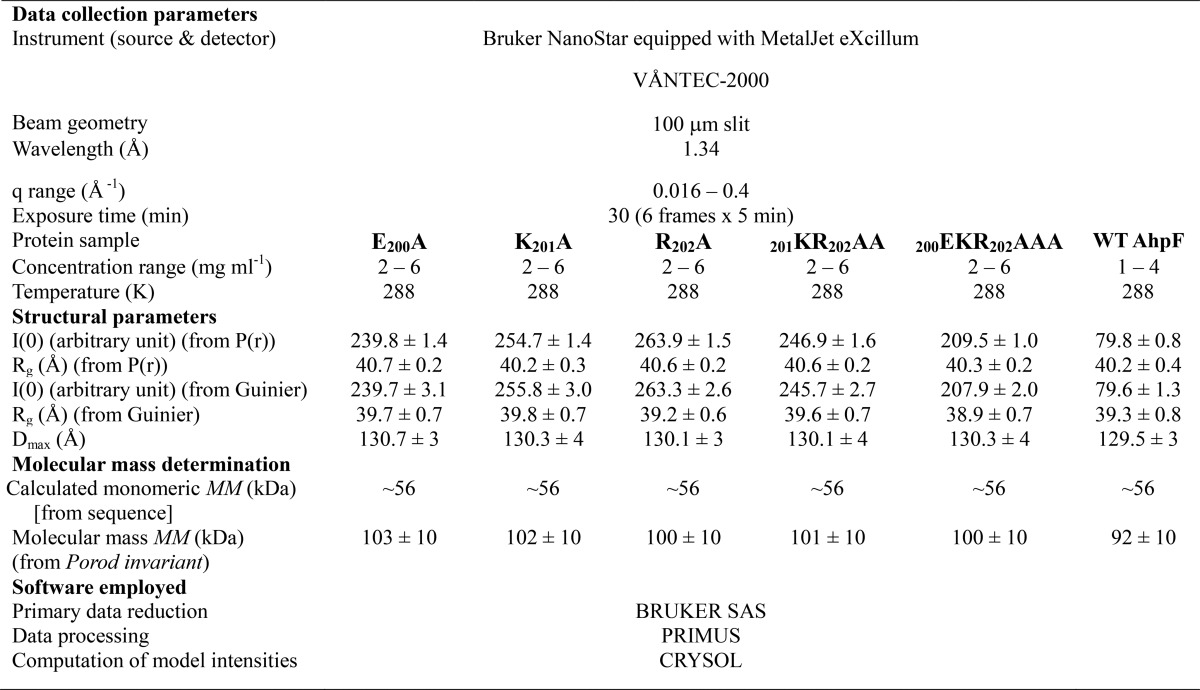
To unravel the effect of the linker on the conformation flexibility of EcAhpF in solution, SAXS experiments were performed with the genetically engineered, expressed, and purified linker mutants (Fig. 2A). SAXS patterns of the various linker mutants were recorded at three different concentrations between 2 and 6 mg/ml (Fig. 2, B–F). The Guinier plots at low angles appeared linear and confirmed good data quality with no indication of protein aggregation (21) (Fig. 2, B–F, inset). The derived Rg values were 39.7, 39.8, 39.2, 39.6, and 39.9 Å for the EcAhpF(E200A), EcAhpF(K201A), EcAhpF(R202A), EcAhpF(K201A/R202A), and EcAhpF(E200A/K201A/R202A) mutants, respectively (Table 1). The P(r) function showed a single peak with a small tail (Fig. 2G) and the maximum particle dimension (Dmax) values were calculated to be 130.7, 130.3, 130.1, 130.1, and 130.3 Å for EcAhpF(E200A), EcAhpF(K201A), EcAhpF(R202A), EcAhpF(K201A/R202A), and EcAhpF(E200A/K201A/R202A), respectively (Table 1) (22). The Rg values extracted from the P(r) function are in agreement with the Rg values extracted from the Guinier region. The estimated Rg, Dmax, and molecular mass of the linker mutants are comparable with the ones of WT EcAhpF (39.3 and 129.5 Å; Table 1) (15), indicating that the linker mutants do not affect the overall structural integrity and dimeric form of the protein in solution.
Figure 2.
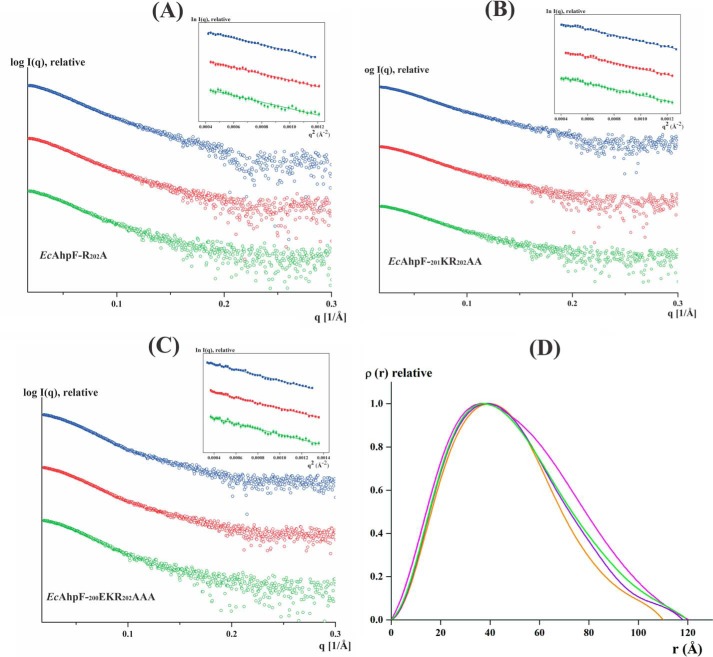
Solution X-ray scattering studies of EcAhpF linker mutants without NADH. A, the 17% SDS gel shows the high purity of the recombinant: lane 1, EcAhpF(E200A/K201A/R202A); lane 2, EcAhpF(K201A/R202A); lane 3, EcAhpF(E200A); lane 4, EcAhpF(K201A); and lane 5, EcAhpF(R202A). Lane M contains a molecular mass protein marker. Small angle X-ray scattering profiles with their corresponding Guinier plots (insets) are shown for the mutants EcAhpF(E200A) (B), EcAhpF(K201A) (C), EcAhpF(R202A) (D), EcAhpF(K201A/R202A) (E), and EcAhpF(E200A/K201A/R202A) (F) measured at various concentrations ranging from 2 to 6 mg/ml in buffer containing 50 mm Tris-HCl, pH 7.5, 200 mm NaCl. G, pair-distance distribution function P(r) shown for EcAhpF(E200A) (yellow), EcAhpF(K201A) (magenta), EcAhpF(R202A) (green), EcAhpF(K201A/R202A) (violet), and EcAhpF(E200A/K201A/R202A) (orange) along with the WT EcAhpF (cyan).
Table 1.
Data collection and scattering derived parameters for EcAhpF linker mutants
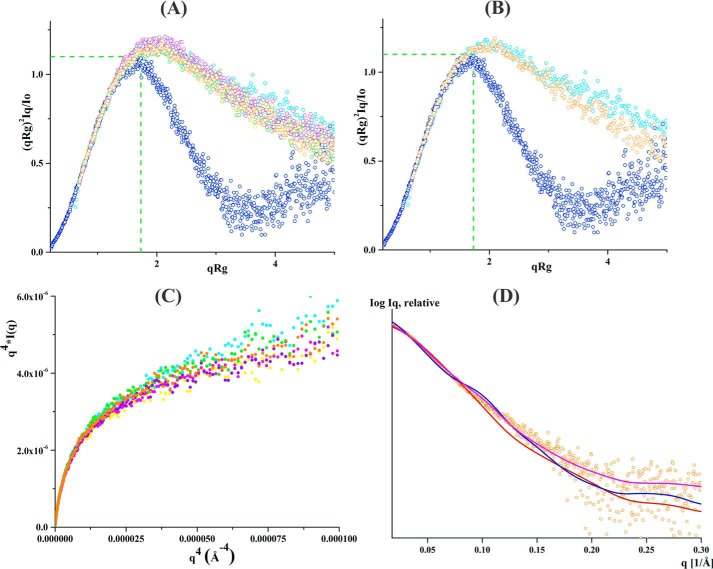
To further elucidate the role of the linker mutants on protein flexibility, the normalized Kratky- and Porod-Debye plots were derived and compared with that of WT EcAhpF as well as to the compact protein lysozyme (23, 24) (Fig. 3A). For clarity, the comparison between the WT EcAhpF, EcAhpF(E200A/K201A/R202A), and lysozyme, is shown in Fig. 3B. Visual inspection of the normalized Kratky plot reveals a bell-shaped profile for lysozyme, which vanishes for both the WT EcAhpF and the linker mutants demonstrating their flexibility in solution (23, 15) (Fig. 3A). Moreover, no significant difference was noticed in the normalized Kratky profiles for the linker mutants (Fig. 3A). However, the profile of the mutants showed a slight inwards shift when compared with WT EcAhpF (Fig. 3, A and B), suggesting an alteration in structural flexibility of the mutants. In addition, the absence of a plateau at low q-angles in the Porod-Debey plot supports the continued flexibility of the linker mutants (Fig. 3C). As in the normalized Kratky plot, the Porod-Debey plots confirm a slightly higher flexibility of WT EcAhpF compared with the mutants (Fig. 3C).
Figure 3.
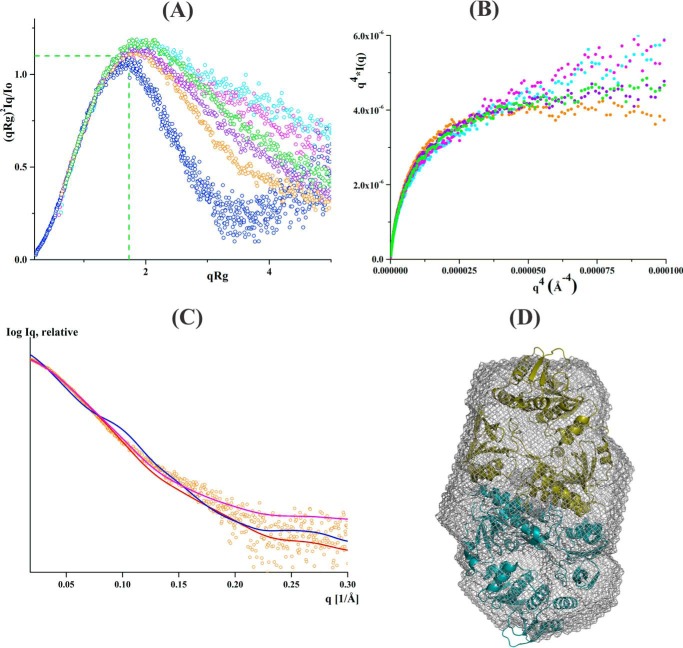
SAXS data analysis of linker mutants without NADH. A, normalized Kratky plot of lysozyme (blue) shows the bell-shaped profile. The bell-shapes profile vanishes for the EcAhpF(E200A) (yellow), EcAhpF(K201A) (magenta), EcAhpF(R202A) (green), EcAhpF(K201A/R202A) (violet), EcAhpF(E200A/K201A/R202A) (orange), and WT EcAhpF (cyan). B, normalized Kratky plots of EcAhpF(E200A/K201A/R202A) (orange) show a slightly altered profile compared with WT EcAhpF (cyan). C, the absence of plateau at low q angles of the Porod-Debye plots for EcAhpF(E200A) (yellow), EcAhpF(K201A) (magenta), EcAhpF(R202A) (green), EcAhpF(K201A/R202A) (violet), EcAhpF(E200A/K201A/R202A) (orange), and WT EcAhpF (cyan) supports the presence of flexibility. D, the fits between the experimental scattering data of EcAhpF(E200A/K201A/R202A) (○) and the theoretical scattering pattern (—) calculated using the dimers of compact (red) and extended (blue) crystal conformations. The mixture of both conformations (magenta) fits better to the experimental data.
To investigate the content of a compact or extended form of the mutants, the theoretical scattering curves were computed and compared with the experimental data using CRYSOL (25). In the case of a 100% compact dimer of EcAhpF(E200A), EcAhpF(K201A), EcAhpF(R202A), EcAhpF(K201A/R202A), and EcAhpF(E200A/K201A/R202A), χ2 values of 1.92, 2.44, 2.08, 1.98, and 1.77, respectively, were computed. In addition, χ2 values of 2.18, 2.53, 2.09, 2.38, and 2.01 were calculated for the respective mutants for an extended dimer (Table 2). However, a best fit with χ2 values of 1.25, 1.25, 0.89, 1.12, and 0.84 were achieved for EcAhpF(E200A), EcAhpF(K201A), EcAhpF(R202A), EcAhpF(K201A/R202A), and EcAhpF(E200A/K201A/R202A), respectively, when a mixture of a compact (∼60%) and extended (∼40%) conformation were present using OLIGOMER (26) (Table 2, Fig. 3D). The result is in agreement with the one determined for WT EcAhpF, which showed a best fit (χ2 = 0.63) in the presence of an equal mixture of compact (∼49%) and extended (∼51%) conformations (Table 2) (15). When compared with WT EcAhpF, the compact conformation of the linker EcAhpF mutants is slightly preferred over the extended conformation.
Table 2.
Fitting between the experimental scattering data of various EcAhpF forms and the calculated scattering data from the atomic structure of the compact dimeric StAhpF and extended dimeric EcAhpF structures using CRYSOL
The volume fraction of each components in a mixture estimated using OLIGOMER.
| Sample | Compact StAhpF | Extended EcAhpF | Mixture (χ2) (closed:extended) |
|---|---|---|---|
| χ2 | % | ||
| WT AhpF | 1.78 | 1.64 | 0.63 (49:51) |
| E200A | 1.92 | 2.18 | 1.25 (61:39) |
| K201A | 2.44 | 2.53 | 1.25 (59:41) |
| R202A | 2.08 | 2.09 | 0.89 (58:42) |
| K201A/R202A | 1.98 | 2.38 | 1.12 (61:38) |
| E200A/K201A/R202A | 1.77 | 2.01 | 0.84 (58:42) |
Substrate-NADH induced conformation flexibility analysis
To understand the effect of NADH on the EcAhpF linker mutants, SAXS data have been collected for EcAhpF(E200A/K201A/R202A), EcAhpF(K201KA/R202A), and EcAhpF(R202A) at three protein concentrations in the presence of 1 mm NADH. The scattering profiles of the three above mentioned mutants in the presence of NADH are shown in Fig. 4, A–C. The Rg and Dmax values of the single EcAhpF(R202A) mutant were 37.8 and 120 Å, respectively, which is comparable with that of NADH-bound EcAhpF (37.3 and 120 Å; Table 3 (15)). However, a significant decrease in the Rg and Dmax values were observed for the double and triple mutants EcAhpF(K201A/R202A) (Rg = 36.5 Å and Dmax = 118 Å) and EcAhpF(E200A/K201A/R202A) (Rg = 35.5 Å and Dmax = 110 Å), respectively (Table 3; Fig. 4D).
Figure 4.
SAXS studies of linker mutants in the presence of NADH. Experimental scattering profile of linker mutants in the presence of 1 mm NADH for: A, EcAhpF(R202A); B, EcAhpF(K201A/R202A); and C, EcAhpF(E200A/K201A/R202A) and their corresponding Guinier plots (insets) demonstrates linearity, indicating no aggregation. D, pair-distance distribution function P(r) for EcAhpF(R202A) (green), EcAhpF(K201A/R202A) (violet), and EcAhpF(E200A/K201A/R202A) (orange) is shown along with WT EcAhpF (magenta).
Table 3.
The SAXS derived parameters for the WT EcAhpF, EcAhpF(R202A), EcAhpF(K201A/R201A), and EcAhpF(E200A/K201A/R202A) in the presence of NADH
| Sample (1 mm NADH) | Guinier Rg | Real space Rg | Dmax | Calculated molecular mass (from Porod invariant) |
|---|---|---|---|---|
| Å | kDa | |||
| R202A | 37.8 ± 0.5 | 38.1 ± 0.2 | 120 ± 4 | 98 ± 10 |
| K201A/R202A | 36.5 ± 0.4 | 37.3 ± 0.2 | 118 ± 3 | 100 ± 10 |
| E200A/K201A/R202A | 35.5 ± 0.3 | 35.9 ± 0.1 | 110 ± 3 | 100 ± 10 |
| WT AhpF | 37.2 ± 1 | 38.9 ± 0.3 | 120 ± 2 | 99 ± 10 |
In addition, the linker mutants showed a significant difference in their normalized Kratky curves, which started to shift progressively inwards from a single to the triple mutant (Fig. 5A). The Porod-Debey plot also demonstrated the gradual decrease in the slope as q increases, for EcAhpF(R202A) and EcAhpF(K201A/R202A). Interestingly, a plateau was observed for the EcAhpF(E200A/K201A/R202A) triple mutant (Fig. 5B). These results clearly demonstrate a significant decrease in the structural flexibility of linker mutants in the presence of NADH. Furthermore, the flexibility of the EcAhpF(E200A/K201A/R202A) is affected considerably followed by the EcAhpF(K201A/R202A) and EcAhpF(R202A) mutants. In comparison, the substrate-free and NADH-bound forms of WT EcAhpF showed a similar normalized Kratky- and Porod-Debye profile, revealing no alteration in their protein flexibility (15) (Fig. 5A).
Figure 5.
SAXS flexibility analysis of linker mutants in the presence of NADH. A, normalized Kratky plots of EcAhpF(R202A) (green), EcAhpF(K201A/R202A) (violet), EcAhpF(E200A/K201A/R202A) (orange), and WT EcAhpF (magenta) measured in the presence of NADH along with the substrate-free WT EcAhpF (cyan) and lysozyme (blue). B, Porod-Debye plots of EcAhpF(R202A) (green), EcAhpF(K201A/R202A) (violet), and EcAhpF(E200A/K201A/R202A) (orange) and WT EcAhpF (magenta) measured in the presence of NADH along with the substrate-free WT EcAhpF (cyan). C, the fits of the experimental scattering data of the NADH-bound form of EcAhpF(E200A/K201A/R202A) (○) with the theoretical scattering pattern (—) calculated using the compact (red), extended (blue) dimers and the mixture of both conformations. D, the averaged ab initio model (mesh) is overlaid with the compact dimer structure of StAhpF (16).
The CRYSOL (25) and OLIGOMER (26) analysis demonstrated that the data of NADH-bound EcAhpF(K201A/R202A) and EcAhpF(R202A) had a much better fit with the compact conformation (χ2 values of 1.37 and 1.44, respectively) than the extended conformation (χ2 values of 8.76 and 6.31, respectively). The fit marginally increased with χ2 values of 1.30 and 1.01, respectively, considering a mixture of compact (about 80%) and extended (about 20%) conformations present in solution (Table 4). Importantly, EcAhpF(E200A/K201A/R202A) had a best fit with a 100% compact form in solution (χ2 = 1.1) (Fig. 5C). Because the scattering data of EcAhpF(E200A/K201A/R202A) suggested the presence of a single compact component, the solution shape was ab initio reconstructed. The averaged model (DAMMIF, Fig. 5D) had a normalized spatial discrepancy value of 0.81 ± 0.07, and revealed a compact shape, which superimposed nicely with the compact structure (16) with a calculated normal spatial discrepancy value of 1.4 (Fig. 5D).
Table 4.
Fitting the experimental data of WT EcAhpF, EcAhpF(R202A), EcAhpF(K201A/R202A), and EcAhpF(E200A/K201A/R202A) measured in the presence of NADH with the calculated scattering data from the compact dimeric StAhpF, extended dimeric EcAhpF structures using CRYSOL
The volume fraction of each components in a mixture is estimated using OLIGOMER.
| Sample (1 mm NADH) | Compact StAhpF | Extended EcAhpF | Mixture (χ2) (closed:extended) |
|---|---|---|---|
| χ2 | % | ||
| WT AhpF | 1.30 | 1.43 | 0.51 (52:48) |
| R202A | 1.44 | 6.31 | 1.01 (77:23) |
| K201A/R202A | 1.37 | 8.76 | 1.30 (84:16) |
| E200A/K201A/R202A | 1.10 | 8.96 | 1.18 (94:6) |
The flexibility of EcAhpF is essential for peroxidase activity
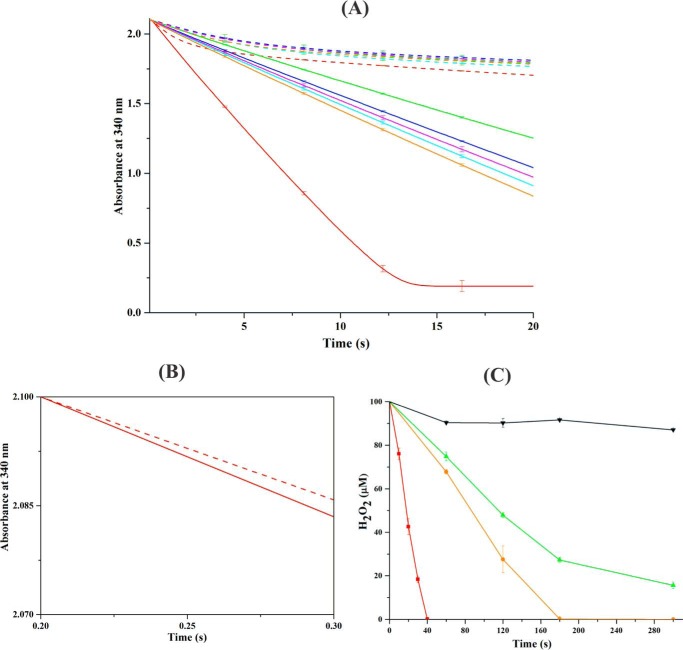
To evaluate the essential role of structural flexibility in EcAhpF on the peroxidase activity, NADH-dependent peroxidase activity was measured in the presence of EcAhpC, H2O2, and WT EcAhpF or the linker mutants by observing the decrease in NADH absorbance at 340 nm (Fig. 6A) (12, 14). NADH absorbance dropped significantly for WT EcAhpF in the presence of EcAhpC and 1 mm H2O2, resulting in a reaction rate constant of 170 ± 2 s−1 (Fig. 6A). Importantly, the linker mutants showed a lower level of NADH-oxidation in the presence of EcAhpC and 1 mm H2O2 compared with WT EcAhpF, demonstrating that the linker mutants affect the peroxidase activity. Among the linker mutants, EcAhpF(E200A/K201A/R202A) revealed the lowest reaction rate of 47 ± 1 s−1, followed by EcAhpF(K201A/R202) with a value of 58 ± 2 s−1. A similar rate constant of 64 ± 2, 62 ± 2, and 66 ± 2 s−1 was observed for the three single mutants EcAhpF(E200A), EcAhpF(K201A), and EcAhpF(R202A), respectively (Fig. 6A). Interestingly, as shown in Fig. 6B, NADH oxidation in the initial phase of 0.28 s for EcAhpC and WT EcAhpF in the presence and absence of H2O2 are similar (Fig. 6B), whereas it is continuous in the presence of H2O2 afterward due to multiple turnover with peroxide. These results indicate that the EcAhpC turnover with peroxide is at least as fast as the transfer of electron to EcAhpC by the WT EcAhpF. In addition, using a ferrous oxidation xylenol assay we confirmed that WT EcAhpC was able to efficiently decompose H2O2, and that the amount of reduced H2O2 was lower in case of the EcAhpF(R202A) and even more in EcAhpF(E200A/K201A/R202A) (Fig. 6C). The difference in peroxidase activity observed for WT EcAhpF and its linker mutants might arise due to the inability of the linker mutants to shuttle the electron from the AhpF-CTD to AhpC for efficient regeneration during the peroxidase cycle (Fig. 6, A and C).
Figure 6.
Peroxidase activity of the AhpF-AhpC complex. A, NADH-dependent peroxidase activity was measured for 0.4 μm WT EcAhpF (red) or the different linker mutants, EcAhpF(E200A) (cyan), EcAhpF(K201A) (magenta), EcAhpF(R202A) (orange), EcAhpF(K201A/R202A) (blue), and EcAhpF(E200A/K201A/R202A) (green) in the presence of 30 μm EcAhpC, 300 μm NADH with (straight lines) or without (dashed lines) 1 mm H2O2 in a reaction mixture containing 50 mm phosphate buffer, pH 7.0, 100 mm ammonium sulfate, and 0.5 mm EDTA. B, the initial rate of NADH oxidation for WT EcAhpF and EcAhpC with (straight lines) or without (dashed lines) 1 mm H2O2. C, to determine the H2O2 decomposition, 0.4 μm (■) WT EcAhpF, (●) EcAhpF(R202A), and (▴) EcAhpF(E200A/K201A/R202A) were added to the reaction mixture containing 30 μm EcAhpC, 750 μm NADH, and 100 μm H2O2. The residual H2O2 in the reaction mixtures was measured at the indicated times using ferrous oxidation xylenol reagent. The control was measured in the absence of EcAhpF (▾). The linker mutants significantly affect the EcAhpC-dependent peroxidase activity. The data presented were averaged of three independent measurements, and error bars represent the standard deviation.
EcAhpC is not effectively reduced by EcAhpF(E200A/K201A/R202A)
To specifically evaluate the ability of the WT EcAhpF and EcAhpF(E200A/K201A/R202A) to reduce oxidized EcAhpC, non-reducing SDS-PAGE experiments were conducted (10). The oxidized EcAhpC (20 μm), running as a band corresponding to a dimer in a non-reducing SDS gel, was added to 50 nm WT EcAhpF and 100 μm NADH and the reduction of EcAhpC was observed over 10 min (Fig. 7, A and B). As shown in Fig. 7A, the intensity of the dimeric band of EcAhpC decreased, whereas the intensity of the monomeric band increased gradually, reflecting disulfide reduction of EcAhpC by WT EcAhpF. In comparison, when the mutant EcAhpF(E200A/K201A/R202A) was used for EcAhpC reduction, only a very faint monomeric band of EcAhpC was observed, demonstrating that the EcAhpF(E200A/K201A/R202A) failed to reduce EcAhpC as effectively as WT EcAhpF (Fig. 7B).
Figure 7.
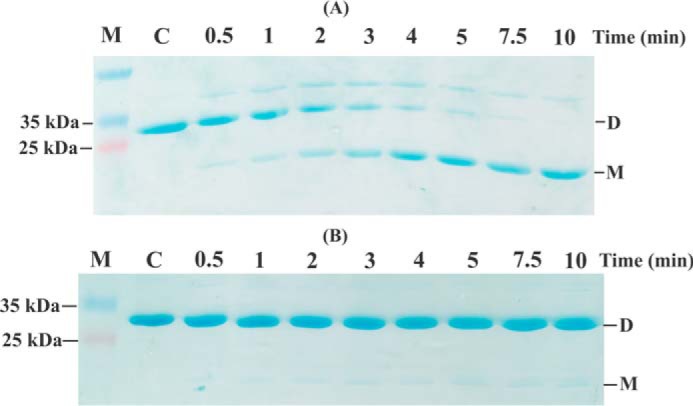
EcAhpC reduction assay. Non-reducing SDS-PAGE analysis for the reduction of 20 μm oxidized EcAhpC by 50 nm WT EcAhpF (A) and EcAhpF(E200A/K201A/R202A) (B) in the presence of 100 μm NADH for different time periods (0.5 to 10 min) in 50 mm HEPES buffer, pH 7.0. The reaction without NADH was measured as control (denoted as C). Reduced AhpC runs as a monomer (M) and oxidized AhpC forms a dimer (D).
Discussion
The flexible linker is critical for electron transfer reaction
Electron transfer reactions are key biochemical events, which underpin fundamental processes like respiration and photosynthesis. Many metabolic pathways as well as the detoxification reactions involve electron transfers (27, 28). Inter-protein electron transfer between PrxR and Prxs is essential for the detoxification of peroxides and peroxinitrite. Particularly, the dithiol-disulfide interchange between the NTD of AhpF and AhpC constitutes one of the most efficient Prx recycling reactions reported, highlighting the advantage gained by expression of a specialized AhpC reductase in bacteria (29). The inter-protein electron transfer demands transient complex formation, which is a dynamic process and involves many intriguing factors. The determined redox potential of the NTD (E′0 = −264 mV) and AhpC (E′0 = −178 mV) redox centers favor the dithiol-disulfide exchange (30). In addition, previous work established that the interaction between NTD and AhpC is highly specific with a Km(NTD) of about 6 μm (30). The measured thermodynamics of the NTD-AhpC interaction by isothermal titration calorimetry revealed a dissociation constant of Kd = 10.5 μm (13). Furthermore, we demonstrated that the assembly and disassembly of the transient complex is favored by the redox state of AhpC (13, 17).
Besides the factors mentioned above, the efficient regeneration of AhpC is facilitated by the efficient intramolecular electron transfer among multiple redox centers of AhpF. The estimated rate constant (170 s−1) for electron transfer through all the redox centers inside WT EcAhpF is comparable with that of the reduction rate of enzyme-bound FAD (200 s−1) by NADH (31). This clearly indicates that electron transfer from the CTD via the NTD to AhpC should progress rapidly, and requires conformational changes (32, 33). The determined rate constant of 64, 62, 66, 58, and 47 s−1 of the linker region mutants, EcAhpF(E200A), EcAhpF(K201A), EcAhpF(R202A), EcAhpF(K201A/R202A), and EcAhpF(E200A/K201A/R202A), respectively, underline the essential role of the flexible linker in the conformational alteration of the NTD. The SAXS analysis of the EcAhpF linker mutants presented demonstrate a similar flexibility of the linker mutants compared with that of WT EcAhpF (15), suggesting that the introduced point mutations did not significantly affect the flexibility of the substrate-free enzyme (Table 1, Fig. 3, A and C). However, linker mutants in the presence of NADH showed remarkable differences in structural flexibility. Interestingly, the normalized Kratky and Porod-Debye profiles illustrated the different levels of reduced flexibility among the mutants, with the mutant EcAhpF(E200A/K201A/R202A) being more rigid than EcAhpF(K201A/R202A) and EcAhpF(R202A) (Fig. 5, A and B). Moreover, the reduced flexibility seems to affect equilibrium of the NTD motion, which is highlighted by the significant decrease in Rg and Dmax values and by the increasing amount of the compact conformation in solution (Tables 3 and 4). The significant difference in the structural flexibility between WT EcAhpF and the EcAhpF(E200A/K201A/R202A) mutant reflects the importance of the proper linker residue composition to effectively reduce EcAhpC for peroxidation (Fig. 6, A and C). Furthermore, the NADH-bound forms of EcAhpF(R202A), EcAhpF(K201A/R202A), and EcAhpF(E200A/K201A/R202A) show a progressive reduce in structural flexibility, which is reflected in the decrease in the peroxide reduction rates of 66, 58, and 47 s−1, respectively.
The proposed mode of inter-domain motion for efficient regeneration of AhpC
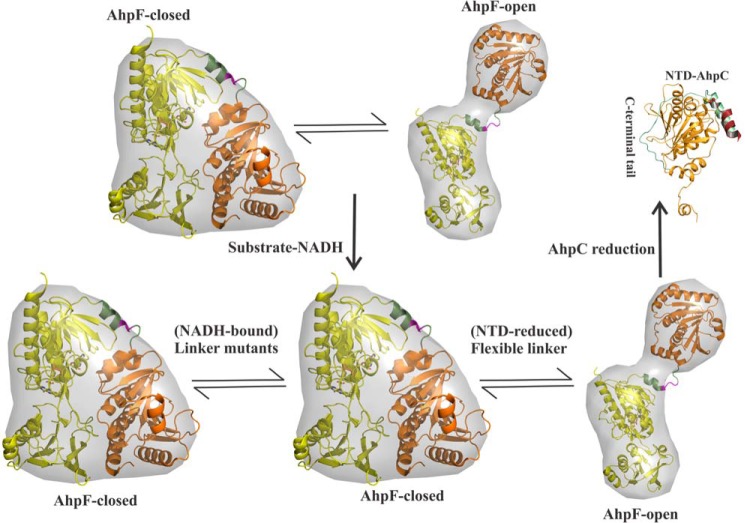
The large-scale conformational flexibility of EcAhpF demonstrates that the NTD is present in an equilibrium between two alternative conformations, termed the closed and open states, enabling the NTD to interact with the CTD and AhpC, respectively (Fig. 8) (14, 15). In addition, the linker mutations upon NADH-binding significantly affect the structural flexibility, indicating that NADH-binding induces conformational changes for NTD movements, leading to an AhpF closed conformation, which enables the dithiol-disulfide exchange with the CTD (Fig. 8). Nevertheless, the flexible linker assists the conformational equilibrium of the NTD motion as shown for the NADH-bound WT EcAhpF in solution (Table 4) (15). In comparison, the linker mutants, perturbing the flexibility, are unable to provide the desired conformational alterations and consequently affect the conformational equilibrium of NTD movement, represented by a prevalent closed AhpF conformation in solution (Fig. 8).
Figure 8.
NTD conformational selection. The NTD (orange) of AhpF is proposed to be present in a rapid conformational equilibrium between the closed and open states. The flexible linker (green) enables the rapid equilibria of NTD motion, wherein the closed state enables the NTD to interact with the CTD (yellow) and the open conformation allows the NTD to bind the oxidized AhpC for regeneration. The 200EKR202 mutants (magenta) affect the conformational equilibria of NTD movement in solution. For clarity, AhpF is shown as a monomer.
Besides the rotation and translation rearrangements observed for the NTD (Fig. 1A) (14), the substrate (NADH)- and product (NAD+)-bound crystal structures revealed that the NADH domain has to rotate back-and-forth by about 65° (15). The comparison between the compact and extended AhpF structures showed that the NADH domain in the compact form rotates about 9° and shifts about 0.79 Å along a screw axis running parallel to the center of the molecule (14). Therefore, a model was suggested in which the NTD would bend down toward the NADH-domain redox center once the dithiol-disulfide exchange with the AhpC catalytic center is completed (14). In a coordinated manner, the NADH-domain would twist to bring the electron donor NADH close to the flavin, followed by the electron transfer from the reduced flavin to the CTD redox-active center (Cys345/Cys348), which finally transfers the electrons to the redox-active center of the NTD. The reduced NTD would move up for a new cycle of H2O2 reduction in the AhpC oligomer. Because the NADH-bound triple mutant EcAhpF(E200A/K201A/R202A) becomes “trapped” into a closed conformation, we speculate that the transient elastic energy inside the 200EKR202-swivel may be sufficient to overcome the interaction of the NADH domain and the NTD during electron transfer, and allows the upward movement of the NTD. In comparison, the upwards movement of the NTD is gradually affected, allowing the gradual increase in the NTD-closed conformation for the NADH-bound forms of EcAhpF(R202A), EcAhpF(K201A/R202A), and EcAhpF(E200A/K201A/R202A), respectively (Table 4).
Furthermore, the studies also demonstrate that the existence of rapid equilibria of NTD motion, specifically the presence of an open conformation, enhances the NTD-AhpC ensemble formation for an effective regeneration of AhpC (Fig. 7), resulting in maximum peroxidase activity as shown by the WT EcAhpF (Fig. 6, A and C). The rigid triple mutant EcAhpF(E200A/K201A/R202A), which lacks the open conformation, shows low AhpC reduction and peroxidase activity (Figs. 6 and 7). Moreover, we suggest that the open conformation of AhpF explicitly selects the oxidized AhpC through interactions with the flexible C-terminal tail present in the oxidized AhpC (Fig. 8). This enhances the ability of the NTD to dock with AhpC in the appropriate redox state prior to each electron transfer. Furthermore, the disassembly of the transient complex is mediated by the conformational changes at the active site and the C-terminal tail of AhpC after reduction (13). Overall, the studies demonstrate that the linker with its 200EKR202 swivel provides an optimal cross-talk between the redox centers inside AhpF and a robust design principle for efficient shuttling of electrons to regenerate AhpC during the peroxidation cycle.
Experimental procedures
Cloning, overexpression, and purification of proteins
Single EcAhpF mutants EcAhpF(E200A), EcAhpF(K201A), and EcAhpF(R202A), a double mutant EcAhpF(K201A/R202A), and a triple mutant EcAhpF(E200A/K201A/R202A), were generated by overlap extension PCR using forward primer 5′-CAT GCC ATG GCA ATG CTC GAC ACA AAT ATG AAA CTC AAC TCA AGG-3′ and the reverse primer 5′-GCG AGC TCG TTA TGC AGT TTT GGT GCG AAT CAG GTAG-3′ with NcoI and SacI restriction site (bold), respectively, as flanking primers complementary to the ends of the target sequence. In addition, the following internal forward primers 5′-ACT GGC GCG GCT AAA CGT GCG GCA GAA GAG CTG-3′, 5′-ACT GGC GCG GAA GCA CGT GCG GCA GAA GAG CTG-3′, 5′-ACT GGC GCG GAA AAA GCG GCG GCA GAA GAG CTG-3′, 5′-ACT GGC GCG GAA GCA GCG GCG GCA GAA GAG CTG-3′, and 5′-ACT GGC GCG GCA GCA GCA GCG GCA GAA GAG CTG-3′, and the following internal reverse primers 5′-CAG CTC TTC TGC CGC ACG TTT AGC CGC GCC AGT-3′, 5′-CAG CTC TTC TGC CGC ACG TGC TTC CGC GCC AGT-3′, 5′-CAG CTC TTC TGC CGC CGC TTT TTC CGC GCC AGT-3′, 5′-CAG CTC TTC TGC CGC CGC TGC TTC CGC GCC AGT-3′, and 5′-CAG CTC TTC TGC CGC TGC TGC TGC CGC GCC AGT-3′, which contains the desired mutations to generate the EcAhpF(E200A), EcAhpF(K201A), EcAhpF(R202A), EcAhpF(K201A/R202A), and EcAhpF(E200A/K201A/R202A), respectively, were used for the amplification of the gene. The amplified products were ligated into the pET9-d1-His6 vector (34). The coding sequences for all constructs were verified by DNA sequencing. The final plasmids were subsequently transformed into E. coli BL21 (DE3) cells (Stratagene). To express the respective proteins, liquid cultures were shaken in LB medium containing kanamycin (30 μg/ml) at 310 K until an optical density A600 of 0.6–0.7 was reached. To induce the production of proteins, cultures were supplemented with isopropyl β-d-1-thiogalactopyranoside to a final concentration of 1 mm, followed by incubation for 4 h at 310 K.
E. coli cells containing recombinant EcAhpF linker mutant proteins were harvested from 1-liter cultures by centrifugation at 8,000 × g for 10 min at 279 K. The cells producing the respective recombinant proteins were lysed on ice by sonication with an ultrasonic homogenizer (Bandelin, KE76 tip) for 3 × 1 min in buffer A (50 mm Tris-HCl, pH 7.5, 200 mm NaCl, 2 mm PMSF, 1 mm Pefabloc SC, 0.8 mm DTT). After sonication, the cell lysate was centrifuged at 10,000 × g for 35 min at 277 K. The resulting supernatant was passed through a filter (0.45 μm; Millipore) and supplemented with nickel-nitrilotriacetic acid resin pre-equilibrated in buffer A. The His-tagged proteins were allowed to bind to the matrix for 1.5 h at 277 K by mixing on a sample rotator (Neolab). To avoid remaining DTT from the lysis buffer A, nickel-nitrilotriacetic acid was initially washed with 10 column volumes of buffer A without DTT and subsequently eluted with an imidazole gradient (0–500 mm). Fractions containing the required proteins were identified by SDS-PAGE (35), pooled, and concentrated using a Millipore spin concentrator with a molecular mass cutoff of 10 kDa. The samples were further purified by gel filtration chromatography using a Superdex 75 HR 10/30 column (GE Healthcare). The final purity and homogeneity of the protein samples were analyzed by SDS-PAGE and the gels were stained with Coomassie Brilliant Blue G-250 (35). Wild-type EcAhpF and -AhpC were purified by the method described earlier (14).
Small angle X-ray scattering data collection
SAXS data of the EcAhpF linker mutants were measured with the BRUKER NANOSTAR SAXS instrument equipped with a Metal-Jet X-ray source (Excillum, Germany) and VÅNTEC 2000-detector system. The X-ray radiation was generated from a liquid gallium alloy with microfocus electron source (Kα = 0.13414 nm with a potential of 70 kV and a current of 2.857 mA). The X-ray beam was focused and monochromated using Montel mirrors. The parasitic scattering is eliminated using two pinhole collimation systems. Liquid samples are placed in a sealed quartz capillary, whereas the solid glassy carbon that is used as a standard is fixed into the path of the beam after the sample. The sample chamber and beam path are evacuated at around 1 pascal and all measurements were performed at 15 °C. The sample to detector distance was set at 0.67 m, which covers a range of momentum transfer of 0.16 < q < 4 nm−1 (q = 4π sin(θ)/λ, where 2θ is the scattering angle) (36, 37).
SAXS experiments were carried out at 15 °C in a concentration series ranging from 2 to 6 mg/ml in buffer containing 50 mm Tris-HCl, pH 7.5, 200 mm NaCl for a sample volume of 40 μl in a vacuum tight quartz capillary. The data were collected for 30 min and for each measurement a total of six frames at 5-min intervals were recorded for both the sample and the corresponding buffer. SAXS measurements with the standard globular protein, lysozyme, were performed at 5 mg/ml in buffer containing 100 mm acetic acid, pH 4.5, employing the same strategy as above for comparison. The scattered X-rays, detected by a two-dimensional area detector, were flood-field and spatially corrected. The flood-field correction rectifies the intensity distortions arising due to the non-uniformity in the pixel to pixel sensitivity differences in the detector using a radioactive source (55Fe) supplied by Bruker AXS, Germany. The spatial correction fixes the inherent geometrical pincushion distortion by placing a mask with regular pattern before the detector and measuring the deviation from regularity in the detected image. The data were converted to one-dimensional scattering as a function of momentum transfer by radial averaging using the built-in SAXS software (Bruker AXS, Germany). The data were normalized by the incident intensity and transmission of the sample using a strongly scattering glassy carbon of known X-ray transmission. The transmission of X-rays through the sample is determined by an indirect method, in which the integrated counts per unit time for glassy carbon is related to the relative intensity of the direct beam and hence the transmission of the sample (38). The data were then tested for possible radiation damage by comparing the six data frames and no changes were observed. The scattering of the buffer was subtracted from the scattering of the sample, and all the scattering data were normalized by the concentration.
Similarly, SAXS data for the EcAhpF linker mutants in the presence of substrate NADH were measured for each protein sample incubated with 1 mm NADH for 30 min. Corresponding to each protein sample, data were collected for a buffer under identical experimental conditions, providing a background scattering curve.
SAXS data analysis
All the data processing steps were performed using the program package PRIMUS (26). The experimental data obtained for all protein samples were analyzed for aggregation using the Guinier region (21). The forward scattering I(0) and the radius of gyration, Rg, were computed using the Guinier approximation assuming that at very small angles (q < 1.3/Rg) the intensity is represented as I(q) = I(0)·exp(−(qRg)2/3) (21). The Rg is the root mean square distance of all atoms to the center of the scattering volume of the particles in solution, weighted by their scattering length density. Rg provides a measure of the overall size of the macromolecule. The distance distribution function, P(r), was computed from the extended scattering patterns using the indirect transform package GNOM (22). P(r) is a histogram of distances between all possible pairs of atoms within a particle. The maximum dimension of the particle, Dmax, estimated from the P(r) function satisfying the condition P(r) = 0 for r > Dmax. In addition, the radius of gyration, Rg, and forward scattering I(0) were also estimated from P(r) function. SAXS data of good quality should exhibit a close agreement between the I(0) and Rg values extracted from Guinier and P(r) analyses. To assess compactness and molecular flexibility, the SAXS data were analyzed by using the normalized Kratky plot ((qRg)2(I(q)/I(0)) versus qRg) (39). The scattering pattern of globular proteins in a normalized Kratky plot exhibits a bell-shaped profile with a clear maximum value of 1.104 for qRg = √3. The Porod-Debye plot was generated by transforming scattering data as q4·I(q) versus q4, and the Porod-Debye law approximates the scattering for well folded compact particle decays as q−4 scaled by the surface area, concentration, and electron density contrast of the particle within a limited angle range. The compact particle will produce a sharp scattering contrast and subsequently, display a plateau in Porod-Debye plot, which is absent in the case of increasing flexibility of the particle, due to the diffuse in scattering contrast (24). Ab initio low-resolution models of the proteins were built by the program DAMMIF (40) considering low angle data (q < 2 nm−1). The algorithm constructs bead models yielding a scattering profile with the lowest possible discrepancy (χ2) to the experimental data whereas keeping beads interconnected and the model compact. Ten independent ab initio reconstructions were performed for each protein and then averaged using DAMAVER (41). Superimposition between ab initio reconstruction and atomic model was performed using the software SUPCOMB (42).
The theoretical scattering curves from atomic structures were generated and evaluated against experimental scattering curves using CRYSOL (25). Utilizing the intensities from each component, the volume fraction of each component can be determined by OLIGOMER (26).
NADH-dependent peroxidase activity
The peroxidase activity of recombinant EcAhpC was measured by coupling its activity with consumption of NADH (ϵ340 = 6220 m−1 cm−1) catalyzed by WT EcAhpF and the various linker mutants (43). The peroxidase activity was measured at 25 °C by monitoring the decrease in NADH absorbance at 340 nm using a stopped-flow spectrophotometer SX20 (Applied Photophysics, UK). The reaction mixture containing 50 mm phosphate buffer, pH 7.0, 100 mm ammonium sulfate, 0.5 mm EDTA, 300 μm NADH, 30 μm EcAhpC, and 0.4 μm WT EcAhpF or the linker mutant proteins were mixed with or without 1 mm H2O2 and monitored at 340 nm.
Ferrous oxidation xylenol orange assay
Peroxide decomposition catalyzed by EcAhpC in the presence of WT EcAhpF or linker mutants were measured using the ferrous oxidation xylenol assay (44). The reaction mixture (100 μl total volume) contained 50 mm phosphate buffer, pH 7.0, 30 μm EcAhpC, 100 μm H2O2, 750 μm NADH, and 0.4 μm WT EcAhpF or EcAhpF(R202A) or EcAhpF(E200A/K201A/R202A). At several time intervals, 10-μl aliquots were removed from the reaction mixture, added to 190 μl of ferrous oxidation xylenol reagent, and incubated at room temperature for 30 min. The sample absorbance was measured at 560 nm to quantitate the residual peroxide.
AhpC reduction assay using non-reducing SDS-PAGE
This assay is based on the observations that the reduced and oxidized EcAhpC run as a monomer and dimer, respectively, in a non-reducing SDS-PAGE (10). The 200 μl of total reaction volume containing 50 mm HEPES buffer, pH 7.0, 20 μm oxidized AhpC, 50 nm WT EcAhpF, or EcAhpF(E200A/K201A/R202A) were added with 100 μm NADH to initiate the reaction. The 10 μl of sample was taken out at the desired time interval (0.5–10 min) and added to 50 mm N-ethylmaleimide in a sample buffer (4% SDS, 10% glycerol, and 62.5 mm Tris-HCl, pH 6.8) to avoid artifactual disulfides. The incubation of two proteins in the absence of NADH was taken as control. The reduction of oxidized EcAhpC by EcAhpF or the EcAhpF(E200A/K201A/R202A) mutant was monitored by transition from a dimeric to a monomeric band in the non-reducing 17% SDS gel.
Author contributions
N. K., B. E., F. E., and G. G. designed the experiments. N. K. performed the experiments. N. K. and G. G. analyzed the data. N. K., B. E., F. E., and G. G. wrote the paper.
This work was supported in part by NTU-fund (Nanyang Technological University) Grant M4080811.080 (to G. G.). The authors declare that they have no conflicts of interest with the contents of this article.
- ROS
- reactive oxygen species
- Prxs
- peroxiredoxin
- PrxRs
- peroxiredoxin reductases
- AhpF
- alkyl hydroperoxide reductase subunit F
- AhpC
- alkyl hydroperoxide reductase subunit C
- NTD
- N-terminal domain
- CTD
- C-terminal domain
- CP
- peroxidatic cysteine
- CR
- resolving cysteine
- SAXS
- small angle X-ray scattering
- PDB
- Protein Data Bank.
References
- 1. Imlay J. A. (2013) The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 11, 443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. D'Autréaux B., and Toledano M. B. (2007) ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 8, 813–824 [DOI] [PubMed] [Google Scholar]
- 3. Winterbourn C. C. (2008) Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 4, 278–286 [DOI] [PubMed] [Google Scholar]
- 4. Rhee S. G., Woo H. A., Kil I. S., and Bae S. H. (2012) Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J. Biol. Chem. 287, 4403–4410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jacobson F. S., Morgan R. W., Christman M. F., and Ames B. N. (1989) An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage: purification and properties. J. Biol. Chem. 264, 1488–1496 [PubMed] [Google Scholar]
- 6. Poole L. B., and Ellis H. R. (1996) Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium: 1. purification and enzymatic activities of overexpressed AhpF and AhpC proteins. Biochemistry 35, 56–64 [DOI] [PubMed] [Google Scholar]
- 7. Seaver L. C., and Imlay J. A. (2001) Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183, 7173–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bryk R., Griffin P., and Nathan C. (2000) Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407, 211–215 [DOI] [PubMed] [Google Scholar]
- 9. Hall A., Nelson K., Poole L. B., and Karplus P. A. (2011) Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid. Redox Signal. 15, 795–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kamariah N., Sek M. F., Eisenhaber B., Eisenhaber F., and Grüber G. (2016) Transition steps in peroxide reduction and a molecular switch for peroxide robustness of prokaryotic peroxiredoxins. Sci. Rep. 6, 37610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Poole L. B. (2005) Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch. Biochem. Biophys. 433, 240–254 [DOI] [PubMed] [Google Scholar]
- 12. Dip P. V., Kamariah N., Nartey W., Beushausen C., Kostyuchenko V. A., Ng T. S., Lok S. M., Saw W. G., Eisenhaber F., Eisenhaber B., and Grüber G. (2014) Key roles of the Escherichia coli AhpC C-terminus in assembly and catalysis of alkylhydroperoxide reductase, an enzyme essential for the alleviation of oxidative stress. Biochim. Biophys. Acta 1837, 1932–1943 [DOI] [PubMed] [Google Scholar]
- 13. Nartey W., Basak S., Kamariah N., Manimekalai M. S., Robson S., Wagner G., Eisenhaber B., Eisenhaber F., and Grüber G. (2015) NMR studies reveal a novel grab and release mechanism necessary for efficient catalysis of the bacterial 2-Cys peroxiridoxin machinery. FEBS J. 282, 4620–4638 [DOI] [PubMed] [Google Scholar]
- 14. Dip P. V., Kamariah N., Subramanian Manimekalai M. S., Nartey W., Balakrishna A. M., Eisenhaber F., Eisenhaber B., and Grüber G. (2014) Structure, mechanism and ensemble formation of the alkylhydroperoxide reductase subunits AhpC and AhpF from Escherichia coli. Acta Crystallogr. D Biol. Crystallogr. 70, 2848–2862 [DOI] [PubMed] [Google Scholar]
- 15. Kamariah N., Manimekalai M. S., Nartey W., Eisenhaber F., Eisenhaber B., and Grüber G. (2015) Crystallographic and solution studies of NAD+- and NADH-bound alkyl hydroperoxide reductase subunit F (AhpF) from Escherichia coli provide insight into sequential enzymatic steps. Biochim. Biophys. Acta 1847, 1139–1152 [DOI] [PubMed] [Google Scholar]
- 16. Wood Z. A., Poole L. B., and Karplus P. A. (2001) Structure of intact AhpF reveals a mirrored thioredoxin-like active site and implies large domain rotations during catalysis. Biochemistry 40, 3900–3911 [DOI] [PubMed] [Google Scholar]
- 17. Kamariah N., Nartey W., Eisenhaber B., Eisenhaber F., and Grüber G. (2016) Low resolution solution structure of an enzymatic active AhpC10:AhpF2 ensemble of the Escherichia coli Alkyl hydroperoxide reductase. J. Struct. Biol. 193, 13–22 [DOI] [PubMed] [Google Scholar]
- 18. Papaleo E., Saladino G., Lambrughi M., Lindorff-Larsen K., Gervasio F. L., and Nussinov R. (2016) The role of protein loops and linkers in conformational dynamics and allostery. Chem. Rev. 116, 6391–6423 [DOI] [PubMed] [Google Scholar]
- 19. Gokhale R. S., and Khosla C. (2000) Role of linkers in communication between protein modules. Curr. Opin. Chem. Biol. 4, 22–27 [DOI] [PubMed] [Google Scholar]
- 20. Poole L. B., Godzik A., Nayeem A., and Schmitt J. D. (2000) AhpF can be dissected into two functional units: tandem repeats of two thioredoxin-like folds in the N-terminus mediate electron transfer from the thioredoxin reductase-like C-terminus to AhpC. Biochemistry 39, 6602–6615 [DOI] [PubMed] [Google Scholar]
- 21. Guinier A. (1939) La diffraction des rayons X aux tres petits angles; application a l'etude de phenomenes ultramicroscopiques. Ann. Phys. (Paris) 12, 161–237 [Google Scholar]
- 22. Svergun D. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 23. Kikhney A. G., and Svergun D. I. (2015) A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett. 589, 2570–2577 [DOI] [PubMed] [Google Scholar]
- 24. Rambo R. P., and Tainer J. A. (2011) Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 95, 559–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Svergun D. I., Barberato C., and Koch M. H. J. (1995) CRYSOL: a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 26. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., and Svergun D. I. (2003) PRIMUS: Windows-PC based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 27. Moser C. C., Keske J. M., Warncke K., Farid R. S., and Dutton P. L. (1992) Nature of biological electron transfer. Nature 355, 796–802 [DOI] [PubMed] [Google Scholar]
- 28. Leys D., and Scrutton N. S. (2004) Electrical circuitry in biology: emerging principles from protein structure. Curr. Opin. Struct. Biol. 14, 642–647 [DOI] [PubMed] [Google Scholar]
- 29. Parsonage D., Nelson K. J., Ferrer-Sueta G., Alley S., Karplus P. A., Furdui C. M., and Poole L. B. (2015) Dissecting peroxiredoxin catalysis: separating binding, peroxidation, and resolution for a bacterial AhpC. Biochemistry 54, 1567–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parsonage D., Karplus P. A., and Poole L. B. (2008) Substrate specificity and redox potential of AhpC, a bacterial peroxiredoxin. Proc. Natl. Acad. Sci. U.S.A. 105, 8209–8214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Niimura Y., and Massey V. (1996) Reaction mechanism of Amphibacillus xylanus NADH oxidase/alkyl hydroperoxide reductase flavoprotein. J. Biol. Chem. 271, 30459–30464 [DOI] [PubMed] [Google Scholar]
- 32. Poole L. B., Higuchi M., Shimada M., Calzi M. L., and Kamio Y. (2000) Streptococcus mutans H2O2-forming NADH oxidase is an alkyl hydroperoxide reductase protein. Free Radic. Biol. Med. 28, 108–120 [DOI] [PubMed] [Google Scholar]
- 33. Poole L. B., Reynolds C. M., Wood Z. A., Karplus P. A., Ellis H. R., and Li Calzi M. (2000) AhpF and other NADH:peroxiredoxin oxidoreductases, homologues of low Mr thioredoxin reductase. Eur. J. Biochem. 267, 6126–6133 [DOI] [PubMed] [Google Scholar]
- 34. Grüber G., Godovac-Zimmermann J., Link T. A., Coskun U., Rizzo V. F., Betz C., and Bailer S. M. (2002) Expression, purification, and characterization of subunit E, an essential subunit of the vacuolar ATPase. Biochem. Biophys. Res. Commun. 298, 383–391 [DOI] [PubMed] [Google Scholar]
- 35. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 36. Balakrishna A. M., Basak S., Manimekalai M. S., and Grüber G. (2015) Crystal structure of subunits D and F in complex give insight into energy transmission of the eukaryotic V-ATPase from Saccharomyces cerevisiae. J. Biol. Chem. 290, 3183–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saw W. G., Tria G., Grüber A., Subramanian Manimekalai M. S., Zhao Y., Chandramohan A., Srinivasan Anand G., Matsui T., Weiss T. M., Vasudevan S. G., and Grüber G. (2015) Structural insight and flexible features of NS5 proteins from all four serotypes of Dengue virus in solution. Acta Crystallogr. D Biol. Crystallogr. 71, 2309–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dreiss C. A., Jack K. S., and Parker A. P. (2006) On the absolute calibration of bench-top small-angle X-ray scattering instruments: a comparison of different standard methods. J. Appl. Crystallogr. 39, 32–38 [Google Scholar]
- 39. Durand D., Vivès C., Cannella D., Pérez J., Pebay-Peyroula E., Vachette P., and Fieschi F. (2010) NADPH oxidase activator p67phox behaves in solution as a multidomain protein with semi-flexible linkers. J. Struct. Biol. 169, 45–53 [DOI] [PubMed] [Google Scholar]
- 40. Franke D., and Svergun D. I. (2009) DAMMIF, a program for rapid ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Volkov V. V., and Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kozin M. B., and Svergun D. I. (2001) Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34, 33–41 [Google Scholar]
- 43. Niimura Y., Poole L. B., and Massey V. (1995) Amphibacillus xylanus NADH oxidase and Salmonella typhimurium alkyl-hydroperoxide reductase flavoprotein components show extremely high scavenging activity for both alkyl hydroperoxide and hydrogen peroxide in the presence of S. typhimurium alkyl-hydroperoxide reductase 22-kDa protein component. J. Biol. Chem. 270, 25645–25650 [DOI] [PubMed] [Google Scholar]
- 44. Wolff S. P. (1994) Ferrous ion oxidation in presence of ferric ion indicator xylenol orange for measurement of hydroperoxides. Methods Enzymol. 233, 182–189 [Google Scholar]