Abstract
Microtubule-associated protein 2c (MAP2c) is involved in neuronal development and is less characterized than its homolog Tau, which has various roles in neurodegeneration. Using NMR methods providing single-residue resolution and quantitative comparison, we investigated molecular interactions important for the regulatory roles of MAP2c in microtubule dynamics. We found that MAP2c and Tau significantly differ in the position and kinetics of sites that are phosphorylated by cAMP-dependent protein kinase (PKA), even in highly homologous regions. We determined the binding sites of unphosphorylated and phosphorylated MAP2c responsible for interactions with the regulatory protein 14-3-3ζ. Differences in phosphorylation and in charge distribution between MAP2c and Tau suggested that both MAP2c and Tau respond to the same signal (phosphorylation by PKA) but have different downstream effects, indicating a signaling branch point for controlling microtubule stability. Although the interactions of phosphorylated Tau with 14-3-3ζ are supposed to be a major factor in microtubule destabilization, the binding of 14-3-3ζ to MAP2c enhanced by PKA-mediated phosphorylation is likely to influence microtubule-MAP2c binding much less, in agreement with the results of our tubulin co-sedimentation measurements. The specific location of the major MAP2c phosphorylation site in a region homologous to the muscarinic receptor-binding site of Tau suggests that MAP2c also may regulate processes other than microtubule dynamics.
Keywords: 14-3-3 protein, mass spectrometry (MS), microtubule-associated protein (MAP), nuclear magnetic resonance (NMR), protein kinase A (PKA)
Introduction
Cytoskeletal microtubule-associated proteins (MAPs)3 are proteins of critical importance for regulating the stability and dynamics of microtubules (1). MAP2 and Tau represent MAP subfamilies expressed in neurons; MAP2 is localized in dendrites, whereas Tau is found mainly in axons (2). Tau and MAP2 belong to the class of intrinsically disordered proteins (IDPs), which lack a unique structure and which exist in multiple, quickly interconverting conformations (3–7). NMR is the method of choice for structural investigation of this type of protein. Tau and MAP2 differ in their N-terminal projection domains, which contain acidic and proline-rich subdomains, whereas the C-terminal parts, containing the microtubule-binding domain (MTBD) and the C-terminal region, are homologous (8). Tau is expressed in several splice variants. The human brain isoforms differ in the number of microtubule-binding regions (MTBRs) in the C-terminal portion and in the presence of two inserts near the N terminus. For the sake of simplicity, only the 441-residue variant lacking exons 6, 8, and 10 (9) is discussed in this paper. This variant has been studied in detail and was shown to form paired helical filaments and neurofibillary tangles in brains of patients suffering from Alzheimer's disease (10). The MAP2 family is composed of two high-molecular-weight proteins, MAP2a and MAP2b, each consisting of 1830 amino acids, and two low-molecular-weight proteins, MAP2c and MAP2d, consisting of 467 and 498 amino acids, respectively. The MAP2 isoforms differ mainly in their projection domains (8), with MAP2c being the shortest functional isoform. MAP2c mainly is expressed perinatally (2). Postnatally, its expression is restricted to regions exhibiting postnatal plasticity, such as the olfactory bulb (11), suggesting a role in neuronal development.
The phosphorylation of MAPs, which regulates their binding to microtubules (12–17) and consequently the microtubule dynamics (18), is implicated in neuronal development and plasticity (19). In vivo MAPs are substrates of various protein kinases, such as cAMP-dependent protein kinase A (PKA), protein kinase C (PKC), cdc2 kinase, and the like (16, 17, 20, 21). Phosphorylation of Tau weakens its interaction with microtubules and increases its affinity to regulatory 14-3-3 proteins (vide infra).
The 14-3-3 family includes highly conserved ubiquitous proteins (22) mostly expressed in the brain, and in particular in regions exhibiting neuroplasticity, and having important roles in neuronal development (23). The seven 14-3-3 isoforms present in mammals are β, γ, ϵ, ζ, τ, σ, and η. The 14-3-3ζ isoform was reported to be associated with microtubules in the brain (24). In vitro, 14-3-3ζ interacts with unphosphorylated Tau; however the phosphorylation of Tau increases its affinity for 14-3-3ζ (24–27).
Our goal was to compare the phosphorylation of Tau and MAP2c by PKA and the interactions of Tau and MAP2c with 14-3-3ζ. To the best of our knowledge, no data have been published on the interaction of 14-3-3ζ with MAP2c until now. A prerequisite for such studies is the knowledge of the phosphorylated residues of MAP2c. MAP2c phosphorylation by PKA has been studied previously, but the reported data are partially contradictory (28, 29). Therefore, we re-addressed this issue and determined the PKA phosphorylation sites of MAP2c by NMR and mass spectrometry (MS). We then characterized the interaction of MAP2c with 14-3-3ζ at a molecular level, and we studied the effect of 14-3-3ζ on tubulin polymerization induced by MAP2c. We found that the same regions of unphosphorylated MAP2c and Tau interact with 14-3-3ζ and that the binding affinity of MAP2c is greatly increased by phosphorylation, as described previously for Tau (25, 39). However, NMR analysis of the phosphorylation kinetics revealed that, despite their highly homologous sequences, PKA phosphorylates MAP2c and Tau in a different manner. Consequently, the high-affinity binding sites of phosphorylated MAP2c and Tau differ substantially.
Results
NMR assignment of the phosphorylated MAP2c
Chemical shifts of unphosphorylated MAP2c were assigned in our previous study (30). The same assignment strategy for PKA-phosphorylated MAP2c was used in this study. The resonance frequencies of the phosphorylated MAP2c were assigned using 5D CACONCACO, 3D (H)CANCO, and 5D HC(CC-TOCSY)CACON NMR experiments. The spectra were measured on a 1.1 mm [13C,15N]MAP2c sample phosphorylated by PKA for 24 h. The assigned backbone chemical shifts were similar to those obtained for unphosphorylated MAP2c other than the phosphorylated residues and their neighbors. The major changes in the CACONCACO spectrum were observed near Ser-435, where six neighboring residues showed a significant change in backbone chemical shifts, up to 1 ppm for 15N and 0.3 ppm for protons.
Identification of phosphorylation sites
The phosphorylated MAP2c residues were first detected by NMR spectroscopy. A 1H,15N HSQC spectrum of [15N,13C] MAP2c was measured after 24 h of phosphorylation. Four new intense peaks and several weaker signals appeared at proton frequencies downfield from 8.6 ppm, corresponding to a chemical shift of the NH groups influenced by phosphorylation (Fig. 1). The 5D CACONCACO spectra of phosphorylated MAP2c provided an unambiguous assignment of the major new peaks in the 1H,15N HSQC spectrum to pSer-435, pSer-184, pThr-220, and to Arg-221 following pThr-220. The 3D HNCACB spectrum allowed us to classify the types of amino acids preceding the four additional phosphoserines/phosphothreonines detected in the 1H,15N HSQC spectrum.
Figure 1.
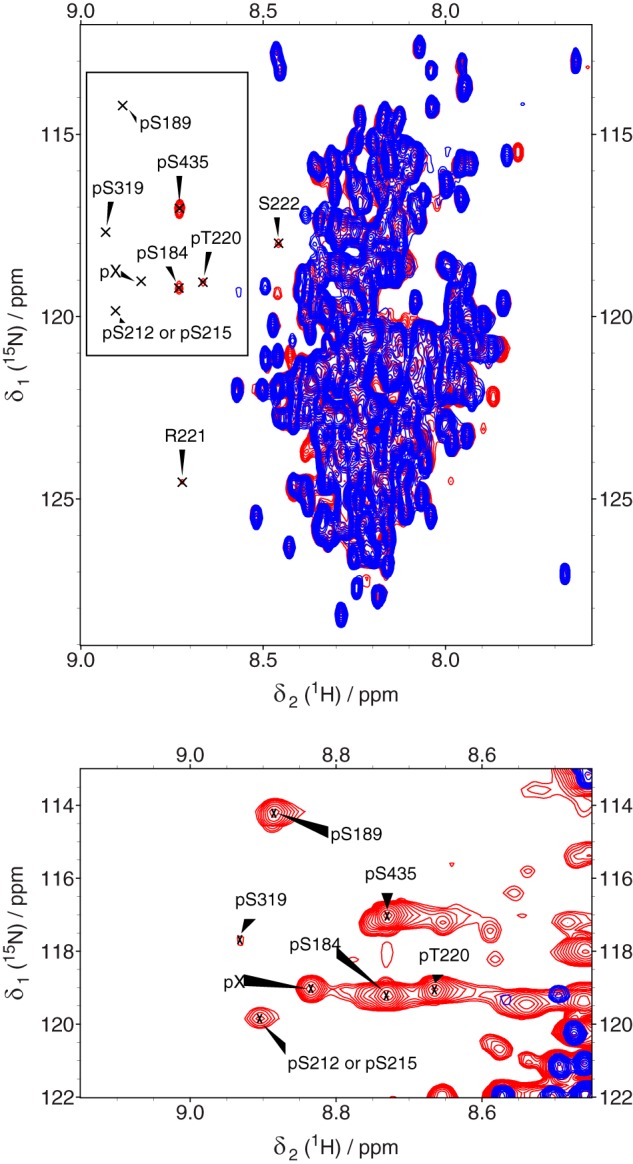
Overlaid 1H,15N HSQC spectra of unphosphorylated [15N,13C]MAP2c (blue), and [15N,13C]MAP2c phosphorylated by PKA for 24 h (red). The well-resolved peaks of phosphorylated residues (pSer-184, pSer-189, pThr-220, and pSer-435) at proton frequencies downfield from 8.6 ppm and of their neighbors (Arg-221 and Ser-222) are labeled. Bottom, close-up of the region of phosphorylated residues plotted with a lowered signal threshold level to show minor peaks. pX, this peak could not be assigned.
The phosphorylation sites were also identified by MS (Table 1). After incubation with PKA for 24 h, MAP2c was digested with trypsin. The phosphorylated peptides were separated from the unphosphorylated ones by TiO2 fractionation, and the phosphopeptides were subjected to LC-MS/MS analysis. In agreement with the NMR data, the most intense peaks corresponded to peptides containing pSer-184 and pSer-435. Phosphopeptides containing pThr-220 were not detected in our tryptic digests (possibly because of the high frequency of tryptic cleavage sites in the vicinity of Thr-220 resulting in peptides too short for analysis under the conditions used in our study) but were unambiguously identified previously (29). The peak areas of the obtained phosphopeptides relative to the sum of the areas of peaks containing pSer-435 are presented in Fig. 2. The phosphopeptides identified by MS are presented in Table 1.
Table 1.
Phosphopeptides identified by MS in percentage compared with the most intense peak, Ser-435
Only the phosphopeptides that have an intensity higher than 1% of the intensity of the most intense peak are presented here. In the absence of PKA, none of the phosphopeptides identified has an intensity higher than 1% of the most intense peak. The phosphorylated residues are indicated in bold.
| Residue | Peptide | % Intensity |
|---|---|---|
| Ser-184 | 183SSLPRPSSILPPR195 | 65 |
| Ser-199 | 196RGVSGDREENSFSLNSSISSAR217 | 5 |
| Ser-212 | 197GVSGDREENSFSLNSSISSAR217 | 1 |
| Ser-214 | 197GVSGDREENSFSLNSSISSAR217 | 2 |
| Ser-231 | 228AGKSGTSTPTTPGSTAITPGTPPSYSSR255 | 2 |
| Ser-319 | 315SKIGSTDNIK324 | 7 |
| Ser-367 | 363VKIESVKLDFK373 | 2 |
| Ser-435 | 433RLSNVSSSGSINLLESPQLATLAEDVTAALAK464 | 100 |
| Ser-435, Ser-438 | 433RLSNVSSSGSINLLESPQL451 | 3 |
| Ser-435, Ser-442 | 433RLSNVSSSGSIN444 | 1 |
Figure 2.
Phosphorylation of MAP2c. The dots represent the ratio Uu/Up, where Uu and Up are peak intensities of unphosphorylated residues in the unphosphorylated and phosphorylated MAP2c samples, respectively. Triangles represent the ratio Pp/(Up + Pp), where Pp are peak intensities of phosphorylated residues in the phosphorylated MAP2c sample. The bars represent the sum of areas of peptide peaks containing the given phosphorylated residue in mass spectra (relative to the sum of areas of peptides peaks containing Ser-435). Letter P (above the plot), indicates proline residues.
Verification of NMR assignment of phosphorylated residues by site-directed mutagenesis
Because the sensitivity of the 5D CACONCACO spectra was not sufficient to assign minor phosphorylation sites, we designed several mutants (S184D, S189D/S435D, S199D/S435D, T220E, S319D/S435D, T320E/S435D, S342D/S435D, S350D/S435D, S367D/S435D, S382D/S435D, and S435D) based on our MS data and on phosphorylation sites reported in the literature (28). The mutants were uniformly 15N-labeled and phosphorylated, and the 1H,15N HSQC spectra were recorded. In the spectra of the S184D, T220E, S435D, S189D/S435D, and S319D/S435D mutants, the peaks of the mutated phosphorylated residues disappeared, confirming our previous assignment and allowing us to assign two minor peaks to pSer-189 and pSer-319. No significant changes were observed in the region of the phosphorylated serines and threonines in the HSQC spectra of the other mutants, suggesting that their degree of phosphorylation is low.
Sequence-based prediction of phosphorylation sites
In principle, the target sites for specific kinases are encoded in the amino acid sequence and by their accessibility to the kinase. However, currently available Web server predictors of phosphorylation sites have been optimized for structured proteins, whereas their reliability for IDPs is not well documented. Fig. 3 compares the PKA phosphorylation sites of MAP2c determined by NMR in this study with three popular Web server predictors. One can see that the PKA phosphorylation of Ser-435 was predicted by all three predictors. Sites Ser-184, Ser-189, and Thr-220 were predicted by Scansite3 (31) and GPS3 (32) but not by Kinasephos2 (33). This limited comparison indicates that Scansite3 and GPS3 can identify the most important PKA phosphorylation sites within the intrinsically disordered protein MAP2c. On the other hand, all three predictors generated significant number of false-positive predictions with respect to the NMR data (Fig. 3). Interestingly, the same type of prediction for Tau protein shows better agreement between GPS3 and Kinasephos2 in contrast to Scansite3 (Fig. 3). New experimental results are therefore needed to improve the reliability of prediction of phosphorylated sites within IDPs.
Figure 3.
Comparison of sequences, phosphorylation sites, and electrostatic properties of Tau and MAP2c. Experimentally determined phosphorylation sites are indicated by background colors: red, class I; orange, class II; green, class III (according to the classification used by Laudrieu et al. (43)). The predicted phosphorylation sites are indicated by dots below the sequences: red, prediction by Scansite3; green, prediction by GPS3; blue, prediction by Kinasephos2. Residues predicted by 1433pred (48) to interact with 14-3-3ζ are underlined. Regions with a propensity to form helical, β-strand, and polyproline II structures according to Mukrasch et al. (38) and Nováček et al. (30) are indicated by magenta, cyan, and gray bars above the sequences, respectively. The relative electrostatic potential is shown as color-coded boxes below the sequences for unphosphorylated (upper row) and phosphorylated (lower row) form of each protein. The potential is approximated by ΣjCQi/(d0 + d1|ni − nj|), where Qi and nj are the charge and sequential number of the i-th residue, C is a constant including the electric permittivity, and dk are distance constants. The ratio d1/d0 was set to 2.0, and the colors were chosen so that red and blue correspond to the highest negative and positive potential, respectively, which makes the color code independent of C/d0. Gaps in the sequences were inserted manually to optimize the alignment of regions with similar trends to form transient secondary structures. The annotation of functionally important regions of Tau, defined by the double-headed arrows above the sequences, was taken from the literature (50, 38, 9).
Quantitative determination of phosphorylation
Strong signals indicating a high degree of phosphorylation at Ser-184 and Ser-435 were observed using both NMR and MS methods. NMR was used to determine the degree of phosphorylation quantitatively. A comparison of the peak intensities in the 3D HNCO spectra of phosphorylated and unphosphorylated MAP2c revealed three regions showing a significant decrease in the intensity of the peaks, corresponding to unphosphorylated residues in the phosphorylated sample (Fig. 2). The intensity levels decreased to ∼40% in the vicinity of Thr-220, to less than 10% in the vicinity of Ser-184 and below the detection limit in the vicinity of Ser-435. Such a comparison provided an overall quantitative picture of the phosphorylation and allowed us to estimate the degree of phosphorylation of individual amino acids (with the exception of residues close in sequence to Ser-184, Thr-220, and Ser-435, where the effect of possible minor phosphorylation was obscured by the decrease in the signal intensity due to a major phosphorylation site). In summary, both NMR and MS identified Ser-184 and Ser-435 as major PKA phosphorylation sites of MAP2c, and significant phosphorylation was observed also at Thr-220 (Fig. 2).
Kinetics of phosphorylation
The kinetics of phosphorylation was followed by real-time NMR spectroscopy from 2 min to 30 h after the addition of PKA. The signal of the first phosphorylated residue, pSer-435, already appeared in the first spectrum recorded after the addition of PKA and reached saturation after 1.5 h of incubation (Fig. 4). The kinetic curve was exponential even for short reaction times, indicating that PKA was not saturated by MAP2c. The other residues were phosphorylated much more slowly, and their buildup curves exhibited lag phases documenting competition of the phosphorylation sites for PKA. After the lag phase, the ratio of phosphorylation rates of Ser-184, Ser-189, and Thr-220 was almost constant (∼4:1:2).
Figure 4.
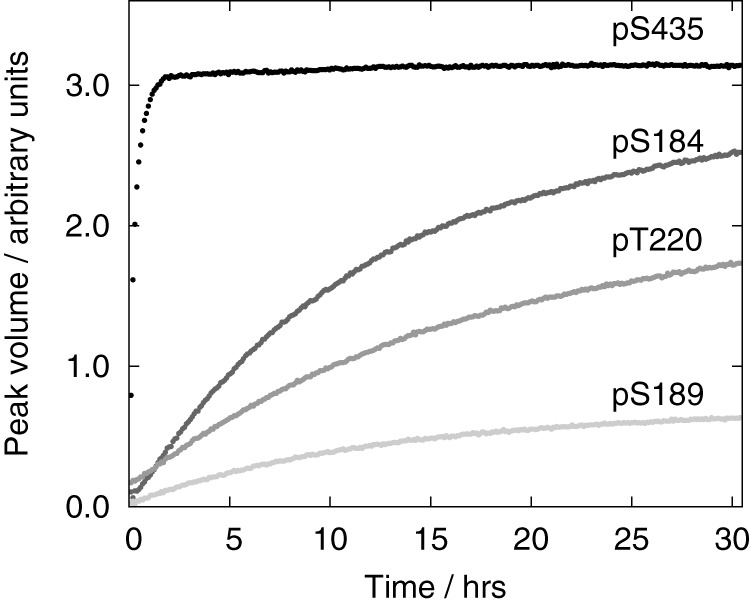
Volumes of pSer-435 (black), pSer-184 (dark gray), pThr-220 (medium gray), and pSer-189 (light gray) peaks in the SOFAST-HMQC spectra recorded during MAP2c phosphorylation by PKA.
To compare the phosphorylation of individual sites quantitatively, apparent rate constants (kobs) were estimated as described under “Experimental procedures.” The evaluation of kobs was facilitated by the kinetic separation of Ser-435, allowing us to assume that the other sites were unphosphorylated during phosphorylation of Ser-435 by the aforementioned kinetic partitioning of Ser-184, Ser-189, and Thr-220. The estimated kobs values were 9700 ± 1500 m−1s−1 for Ser-435, 200 ± 14 m−1s−1 for Ser-184, 50 ± 6 m−1s−1 for Ser-189, and 95 ± 21 m−1s−1 for Thr-220. In summary, the real-time NMR measurements revealed a dramatic difference between phosphorylation of Ser-435 and other residues.
Effect of phosphorylation on the secondary structure propensities of MAP2c
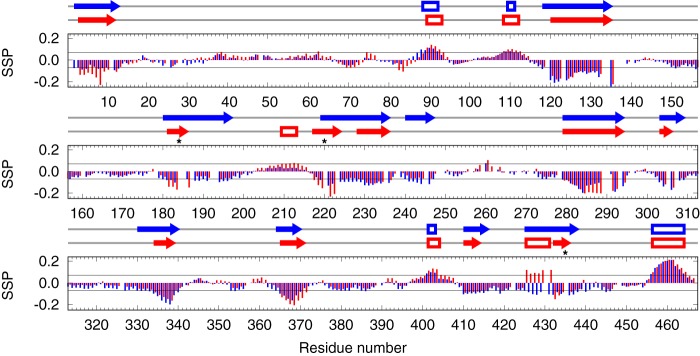
To determine the effect of phosphorylation on the secondary structure, secondary structure propensities (SSP) of unphosphorylated and phosphorylated MAP2c were calculated. To identify SSP changes close to the phosphorylation sites, the SSP was calculated from the chemical shifts of 13Cα, 13Cβ, and 1Hα, recently found to be insensitive to the presence of the phosphate group attached to neighboring residues (34). The most significant changes were observed in two regions close to the major phosphorylation sites. In the vicinity of pSer-184 and especially pThr-220, the SSP value indicates that phosphorylation stabilizes the already existing secondary structures (Fig. 5). The opposite effect was observed in a region preceding pSer-435, where phosphorylation alters the SSP of residues Arg-425–Pro-431 from the propensity to form an extended secondary structure in the unphosphorylated state to a helical propensity in the phosphorylated state (Fig. 5). The less pronounced changes observed for residues distant from the phosphorylation sites (including the N-terminal region) indicate that phosphorylation by PKA also influences the intramolecular interactions of MAP2c.
Figure 5.
Effects of phosphorylation on the secondary structure propensity of unphosphorylated MAP2c (blue) and phosphorylated MAP2c (red). The symbols above the graphs indicate continuous regions of unphosphorylated (blue) and phosphorylated (red) with the propensity to form helical (empty boxes) or extended (arrows) structures (see “Experimental procedures” for details). Asterisks indicate the major PKA phosphorylation sites.
Apparent dissociation constants of MAP2c-14-3-3ζ complexes
The affinities of 14-3-3ζ to MAP2c in phosphorylated and non-phosphorylated forms were compared by microscale thermophoresis (MST). To achieve a sufficient sensitivity, MAP2c was labeled fluorescently. MAP2c contains a single cysteine residue (Cys-348), but it is located in a potential binding region (MTBR3). Therefore, an E52C/C348S MAP2c construct was prepared and labeled specifically by the fluorescent dye Alexa Fluor 647 at the introduced Cys-52.
Titration of E52C/C348S MAP2c with 14-3-3ζ revealed that both unphosphorylated and phosphorylated MAP2c bind 14-3-3ζ, but phosphorylation greatly enhances the affinity. The titration curve of phosphorylated E52C/C348S MAP2c exhibited a well-defined inflection point in the low micromolar range of 14-3-3ζ concentrations and another less-defined inflection point at a concentration 2 orders of magnitude higher (Fig. 6). Fitting the first sigmoidal region separately (for 14-3-3ζ monomer concentrations lower than 100 μm) resulted in an apparent dissociation constant (expressed for the 14-3-3ζ monomer concentration as described under “Experimental procedures”) of 0.57 ± 0.37 μm and stoichiometry of n = 1.53 ± 0.14 (14-3-3ζ monomer/MAP2c molecule). The fractional stoichiometry may indicate the contribution of several binding modes, but the size of the experimental errors made a precise determination of the stoichiometry difficult. Fitting the second region gives K′D ≈ 280 ± 120 μm, but full saturation could not be achieved.
Figure 6.
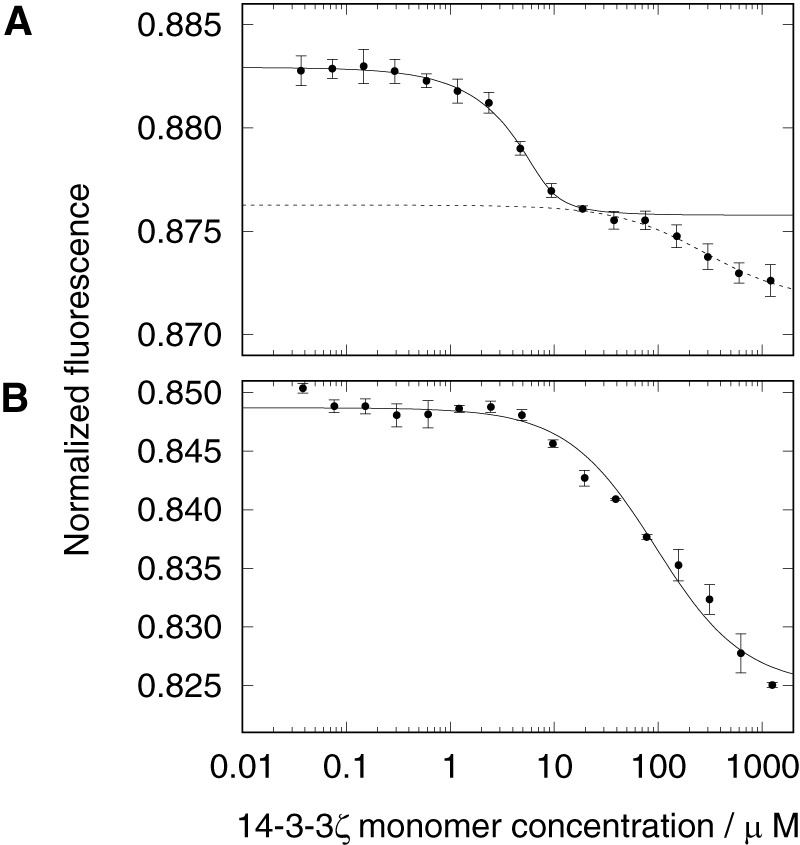
Microscale thermophoretic analysis of interaction between 5μm E52C/C348S MAP2c and 14-3-3ζ in 50 mm Tris buffer. A, unphosphorylated MAP2c; B, phosphorylated MAP2c. The plots show interactions in the 14-3-3ζ monomer concentration range from 36.6 nm to 1.2 mm. The mean values ± S.D. for each concentration point were calculated from triplicate measurements.
The binding curve of unphosphorylated E52C/C348S MAP2c showed an interaction at least 1 order of magnitude weaker than observed for the phosphorylated form. Fitting the data provided K′D = 92 ± 12 μm, but the shape of the titration curve indicates that this number is probably affected by additional binding events that occur at a higher 14-3-3ζ concentration and cannot be fully separated.
The obtained apparent dissociation constants are useful for a rough comparison of the binding affinities. However, they should not be overinterpreted because the MAP2c-14-3-3ζ interaction is more complex than the simple binding model used for the data fitting (35).
Identification of 14-3-3ζ-binding sites in MAP2c
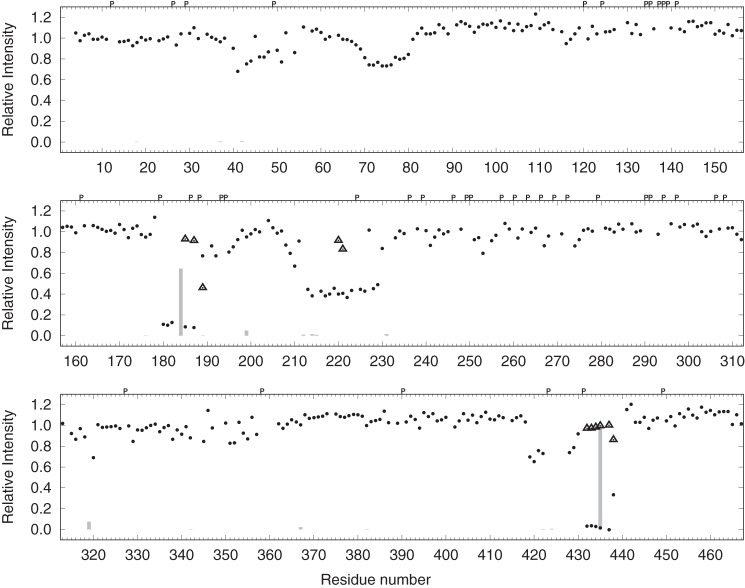
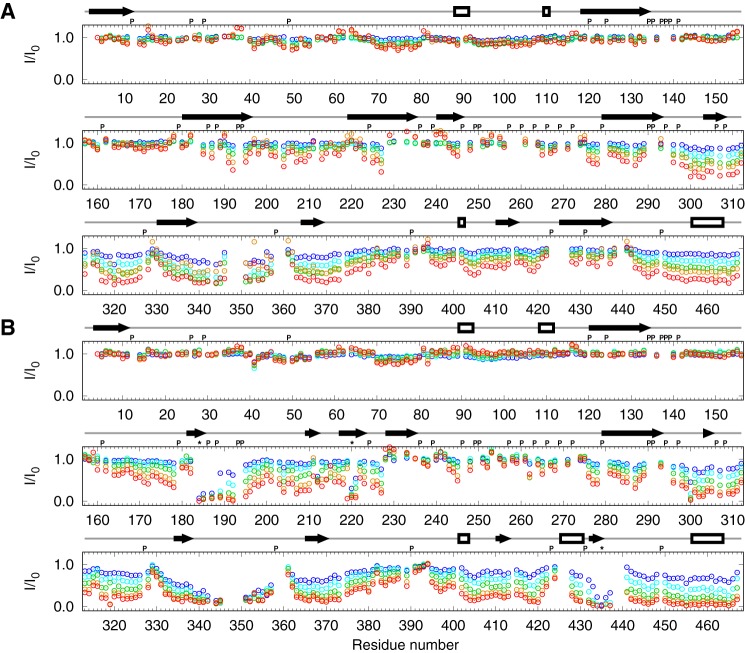
NMR spectroscopy was used to identify individual residues of MAP2c that interact with 14-3-3ζ. Unphosphorylated [13C,15N]MAP2c and [13C,15N]MAP2c phosphorylated for 24 h by PKA were mixed with 14-3-3ζ in ratios of 1:0.125, 1:0.25, 1:0.5, 1:1, and 1:2 (MAP2c:14-3-3ζ monomer). A 3D HNCO spectrum was recorded before titration and after each addition of 14-3-3ζ. When titrating unphosphorylated MAP2c with 14-3-3ζ (Fig. 7), we observed a gradual diminution of the intensity of the peaks of the residues in the MTBD (Thr-296–Ser-380) and in the C-terminal region with a strong helical propensity (Ile-443–Leu-467), indicating that these regions bind 14-3-3ζ and become less flexible. Note that the broad region of residues with reduced peak intensity due to the interaction with 14-3-3ζ is interrupted by several short stretches of amino acids already exhibiting significant line broadening in free MAP2c, and therefore it is little affected by 14-3-3ζ binding. This effect is particularly notable for the PXGG motifs immediately preceding the regions with high β-strand propensity in the MTBD. These regions initiate aggregation in Tau (36, 37). On the other hand, a decrease in the peak intensity was also observed for several residues in the N-terminal portion of MAP2c, indicating that interdomain interactions exist in MAP2c as in Tau (38).
Figure 7.
Intensities of the peaks in MAP2c HNCO spectra upon addition of 14-3-3ζ compared with intensities of the peaks in free MAP2c with the MAP2c:14-3-3ζ monomer ratios of 1:0.125 (blue), 1:0.25 (cyan), 1:0.5 (green), 1:1 (orange), and 1:2 (red). A, unphosphorylated MAP2c; B, phosphorylated MAP2c. The empty boxes and arrows above the graphs indicate propensity to form helical and extended structures, respectively, according to the data presented in Fig. 5. Letter P and asterisks (above the plot), indicate proline residues and major phosphorylation sites, respectively.
The addition of 14-3-3ζ to phosphorylated MAP2c resulted, in addition to the changes described above, in a strong decrease of peak heights of amino acids in the vicinity of pSer-184 and pThr-220 in the proline-rich domain and of pSer-435 in the region corresponding to the muscarinic receptor-binding site of Tau (Fig. 3). The fact that the peak intensities were substantially reduced already in the presence of substoichiometric amounts of 14-3-3ζ suggests that the spectra of the 14-3-3ζ-bound MAP2c are also influenced by an intermediate chemical exchange. Therefore, we did not use our NMR data for quantitative determination of the binding affinity.
Effect of 14-3-3ζ on MAP2c-induced tubulin polymerization
The effect of 14-3-3ζ on the polymerization of tubulin induced by unphosphorylated and phosphorylated MAP2c was measured by co-sedimentation assays (Fig. 8). To measure the direct effect of MAP2c and 14-3-3ζ on tubulin polymerization, tubulin was not stabilized by Taxol in the assay. 14-3-3ζ alone was not able to induce the polymerization of microtubules (data not shown). About 20% of the tubulin polymerized spontaneously. In the presence of 2 μm unphosphorylated and phosphorylated MAP2c, tubulin polymerization increased by 13 ± 2% and 12 ± 1%, respectively. The addition of up to 2 μm 14-3-3ζ (monomer concentration) had a negligible effect, but 20 μm 14-3-3ζ reduced the amount of tubulin in the pellet to a level comparable with the background of its spontaneous polymerization. The effect of MAP2c phosphorylation was negligible. Thus the data show that phosphorylation of MAP2c by PKA does not influence its affinity to tubulin under the conditions used and that 14-3-3ζ competes with tubulin for binding unphosphorylated and PKA-phosphorylated MAP2c with a comparable efficiency.
Figure 8.
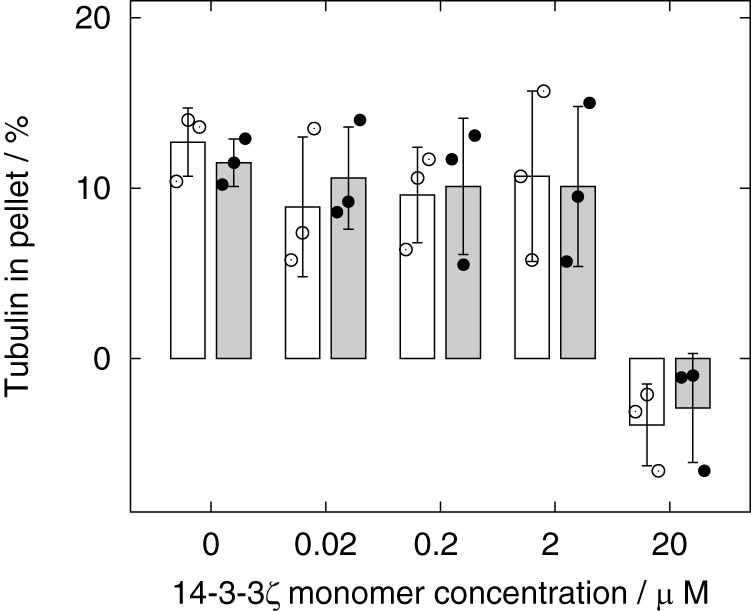
Results of the tubulin-MAP2c co-sedimentation assay. The relative amount of tubulin in the pellet (after substraction of the background of spontaneous tubulin polymerization) with increasing concentrations of 14-3-3ζ in the presence of unphosphorylated MAP2c and phosphorylated MAP2c is shown as white and black boxes, respectively. The error bars correspond to the standard deviations calculated from triplicate measurements. Only the effect of 20 μm 14-3-3ζ is statistically significant at α = 0.05.
Discussion
The key role of phosphorylation in regulating the interactions of MAPs with microtubules has been well-known for decades (12–17). 14-3-3 proteins are proposed to be involved in the phosphorylation-dependent control of microtubule dynamics by competing for phosphorylated Tau with tubulin (24, 39, 40). PKA seems to be the key kinase in this mechanism. Phosphorylation of Tau by PKA or PKB, but not by GSK-3β, CDK2, or CK1, enhances the interaction with 14-3-3ζ (25), whereas Tau phosphorylated by PKA binds to tubulin with 7-fold lower affinity (41, 20).
In our study, we searched for possible differences between Tau and its homolog, MAP2c, that would implicate distinct regulatory roles for these proteins. As phosphorylation by PKA is sufficient to form the sites critical for 14-3-3ζ binding in Tau, we also used PKA in our studies of MAP2c.
Phosphorylation of MAP2c by PKA has been studied already in the past. However, early studies of MAP2c phosphorylation resulted in partially contradictory conclusions. Using two-dimensional electrophoresis combined with mass spectrometry and site-directed mutagenesis, Ozer and Halpain (28) identified Ser-319, Ser-350, and Ser-382 as early phosphorylation sites. Later, Ser-184, Thr-220, and Ser-435 were reported by Alexa et al. (29) to be the major phosphorylation sites in MAP2c, in agreement with the prediction based on the sequence (42). A single completely phosphorylated residue, Thr-220, was found in a 20-kDa peptide (Asn-205–Glu-366) of MAP2c phosphorylated by PKA (29). Although the Asn-205–Glu-366 peptide also contains Ser-319, Ser-350, and Ser-382, phosphorylation of these serines was not observed by Alexa et al. (29). Therefore, we revisited the topic of phosphorylation of MAP2c by PKA and, taking advantage of the recent methodological progress in MS and NMR spectroscopy, clarified which amino acids are predominantly phosphorylated under the given conditions. Our results confirm that Ser-184, Thr-220, and Ser-435 are phosphorylated most efficiently. The Web server predictors also identified Ser-184, Thr-220, and Ser-435 as phosphorylation sites, but the number of false positive predictions documents that experimental determination of phosphorylation sites is still necessary (Fig. 3).
The locations of the phosphorylation sites in MAP2c, and especially the kinetics of phosphorylation, differ significantly from Tau. Our results can be compared quantitatively with the data published by Landrieu et al. (43), which were obtained under similar conditions.
The major phosphorylation site of Tau (class I according to the classification introduced by Landrieu et al. (43), corresponding to kobs ≈ 104 m−1s−1 and highlighted by the red background in Fig. 3) is 211RTPpSLP216 in the P2 region of the proline-rich domain (Fig. 3), representing a typical 14-3-3ζ-binding motif, RS/TXpSXP (25). The P2 region is involved in interactions with tubulin (38, 44), and phosphorylation of Ser-214 reduces tubulin binding (44) and the ability of Tau to promote microtubule assembly (45). The corresponding sequence in MAP2c, 255RTPGTP260, does not contain a phosphorylatable serine or threonine in the position of pSer-214 as in Tau. This correlates with the fact that the phosphorylation by PKA inhibits the microtubule-stabilizing (growth promoting) activity of Tau (46) but not of MAP2 (47). However, the proline-rich region of MAP2c also contains a similar phosphorylated 14-3-3ζ-binding site (48) but in a different position (181KRSpSLP186), in a region exhibiting a certain sequence homology with the P1 region (see Fig. 3) of Tau. Ser-184 of MAP2c is phosphorylated more slowly than Ser-214 of Tau, exhibiting the second highest phosphorylation rate among the MAP2c phosphorylation sites (class II, with kobs ≈ 102 m−1s−1, yellow background in Fig. 3). Both Ser-214 of Tau and Ser-184 of MAP2c are accompanied by weaker phosphorylation sites (Ser-208 and Ser-189, respectively) in close proximity.
The proline-rich domain of MAP2c contains another class II phosphorylation site, Thr-220, in a region that has little homology with Tau (Fig. 3). The SSP calculated from the chemical shifts indicates that phosphorylation by PKA stabilizes the secondary structure in the vicinity of Thr-220 (Fig. 5). This site has been studied extensively by Alexa et al. (29), who found that pThr-220 is extraordinarily sensitive to phosphatases and that MAP2c phosphorylated at Ser-184 and Ser-435, but not at Thr-220, stabilizes microtubules significantly more strongly than the other combinations of phosphorylation.
The remaining phosphorylation sites are located in a C-terminal region with a high sequence homology between MAP2c and Tau. The site with the second highest phosphorylation rate (class II) in Tau is 321KCGpS324 in MTBR3 (Fig. 3), representing the typical KXGS motif responsible for microtubule binding (16, 17). Ozer and Halpain (28) detected a rapid PKA phosphorylation of the corresponding residue Ser-350 (and Ser-319 and Ser-382 in other KXGS motifs) in MAP2c and found that the mutation of serines in the KXGS motifs of MAP2c has a great impact on interactions with microtubules. However, our quantitative data clearly show that the degree of phosphorylation at Ser-350 is low under our conditions. On the contrary, PKA in our study preferentially phosphorylated Ser-435 in the C-terminal region of MAP2c, corresponding to the muscarinic receptor-binding site of Tau (Fig. 3). The rate of Ser-435 phosphorylation was comparable with that of Ser-214 of Tau (class I). Despite the high sequence homology, the corresponding serine (Ser-409) is only a minor phosphorylation site (class III, indicated by the green background in Fig. 3) of Tau.
The listed striking differences between phosphorylation patterns and kinetics of MAP2c and Tau, determined by the same experimental approach (real-time NMR spectroscopy), are surprising considering the high sequence homology (especially in MTBDs) and have an important implication for 14-3-3ζ binding. Our interpretation is that these highly homologous regions have different local structures within MAP2c and Tau with a direct consequence: differing accessibility for PKA. This explanation is in agreement with the current view of IDPs as proteins that exhibit structural features significantly different from a random coil organization.
A recent study by Joo et al. (39) investigating the interaction of phosphorylated Tau with 14-3-3σ allowed us to make a direct comparison of the interactions of 14-3-3 with MAP2c and Tau. Unphosphorylated forms of MAP2c and Tau bind the 14-3-3 proteins, but phosphorylation by PKA significantly increases the binding affinities. The phosphorylation-independent 14-3-3ζ-binding sites of MAP2c are located in the MTBD and in the C-terminal domain. The same regions have been shown to bind 14-3-3ζ in unphosphorylated Tau (24, 25). Residue-specific data for unphosphorylated Tau are not reported by Joo et al. (39), but their analysis of the set of all residues of phosphorylated Tau influenced by 14-3-3σ binding leads to the same conclusion.
Phosphorylated forms of both MAP2c and Tau seem to interact with 14-3-3 proteins predominantly via two phosphoserines, one located in the proline-rich domain (25, 27, 39) and the other in the C-terminal portion (27, 39). As discussed above, the phosphorylation sites of MAP2c and Tau differ significantly even in regions of high sequential homology, which has a direct impact on phosphorylation-dependent 14-3-3 binding.
Charge distribution along the MAP2c and Tau sequences provides a physicochemical explanation of the observed differences in 14-3-3ζ binding. Positively charged regions of Tau are known to bind to acidic residues of tubulin and polyanions (49, 50), and MAP2c is likely to behave in a similar manner. We propose that the stretches of positively charged residues in MAP2c and Tau are also involved in the interaction with electronegative regions within the highly acidic 14-3-3ζ protein (pI ≈ 4.7). To explain the observed differences in the interactions of MAP2c and Tau, one should search for regions of the proteins that differ in charge distributions.
Fig. 3 shows that the electrostatic potential in the compared MAPs is similar in MTBR1 and MTBR4 but very different in MTBR3. A long stretch of positively charged residues is present in this region of MAP2c. As PKA did not phosphorylate Ser-350 under the conditions of our study, phosphorylation did not influence the charge distribution in that region of MAP2c. In contrast, the corresponding MTBR3 region in Tau is efficiently phosphorylated at Ser-324 (class II kinetics), which interrupts a stretch of positively charged residues with a negative patch. Therefore, the possible electrostatic interaction with acidic regions of 14-3-3 proteins is likely to be greatly suppressed by phosphorylation in the case of Tau but is little affected by phosphorylation in the case of MAP2c. This working hypothesis is consistent with our finding that 14-3-3ζ competes with MAP2c-induced tubulin polymerization regardless of MAP2c phosphorylation (Fig. 8), in contrast to the model of the Tau-regulated microtubule polymerization (51).
Another region with differing electrostatic potential between MAP2c and Tau is located 30 to 40 residues downstream of MTBR4. Phosphorylation of Ser-435, representing the major PKA target in MAP2c (but not in Tau), significantly reduces the positive charge in the mentioned region of MAP2c. This phosphorylation also induced the propensity to form a helical structure in the region Arg-425–Pro-431 (Fig. 5). The specific phosphorylation of MAP2c Ser-435 and its physicochemical consequences are particularly interesting because this region is responsible for the ability of Tau to act as a muscarinic agonist (52).
The observed phosphorylation patterns and interactions with 14-3-3 are related to the biological functions of Tau and MAP2c. Phosphorylation of Tau and MAP2c is controlled by various neurotransmitter receptors (53–55). PKA plays a key role in the signaling cascades triggered by receptors activating adenyl cyclase directly but is also involved, via Ca2+-stimulated adenyl cyclases, in pathways employing Ca2+ as a second messenger (56). PKA phosphorylates Tau and MAP2c directly but also activates downstream kinases able to phosphorylate residues not targeted by PKA (57–60). The effects of direct phosphorylation by PKA addressed in this study differ not only between Tau and MAP2 isoforms but also between different activities of the MAPs. PKA moderately decreases the binding of Tau and MAP2 isoforms to microtubules (29, 41, 47), suppresses the microtubule-nucleating activity of both Tau and high-molecular-weight MAP2 (46, 47), and inhibits the microtubule-stabilizing activity of Tau (46), but it does not affect the microtubule-stabilizing activity of high-molecular-weight MAP2 (47). Differences in the phosphorylation of Tau and MAP2c also influence the interactions with 14-3-3 proteins, which are proposed to regulate interactions with microtubules and modulate phosphorylation of Tau by various kinases (for review, see Ref. 51). Moreover, binding to microtubules is in equilibrium with the interactions of MAPs with other components of the cytoskeleton. Phosphorylation in MTBRs promotes MAP2c localization to actin-rich regions of neurons (28), with a direct impact on neuronal development and plasticity (19, 61, 62). It is evident that the kinetics of phosphorylation is an important factor in determining the balance of the aforementioned effects and contributes to the specificity of the control of microtubule stability by Tau and MAP2c under different physiological conditions and in different regions of neurons (8). Some of the functional differences may be directly related to the observed distinct kinetics of phosphorylation: PKA phosphorylation of Ser-214 in Tau inhibits the microtubule-stabilizing activity of Tau but does not affect the microtubule-stabilizing activity of MAP2 (46, 47); and the kinetics of phosphorylation and dephosphorylation of Thr-220 in MAP2c should be comparable if Thr-220 dephosphorylation plays a regulatory role (29).
In conclusion, PKA phosphorylates Tau and MAP2c differently even in highly homologous regions, which is reflected by the interactions of different sites of Tau and MAP2c with 14-3-3 proteins. The involvement of two proteins responding differently to the same signal (phosphorylation by PKA) may represent a branching point in the signaling pathways controlling microtubule stability and other important events such as activation of cholinergic receptors.
Experimental procedures
Preparation of recombinant proteins
MAP2c expression and purification was performed as described previously (1, 30). The protein was then phosphorylated, or dialyzed, against NMR buffer consisting of 50 mm MOPS, pH 6.9, 150 mm NaCl, and 0.7 mm TCEP. MAP2c was expressed in M9 medium containing 15N NH4Cl and/or 13C glucose for NMR measurement.
MAP2c was phosphorylated at 30 °C with 650 units of the catalytic subunit of PKA (New England Biolabs)/mg of MAP2c in a buffer containing 50 mm Tris-HCl, 10 mm MgCl2, 0.1 mm EDTA, 2 mm DTT, pH 7.5, and 20 mm ATP. After 24 h of incubation, PKA was deactivated by heating to 95 °C for 20 min. For NMR measurement, the protein was dialyzed against the NMR buffer.
14-3-3ζ was purified as described previously (35). Two surface-exposed cysteines were mutated for alanine (C25A and C189A). This allowed us to work with highly concentrated samples over longer times without the risk of disulfide bond formation. We showed earlier that the mutation of these two exposed cysteines does not change the fold or stability of the protein (35). The purity of the final protein samples, including phosphorylated and labeled samples, was checked by MALDI-MS.
Mutagenesis
Single-point mutants of MAP2c (S435D, S184D, T220E, and C348S) were produced using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) following the manufacturer's protocol and using MAP2c in the pET3a vector as a template. To prepare the double mutants (S189D/S435D, S199D/S435D, S319D/S435D, T320E/S435D, S342D/S435D, S350D/S435D, S367D/S435D, and S382D/S435D), S435D MAP2c in pET3a was used as a template, and C348S MAP2c was used as template for the double mutant E52C/C348S. The results of the mutations were confirmed by sequencing. For NMR measurement, the MAP2c mutants were expressed in 1 liter of 15N M9 medium. Phosphorylation was performed as for the wild-type MAP2c.
Mass spectrometry
MAP2c at 0.2 mm concentration was phosphorylated for 24 h with 650 units of PKA/mg of MAP2c. Phosphorylated MAP2c and 0.16 mm unphosphorylated MAP2c were processed by the filter-aided sample preparation (FASP) method (63, 64). Proteins were alkylated and digested by trypsin on the filter unit membrane, and the resulting peptides were eluted by ammonium bicarbonate. One-tenth of the peptide mixture was analyzed directly, and the rest of the sample was used for phosphopeptide enrichment. Both peptide mixtures were separately analyzed on a LC-MS/MS system (RSLCnano connected to Orbitrap Elite, Thermo Fisher Scientific).
The MS data were acquired using a data-dependent strategy by selecting up to the top six precursors based on precursor abundance in the survey scan (350–2000 m/z). High-resolution HCD or ETD MS/MS spectra were acquired in the Orbitrap analyzer. The analysis of the mass spectrometric RAW data files was carried out using Proteome Discoverer software (version 1.4, Thermo Fisher Scientific) with an in-house Mascot (version 2.4.1, Matrix Science) search engine utilization. Peptides with a false discovery rate lower than 1%, rank 1, and search engine rank 1 and with at least 6 amino acids were considered. Quantitative information assessment was performed using Skyline (Skyline Software Systems).
NMR spectroscopy
NMR experiments were performed using a 950-MHz Bruker Avance III spectrometer equipped with a 1H-13C/15N/D TCI cryogenic probe head with z axis gradients and a 700-MHz Bruker Avance III spectrometer equipped with a 1H/13C/15N TXO cryogenic probe head with z axis gradients. All experiments were performed at 27 °C with the temperature calibrated according to the chemical shift differences of pure methanol peaks. The indirect dimensions in 3D and 5D experiments were acquired in a non-uniformly sampled manner. On-grid Poisson disk sampling with a Gaussian probability distribution (65) was applied.
The 1H-15N HSQC (66, 67) spectrum of wild-type MAP2c was recorded with spectral widths set to 11,904 Hz in the direct dimension and to 2500 Hz in the indirect dimension. 2048 and 128 complex points were acquired in the direct and the indirect dimensions, respectively. The 1H-15N HSQC spectra of the MAP2c mutants were recorded with spectral widths set to 17,045 Hz in the direct dimension and to 1000 Hz in the indirect dimension. 2048 and 256 complex points were acquired in the direct and the indirect dimensions, respectively. 16 scans with recycle delay set to 1 s were recorded.
The 1H-15N SOFAST-HMQC (68) spectra were recorded with spectral widths set to 13,297 Hz in the direct dimension and to 2500 Hz in the indirect dimension. 2048 and 64 complex points were acquired in the direct and the indirect dimensions, respectively. 16 scans with the recycle delay set to 228 ms were recorded in each experiment.
The 3D (CACO)NCACO spectrum (30) was acquired with spectral widths set to 7042 (acquired dimension (aq)) × 2000 (15N) × 4000 (13Cα) Hz and with maximal evolution times of 46 ms (15N) and 26 ms (13Cα) in the indirectly detected dimensions. The overall number of 1024 complex points was acquired in the acquisition dimension, and 3000 hypercomplex points were randomly distributed over the indirectly detected dimensions. The 5D CACONCACO spectrum (30) was acquired with spectral widths set to 7042 (aq) × 4000 (13Cα) × 2000 (15N) × 2000 (13C′) × 4000 (13Cα) Hz. The maximal evolution times in the indirectly detected dimensions were set to 26 ms for the 13Cα dimensions, 46 ms for the 15N dimension, and 28 ms for the 13C′ dimension. The overall number of 1024 complex points was acquired in the acquisition dimension, and 3000 hypercomplex points were distributed over the indirectly detected dimensions.
The 3D (H)CANCO spectrum (69) was acquired with spectral widths set to 7042 (aq) × 2000 (15N) × 4000 (13Cα) Hz and with maximal evolution times of 32 ms (15N) and 26 ms (13Cα) in the indirectly detected dimensions. The overall number of 1024 complex points was acquired in the acquisition dimension, and 1500 hypercomplex points were distributed over the indirectly detected dimensions.
The 5D HC(CC-TOCSY)CACON (30) experiment was measured with the spectral widths set to 7042 (aq) × 2500 (15N) × 4000 (13Cα) × 12500 (13Cali) × 5000 (1Hali) Hz where “ali” indicates nuclei in the aliphatic side chain. The maximal evolution times in the indirectly detected dimensions were set to 46 ms for the 15N dimension, 26 ms for the 13Cα dimension, 8 ms for the 13Cali dimension, and 10 ms for the 1Hali dimension. The overall number of 1024 complex points was acquired in the acquisition dimension, and 2000 hypercomplex points were distributed over the indirectly detected dimensions. The 3D (HC(CC-TOCSY))CACON (30) experiment was acquired with spectral widths set to 7042 (aq) × 1956 (15N) × 4000 (13Cα) Hz and maximal evolution times of 46 ms for 15N and 26 ms for 13Cα indirectly detected dimensions. The overall number of 1024 complex points was acquired in the acquisition dimension, and 2000 hypercomplex points were distributed over the indirectly detected dimensions.
The 3D HNCO (70) spectra were acquired with spectral widths set to 18939 (aq) × 2000 (15N) × 2000 (13C′) Hz and maximal evolution times of 120 ms for 15N and 80 ms for 13C′ indirectly detected dimensions. The overall number of 2048 complex points was acquired in the acquisition dimension, and 2000 hypercomplex points were distributed over the indirectly detected dimensions.
The 3D HNCACB experimental data (71) were acquired with spectral widths set to 18939 (aq) × 2500 (15N) × 17921 (13C′) Hz. The number of recorded complex points was 2048, 40, and 128 for the 1H, 15N, and 13C dimensions, respectively.
Uniformly sampled data processing and direct dimension processing of non-uniformly sampled data were done using NMRPipe software (72). Multidimensional Fourier transform with iterative algorithm for artifact suppression (73) was employed to process indirect dimensions in three-dimensional experiments. Indirect dimensions in five-dimensional experiments were processed using the sparse multidimensional Fourier transform (74). Spectral analysis was done using the software Sparky 3.115 (T. D. Goddard and D. G. Kneller, University of California, San Francisco).
The transient secondary structure propensities of phosphorylated and unphosphorylated MAP2c were calculated using the program SSP (75) as described previously (30). To obtain values not biased by the direct chemical effect of the presence of the phosphate group, SSP values were calculated only from the chemical shifts of 13Cα, 13Cβ, and 1Hα, which are insensitive to the presence of the phosphate group attached to neighboring residues, with the exception of 13Cβ of the phosphorylated side chain (34). Residues pSer-184, pThr-220, and pSer-435 were excluded from the analysis. Regions of propensity to form helical structures were defined as stretches of at least three residues with SSP higher than 0.07 (interrupted by no more than two residues with SSP lower than 0.07). Regions of propensity to form extended structures were defined as stretches of at least three residues with SSP lower than −0.07 (interrupted by no more than two residues with SSP higher than −0.07).
Phosphorylation kinetics
Real-time phosphorylation was followed by recording 1H,15N-SOFAST-HMQC spectra at 27 °C in 5-min intervals for 30 h after the addition of PKA. [15N]MAP2c at a 0.65 mm concentration in 50 mm MOPS, pH 6.9, 150 mm NaCl, 0.7 mm TCEP, 10 mm MgCl2, 20 mm ATP, and 0.1 mm EDTA was mixed with 12,500 units of PKA directly in the NMR tube. According to the manufacturer, the PKA concentration was ∼0.1 μm. The first SOFAST-HMQC experiment was started 2 min after the addition of PKA. The relative concentrations of individual phosphorylation sites were assumed to be proportional to the corresponding peak volumes, and PKA was assumed to be saturated by ATP and magnesium. Phosphorylation rates were modeled as
| (Eq. 1) |
where t is time, [S]i and [P]i are concentrations of the i-th unphosphorylated and phosphorylated phosphorylation sites, respectively, kcat,i and Km,i are catalytic and Michaelis constants, respectively, and cPKA is the total concentration of PKA. The apparent rate constants, kobs,i, were defined as
| (Eq. 2) |
where cMAP2c is the total concentration of MAP2c.
The kobs value for Ser-435 was estimated by fitting the peak volumes to the integrated form of Equation 1, assuming that concentrations of other phosphorylation sites were equal to cMAP2c. The value of kcat could not be reliably separated from kobs for the obtained data. To estimate kobs for Ser-184, Ser-189, and Thr-220, the ratios of peak volumes of pSer-184, pSer-189, and pThr-220 were assumed to be constant, and peak volumes, V, were fitted to the equation
| (Eq. 3) |
where a < b, a = kobscPKA at [Ser-435] = 0, and b approximates the rate of the preceding phosphorylation of Ser-435.
Interaction with 14-3-3ζ by NMR
A sample containing 550 μl of 0.69 mm unphosphorylated [15N,13C]MAP2c was titrated with 17, 34, 69, 139, and 280 μl of 2.5 mm 14-3-3ζ (monomer concentration). After each addition, a 3D HNCO spectrum was recorded. 500 μl of 0.61 mm phosphorylated [15N,13C]MAP2c was titrated with 17, 34, 68, 136, and 272 μl of 2.2 mm 14-3-3ζ. For each titration point, 3D HNCO and 2D 1H,15N HSQC spectra were measured.
Microscale thermophoresis
Binding experiments between 14-3-3ζ and MAP2c in the unphosphorylated and phosphorylated forms were measured by MST at 20 °C. E52C/C348S MAP2c was specifically labeled at position Cys-52 with the fluorophore Alexa Fluor 647 C2 maleimide (AF647, Thermo Fisher Scientific). Titration experiments were performed in buffer containing 50 mm Tris, pH 7.5. In addition, 0.5 mg/ml BSA and 0.05% Tween 20 were added to prevent aggregation of the studied proteins on standard capillary walls. Binding studies were performed at 30% laser power with a Monolith NT.115 device (NanoTemper Technologies, Munich, Germany) in combination with three different MST power setups (at 40, 60, and 80%). The data were fitted to the following model,
| (Eq. 4) |
where Φ = F/Fmax is the normalized fluorescence, c is the total concentration of MAP2c, x is the total monomer concentration of 14-3-3ζ, and K′D is the apparent dissociation constant expressed for the 14-3-3ζ monomer concentration.
Microtubular co-sedimentation assay
Co-sedimentation assays were adapted from Valencia et al. (76). Samples of 5 μm porcine brain tubulin in 30 μl of tubulin buffer (80 mm PIPES, pH 6.9, 2 mm MgCl2, and 0.5 mm EGTA) were polymerized by the addition of 2 μm MAP2c and incubated for 15 min at 37 °C. 20 nm, 200 nm, 2 μm, and 20 μm concentrations of 14-3-3ζ (monomer concentrations) in tubulin buffer were added to the MAP2c-tubulin samples before the incubation. Microtubule bundles were pelleted by centrifugation (50,000 × g, 30 min, 37 °C). The pellet was washed by tubulin buffer and adjusted to the same volume as the supernatant with the buffer. Both supernatant and pellet were mixed with the SDS gel loading buffer. Equal volumes of supernatant and pellet were separated by SDS-PAGE. The Coomassie Brilliant Blue-stained protein bands were analyzed using the densitometer software QuantiScan 3.0.
Author contributions
S. J., L. Z., J. H., J. N., and K. M. conceived and designed the research. S. J., E. Z., K. M., and Z. T. prepared the protein samples. Z. T. and K. M., respectively, performed the MST and co-sedimentation assays. L. Z., M. Z., S. J., and K. M. performed the NMR experiments, and K. H. and Z. Z. performed the MS experiments. S. J., L. Z., and J. H. wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
CIISB research infrastructure project LM2015043, funded by Ministry of Education, Youth and Sports of the Czech Republic (MEYS CR), is gratefully acknowledged for partial financial support of the measurements at the Josef Dadok National NMR Centre, Biomolecular Interactions and Crystallization and Proteomics Core Facilities, Central European Institute of Technology (CEITEC), Masaryk University. We thank Tanvir Shaikh for language corrections and Arnošt Mládek and Irmgard Fischer for technical help.
This work was supported by Grants GA15-14974S (to S. J., K. M., K. H., M. Z., and L. Z.) and GF15-34684L (Z. T., E. Z., and J. H.) from the Czech Science Foundation. The authors declare that they have no conflicts of interest with the contents of this article.
The chemical shifts were deposited in the Biological Magnetic Resonance Bank under BMRB accession no. 26960.
- MAP
- microtubule-associated protein
- IDP
- intrinsically disordered protein
- MTBD
- microtubule-binding domain
- MTBR
- microtubule-binding region
- SSP
- secondary structure propensity
- MST
- microscale thermophoresis
- TCEP
- tris(2-carboxyethyl)phosphine.
References
- 1. Gamblin T. C., Nachmanoff K., Halpain S., and Williams R. C. (1996) Recombinant microtubule-associated protein 2c reduces the dynamic instability of individual microtubules. Biochemistry 35, 12576–12586 [DOI] [PubMed] [Google Scholar]
- 2. Jalava N. S., Lopez-Picon F. R., Kukko-Lukjanov T. K., and Holopainen I. E. (2007) Changes in microtubule-associated protein-2 (MAP2) expression during development and after status epilepticus in the immature rat hippocampus. Int. J. Dev. Neurosci. 25, 121–131 [DOI] [PubMed] [Google Scholar]
- 3. Dunker A. K., Obradovic Z., Romero P., Garner E. C., and Brown C. J. (2000) Intrinsic protein disorder in complete genomes. Genome Inform. 11, 161–171 [PubMed] [Google Scholar]
- 4. Dunker A. K., Oldfield C. J., Meng J., Romero P., Yang J. Y., Chen J. W., Vacic V., Obradovic Z., and Uversky V. N. (2008) The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics 9, S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dyson H. J., and Wright P. E. (2005) Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 6. Tompa P. (2005) The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 579, 3346–3354 [DOI] [PubMed] [Google Scholar]
- 7. Fink A. L. (2005) Natively unfolded proteins. Curr. Opin. Struct. Biol. 15, 35–41 [DOI] [PubMed] [Google Scholar]
- 8. Dehmelt L., and Halpain S. (2005) The MAP2/Tau family of microtubule-associated proteins. Genome Biol. 6, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sündermann F., Fernandez M. P., and Morgan R. O. (2016) An evolutionary roadmap to the microtubule-associated protein MAP Tau. BMC Genomics 17, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grundke-Iqbal I., Iqbal K., Quinlan M., Tung Y. C., Zaidi M. S., and Wisniewski H. M. (1986) Microtubule-associated protein Tau: a component of Alzheimer paired helical filaments. J. Biol. Chem. 261, 6084–6089 [PubMed] [Google Scholar]
- 11. Viereck C., Tucker R. P., and Matus A. (1989) The adult rat olfactory system expresses microtubule-associated proteins found in the developing brain. J. Neurosci. 9, 3547–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vallee R. (1980) Structure and phosphorylation of microtubule-associated protein 2 (MAP2). Proc. Natl. Acad. Sci. U.S.A. 77, 3206–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamauchi T., and Fujisawa H. (1983) Disassembly of microtubules by the action of calmodulin-dependent protein kinase (kinase II) which occurs only in the brain tissues. Biochem. Biophys. Res. Commun. 110, 287–291 [DOI] [PubMed] [Google Scholar]
- 14. Burns R. G., Islam K., and Chapman R. (1984) The multiple phosphorylation of the microtubule-associated protein MAP2 controls the MAP2:tubulin interaction. Eur. J. Biochem. 141, 609–615 [DOI] [PubMed] [Google Scholar]
- 15. Hoshi M., Akiyama T., Shinohara Y., Miyata Y., Ogawara H., Nishida E., and Sakai H. (1988) Protein-kinase-C-catalyzed phosphorylation of the microtubule-binding domain of microtubule-associated protein 2 inhibits its ability to induce tubulin polymerization. Eur. J. Biochem. 174, 225–230 [DOI] [PubMed] [Google Scholar]
- 16. Ainsztein A. M., and Purich D. L. (1994) Stimulation of tubulin polymerization by MAP-2: control by protein kinase C-mediated phosphorylation at specific sites in the microtubule-binding region. J. Biol. Chem. 269, 28465–28471 [PubMed] [Google Scholar]
- 17. Illenberger S., Drewes G., Trinczek B., Biernat J., Meyer H. E., Olmsted J. B., Mandelkow E. M., and Mandelkow E. (1996) Phosphorylation of microtubule-associated proteins MAP2 and MAP4 by the protein kinase p110mark: phosphorylation sites and regulation of microtubule dynamics. J. Biol. Chem. 271, 10834–10843 [DOI] [PubMed] [Google Scholar]
- 18. Drewes G., Ebneth A., and Mandelkow E. M. (1998) MAPs, MARKs and microtubule dynamics. Trends Biochem. Sci. 23, 307–311 [DOI] [PubMed] [Google Scholar]
- 19. Sánchez C., Díaz-Nido J., and Avila J. (2000) Phosphorylation of microtubule-associated protein 2 (MAP2) and its relevance for the regulation of the neuronal cytoskeleton function. Prog. Neurobiol. 61, 133–168 [DOI] [PubMed] [Google Scholar]
- 20. Illenberger S., Zheng-Fischhöfer Q., Preuss U., Stamer K., Baumann K., Trinczek B., Biernat J., Godemann R., Mandelkow E. M., and Mandelkow E. (1998) The endogenous and cell cycle-dependent phosphorylation of Tau protein in living cells: implications for Alzheimer's disease. Mol. Biol. Cell 9, 1495–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Avila J., Domínguez J., and Díaz-Nido J. (1994) Regulation of microtubule dynamics by microtubule-associated protein expression and phosphorylation during neuronal development. Int. J. Dev. Biol. 38, 13–25 [PubMed] [Google Scholar]
- 22. Aitken A., Collinge D. B., van Heusden B. P., Isobe T., Roseboom P. H., Rosenfeld G., and Soll J. (1992) 14-3-3 proteins: a highly conserved, widespread family of eukaryotic proteins. Trends Biochem. Sci. 17, 498–501 [DOI] [PubMed] [Google Scholar]
- 23. Skoulakis E. M., and Davis R. L. (1998) 14-3-3 proteins in neuronal development and function. Mol. Neurobiol. 16, 269–284 [DOI] [PubMed] [Google Scholar]
- 24. Hashiguchi M., Sobue K., and Paudel H. K. (2000) 14-3-3ζ is an effector of Tau protein phosphorylation. J. Biol. Chem. 275, 25247–25254 [DOI] [PubMed] [Google Scholar]
- 25. Sadik G., Tanaka T., Kato K., Yamamori H., Nessa B. N., Morihara T., and Takeda M. (2009) Phosphorylation of Tau at Ser214 mediates its interaction with 14-3-3 protein: implications for the mechanism of tau aggregation. J. Neurochem. 108, 33–43 [DOI] [PubMed] [Google Scholar]
- 26. Sluchanko N. N., Seit-Nebi A. S., and Gusev N. B. (2009) Effect of phosphorylation on interaction of human Tau protein with 14-3-3ζ. Biochem. Biophys. Res. Commun. 379, 990–994 [DOI] [PubMed] [Google Scholar]
- 27. Sluchanko N. N., Seit-Nebi A. S., and Gusev N. B. (2009) Phosphorylation of more than one site is required for tight interaction of human Tau protein with 14-3-3ζ. FEBS Lett. 583, 2739–2742 [DOI] [PubMed] [Google Scholar]
- 28. Ozer R. S., and Halpain S. (2000) Phosphorylation-dependent localization of microtubule-associated protein MAP2c to the actin cytoskeleton. Mol. Biol. Cell 11, 3573–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alexa A., Schmidt G., Tompa P., Ogueta S., Vázquez J., Kulcsár P., Kovács J., Dombrádi V., and Friedrich P. (2002) The phosphorylation state of threonine-220, a uniquely phosphatase-sensitive protein kinase A site in microtubule-associated protein MAP2c, regulates microtubule binding and stability. Biochemistry 41, 12427–12435 [DOI] [PubMed] [Google Scholar]
- 30. Nováček J., Janda L., Dopitová R., Žídek L., Sklenář V. (2013) Efficient protocol for backbone and side-chain assignments of large, intrinsically disordered proteins: transient secondary structure analysis of 49.2-kDa microtubule-associated protein 2c. J. Biomol. NMR 56, 291–301 [DOI] [PubMed] [Google Scholar]
- 31. Obenauer J. C., Cantley L. C., and Yaffe M. (2003) Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs, Nucleic Acids Res. 31, 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xue Y., Liu Z., Cao J., Ma Q., Gao X., Wang Q., Jin C., Zhou Y., Wen L., and Ren J. (2011) GPS 2.1: enhanced prediction of kinase-specific phosphorylation sites with an algorithm of motif length selection. Protein Eng. Des. Sel. 24, 255–260 [DOI] [PubMed] [Google Scholar]
- 33. Wong Y. H., Lee T. Y., Liang H. K., Huang C., Yang Y. H., Chu C. H., Huang H. D., Ko M. T., and Hwang J. K. (2007) KinasePhos 2.0: a Web server for identifying protein kinase-specific phosphorylation sites based on sequences and coupling patterns. Nucleic Acids Res. 35, 588–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Louša P., Nedozrálová H., Župa E., Nováček J., and Hritz J. (2017) Phosphorylation of the regulatory domain of human tyrosine hydroxylase 1 monitored using non-uniformly sampled NMR. Biophys. Chem. 223, 25–29 [DOI] [PubMed] [Google Scholar]
- 35. Hritz J., Byeon I. J., Krzysiak T., Martinez A., Sklenar V., and Gronenborn A. M. (2014) Dissection of binding between a phosphorylated tyrosine hydroxylase peptide and 14-3-3ζ: a complex story elucidated by NMR. Biophys. J. 107, 2185–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. von Bergen M., Friedhoff P., Biernat J., Heberle J., Mandelkow E. M., and Mandelkow E. (2000) Assembly of Tau protein into Alzheimer paired helical filaments depends on a local sequence motif ((306)VQIVYK(311)) forming beta structure. Proc. Natl. Acad. Sci. U.S.A. 97, 5129–5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xie C., Miyasaka T., Yoshimura S., Hatsuta H., Yoshina S., Kage-Nakadai E., Mitani S., Murayama S., and Ihara Y. (2014) The homologous carboxyl-terminal domains of microtubule-associated protein 2 and Tau induce neuronal dysfunction and have differential fates in the evolution of neurofibrillary tangles. PLoS One 9, e89796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mukrasch M. D., Bibow S., Korukottu J., Jeganathan S., Biernat J., Griesinger C., Mandelkow E., and Zweckstetter M. (2009) Structural polymorphism of 441-residue Tau at single residue resolution. PLOS Biol. 7, e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Joo Y., Schumacher B., Landrieu I., Bartel M., Smet-Nocca C., Jang A., Choi H. S., Jeon N. L., Chang K. A., Kim H. S., Ottmann C., and Suh Y. H. (2015) Involvement of 14-3-3 in tubulin instability and impaired axon development is mediated by Tau. FASEB J. 29, 4133–4144 [DOI] [PubMed] [Google Scholar]
- 40. Sluchanko N. N., and Gusev N. B. (2010) 14–3-3 proteins and regulation of cytoskeleton. Biochemistry (Mosc.) 75, 1528–1546 [DOI] [PubMed] [Google Scholar]
- 41. Ackmann M., Wiech H., and Mandelkow E. (2000) Nonsaturable binding indicates clustering of Tau on the microtubule surface in a paired helical filament-like conformation. J. Biol. Chem. 275, 30335–30343 [DOI] [PubMed] [Google Scholar]
- 42. Kennelly P. J., and Krebs E. G. (1991) Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J. Biol. Chem. 266, 15555–15558 [PubMed] [Google Scholar]
- 43. Landrieu I., Lacosse L., Leroy A., Wieruszeski J. M., Trivelli X., Sillen A., Sibille N., Schwalbe H., Saxena K., Langer T., and Lippens G. (2006) NMR analysis of a Tau phosphorylation pattern. J. Am. Chem. Soc. 128, 3575–3583 [DOI] [PubMed] [Google Scholar]
- 44. Sillen A., Barbier P., Landrieu I., Lefebvre S., Wieruszeski J. M., Leroy A., Peyrot V., and Lippens G. (2007) NMR investigation of the interaction between the neuronal protein Tau and the microtubules. Biochemistry 46, 3055–3064 [DOI] [PubMed] [Google Scholar]
- 45. Yoshida H., and Goedert M. (2006) Sequential phosphorylation of tau protein by cAMP-dependent protein kinase and SAPK4/p38delta or JNK2 in the presence of heparin generates the AT100 epitope. J. Neurochem. 99, 154–164 [DOI] [PubMed] [Google Scholar]
- 46. Brandt R., Lee G., Teplow D. B., Shalloway D., and Abdel-Ghany M. (1994) Differential effect of phosphorylation and substrate modulation on Tau's ability to promote microtubule growth and nucleation. J. Biol. Chem. 269, 11776–11782 [PubMed] [Google Scholar]
- 47. Itoh T. J., Hisanaga S., Hosoi T., Kishimoto T., and Hotani H. (1997) Phosphorylation states of microtubule-associated protein 2 (MAP2) determine the regulatory role of MAP2 in microtubule dynamics. Biochemistry 36, 12574–12582 [DOI] [PubMed] [Google Scholar]
- 48. Madeira F., Tinti M., Murugesan G., Berrett E., Stafford M., Toth R., Cole C., MacKintosh C., and Barton G. J. (2015) 14-3-3-Pred: improved methods to predict 14–3-3-binding phosphopeptides. Bioinforma 31, 2276–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mukrasch M. D., Biernat J., von Bergen M., Griesinger C., Mandelkow E., and Zweckstetter M. (2005) Sites of Tau important for aggregation populate β-structure and bind to microtubules and polyanions. J. Biol. Chem. 280, 24978–24986 [DOI] [PubMed] [Google Scholar]
- 50. Mukrasch M. D., von Bergen M., Biernat J., Fischer D., Griesinger C., Mandelkow E., and Zweckstetter M. (2007) The “jaws” of the Tau-microtubule interaction. J. Biol. Chem. 282, 12230–12239 [DOI] [PubMed] [Google Scholar]
- 51. Sluchanko N. N., and Gusev N. B. (2011) Probable participation of 14–3-3 in Tau protein oligomerization and aggregation. J. Alzheimers Dis. 27, 467–476 [DOI] [PubMed] [Google Scholar]
- 52. Gómez-Ramos A., Díaz-Hernández M., Rubio A., Miras-Portugal M. T., and Avila J. (2008) Extracellular Tau promotes intracellular calcium increase through M1 and M3 muscarinic receptors in neuronal cells. Mol. Cell. Neurosci. 37, 673–681 [DOI] [PubMed] [Google Scholar]
- 53. Gardiner J., Overall R., and Marc J. (2011) The microtubule cytoskeleton acts as a key downstream effector of neurotransmitter signaling. Synapse 65, 249–256 [DOI] [PubMed] [Google Scholar]
- 54. Ovsepian S. V., O'Leary V. B., and Zaborszky L. (2016) Cholinergic mechanisms in the cerebral cortex: beyond synaptic transmission. Neuroscientist 22, 238–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Busceti C. L., Di Pietro P., Riozzi B., Traficante A., Biagioni F., Nisticò R., Fornai F., Battaglia G., Nicoletti F., and Bruno V. (2015) 5-HT(2C) serotonin receptor blockade prevents tau protein hyperphosphorylation and corrects the defect in hippocampal synaptic plasticity caused by a combination of environmental stressors in mice. Pharmacol. Res. 99, 258–268 [DOI] [PubMed] [Google Scholar]
- 56. Wang H., and Zhang M. (2012) The role of Ca2+-stimulated adenylyl cyclases in bidirectional synaptic plasticity and brain function. Rev. Neurosci. 23, 67–78 [DOI] [PubMed] [Google Scholar]
- 57. Vossler M. R., Yao H., York R. D., Pan M. G., Rim C. S., and Stork P. J. (1997) cAMP activates MAP kinase and Elk-1 through a B-Raf- and Rap1-dependent pathway. Cell 89, 73–82 [DOI] [PubMed] [Google Scholar]
- 58. Kim H. A., DeClue J. E., and Ratner N. (1997) cAMP-dependent protein kinase A is required for Schwann cell growth: interactions between the cAMP and neuregulin/tyrosine kinase pathways. J. Neurosci. Res. 49, 236–247 [PubMed] [Google Scholar]
- 59. Blanco-Aparicio C., Torres J., and Pulido R. (1999) A novel regulatory mechanism of MAP kinases activation and nuclear translocation mediated by PKA and the PTP-SL tyrosine phosphatase. J. Cell Biol. 147, 1129–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ambrosini A., Tininini S., Barassi A., Racagni G., Sturani E., and Zippel R. (2000) cAMP cascade leads to Ras activation in cortical neurons. Brain Res. Mol. Brain Res. 75, 54–60 [DOI] [PubMed] [Google Scholar]
- 61. Mohan R., and John A. (2015) Microtubule-associated proteins as direct crosslinkers of actin filaments and microtubules. IUBMB Life 67, 395–403 [DOI] [PubMed] [Google Scholar]
- 62. Elie A., Prezel E., Guérin C., Denarier E., Ramirez-Rios S., Serre L., Andrieux A., Fourest-Lieuvin A., Blanchoin L., and Arnal I. (2015) Tau co-organizes dynamic microtubule and actin networks. Sci. Rep. 5, 9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wiśniewski J. R., Ostasiewicz P., and Mann M. (2011) High recovery FASP applied to the proteomic analysis of microdissected formalin fixed paraffin embedded cancer tissues retrieves known colon cancer markers. J. Proteome Res. 10, 3040–3049 [DOI] [PubMed] [Google Scholar]
- 64. Wiśniewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 65. Kazimierczuk K., Zawadzka A., and Koźmiński W. (2008) Optimization of random time domain sampling in multidimensional NMR. J. Magn. Reson. 192, 123–130 [DOI] [PubMed] [Google Scholar]
- 66. Bodenhausen G., and Ruben D. J. (1980) Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 69, 185–189 [Google Scholar]
- 67. Sklenar V., Piotto M., Leppik R., and Saudek V. (1993) Gradient-tailored water suppression for 1H-15N HSQC experiments optimized to retain full sensitivity. J. Magn. Reson. 102, 241–245 [Google Scholar]
- 68. Schanda P., and Brutscher B. (2005) Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of second. J. Am. Chem. Soc. 127, 8014–8015 [DOI] [PubMed] [Google Scholar]
- 69. Bermel W., Bertini I., Felli I. C., and Pierattelli R. (2009) Speeding up (13)C direct detection biomolecular NMR spectroscopy. J. Am. Chem. Soc. 131, 15339–15345 [DOI] [PubMed] [Google Scholar]
- 70. Kay L. E., Ikura M., Tschudin R., and Bax A. (1990) Three-dimensional triple-resonance NMR spectroscopy of isotopically enriched proteins. J. Magn. Reson. 89, 496–514 [DOI] [PubMed] [Google Scholar]
- 71. Sattler M., Schleucher J., and Griesinger C. (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucleic Magn. Reson. Spectrosc. 34, 93–158 [Google Scholar]
- 72. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 73. Stanek J., and Koźmiński W. (2010) Iterative algorithm of discrete Fourier transform for processing randomly sampled NMR data sets. J. Biomol. NMR 47, 65–77 [DOI] [PubMed] [Google Scholar]
- 74. Kazimierczuk K., Zawadzka A., and Koźmiński W. (2009) Narrow peaks and high dimensionalities: exploiting the advantages of random sampling. J. Magn. Reson. 197, 219–228 [DOI] [PubMed] [Google Scholar]
- 75. Marsh J. A., Singh V. K., Jia Z., and Forman-Kay J. D. (2006) Sensitivity of secondary structure propensities to sequence differences between α- and γ-synuclein: implications for fibrillation. Protein Sci. 15, 2795–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Valencia R. G., Walko G., Janda L., Novacek J., Mihailovska E., Reipert S., Andrä-Marobela K., and Wiche G. (2013) Intermediate filament-associated cytolinker plectin 1c destabilizes microtubules in keratinocytes. Mol. Biol. Cell 24, 768–784 [DOI] [PMC free article] [PubMed] [Google Scholar]