Abstract
Tumor-specific alternative splicing is implicated in the progression of cancer, including clear-cell renal cell carcinoma (ccRCC). Using ccRCC RNA sequencing data from The Cancer Genome Atlas, we found that epithelial splicing regulatory protein 2 (ESRP2), one of the key regulators of alternative splicing in epithelial cells, is expressed in ccRCC. ESRP2 mRNA expression did not correlate with the overall survival rate of ccRCC patients, but the expression of some ESRP-target exons correlated with the good prognosis and with the expression of Arkadia (also known as RNF111) in ccRCC. Arkadia physically interacted with ESRP2, induced polyubiquitination and modulated its splicing function. Arkadia and ESRP2 suppressed ccRCC tumor growth in a coordinated manner. Lower expression of Arkadia correlated with advanced tumor stages and poor outcomes in ccRCC patients. This study thus reveals a novel tumor-suppressive role of the Arkadia-ESRP2 axis in ccRCC.
Introduction
Alternative mRNA splicing is a crucial event during normal development and, therefore, tightly regulated in various biological processes.1, 2 Aberrant alternative splicing may occur in certain pathophysiological conditions, including tumor progression and cancer metastasis.1, 3 Alternative splicing is modulated by various splicing regulatory proteins, and abnormal splicing profiles in cancer cells are caused by altered activity or expression levels or mutations in these proteins, rather than mutations in target genes.4
For example, splicing occurs in the second half of the third immunoglobulin-like domain of fibroblast growth factor receptor 1–3 (FGFR1–3), leading to differential binding specificities to FGF family ligands.5 FGF2 (also known as basic FGF) and some FGF ligands preferentially bind to the IIIc isoform of FGFRs, whereas FGF7 (also known as keratinocyte growth factor) and other FGFs preferentially bind to the IIIb isoform of FGFRs.5, 6 Recently, FGFR2 IIIc (the IIIc isoform of FGFR2), but not FGFR2 IIIb, was shown to be expressed in clear-cell renal cell carcinoma (ccRCC), accompanied by decreased expression of epithelial splicing regulatory protein 1 (ESRP1) mRNA, resulting in the acquisition of a mesenchymal and malignant phenotype of ccRCC cells.7
ESRP1 and ESRP2 are splicing regulators specifically expressed in epithelial cells, and promote epithelial type splicing.8 In addition to FGFRs, ESRP1 and ESRP2 are known to regulate the splicing of numerous genes, including STE20-like kinase (SLK), integrin α6 (ITGA6), tuberous sclerosis 2 (TSC2), enabled homolog (ENAH, also known as Mena), CD44 and catenin δ1 (CTNND1, also known as p120-catenin), some of which are involved in regulation of the function of cytoskeleton and cell motility.9 Expression of ESRP1 and ESRP2 is transcriptionally decreased during the process of epithelial-to-mesenchymal transition (EMT), accompanied by malignant progression in epithelial-type cancers.10 Transforming growth factor-β (TGF-β) is a potent inducer of EMT11 and suppresses the expression of ESRP2 in mouse mammary epithelial NMuMG cells through induction of zinc-finger transcription factors ZEB1 and ZEB2.12
Arkadia, also known as RNF111, is a RING-type E3 ligase that has a key role in vertebrate development and pathophysiology by enhancing TGF-β signaling activities. Arkadia induces ubiquitin-dependent degradation of negative regulators of the TGF-β signaling pathways, including Smad7, c-Ski and SnoN (also known as Skil).13, 14, 15 Arkadia exhibits both pro- and antitumorigenic functions,16, 17 consistent with TGF-β signaling having bidirectional roles in the progression of cancer in a context- and cell type-dependent manner.11, 18 Moreover, recent data have revealed that Arkadia has important functions independent of TGF-β signaling, including the stimulation of endocytosis of epidermal growth factor receptor,19 assistance with the DNA damage response20 and stimulation of arsenic-induced degradation of polysumoylated PML protein.21
In the present study, we used ccRCC RNA-sequencing (RNA-seq) data from The Cancer Genome Atlas (TCGA) and found that ESRP2 is expressed in ccRCC. We also confirmed that ESRP1 expression is strongly decreased in most ccRCC patients.7 Interestingly, the function of ESRP2, not its mRNA expression, appeared to correlate with the prognosis of ccRCC patients. On the basis of analyses of the TCGA ccRCC data, we determined Arkadia as a candidate for regulating ESRP2-splicing function. Here, we report that Arkadia is a tumor suppressor in ccRCC through modulation of the splicing function of ESRP2.
Results
ESRP2, but not ESRP1, expression is maintained in ccRCC
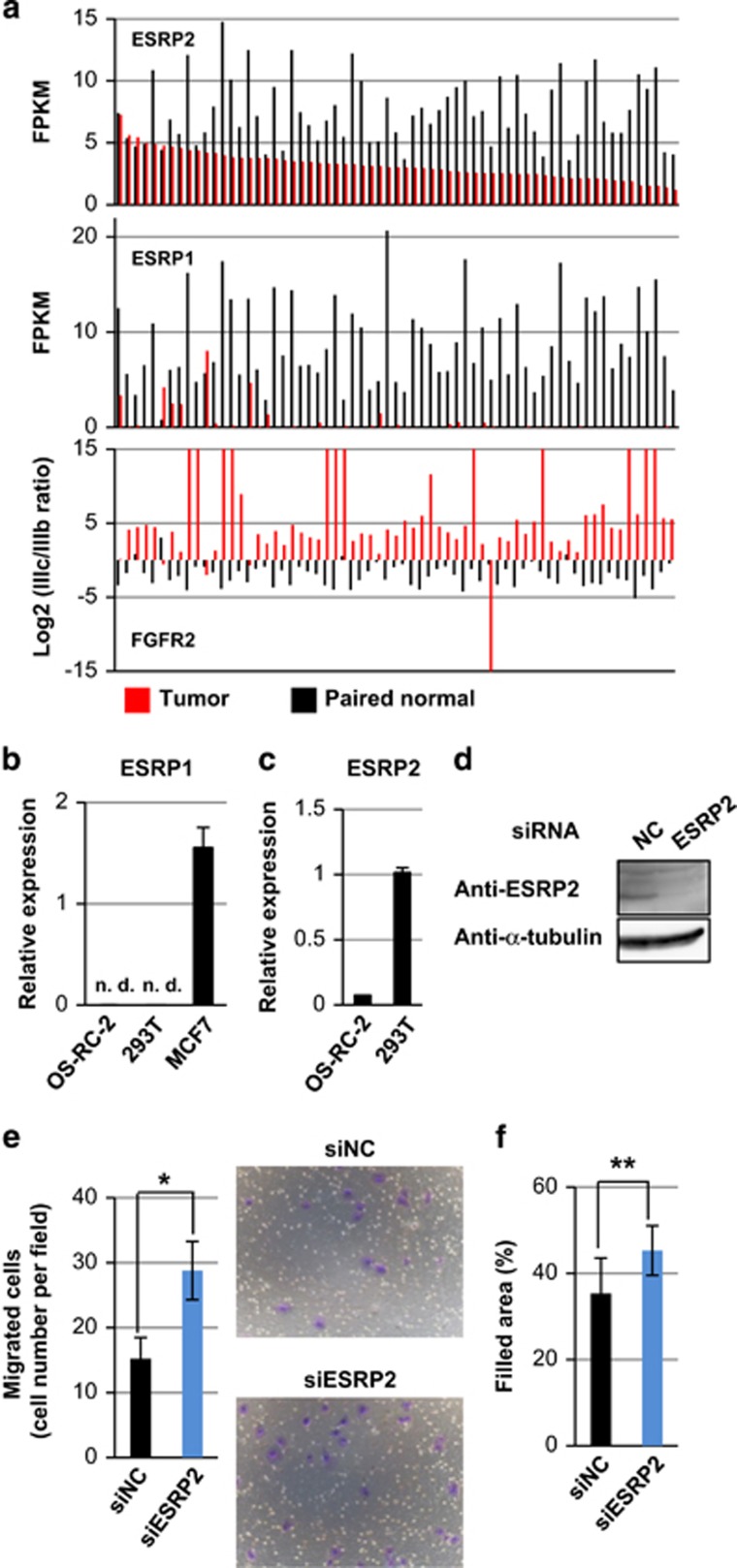
According to TCGA RNA-seq data analyses, tumor samples from ccRCC express the FGFR2 IIIc isoform, whereas paired normal samples express the FGFR2 IIIb isoform.7 In addition, the previous study showed that the expression of ESRP1 is decreased in ccRCC tumor samples compared with paired normal samples. We also analyzed the ccRCC RNA-seq data obtained from TCGA and found that the expression of ESRP2, but not ESRP1, was maintained in ccRCC tumor samples, although the levels of ESRP2 were lower in ccRCC tumor samples compared with paired normal samples (Figure 1a and Supplementary Figure S1). FGFR2 IIIc/IIIb expression ratios did not correlate with the ESRP2 mRNA expression levels (Figure 1a).
Figure 1.
Expression of ESRPs in ccRCC. (a) Expression of ESRP2 (top) and ESRP1 (middle) in 65 tumor tissues and 65 paired normal tissues using the RNA-seq data from TCGA. The log 2 values of FGFR2 IIIc/IIIb ratios are shown in the bottom panel. FPKM: fragments per kilobase of exon per million mapped sequence reads. (b and c) qRT–PCR analysis of ESRP1 (b) and ESRP2 (c) expression in OS-RC-2 and HEK293T cells. MCF7 breast cancer cells were used as a positive control in (b). ND, not detected. (d) Immunoblot analysis to determine ESRP2 protein expression in HEK293T cells treated with ESRP2 siRNA. NC, negative control. (e and f) Transwell cell migration assay (e) and wound healing assay (f) in OS-RC-2 cells treated with siRNA for ESRP2. *P<0.01 and **P<0.05 by two-sided Student's paired t-test. Representative pictures are shown in (e). siNC: negative control siRNA.
Next, we performed quantitative real-time PCR (qRT–PCR) to examine the expression of ESRP1 and ESRP2 mRNA in cultured renal cells. Although we failed to detect ESRP1 expression in the ccRCC cell line OS-RC-2 and human embryonic kidney cell line HEK293T, we detected ESRP2 mRNA in both OS-RC-2 and HEK293T cells (Figures 1b and c). We also detected the expression of ESRP2 protein in HEK293T cells by immunoblot analysis (Figure 1d). To determine whether residual ESRP2 in ccRCC cells is functional, we performed a transwell cell migration assay and wound healing assay after knocking down ESRP2 in OS-RC-2 cells. ESRP1/2 have been reported to be involved in the splicing of molecules that regulate the function of cytoskeleton and cell motility.9, 22 We found that knocking down ESRP2 expression promoted transwell cell migration and two-dimensional cell migration (Figures 1e and f), suggesting that residual ESRP2 is functional in ccRCC.
Correlation of the expression of ESRP-target exons with Arkadia expression in ccRCC
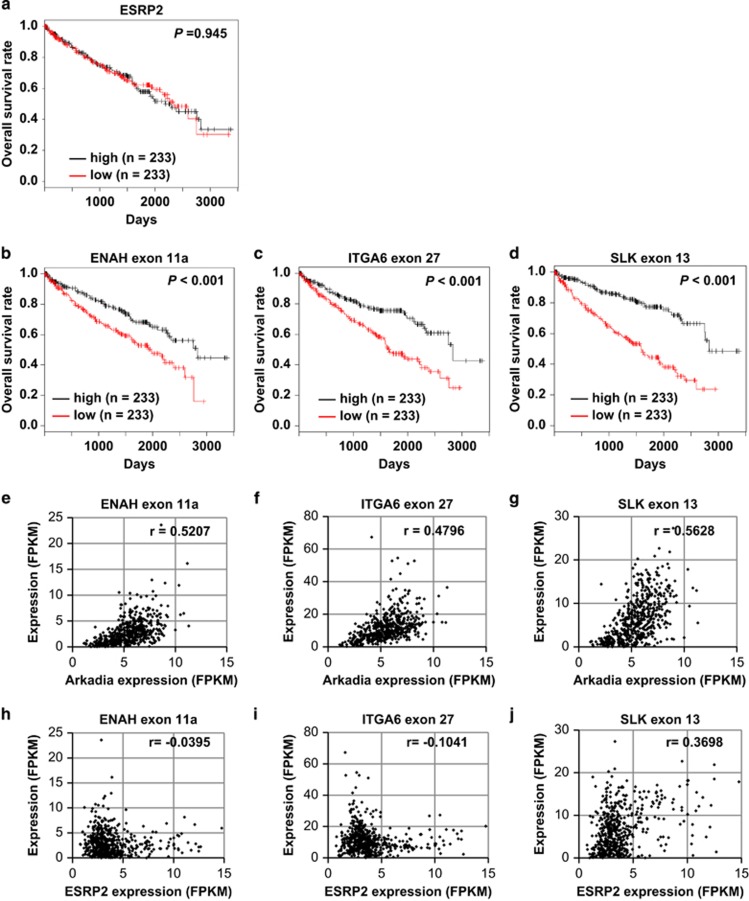
We examined the relationship between the expression of ESRP2 mRNA and the prognosis of ccRCC patients using RNA-seq data and clinical data obtained from TCGA. The ESRP2 mRNA expression levels did not correlate with the overall survival rate of ccRCC patients (Figure 2a). However, high expression levels of some ESRP-target exons, including ENAH exon 11a, ITGA6 exon 27 and SLK exon 13, were significantly correlated with longer overall survival rates (Figures 2b–d), suggesting that the splicing function, not mRNA expression, of ESRP2 is important for prognosis in ccRCC. Therefore, we hypothesized that certain factors regulate splicing function of ESRP2. To determine the regulators of the splicing function of ESRP2, we analyzed the ccRCC RNA-seq data obtained from TCGA using ESRP-target exon expression as the ESRP functional index. We found that the expression of ESRP-target exons shown in Figures 2b–d correlated with the expression of Arkadia mRNA (Figures 2e–g), but not that of ESRP2 mRNA (Figures 2h–j). Taken together, these findings suggest that Arkadia is a functional regulator of ESRP2.
Figure 2.
ESRP-splicing function and prognosis in ccRCC. (a–d) Kaplan–Meier analysis of overall survival in ccRCC patients enrolled in the TCGA database, with classifications based on the expression of ESRP2 mRNA (a), ENAH exon 11a (b), ITGA6 exon 27 (c) or SLK exon 13 (d) in TCGA ccRCC RNA-seq data. Patients were divided into two equally sized groups based on the expression of ESRP2 mRNA or ESRP-target exons. The half of the patients with higher expression of ESRP2 mRNA or each ESRP-target exon are shown in black, and the half of the patients with lower expression are shown in red; n=466. P-values are based on the log-rank significance tests. (e–g) Scatter plots of Arkadia expression and ENAH exon 11a (e), ITGA6 exon 27 (f) or SLK exon 13 (g) expression in TCGA ccRCC RNA-seq data; n=542. (h–j) Scatter plots of ESRP2 expression and ENAH exon 11a (h), ITGA6 exon 27 (i) or SLK exon 13 (j) expression in TCGA ccRCC RNA-seq data; n=542. The r-values were determined using Pearson's correlation. FPKM, fragments per kilobase of exon per million mapped sequence reads.
Arkadia interacts with ESRP2
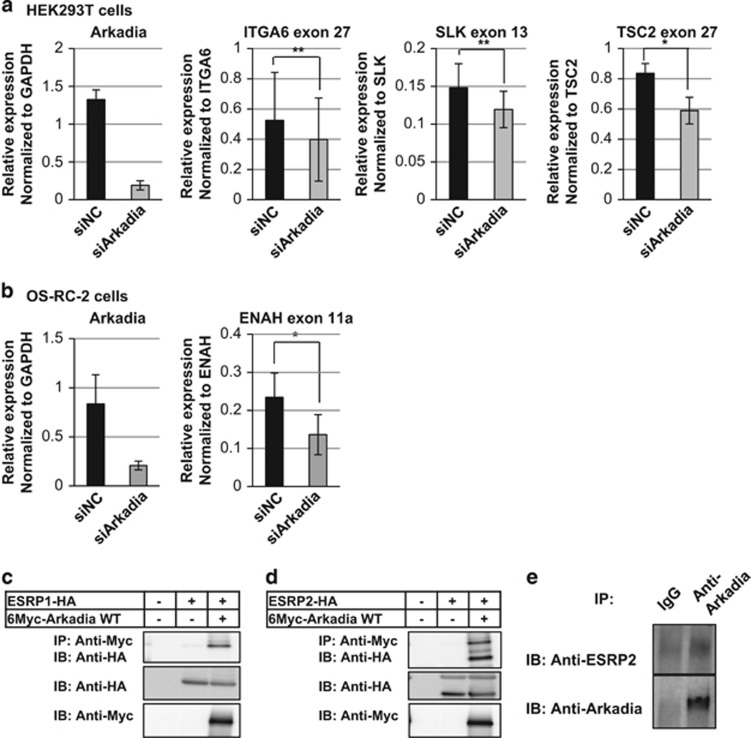
To examine whether the expression of ESRP-target exons correlate with that of Arkadia in cellulo, we knocked down Arkadia in HEK293T and OS-RC-2 cells and measured the expression of ESRP-target exons. We found that the expression of some ESRP-target exons decreased upon knockdown of Arkadia (Figures 3a and b), supporting the notion that Arkadia is an ESRP2 regulator. In contrast, Arkadia showed varied effects on the total mRNA expression levels of ITGA6, SLK and TSC2 in HEK293T cells and ENAH in OS-RC-2 cells (Supplementary Figure S2). To investigate whether Arkadia is a direct modulator of ESRP2, we performed immunoprecipitation studies using HEK293T cells co-transfected with Myc-tagged Arkadia and HA-tagged ESRP1 or ESRP2, and found that Arkadia physically interacts with ESRP1 and ESRP2 (Figures 3c and d). The interaction between endogenous Arkadia and endogenous ESRP2 was also observed in HEK293T cells (Figure 3e), suggesting that Arkadia interacts with ESRP2 and directly modulates the splicing function of ESRP2. Arkadia is known to enhance TGF-β signaling through the degradation of Smad7, c-Ski and SnoN,14, 15, 23, 24, 25 but in HEK293T kidney cells and OS-RC-2 ccRCC cells, both Arkadia and ESRP2 mRNA levels decreased after TGF-β stimulation (Supplementary Figure S3).
Figure 3.
Functional interaction between Arkadia and ESRP2. (a) qRT–PCR analyses of the expression of Arkadia, ITGA6 exon 27, SLK exon 13 and TSC2 exon 27 in HEK293T cells in the presence of negative control siRNA (siNC) or siArkadia. Values were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH), total ITGA6, SLK and TSC2, respectively. (b) qRT–PCR analysis of Arkadia and ENAH exon 11a expression in OS-RC-2 cells in the presence of siNC or siArkadia. Values were normalized to GAPDH and total ENAH, respectively; n=3 in (a) and (b). *P<0.01 and **P<0.05 by two-sided Student's paired t-test. (c and d) Immunoprecipitation assay using HEK293T cells transiently transfected with ESRP1 (c) or ESRP2 (d) and Arkadia. IB, immunoblotting; IP, immunoprecipitation; WT, wild-type. (e) Immunoprecipitation assay using HEK293T cells to determine the interaction between endogenous ESRP2 and Arkadia proteins.
Arkadia modulates ESRP2 function through ubiquitination
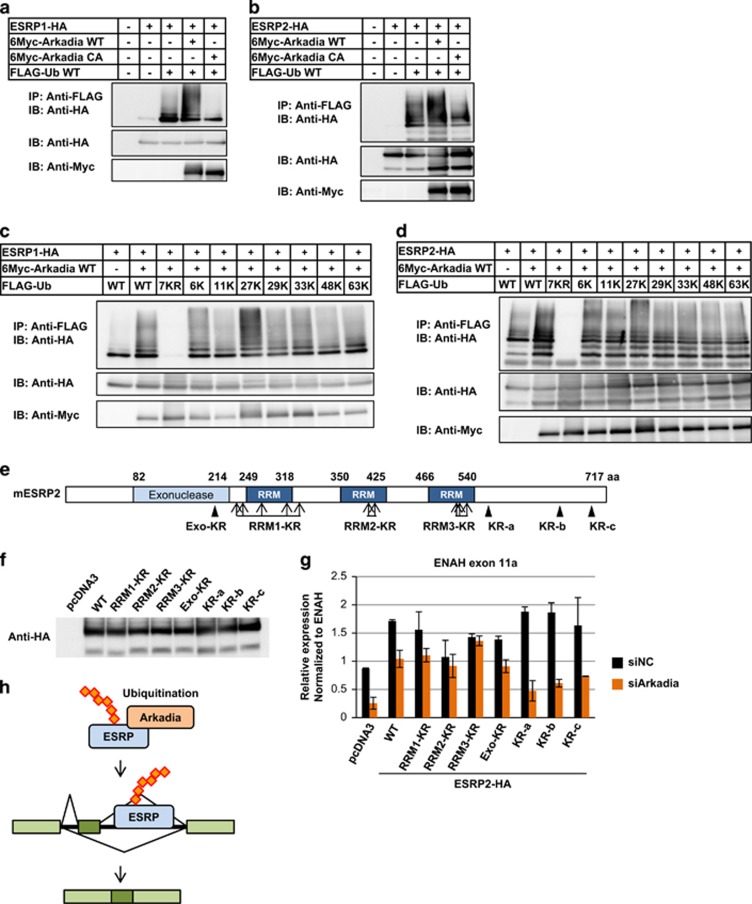
As Arkadia is an E3 ligase, we hypothesized that Arkadia ubiquitinates ESRPs to modulate their splicing function. To examine whether Arkadia ubiquitinates ESRP1/2, cDNAs for Arkadia, ubiquitin and ESRP1 or ESRP2 were transiently transfected in HEK293T cells, and immunoprecipitation assays were performed. We detected ubiquitinated ESRP1 or ESRP2 in the presence of Arkadia, whereas the ubiquitination of ESRP1/2 was strongly decreased when an Arkadia CA mutant with lower E3 ligase activity was co-transfected (Figures 4a and b). Ubiquitin molecules form polyubiquitin chains covalently linked through one of the seven lysine residues (i.e., Lys6, Lys11, Lys27, Lys29, Lys33, Lys48 or Lys63), or through an amino-terminal Met1 residue.26 To determine which lysine residues form polyubiquitin chains, we prepared ubiquitin mutants in which lysine residues were substituted by arginine residues. In the 7KR mutant, all seven lysine residues were substituted by arginine residues; in the other ubiquitin mutants, one lysine residue (6, 11, 27, 29, 33, 48 or 63K) was intact, but the other lysine residues were substituted by arginine residues. The ubiquitin mutants were co-transfected with Arkadia and ESRP1 or ESRP2 in HEK293T cells, followed by immunoprecipitation. Polyubiquitinated ESRP1 and ESRP2 were not observed when the 7KR mutant was co-transfected (Figures 4c and d). Interestingly, polyubiquitination of ESRP1 and ESRP2 was observed when the 27K mutant was co-transfected, but it was attenuated when the other ubiquitin mutants were co-transfected (Figures 4c and d). These findings suggest that polyubiquitination of ESRP by Arkadia occurs on Lys27 of ubiquitin molecules, and imply that it may have other functions than orientation to degradation.26
Figure 4.
Modulation of the splicing function of ESRP2 by Arkadia through ubiquitination. (a and b) Immunoprecipitation assay using HEK293T cells transiently transfected with ESRP1 (a) or ESRP2 (b), Arkadia wild-type or CA mutant, and ubiquitin. IP, immunoprecipitation; IB, immunoblotting; Ub, ubiquitin; WT, wild-type. (c and d) Immunoprecipitation assay using HEK293T cells transiently transfected with ESRP1 (c) or ESRP2 (d), WT Arkadia, and ubiquitin WT or its mutants. 7KR: all seven lysine residues are substituted by arginine residues; 6, 11, 27, 29, 33, 48 and 63K: one lysine residue is intact, but the others are substituted by arginine residues. KR, lysine-to-arginine mutation. (e) Schematic representation of ESRP2-KR mutants. (f) Immunoblot analysis to examine the protein expression of WT or ESRP2-KR mutants overexpressed in HEK293T cells. (g) qRT–PCR analysis of ENAH exon 11a expression in HEK293T cells transiently transfected with WT or ESRP2-KR mutants upon Arkadia knockdown. Data were normalized to total ENAH. Error bars indicate s.d. siNC, negative control siRNA. Experiments were repeated, and a representative set of data are shown in (g). (h) Schematic representation of the regulation of ESRP2 function by Arkadia through ubiquitination.
To investigate whether ubiquitination of ESRP2 by Arkadia modifies the splicing function of ESRP2, we made several ESRP2-KR mutants (Figure 4e). The Exo-KR mutant had one lysine residue in the exonuclease domain replaced with arginine residue. We also prepared KR-a, KR-b and KR-c mutants in which lysine residues in the carboxy-terminal portion were replaced with arginine residues, and RRM1-KR, RRM2-KR and RRM3-KR mutants in which lysine residues within or near each RRM domain were replaced with arginine residues. The expression levels of all these mutants were confirmed (Figure 4f). To determine which lysine residues in ESRP2 are targets of Arkadia and important for the splicing function of ESRP2, we examined the expression of ENAH exon 11a as an ESRP2 functional index, using HEK293T cells overexpressing the ESRP2-KR mutants after knocking down Arkadia (Figure 4g). We found that the expression of ENAH exon 11a was not significantly altered upon knockdown of Arkadia when RRM2-KR and RRM3-KR mutants, but not the other mutants, were overexpressed (Figure 4g), indicating that Arkadia ubiquitinates lysine residues in the RRM2 and RRM3 domains of ESRP2 and modulates the ENAH splicing ability of ESRP2. These findings suggest that Arkadia ubiquitinates ESRP2 and potentiates the splicing function of ESRP2 (Figure 4h).
To determine the role of ESRP2 ubiquitination by Arkadia in protein degradation, we performed a cycloheximide chase assay using HEK293T cells overexpressing wild-type Arkadia and wild-type or KR mutants of ESRP2 (Supplementary Figure S4). We did not observe any differences in the half-life periods of wild-type ESRP2 and its mutants in the presence of Arkadia, suggesting that the ubiquitination of ESRP2 by Arkadia does not induce protein degradation, in agreement with the observation that ubiquitination of ESRP by Arkadia may occur on Lys27 of ubiquitin molecules (Figures 4c and d).
The Arkadia-ESRP2 axis suppresses cancer cell proliferation
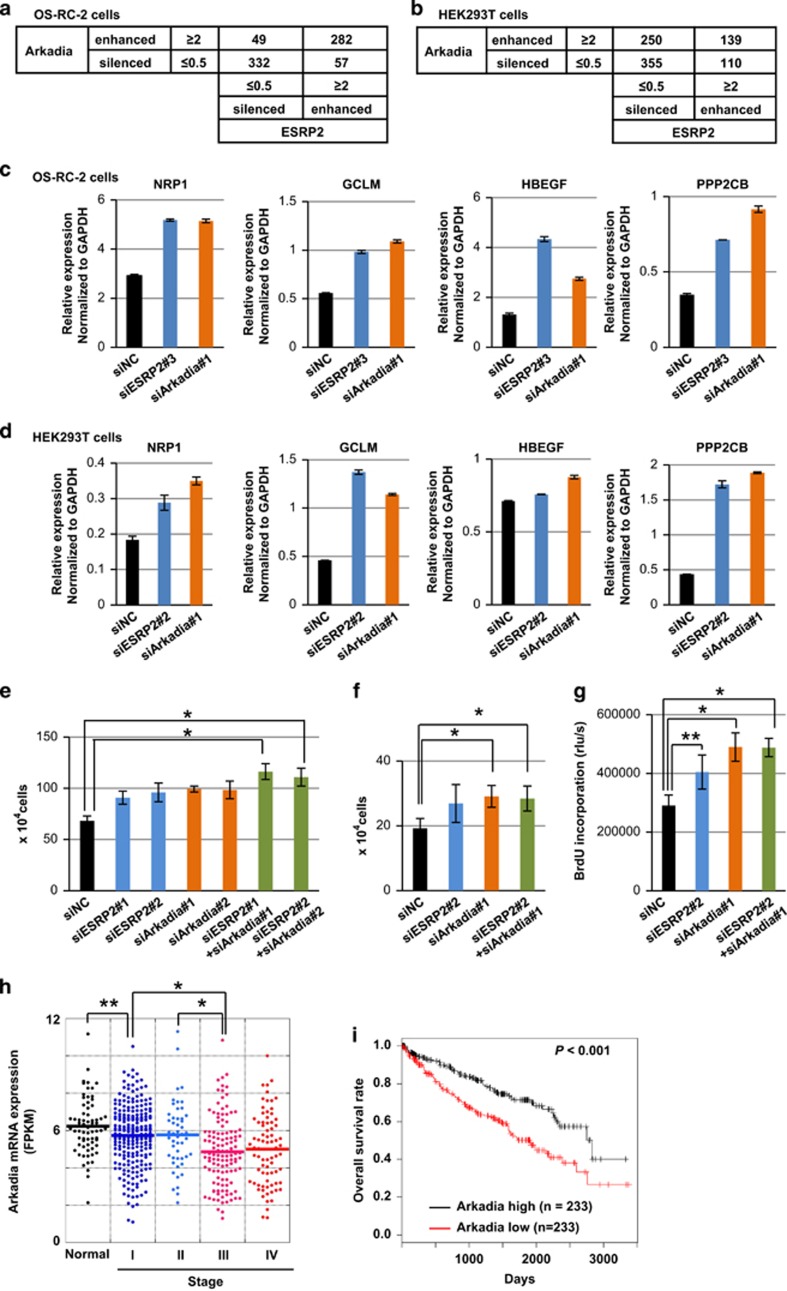
To investigate the roles of Arkadia and ESRP2 in the progression of ccRCC, we performed RNA-seq after knocking down Arkadia or ESRP2 in OS-RC-2 and HEK293T cells. We counted the number of isoforms that were up- or downregulated by knocking down Arkadia or ESRP2. The 2 × 2 contingency matrices showed that Arkadia and ESRP2 act in a significantly coordinated manner in OS-RC-2 and HEK293T cells (Figures 5a and b). To investigate which gene sets are enriched in the genes regulated by both ESRP2 and Arkadia, we performed preranked gene set enrichment analysis using the RNA-seq data in OS-RC-2 cells (Supplementary Tables S1–S4). Analysis using oncogenic signatures c6 gene sets in Molecular signatures database showed that several tumor-promotive gene sets, including the WNT_UP.V1_UP gene set (normalized enrichment score 1.97) and the KRAS.600_UP.V1_UP (normalized enrichment score 1.90), were enriched in genes downregulated by ESRP2 and Arkadia (Supplementary Table S1). Validation of the RNA-seq in OS-RC-2 cells was carried out in OS-RC-2 cells using another small interfering RNA (siRNA) for ESRP2 to exclude the possibility of off-target effects because of the low knockdown efficiency of ESRP2 (Figure 5c and Supplementary Figure S5a). We found the expression of some factors promoting cell proliferation, including NRP1 (encoding neuropillin-1), GCLM (encoding glutamate-cysteine ligase, modifier subunit), HBEGF (encoding heparin-binding epidermal growth factor-like growth factor) and PPP2CB (encoding protein phosphatase 2, catalytic subunit, β-isozyme, PP2Cβ), increased upon knockdown of Arkadia or ESRP2. Similar tendency was observed on the expression of these genes, except for HBEGF, in HEK293T cells by qRT–PCR (Figure 5d and Supplementary Figure S5b).
Figure 5.
Suppression of tumor growth by Arkadia. (a and b) Analysis using the 2x2 contingency matrix of RNA-seq data upon knockdown of Arkadia or ESRP2 in OS-RC-2 cells (a) and in HEK293T cells (b). We select the isoforms showing the maximum FPKM (fragments per kilobase of exon per million mapped sequence reads) values are z⩾5 among the three samples (siNC-treated, siESRP2-treated and siArkadia-treated) in OS-RC-2 or HEK293T cells. ESRP2-enhanced: siNC isoform/siESRP2 isoform is ⩾2. ESRP2-silenced, siNC isoform/siESRP2 isoform is ⩽0.5. Arkadia-enhanced and -silenced were determined using the same cutoff values as ESRP2-enhanced and -silenced, respectively. The number of appropriate isoforms for each category is described in the 2x2 contingency matrix. P<2.2E−16 by Fisher's exact test in (a). P<1.2E−04 by Fisher's exact test in (b). (c and d) qRT–PCR analyses of the expression of NRP1, GCLM, HBEGF and PPP2CB in OS-RC-2 cells (c) and in HEK293T cells (d) upon knockdown of ESRP2 or Arkadia. Values were normalized to GAPDH. Knockdown efficiency of Arkadia or ESRP2 is shown in Supplementary Figure S5. siNC, negative control siRNA; n=2. (e and f) Cell count assays using HEK293T (e) and OS-RC-2 (f) cells upon knockdown of ESRP2 and/or Arkadia. Cells were transfected with siRNA at the same time as seeding and counted after 48 h. Knockdown efficiency of Arkadia and ESRP2 is shown in Supplementary Figure S6. (g) BrdU assay using OS-RC-2 cells after knocking down ESRP2 and/or Arkadia. (h) Arkadia mRNA expression in 68 normal kidney tissues and 474 ccRCC tumors grouped into stages I–IV by TCGA clinical data. (i) Kaplan–Meier analysis of overall survival in ccRCC patients enrolled in the TCGA database, with classifications based on Arkadia mRNA expression in TCGA ccRCC RNA-seq data. Patients were divided into two equally sized groups based on Arkadia mRNA expression. The half of patients with higher Arkadia expression are shown in black, and the half of patients with lower expression are shown in red. In (e–h), *P<0.05 and **P<0.01 by two-sided Student's paired t-test. The P-value in (i) is based on the log-rank significance test.
Preranked gene set enrichment analysis using gene ontology (GO) biological process c5 BP gene sets in Molecular signatures database showed that POSITIVE_REGULATION_OF_CELL_PROLIFERATION gene set was enriched in genes downregulated by ESRP2 and Arkadia (normalized enrichment score 1.80; Supplementary Table S3), indicating that ESRP2 and Arkadia show suppressive effects on cell proliferation. To examine the effect of ESRP2 and Arkadia on cell proliferation, we performed cell count assays in HEK293T cells and OS-RC-2 cells, and a bromodeoxyuridine (BrdU) assay in OS-RC-2 cells (Figures 5e–g and Supplementary Figure S6) after knocking down ESRP2 and Arkadia. In OS-RC-2 cells, ESRP2 mRNA levels decreased after knocking down Arkadia, but this was not observed in HEK293T cells (Supplementary Figures S5 and S6). Knockdown of ESRP2 and Arkadia promoted cell proliferation in both HEK293T and OS-RC-2 cells. BrdU incorporation into OS-RC-2 cells was increased after knocking down ESRP2 and Arkadia, suggesting that ESRP2 and Arkadia decrease cell population at the S phase and regulate cell proliferation.
Arkadia may function as an EMT-promoting factor via enhancing TGF-β signaling, whereas ESRP2 is an EMT-suppressing factor.9, 14 However, many of ccRCC shows mesenchymal phenotypes with low E-cadherin/N-cadherin expression ratios as reported previously7 (Supplementary Figure S7a), and knockdown of Arkadia failed to show significant effects on E- and N-cadherin expression in OS-RC-2 cells (Supplementary Figures S7b). Moreover, while knocking down ESRP2 expression promoted transwell cell migration in OS-RC-2 cells (see Figure 1e), knockdown of Arkadia did not promote transwell cell migration in OS-RC-2 cells (Supplementary Figure S8), possibly because of a wide variety of target genes regulated by Arkadia.
According to the analysis of ccRCC patient data obtained from TCGA, lower Arkadia expression correlated with advanced cancer stage, but Arkadia was expressed in all ccRCC tumors (Figure 5h), in agreement with the findings that Arkadia suppresses ccRCC tumor proliferation. To determine whether Arkadia functions as a tumor suppressor in ccRCC, we performed a Kaplan–Meier survival analysis using the RNA-seq data and clinical data obtained from TCGA. Although Arkadia mRNA expression did not correlate with ESRP2 mRNA expression (Supplementary Figure S9), higher expression of Arkadia mRNA correlated with better patient prognosis (Figure 5i). Taken together, these findings suggest that the splicing function of ESRP2 promoted by Arkadia results in tumor suppression and a better outcome for ccRCC patients.
Discussion
During the process of EMT, the expression of ESRP1 and ESRP2 mRNA is transcriptionally decreased, followed by a switching of the splicing profiles from epithelial type to mesenchymal type, and leading to the acquisition of a more malignant phenotype.9, 27 ESRP1 has been described as a tumor suppressor in pancreatic cancer,28 but reported to promote lung metastasis in breast cancer through CD44 splicing.29 In ccRCC, a major subtype of kidney cancer accounting for 70–80% of all kidney cancer patients,30 ESRP1 expression is lost;7 however, we found in the present study that ESRP2 expression is maintained. Recently, Ishii et al.22 reported that ESRP1 and 2 suppress cancer cell motility through different mechanisms; ESRP1 regulates the expression of Rac1b, whereas ESRP2 attenuates cell motility through regulation of cell–cell adhesion.
Our results show that the splicing function of ESRP2 is important for prognosis in ccRCC patients. In the present study, we chose three exons as indicators of ESRP2 splicing function among the established ESRP-target exons. Our Kaplan–Meier analyses show that these gene products appear to have beneficial effects on the prognosis of ccRCC (Figures 2b–d). However, mechanisms of the beneficial effects of these ESRP-target exons remain to be determined. Interestingly, the ENAH isoform with exon 11a might not bind to actin fiber, which may lead to suppression of the cell motility.31, 32 ITGA6 is a cell adhesion molecule, and SLK reportedly acts as an antiproliferative factor in kidney tissue;33 thus, alternative splicing may affect the function of these molecules, leading to regulation of the progression of ccRCC.
Our present findings demonstrate that ESRP2 is ubiquitinated by Arkadia. Lys27-linked polyubiquitin may act as translocation signaling, and molecular structures may be stabilized by attachment of the Lys27-linked polyubiquitin chain.19, 34, 35 A point mutation in ESRP2 at the second RRM motif has been reported to alter its ability to bind RNA substrates in breast cancer,36 suggesting that the second RRM of ESRP2 is involved in binding to RNA substrates. Our findings show that lysine residues within the second and third RRM motifs in ESRP2 are targets of Arkadia and important for the splicing ability of ESRP2. In agreement with the biochemical data, our analyses of RNA-seq using 2 × 2 contingency matrixes in OS-RC-2 and HEK293T cells show the overall picture for the roles of ESRP2 and Arkadia, suggesting that ESRP2 and Arkadia act in a coordinated manner.
Our analyses of RNA-seq in OS-RC-2 cells indicate that ESRP2 and Arkadia are involved in cell proliferation in ccRCC (Supplementary Tables S1 and S3). The results of preranked gene set enrichment analysis show that several tumor-promotive gene sets, including the Wnt pathway genes, are enriched in genes downregulated by ESRP2 and Arkadia (Supplementary Table S1). It has been reported that the Wnt signaling has oncogenic roles in RCC.37 It should also be noted that the gene products used in validation in OS-RC-2 cells (Figure 5c) stimulate cancer cell proliferation through various mechanisms. Neuropillin-1 binds to vascular endothelial growth factors, and promotes angiogenesis and cell proliferation in various tumors.38 Recently, it has been reported that GCLM functions as a cancer initiation factor through synthesis of the antioxidant glutathione.39 HB-EGF binds to the EGF receptors and promotes tumor malignancy in several types of cancers.40, 41 The PP2Cβ and GAS41 complex dephosphorylate and suppress p53, resulting in increase in cell survival.42 According to our data, ESRP2 and Arkadia suppress the expression of these genes, and exhibit tumor suppressive roles in ccRCC. Notably, TGF-β decreases the expression of both ESRP2 and Arkadia, although these tumor-suppressive roles of ESRP2 and Arkadia appear to be independent of TGF-β signaling.
In contrast to the coordinate function with ESRP2 on cell proliferation, the effects of Arkadia on cell motility and EMT appear to be cell type- and context-dependent. Our results show that residual ESRP2 in ccRCC suppresses cell motility; however, knocking down Arkadia failed to significantly attenuate the cell migration in OS-RC-2 cells, suggesting that Arkadia might have independent and not supportive effect on ESRP2 in the regulation of cell motility. This might be because Arkadia has many targets with various effects on cell motility.14, 19, 20, 21 Arkadia may induce EMT in certain types of cells through enhancing the function of TGF-β however, as ccRCC cells exhibit strong mesenchymal phenotypes, we failed to show significant effects of Arkadia on the regulation of EMT (Supplementary Figure S7). Our present study thus revealed that Arkadia may alter the expression of splicing variants that lead to the regulation of cell proliferation through modulation of the function of ESRP2. Arkadia expression levels correlated with those of ESRP2-target exons, and they significantly correlated with better patient outcome. Thus, our findings suggest that Arkadia has a critical role in suppressing the progression of ccRCC by regulating the function of ESRP2, and that the Arkadia-ESRP2 axis functions as a tumor suppressor in ccRCC.
Materials and methods
TCGA data
Level 3 RNA-seq data containing gene and exon data and clinical data for ccRCC were downloaded from TCGA data portal (http://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm)43 by September 2013. Location information: ENAH exon 11a (hg19, chr1:225 692 755–225 692 693−), ITGA6 exon 27 (hg19, chr2:173 366 500–173 366 629+), SLK exon 13 (hg19, chr10:105 770 574–105 770 666+).
Cell culture
We obtained HEK293T and MCF7 cells from American Type Culture Collection. HEK293T and MCF7 cells were maintained in Dulbecco's modified Eagle's medium (no. 11965; Life Technologies, Waltham, MA, USA) supplemented with 10% fetal bovine serum. OS-RC-2 cells were obtained from RIKEN Cell Bank (Tsukuba, Japan) and maintained in RPMI 1640 (no. 11875; Life Technologies) supplemented with 10% fetal bovine serum. Cells were grown in a humidified atmosphere with 5% CO2 at 37 °C.
RNA interference
siRNAs against human Arkadia/RNF111 (HSS182645, HSS123239), human ESRP2/RBM35B (HSS188321, HSS129250, HSS188322) and control siRNA (cat. no. 12935-200) were purchased from Invitrogen (Waltham, MA, USA). siRNAs were introduced into cells using a reverse transfection method and Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's instructions. The final concentration of siRNA in the culture media was 30 nM.
Transwell assay
OS-RC-2 cells treated with siRNAs were seeded in 24-well 8.0-μm pore size culture inserts (Corning, Corning, NY, USA; 353097) coated with 0.1% gelatin (Millipore, Billerica, MA, USA; ES-006-B), and then placed in 24-well culture plates (Falcon, Corning, NY, USA; 353047) containing culture media. The cells were fixed with Mildform 10N (Wako, Osaka, Japan) and then stained with 0.5% crystal violet. Cells that migrated from inside of the culture insert to outside of that were photographed and counted. Four wells were used for each sample. The assays were performed independently in duplicate.
Wound healing assay
OS-RC-2 cells treated with siRNAs were seeded in 12-well tissue culture plates. The cell monolayers were wounded by scratching with sterile plastic 200-μl micropipette tips and photographed using phase-contrast microscopy immediately and 12 h after scratching. Four wells were used for each sample. The assays were performed independently in duplicate. The migration areas were measured by counting the number of pixels using Image J (NIH, Bethesda, MD, USA; http://imagej.nih.gov/ij/).
Reagents and antibodies
Proteasomal inhibitor MG132 was purchased from Peptide Institute (Osaka, Japan). The following antibodies were used: mouse anti-FLAG (Sigma-Aldrich, St Louis, MO, USA; M2), mouse anti-myc (Oncogene Research Products, San Diego, CA, USA; 9E10), rat anti-HA (Roche, Basel, Switzerland; 3F10), mouse anti-tubulin (Sigma-Aldrich; DM1A), rabbit anti-ESRP2 (Sigma-Aldrich; HPA048597), rabbit anti-RNF111/Arkadia (Sigma-Aldrich; HPA038576), goat anti-Arkadia (Santa Cruz, Dallas, TX, USA; H16 for immunoprecipitation), normal goat IgG (Santa Cruz) and horseradish peroxidase-conjugated secondary antibodies against mouse IgG (Cell Signaling, Cochranville, PA, USA; no. 7076S), rabbit IgG (Cell Signaling; no. 7074S) and rat IgG (Cappel, Cochranville, PA, USA; 56404).
Immunoprecipitation and immunoblotting
HEK293T cells were seeded in 6-well culture plates and transiently transfected with the indicated plasmids using FuGENE6. Cells were harvested with immunoprecipitation buffer (150 mM NaCl, 20 mM Tris-HCl, pH 8.0, 5 mM EDTA, 0.5% Nonidet P-40) containing protease inhibitor cocktail (Roche; 11836145001) at the point of use. For the endogenous binding assay, HEK293T cells were seeded in a 6-cm culture dish. The cells were treated with 10 μM MG132 for 4 h before harvesting with 100 μl (per sample) of RIPA buffer (150 mM NaCl, 50 mM Tris-HCl, pH 8.0, 5 mM EDTA, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate) containing protease inhibitor cocktail at the point of use, and diluted up to 1 ml with immunoprecipitation buffer. Immunoprecipitation was performed as described previously.19 Sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblotting were performed as described previously,19 using a LAS-4000 lumino-image analyzer (Fujifilm, Tokyo, Japan).
cDNA constructs
Mouse Arkadia, human ESRP1, mouse ESRP2 and ubiquitin mutants were described previously.12, 19 ESRP2-KR mutants were produced by PCR-based methods: Exo-KR (K147R), RRM1-KR (K231R, K239R, K268R, K309R, K321R and K329R), RRM2-KR (K413R and K418R), RRM3-KR (K530R, K531R and K534R), KR-a (K568R), KR-b (K678R) and KR-c (K712). In KxR, x denotes the residue number in ESRP2.
RNA isolation and qRT–PCR
Total RNA was extracted as described previously.44 First-strand cDNA was synthesized using PrimeScript2 reverse transcriptase (TakaraBio, Shiga, Japan). qRT–PCR was performed using FastStart Universal SYBR Green Master Mix with ROX (Roche), and the ABI PRISM 7000 Sequence Detection System or StepONE Plus real-time PCR system (Applied Biosystems, Waltham, MA, USA). All samples were run in duplicate and the results averaged. Primer sequences for qRT–PCR are shown in Supplementary Table S5.
RNA-seq and data analysis
RNA-seq was performed as described previously.45 cDNA libraries were prepared using Dynabeads mRNA DIRECT Purification Kit (Life Technologies) and Ion Total RNA-Seq Kit v.2 (Life Technologies). High-throughput sequencing of the cDNA fragments was performed using Ion PROTON, Ion PI Template OT2 200 Kit v.3 and Ion PI Sequencing 200 Kit v.3 or Ion PI IC 200 Kit (Life Technologies) following the manufacturer's protocols. P1 chip v.2 (Life Technologies) was used to sequence three pooled barcoded samples. Sequence reads, whose individual read lengths were determined by evaluating the default sequencing quality using the Torrent Server (Life Technologies), were aligned against the human reference transcriptome (NCBI Build 37, hg19) in TopHat2 (http://ccb.jhu.edu/software/tophat/index.shtml).46 Expression levels were calculated by using the cuffdiff function of Cufflinks (http://cufflinks.cbcb.umd.edu/). Raw sequencing data with FPKM (fragments per kilobase of exon per million mapped sequence reads) calculation results are available at GEO (GSE66741 and GSE60559).
Cell count assay
HEK293T and OS-RC-2 cells treated with siRNA were seeded in 12-well culture plates. The cells were counted 48 h after seeding. Four wells were used for each sample.
BrdU assay
OS-RC-2 cells were seeded in 96-well culture plates. The BrdU assay was performed using the Cell Proliferation ELISA, BrdU Kit (Roche; 11 669 915 001) according to the manufacturer's manual. Four wells were used for each sample.
Kaplan–Meier analysis
From the 470 ccRCC patients, we omitted four patients who had two tumor samples taken. We analyzed the gene and exon expression data and clinical data using the R program (http://www.r-project.org/).
Statistical analyses
Student's t-test was used for two-sample analyses. Differences in survival between groups were evaluated using the log-rank test in the R program. Fisher's exact test in the R program was used for the 2x2 contingency matrices. The statistical software, KaleidaGraph v.4.1 (HULINKS, Tokyo, Japan) was used for dot-blot analysis.
Acknowledgments
We are grateful to Dr Hiroshi I Suzuki (The University of Tokyo) for discussion and advice. We also thank Keiko Yuki for technical assistance, as well as the members of the Miyazono laboratory for discussion and advice. This research was supported by KAKENHI grants-in-aid for scientific research on Innovative Area (Integrative Research on Cancer Microenvironment Network; 22112002, to KM), Young Scientists (B) (24700968 and 26830066, to AM) and Scientific Research (C) (24501311, to DK), from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT). This study was performed, in part, as a research program for the Project for Development of Innovative Research on Cancer Therapeutics (P-Direct), MEXT.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Gamazon ER, Stranger BE. Genomics of alternative splicing: evolution, development and pathophysiology. Hum Genet 2014; 133: 679–687. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet 2011; 12: 715–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Weiss WA. Alternative splicing in cancer: implications for biology and therapy. Oncogene 2014; 34: 1–14. [DOI] [PubMed] [Google Scholar]
- Biamonti G, Catillo M, Pignataro D, Montecucco A, Ghigna C. The alternative splicing side of cancer. Semin Cell Dev Biol 2014; 32C: 30–36. [DOI] [PubMed] [Google Scholar]
- Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 2010; 10: 116–129. [DOI] [PubMed] [Google Scholar]
- Shirakihara T, Horiguchi K, Miyazawa K, Ehata S, Shibata T, Morita I et al. TGF-β regulates isoform switching of FGF receptors and epithelial-mesenchymal transition. EMBO J 2011; 30: 783–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Caballero OL, Davis ID, Jonasch E, Tamboli P, Yung WK et al. Tumor-specific isoform switch of the fibroblast growth factor receptor 2 underlies the mesenchymal and malignant phenotypes of clear cell renal cell carcinomas. Clin Cancer Res 2013; 19: 2460–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell 2009; 33: 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Carstens RP. Complex changes in alternative pre-mRNA splicing play a central role in the epithelial-to-mesenchymal transition (EMT). Semin Cancer Biol 2012; 22: 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S et al. An ESRP-regulated splicing programme is abrogated during the epithelial–mesenchymal transition. EMBO J 2010; 29: 3286–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushima H, Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat Rev Cancer 2010; 10: 415–424. [DOI] [PubMed] [Google Scholar]
- Horiguchi K, Sakamoto K, Koinuma D, Semba K, Inoue A, Inoue S et al. TGF-β drives epithelial-mesenchymal transition through δEF1-mediated downregulation of ESRP. Oncogene 2012; 31: 3190–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Koinuma D. Arkadia—beyond the TGF-β pathway. J Biochem 2011; 149: 1–3. [DOI] [PubMed] [Google Scholar]
- Koinuma D, Shinozaki M, Komuro A, Goto K, Saitoh M, Hanyu A et al. Arkadia amplifies TGF-beta superfamily signalling through degradation of Smad7. EMBO J 2003; 22: 6458–6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano Y, Mavrakis KJ, Lee KL, Fujii T, Koinuma D, Sase H et al. Arkadia induces degradation of SnoN and c-Ski to enhance transforming growth factor-beta signaling. J Biol Chem 2007; 282: 20492–20501. [DOI] [PubMed] [Google Scholar]
- Briones-Orta MA, Levy L, Madsen CD, Das D, Erker Y, Sahai E et al. Arkadia regulates tumor metastasis by modulation of the TGF-β pathway. Cancer Res 2013; 73: 1800–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Antonacopoulou AG, Tanaka S, Panoutsopoulos AA, Bravou V, Kalofonos HP et al. Enhancement of TGF-β signaling responses by the E3 ubiquitin ligase Arkadia provides tumor suppression in colorectal cancer. Cancer Res 2011; 71: 6438–6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierie B, Moses HL. Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer 2006; 6: 506–520. [DOI] [PubMed] [Google Scholar]
- Mizutani A, Saitoh M, Imamura T, Miyazawa K, Miyazono K. Arkadia complexes with clathrin adaptor AP2 and regulates EGF signalling. J Biochem 2010; 148: 733–741. [DOI] [PubMed] [Google Scholar]
- Poulsen SL, Hansen RK, Wagner SA, van Cuijk L, van Belle GJ, Streicher W et al. RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J Cell Biol 2013; 201: 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erker Y, Neyret-Kahn H, Seeler JS, Dejean A, Atfi A, Levy L. Arkadia, a novel SUMO-targeted ubiquitin ligase involved in PML degradation. Mol Cell Biol 2013; 33: 2163–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H, Saitoh M, Sakamoto K, Kondo T, Katoh R, Tanaka S et al. Epithelial Splicing Regulatory Protein 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J Biol Chem 2014; 289: 27386–27399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Scolan E, Zhu Q, Wang L, Bandyopadhyay A, Javelaud D, Mauviel A et al. Transforming growth factor-beta suppresses the ability of Ski to inhibit tumor metastasis by inducing its degradation. Cancer Res 2008; 68: 3277–3285. [DOI] [PubMed] [Google Scholar]
- Levy L, Howell M, Das D, Harkin S, Episkopou V, Hill CS. Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation. Mol Cell Biol 2007; 27: 6068–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzawa H, Koinuma D, Maeda S, Yamamoto K, Miyazawa K, Imamura T. Arkadia represses the expression of myoblast differentiation markers through degradation of Ski and the Ski-bound Smad complex in C2C12 myoblasts. Bone 2009; 44: 53–60. [DOI] [PubMed] [Google Scholar]
- Kulathu Y, Komander D. Atypical ubiquitylation—the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol 2012; 13: 508–523. [DOI] [PubMed] [Google Scholar]
- Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet 2011; 7: e1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda J, Matsuda Y, Yamahatsu K, Uchida E, Naito Z, Korc M et al. Epithelial splicing regulatory protein 1 is a favorable prognostic factor in pancreatic cancer that attenuates pancreatic metastases. Oncogene 2013; 33: 4485–4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yae T, Tsuchihashi K, Ishimoto T, Motohara T, Yoshikawa M, Yoshida GJ et al. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat Commun 2012; 3: 883. [DOI] [PubMed] [Google Scholar]
- Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet 2009; 373: 1119–1132. [DOI] [PubMed] [Google Scholar]
- Di Modugno F, DeMonte L, Balsamo M, Bronzi G, Nicotra MR, Alessio M et al. Molecular cloning of hMena (ENAH) and its splice variant hMena+11a: epidermal growth factor increases their expression and stimulates hMena+11a phosphorylation in breast cancer cell lines. Cancer Res 2007; 67: 2657–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzik M, Kotova TI, Higgs HN, Hazelwood L, Hanein D, Gertler FB et al. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J Biol Chem 2005; 280: 28653–28662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulsky AV, Takano T, Guillemette J, Papillon J, Volpini RA, Di Battista JA. The Ste20-like kinase SLK promotes p53 transactivation and apoptosis. Am J Physiol Renal Physiol 2009; 297: F971–F980. [DOI] [PubMed] [Google Scholar]
- Peng DJ, Zeng M, Muromoto R, Matsuda T, Shimoda K, Subramaniam M et al. Noncanonical K27-linked polyubiquitination of TIEG1 regulates Foxp3 expression and tumor growth. J Immunol 2011; 186: 5638–5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 2010; 12: 119–131. [DOI] [PubMed] [Google Scholar]
- Horvath A, Pakala SB, Mudvari P, Reddy SD, Ohshiro K, Casimiro S et al. Novel insights into breast cancer genetic variance through RNA sequencing. Sci Rep 2013; 3: 2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu RJ, Ho JY, Cha TL, Yu DS, Wu CL, Huang WP et al. WNT10A plays an oncogenic role in renal cell carcinoma by activating WNT/β-catenin pathway. PLoS One 2012; 7: e47649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild JR, Staton CA, Chapple K, Corfe BM. Neuropilins: expression and roles in the epithelium. Int J Exp Pathol 2012; 93: 81–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris IS, Treloar AE, Inoue S, Sasaki M, Gorrini C, Lee KC et al. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell 2015; 27: 211–222. [DOI] [PubMed] [Google Scholar]
- Zhou ZN, Sharma VP, Beaty BT, Roh-Johnson M, Peterson EA, Van Rooijen N et al. Autocrine HBEGF expression promotes breast cancer intravasation, metastasis and macrophage-independent invasion in vivo. Oncogene 2014; 33: 3784–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray KC, Moss ME, Franklin JL, Weaver CJ, Higginbotham J, Song Y et al. Heparin-binding epidermal growth factor-like growth factor eliminates constraints on activated Kras to promote rapid onset of pancreatic neoplasia. Oncogene 2014; 33: 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Smith RJ, Shieh SY, Roeder RG. The GAS41-PP2Cbeta complex dephosphorylates p53 at serine 366 and regulates its stability. J Biol Chem 2011; 286: 10911–10917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani A, Koinuma D, Tsutsumi S, Kamimura N, Morikawa M, Suzuki HI et al. Cell type-specific target selection by combinatorial binding of Smad2/3 proteins and hepatocyte nuclear factor 4alpha in HepG2 cells. J Biol Chem 2011; 286: 29848–29860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogaya K, Koinuma D, Tsutsumi S, Saito RA, Miyazawa K, Aburatani H et al. A Smad3 and TTF-1/NKX2-1 complex regulates Smad4-independent gene expression. Cell Res 2014; 24: 994–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.