Abstract
Biomarker, neuroimaging, and genetic findings implicate the serotonin transporter (SERT) in autism spectrum disorder (ASD). Previously, we found that adult male mice expressing the autism-associated SERT Ala56 variant have altered central serotonin (5-HT) system function, as well as elevated peripheral blood 5-HT levels. Early in gestation, before midbrain 5-HT projections have reached the cortex, peripheral sources supply 5-HT to the forebrain, suggesting that altered maternal or placenta 5-HT system function could impact the developing embryo. We therefore used different combinations of maternal and embryo SERT Ala56 genotypes to examine effects on blood, placenta and embryo serotonin levels and neurodevelopment at embryonic day E14.5, when peripheral sources of 5-HT predominate, and E18.5, when midbrain 5-HT projections have reached the forebrain. Maternal SERT Ala56 genotype was associated with decreased placenta and embryonic forebrain 5-HT levels at E14.5. Low 5-HT in the placenta persisted, but forebrain levels normalized by E18.5. Maternal SERT Ala56 genotype effects on forebrain 5-HT levels were accompanied by a broadening of 5-HT-sensitive thalamocortical axon projections. In contrast, no effect of embryo genotype was seen in concepti from heterozygous dams. Blood 5-HT levels were dynamic across pregnancy and were increased in SERT Ala56 dams at E14.5. Placenta RNA sequencing data at E14.5 indicated substantial impact of maternal SERT Ala56 genotype, with alterations in immune and metabolic-related pathways. Collectively, these findings indicate that maternal SERT function impacts offspring placental 5-HT levels, forebrain 5-HT levels, and neurodevelopment.
INTRODUCTION
The serotonin (5-hydroxytryptamine, 5-HT) system has long been implicated in autism spectrum disorder (ASD), but a clear understanding of how 5-HT contributes to the pathophysiology remains incomplete. Besides its role as a neurotransmitter, 5-HT is now recognized as a critical signaling molecule during neurodevelopment. Several key proteins in the 5-HT system are transiently expressed in the developing forebrain before the arrival of 5-HT projections from the midbrain raphe nuclei (Bonnin et al, 2006; Gaspar et al, 2003), suggesting that exogenous sources of 5-HT have a role in development. Notably, the placenta synthesizes and delivers 5-HT to the fetal forebrain (Bonnin et al, 2011). Although disruptions in this placental 5-HT synthetic pathway appear poised to impact fetal neurodevelopment, this possibility has yet to be examined in genetic animal models of ASD.
Elevated whole blood 5-HT levels have been consistently identified in approximately 30% of children with ASD (Gabriele et al, 2014). Due to its primary role as a regulator of peripheral and central 5-HT homeostasis, the 5-HT transporter (5-HTT, SERT) has been intensively scrutinized (Muller et al, 2015). Linkage studies have implicated the chromosome 17q region harbouring the SERT gene, SLC6A4, with stronger linkage found in families with only affected males (Sutcliffe et al, 2005; Yonan et al, 2003). Although there is modest evidence to connect common SLC6A4 variation to ASD (Devlin et al, 2005), a number of rare, hyperfunctional, dysregulated SERT variants were identified in families with evidence of linkage (Prasad et al, 2009; Sutcliffe et al, 2005). The most common of the rare variants, Gly56Ala, is also found in unaffected carriers (Sakurai et al, 2008; Sutcliffe et al, 2005), suggesting the presence of other genetic or environmental factors that modulate ASD risk. Emerging evidence points to maternal genotype as a potential contributor to offspring risk of neurodevelopmental phenotypes. For example, the prenatal impact of maternal genotype in 5-HT1A receptor null mice confers life-long anxiety-like behavior in offspring, independent of offspring genotype (Gleason et al, 2010). This possibility has recently been explored in ASD, with risk increased in the offspring of mothers with the high-expressing SLC6A4 5-HTTLPR long allele (Kistner-Griffin et al, 2011). It remains unclear how increased SERT function could alter the maternal prenatal environment.
Previously, we found that male SERT Ala56 knock-in mice on the 129S6/S4 inbred background have hyperserotonemia, altered central 5-HT system function, and behavioural abnormalities relevant to ASD (Veenstra-VanderWeele et al, 2012). These findings highlighted clear differences between SERT Ala56 and wild-type littermates, but it remained unknown whether offspring are also sensitive to maternal SERT function. In the current study, we examined whether maternal SERT Ala56 genotype would impact neurodevelopment. As hypothesized, we found maternal effects on the developmental trajectory of the 5-HT system in the fetal brain that suggest altered placental function. Furthermore, we found parallel changes in the development of thalamocortical axons (TCAs), a known 5-HT sensitive brain structure (Bonnin et al, 2007; Cases et al, 1996). RNA sequencing data suggested that altered maternal SERT function has broad effects on placental development. Collectively, these findings suggest that some ASD risk alleles may affect neurodevelopment by altering the maternal prenatal environment.
MATERIALS AND METHODS
Mice
All animal procedures were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Vanderbilt University Institutional Animal Care and Use Committee. SERT Gly56Ala mice used for experiments were maintained on the 129S6/S4 inbred strain background (Veenstra-VanderWeele et al, 2009, 2012).
Timed breeding pairs were composed of wild-type (Gly56/Gly56), heterozygous (Gly56/Ala56), or homozygous mutant (Ala56/Ala56) dams mated with sires of matching genotypes, with presence of vaginal plug considered E0.5. Thus, our experimental groups consisted of wild-type concepti from wild-type dams, wild-type concepti from heterozygous dams, Ala56 concepti from heterozygous dams, and Ala56 concepti from homozygous Ala56 dams. In our primary analysis, wild-type and Ala56 littermates from heterozygous dams assessed embryo genotype effects; whereas samples collected from wild-type and Ala56 dams evaluated the impact of maternal genotype.
For all breeder schemes, four to six litters were harvested per developmental time point analyzed. For wild-type and homozygous Ala56 breeders, two to four samples per litter were randomly chosen for HPLC and tryptophan hydroxylase activity assay analyses, and a single representative sample per litter for all other assays. For heterozygous breeders, multiple wild-type and Ala56 samples, if present, were collected per litter to increase statistical power for HPLC, and a single pair of littermates was chosen for all other assays. Separate cohorts of mice were used for all the assays. As initial homozygous breeder HPLC data indicated no gender effects, the samples were collapsed into single genotype groups regardless of sex for all the subsequent analyses.
High-Performance Liquid Chromatography
Placental and embryonic brain samples were dissected from phosphate buffered saline-perfused dams at E14.5 and E18.5. All tissues were weighed, flash frozen, and stored at −80 °C until HPLC analysis with fluorometric detection of 5-HT, tryptophan (TRP), and 5-hydroxyindole acetic acid (5-HIAA) as previously described, with 5-HIAA/5-HT used as an index of 5-HT turnover (Anderson et al, 1987). 5-HT intra- and inter-assay coefficients of variation were less than 5 and 10%, respectively. At E14.5, forebrain 5-HIAA measurements were below the detection limit. Placental levels of 5-HIAA were less than 1% of 5-HT and could not be consistently measured due to an interfering spectra peak.
Netrin-G1a Immunohistochemistry and Quantification
Embryos were harvested from timed-pregnant dams at E18.5 and collected brains were fixed overnight in 4% paraformaldehyde. After cryoprotection in 30% sucrose, the brains were embedded and sectioned (40 μm) on a cryostat. Peroxidase-quenched sections were blocked in PBS containing 3% donkey serum and 0.1% Triton X-100. The sections were incubated overnight with Netrin-G1a antibody (1 : 100, R&D systems), washed, and incubated with peroxidase-conjugated donkey anti-goat secondary antibody (1 : 800, Jackson Immunoresearch; Bonnin et al, 2007). Per manufacturer's instructions, tyramide signal amplification was used for fluorescein labeling (1 : 100, PerkinElmer), and sections were counterstained with DAPI mounting medium (Vector Labs). Fluorescent images were taken with a Zeiss Axio Imager M2. Using ImageJ software, TCA and cortical thickness were measured, blind to genotype, from three separate locations along the TCA distal–proximal axis in matched rostral and caudal brain sections.
Tryptophan Hydroxylase (TPH) Activity Assay
TPH activity was assayed from in vitro placenta preparations as previously described (Bonnin et al, 2011) with slight modifications. Frozen placental tissue was homogenized in 350 μl of extraction buffer containing 0.05 M Tris-HCL buffer pH 7.5, 1 mM EGTA, and 1 mM DTT. Homogenates were centrifuged at 21 000 g for 15 min at 4 °C. A total 20 μl of supernatant (~70 μg of protein (DC Protein Assay, Bio-Rad)) was added to 80 μl of a reaction master mix, resulting in final concentrations: 0.05 M Tris-HCL buffer pH 7.5, 1 mM EGTA, 100 μM L-tryptophan, 0.05 mg/ml catalase, 100 μM tetrahydrobiopterin, 1 mM ferrous ammonium sulfate. After incubation at 37 °C for 30 min, 100 μl of termination buffer (0.2 M perchloric acid and 100 μM EDTA) was added, followed by incubation on ice for 15 min. After centrifugation at 21 000 g for 15 min at 4 °C, supernatant was flash-frozen for subsequent HPLC with electrochemical detection of 5-hydroxytryptophan (5-HTP).
Blood Collection
Beginning three days before the experimental protocol, female mice were handled daily to minimize stress reactivity during blood draw procedures. Using submandibular bleeds, blood was drawn from unanesthetized females either at a single time point 14.5 days post conception or longitudinally at 7 days before mating, 7.5 days post conception, 14.5 days post conception, 18.5 days post conception, and 7 days post-partum. The blood was collected into K2EDTA-coated tubes (BD Biosciences) and stored at −80 °C until HPLC analysis with fluorometric detection, as previously described (Anderson et al, 1987).
RNA Isolation
RNA from fresh-frozen placental tissue was isolated using Qiagen RNeasy Mini Kit with DNase I treatment per manufacturer's instructions. RNA quality and yield were assessed by NanoDrop spectrophotometer (Thermo Scientific). RNA sequencing sample integrity was evaluated by Agilent Bioanalyzer (Agilent Technologies), with all the samples possessing RNA Integrity Numbers >9.0 (Schroeder et al, 2006).
Quantitative RT-PCR
cDNA was generated using the High Capacity cDNA Reverse Transcription Kit (Life Technologies) per manufacturer's instructions. qPCR was performed in duplicate using Tph1 (Mm00493794_m1) and GusB (Mm00446953_m1) TaqMan gene expression assays on a 7900HT system (Life Technologies), with differential gene expression calculated using the ΔΔCT method.
RNA Sequencing
Isolated RNA from E14.5 placental tissue was submitted to VANTAGE core facilities at Vanderbilt University for cDNA library preparation (Illumina Tru Seq Sample Prep Kit) and whole transcriptome sequencing (30M reads/sample, single-end 50 bp reads) on the Illumina HiSeq 2500 platform. Three biological replicates were sequenced per experimental group, with each replicate consisting of two unique samples pooled together.
RNA Sequencing Analysis
Sequencing reads were mapped to the mouse genome mm10 using TopHat-2.0.10 (Kim et al, 2013) and quantified by samtools-0.1.19 (Li et al, 2009a) and HTSeq-0.5.4p5 (Anders et al, 2014). To further assess the sample quality, we examined intrasample correlation of normalized counts of the top 1000 expressed genes in the placenta. Although intrasample correlation coefficients were generally high (r>0.90), one Ala56 sample from the heterozygous breeding experimental group consistently fell below this threshold and was omitted from further analysis. Count-based differential expression analysis was performed using edgeR_3.4.2 (Anders et al, 2013; McCarthy et al, 2012), which implements general differential analyses based on the negative binomial model. False discovery rate-adjusted p-values were used to evaluate genome-wide statistical significance (<0.05) of individual gene comparisons. For pathway and network analyses, lists of differentially expressed genes with significant unadjusted p-values (p<0.05) were input into Ingenuity Pathway Analysis (IPA, Qiagen, Redwood City, CA) for canonical pathway and upstream regulator analyses.
RESULTS
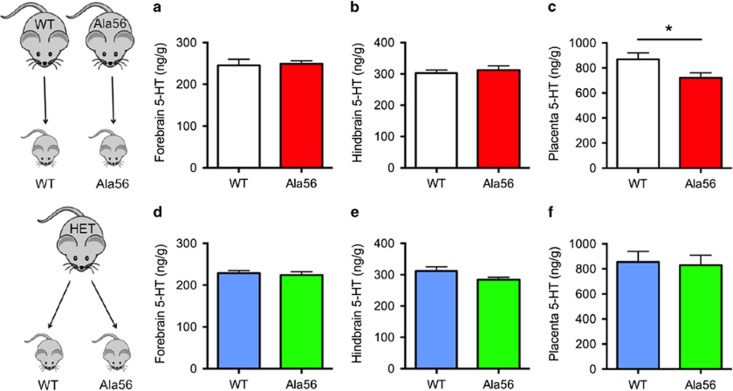
Using HPLC, we measured forebrain, hindbrain, and placenta tissue levels of 5-HT at embryonic day E14.5, when the placenta predominantly supplies 5-HT to the fetal forebrain (Bonnin et al, 2011). Forebrain 5-HT levels were significantly decreased in SERT Ala56 embryos from SERT Ala56 dams compared with wild-type embryos from wild-type dams (t=2.32, p=0.029; Figure 1a); whereas hindbrain levels were not different (t=0.077, p=0.94; Figure 1b). Paralleling these findings, placentas from SERT Ala56 dams also exhibited significant decreases in 5-HT levels (t=2.65, p=0.014, with Welch's correction for unequal variances; Figure 1c). In contrast, SERT Ala56 and wild-type littermate embryos from heterozygous matings exhibited no genotype differences in 5-HT content in any tissue studied (All p>0.1; Figure 1d–f).
Figure 1.
Maternal SERT Ala56 genotype effect on fetal forebrain 5-HT levels at embryonic day 14.5. In homozygous Ala56 or wild-type (WT) dams carrying Ala56 or wild-type embryos, respectively, E14.5 forebrain (a) 5-HT levels were decreased in knock-in embryos compared with wild-type (n=12–15/genotype), with no changes in the hindbrain (b) 5-HT levels. E14.5 placenta (c) 5-HT levels were similarly decreased in Ala56 concepti compared with wild-type within dams of the same genotype. In contrast, there were no significant changes in the forebrain (d), hindbrain (e), or placenta (f) 5-HT levels found between Ala56 and wild-type concepti from heterozygous dams (n=8–11/genotype). *p<0.05.
Tissue HPLC measurements were repeated at E18.5, when the midbrain raphe is the primary source of 5-HT in the fetal forebrain. In contrast to E14.5, forebrain 5-HT levels in E18.5 SERT Ala56 embryos from SERT Ala56 dams were indistinguishable from wild-type embryos from wild-type dams (t=0.20, p=0.84; Figure 2a). In contrast, E18.5 placental 5-HT levels in SERT Ala56 dams remained significantly lower than in wild-type dams (t=2.28, p=0.031; Figure 2c). In E18.5 littermates from heterozygous dams, there were again no significant genotype differences in any tissue studied (All p>0.1; Figure 2d–f). There were no changes in 5-HIAA levels or 5-HT turnover in the hindbrain at E14.5, and levels of 5-HIAA were too low to measure reliably in the forebrain at E14.5. There were no changes in 5-HIAA levels or 5-HT turnover at E18.5 with the exception of decreased 5-HIAA (but not 5-HT turnover) in the hindbrain of Ala56 embryos from heterozygous dams (Supplementary Figures 1 and 2).
Figure 2.
Lack of maternal SERT Ala56 genotype effect on fetal forebrain 5-HT levels at embryonic day 18.5. In homozygous Ala56 or wild-type (WT) dams carrying Ala56 or wild-type embryos, respectively, E18.5 forebrain (a) and hindbrain (b) 5-HT levels were unchanged in knock-in embryos compared with wild-type (n=14 / genotype), despite continued decrease in placenta 5-HT (c). In contrast, there were no significant changes in the (d) forebrain, (e) hindbrain, or (f) placenta 5-HT levels found between Ala56 and wild-type concepti (n=11–12/genotype) from heterozygous dams. *p<0.05.
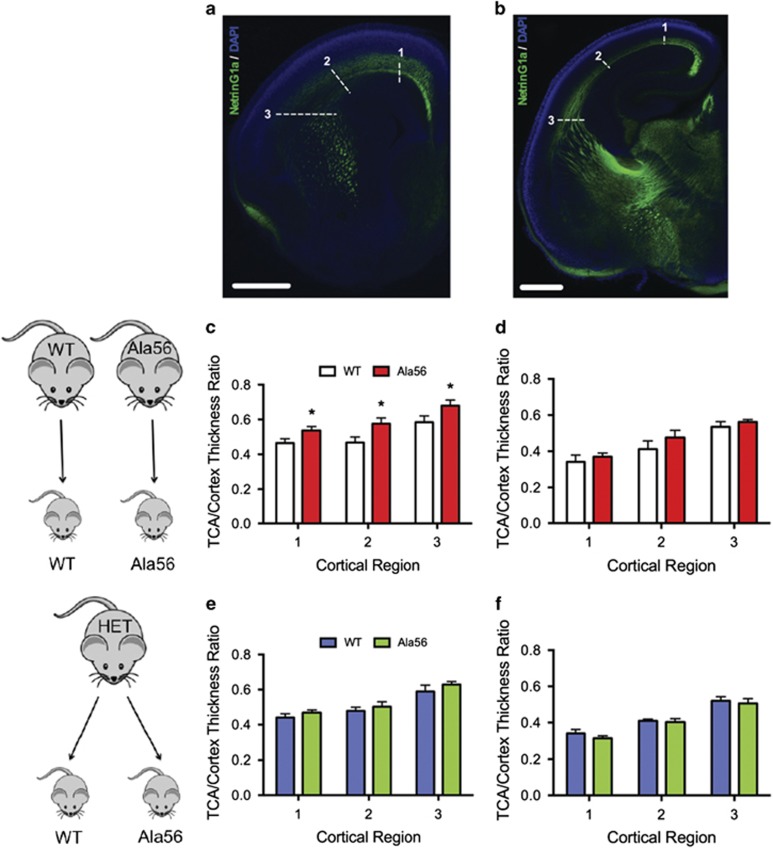
To examine potential downstream effects of altered fetal forebrain 5-HT levels at E14.5, we performed NetrinG1a immunostaining of thalamocortical axons (TCA), since these projections are sensitive to changes in 5-HT signaling during neurodevelopment (Bonnin et al, 2007; Chen et al, 2015). The normalized breadth of TCA projections was significantly increased in rostral brain regions of embryos from Ala56 dams compared with embryos from wild-type dams (repeated measures two-way ANOVA F=10.66, p=0.031; Figure 3c), with no significant difference observed in caudal regions (F=0.83, p=0.41; Figure 3d). Similar to HPLC data, Ala56 and wild-type littermates from heterozygous dams exhibited no genotype differences in normalized TCA trajectory thickness (Both p>0.3; Figure 3e and f).
Figure 3.
Maternal SERT Ala56 genotype effects on thalamocortical axon breadth. NetrinG1a immunostaining (a and b) revealed no gross morphology changes in TCA development at E18.5. In homozygous Ala56 or wild-type (WT) dams carrying Ala56 or wild-type embryos, respectively, however, repeated-measures two-way ANOVA indicated that the TCA/cortex thickness ratio in Ala56 rostral brain sections was significantly increased compared to wild-type, with Bonferroni post hoc test demonstrating significance at each of the three TCA points measured (n=5/genotype group) (c). This phenotype was attenuated in caudal brain sections (d). There were no differences in the TCA/cortex thickness ratio (e and f) between the wild-type (n=5) and Ala56 (n=5) littermates from heterozygous dams. White numbers and dashed lines in (a) and (b) indicate cortical regions measured. Scale bar, 500 μm. TCAs, thalamocortical axons. *p<0.05 relative to WT.
To evaluate placenta synthesis as a possible source of decreased 5-HT levels, we examined TPH activity using an in vitro preparation. 5-HTP production was unchanged in E14.5 placentas from Ala56 dams compared to wild-types (t=0.74, p=0.48; Figure 4a). RT-PCR also indicated no difference in Tph1 expression (t=1.23, p=0.26; Figure 4b). Similarly, no effect of embryo genotype was observed in 5-HTP production or Tph1 expression in placentas from heterozygous dams (both p>0.2; Figure 4d and e).
Figure 4.
Maternal SERT Ala56 genotype effects on placental 5-HT. At E14.5, there were no significant differences in 5-HTP neosynthesis between Ala56 and wild-type (WT) placentas in the homozygous (a) or heterozygous (d) dam comparison (n=6 per genotype group). Likewise, Tph1 expression was unchanged between the Ala56 and wild-type placentas in the homozygous (b) or heterozygous (e) dam comparison (n=5 per genotype group, data presented as mean±upper/lower limit). Two-way ANOVA indicated a maternal genotype effect on placental tissue weight, with placentas harvested from Ala56 dams weighing more than samples from WT dams, with Bonferroni post hoc test significant at E14.5 (c). No significant effects of embryo genotype were seen in placentas from heterozygous dams (f). Maternal blood 5-HT levels were significantly higher in the Ala56 dams compared with the wild-type dams at E14.5 (g). Overall, however, two-way repeated measures ANOVA indicated there were no significant main effects of genotype for whole-blood 5-HT (h) or (i) tryptophan (TRP) levels when measured from 7 days before pregnancy (NP, non-partum), throughout pregnancy, and extending to 7 days postpartum (PP) between the Ala56 (n=7) and wild-type (n=7) dams. *p<0.05 relative to WT.
Although dams were perfused before harvest, placentas inevitably contain some maternal blood, which could potentially contribute to the decreased placenta 5-HT levels. A longitudinal study of SERT Ala56 and WT dams indicated that whole blood 5-HT levels were dynamic during pregnancy (main effect of time point F=15.96, p<0.0001), with no main effect of maternal genotype across pregnancy or interaction effect as determined by two-way repeated-measures ANOVA (main effect of genotype F=0.35, p=0.56; interaction effect F=1.17, p=0.34; Figure 4h). At E14.5, however, maternal blood 5-HT levels were higher in Ala56 dams than in wild-type dams (t=2.14, p=0.042; Figure 4g). Alternatively, decreases in maternally derived tryptophan (TRP) could reduce placental 5-HT neosynthesis. Maternal blood TRP levels were also dynamic during pregnancy (main effect of time point F=8.46, p<0.0001), but no genotype differences or interaction effects were found (main effect of genotype F=0.12, p=0.73; interaction effect F=0.57, p=0.69; Figure 4i), nor were TRP levels different in E14.5 or E18.5 placental tissue (Supplementary Figure 3). Interestingly, we did observe an increase in tissue weight in E14.5 and E18.5 placentas harvested from Ala56 dams compared with placentas from wild-type dams (main effect of genotype F=8.32, p=0.005; Figure 4c); whereas no significant difference in weight was observed in placentas harvested from heterozygous dams (main effect of genotype F=0.18, p=0.67; Figure 4f).
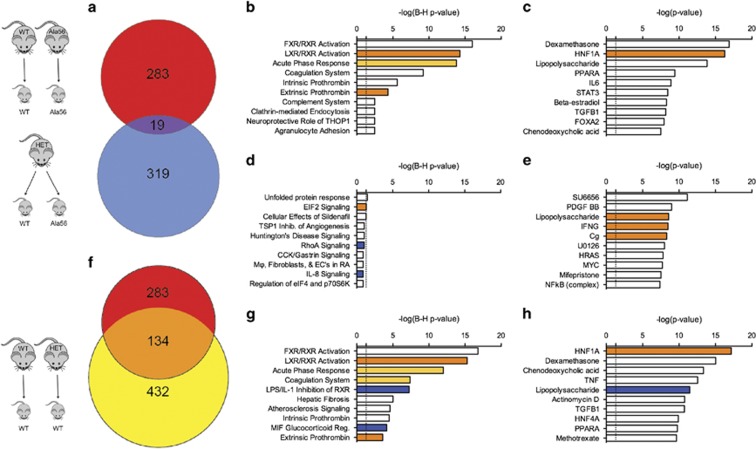
As an unbiased approach to examine maternal and embryo genotype-mediated effects on placental development, we performed whole transcriptome sequencing on E14.5 placentas. In our first comparison of interest, we identified 283 differentially expressed genes (DEGs), with 141 upregulated and 142 downregulated genes noted in placentas from Ala56 dams relative to wild-type (Supplementary Table 1). In the littermate sample comparison, 319 DEGs were identified in Ala56 placentas from heterozygous dams, with 138 upregulated and 181 downregulated genes relative to wild-type samples from the same dams (Supplementary Table 2). No single gene comparison survived false discovery rate analysis. There was significant overlap between the DEGs found in each comparison group, with 2.5-fold enrichment compared with the overlap expected by chance (Fisher's exact test, p=0.0002; Figure 5a). As an alternative strategy to evaluate maternal genotype effects, we compared gene expression changes in wild-type placentas from heterozygous dams compared with wild-type placentas from wild-type dams. Despite possessing genetically identical wild-type SERT, placentas from heterozygous dams had 432 DEGs relative to placentas from wild-type dams (Supplementary Table 3). There was also significant overlap between the 432 DEGs identified in this comparison and the 283 DEGs noted in placentas from Ala56 dams relative to placentas from wild-type dams. The 134 DEGs in common between the two comparisons represents a 9.6-fold enrichment compared with that expected by chance (Fisher's exact p<0.0001 in comparison with 432 genes differentially expressed compared with 13 431 transcripts identified, Figure 5f). The DEGs observed in placentas differing at both maternal and embryo genotypes (Ala56 embryos from Ala56 dams vs WT embryos from WT dams) therefore have much greater overlap with placentas with differing maternal genotypes than with placentas with differing embryo genotypes (5.2-fold greater overlap, Fisher's exact p<0.0001).
Figure 5.
Examination of maternal and embryonic SERT Ala56 genotype effects on E14.5 placental gene expression. In the homozygous and heterozygous dam comparisons, 283 and 319 differentially expressed genes (DEGs) were identified in Ala56 placentas relative to wild-type, respectively. Only 19 DEGs (a) overlapped between the two group comparisons. Conversely, of the 432 DEGs identified when comparing the wild-type placentas from heterozygous and the wild-type dams, 134 DEGs overlapped with the homozygous dam group comparison (f). Ingenuity Pathway Analysis (IPA) identified DEG-enriched canonical pathways and predicted upstream regulators, with the top 10 shown for the homozygous dam (b and c), heterozygous dam (d and e), and wild-type (g and h) data sets. Light and dark orange bars are moderately and strongly predicted upregulated pathways/networks, respectively. Blue bars are strongly predicted downregulated pathways/networks. White bars indicate that the direction of activation could not be predicted based on the direction of regulation of DEGs within the pathways/networks. Vertical dashed line indicates threshold required to meet statistical significance (p<0.05) for pathway/network enrichment.
We used Ingenuity Pathway Analysis (IPA) to identify biological pathways and molecular networks that were significantly impacted by maternal and embryo genotype. Indicative of the distinct DEG populations identified in our initial analysis, IPA highlighted enrichment of genes in canonical pathways that were unique to the dam genotype comparison (Figure 5b) vs those in the embryo-only genotype comparison (Figure 5d). Furthermore, the magnitude of DEG enrichment in canonical pathways found in placentas from Ala56 dams was significantly greater, with only the top three pathways in the littermate sample comparison reaching Benjamini–Hochberg corrected statistical thresholds (Figure 5d). Strikingly, wild-type placentas from heterozygous dams and Ala56 placentas from Ala56 dams exhibited similar DEG enrichment in canonical pathways relative to wild-type placentas from wild-type dams, with the same top four pathways in both comparisons (Figure 5g).
In addition to evaluating DEG enrichment in canonical pathways, we also carried out network-based inquiries, which identify direct and indirect relationships between DEGs. Potential upstream regulators (endogenous or exogenous) can be predicted by examining patterns of gene expression. Mirroring the IPA canonical pathway findings, there were clear differences in the predicted upstream regulators in the dam genotype comparison when compared to the embryo genotype comparison (Figure 5c and e). In contrast, there was substantial overlap between the upstream regulators predicted for gene expression changes in wild-type placentas from heterozygous dams and Ala56 placentas from Ala56 dams relative to placentas from wild-type dams (Figure 5h). Dexamethasone, a synthetic glucocorticoid receptor (GR) agonist, and the transcription factor, hepatocyte nuclear factor 1 alpha (HNF1A), were the top-rated regulators for both data sets.
DISCUSSION
This is the first study to demonstrate that maternal genotype at an autism-associated gene variant affects forebrain 5-HT levels and neurodevelopment. Maternal effects on fetal development have been explored in other mouse models related to autism, but these studies have focused on environmental stressors that impact the prenatal environment (Bronson and Bale, 2014; Malkova et al, 2012). Recent studies related to mood and anxiety have indicated that genetic factors may also influence neurodevelopment by altering maternal physiology during pregnancy (Toth, 2014). Interestingly, maternal genotype effects on fetal development have been reported in offspring of 5-HT1A and TPH1 null mice (Cote et al, 2007; Gleason et al, 2010). Although very useful for understanding these components of the 5-HT system, complete loss of these critical proteins does not occur in the human population. Our findings demonstrate that maternal expression of an autism-associated SERT amino acid variant can perturb embryonic 5-HT homeostasis and alter neurodevelopment.
Based upon the time course of diminished forebrain 5-HT levels in embryos from SERT Ala56 dams, axonal projections from the thalamus into the cortex would be exposed to altered 5-HT levels. These glutamatergic projections transiently express a number of key components of the 5-HT system, most notably SERT and 5-HT1B/1D receptors, and are sensitive to altered extracellular 5-HT in the fetal forebrain (Bonnin et al, 2007; Chen et al, 2015). Decreased forebrain 5-HT levels, as found in embryos from Ala56 dams, may alter TCA 5-HT1B/1D signaling, thereby affecting overall thalamocortical trajectory. The observed rostral-caudal gradient of altered TCA projection breadth aligns with the previously described gradient in 5-HT receptor expression in the anterior and posterior dorsal thalamic axons (Bonnin et al, 2007). Importantly, target regions for these projections include the somatosensory cortex. The original linkage study found association between the SERT Ala56 allele and sensory aversion (Sutcliffe et al, 2005), which is paralleled by recent findings of association between high-expressing SERT genotypes with tactile hyperresponsiveness in ASD (Schauder et al, 2015), potentially quite relevant to the current neurodevelopmental findings.
The impact of maternal SERT genotype on forebrain 5-HT levels is surprising because previous work found that 5-HT does not cross the maternal–fetal barrier (Robson and Senior, 1964), and its synthesis in the placenta is dependent upon embryo genotype (Bonnin et al, 2011). The observed developmental trajectory of forebrain 5-HT, however, suggests that altered maternal 5-HT homeostasis impacts the placenta. At E14.5, during a developmental time period when the placenta provides the main source of forebrain 5-HT (Bonnin et al, 2011), 5-HT levels were specifically decreased in the forebrain of embryos from Ala56 dams. Furthermore, E14.5 placentas from Ala56 dams also showed parallel decreases in 5-HT, suggesting altered placental 5-HT synthesis, handling, or release caused reduced forebrain 5-HT levels. Complementing these findings, forebrain 5-HT levels in embryos from Ala56 dams were normalized at E18.5, when endogenous sources of 5-HT originating from the midbrain raphe nuclei are predominant in the forebrain (Bonnin et al, 2011). In contrast, placenta 5-HT remained decreased at E18.5, indicating that alterations in placental function persist throughout pregnancy in Ala56 dams. Supporting this idea, we did observe a small but significant increase in placenta weight from Ala56 dams compared with wild-type dams.
Serotonin production in the placenta does not appear to account for the maternal genotype effect, as neither tryptophan hydroxylase mRNA expression nor tryptophan hydroxylase activity were altered (Amireault et al, 2013). Within the placenta, SERT is present in the labrynth zone, where cells are of fetal origin, and this expression reaches high levels only later in development (~E17.5; Wu et al, 2016). This suggests that increased maternal SERT function indirectly affects placental handling and storage of 5-HT. Although little is known about the mechanisms that regulate the delivery of placental 5-HT into fetal circulation, future investigations are warranted in ex vivo placenta perfusion preparations that can examine placenta physiology in the context of different environmental and genetic manipulations (Goeden and Bonnin, 2013).
We observed dynamic changes in maternal whole blood 5-HT levels throughout pregnancy. Our previous work identified elevated blood 5-HT levels in male SERT Ala56 mice, but the female mice studied here showed no appreciable difference before or after pregnancy. These findings suggest a sexual dimorphism in SERT's influence on blood 5-HT levels, consistent with a previous study in the human population (Weiss et al, 2005). Interestingly, SERT Ala56 dams exhibited a transient increase in blood 5-HT levels at E14.5. As maternal 5-HT cannot cross the placenta into fetal circulation (Bonnin et al, 2011; Robson and Senior, 1964), it is unlikely that changes in maternal 5-HT blood levels would directly affect placental or fetal forebrain 5-HT levels. Syncytiotrophoblasts, the cellular interface between the maternal and fetal blood supply in the placenta, are thought to tightly regulate free 5-HT via MAOA degradation (Wu et al, 2016). Owing to technical limitations, we were unable to measure 5-HT turnover in the placenta. As a result, the possibility remains that changes in maternal whole blood 5-HT levels could disrupt placental 5-HT degradation mechanisms in syncytiotrophoblasts, which could indirectly affect storage and transfer of 5-HT to the fetus.
Our RNA sequencing data indicate that altered maternal SERT function causes a broad array of placental changes. In contrast, embryo genotype led to minimal enrichment in known canonical signaling pathways and molecular networks, which may reflect the peak in SERT placenta expression later in gestation (Verhaagh et al, 2001). In contrast, wild-type placentas from heterozygous dams and placentas from Ala56 dams exhibited substantial overlap in differentially expressed genes, as well as canonical pathway and network enrichment, suggesting that a single maternal SERT Ala56 allele may be sufficient to impact placental function. The top canonical pathways identified point to potential metabolic abnormalities in placentas from Ala56 dams (Hong and Tontonoz, 2014; Janowski et al, 1996; Kuipers et al, 2014; Parks et al, 1999). We also observed enrichment in gene expression pathways related to coagulation and the innate immune system. These systems have well-documented interactions with one another as critical players in the initial inflammatory response to infectious or noxious stressors (Cray, 2012; Li and Huang, 2009b). These findings were reflected in dexamethasone, a commonly used immunosuppressant, being predicted as an upstream regulator of differentially expressed genes in our data set.
The precise mechanism underlying the maternal genotype effect on fetal forebrain 5-HT levels at E14.5 remains unclear. However, our RNA sequencing data suggests that maternal carriers of the SERT Ala56 variant possess changes in metabolism and the innate immune system, which have known pathological effects on the placenta and offspring (Balsells et al, 2012; Hsiao and Patterson, 2011; Li and Huang, 2009b; Radaelli et al, 2003). Interestingly, evidence has emerged directly connecting these systems to 5-HT homeostasis in the placenta. Impaired insulin signaling, as found in gestational diabetes, decreases 5-HT uptake in trophoblasts by disrupting plasma membrane SERT expression (Li et al, 2014). Maternal immune activation (MIA) increases placental production of 5-HT; although this effect is at least partially owing to increased placental tryptophan hydroxylase activity (Goeden et al, 2016).
Although placental pathology has been suggested in ASD (Anderson et al, 2007; Hsiao and Patterson, 2012; Walker et al, 2013, 2015), the mechanisms supporting these observations remain unclear. Our findings suggest that ASD-associated risk alleles, such as SERT Gly56Ala, could cause placental dysfunction in maternal carriers. Few studies have examined the potential effects of maternal genetic variation in ASD. In the only study to focus on the effects of maternal SERT genotype, Kistner and colleagues found increased risk of ASD in the offspring of mothers with the high-expressing SLC6A4 5-HTTLPR long allele (Kistner-Griffin et al, 2011). Two groups have performed underpowered genome-wide association studies in mothers of affected children (Tsang et al, 2013; Yuan and Dougherty, 2014). Although both studies failed to detect common SNPs that reached genome-wide significance, the number of suggestive associations that did not overlap with their proband analyses indicated that true maternal effects were most likely below their statistical ability to detect.
Our findings have some important limitations. First, we have identified parallel changes in placenta and forebrain 5-HT levels, as well as thalamocortical neuroanatomy, but the underlying mechanisms remain unclear. In addition, we focused on embryonic development, with questions remaining about long-term impact on sensory function and behavior. Further work is necessary to directly connect changes in maternal SERT function to altered thalamic connectivity and sensory behavior, but these findings suggest that altered TCA development observed in SERT Ala56 mice may be relevant to pathophysiology underlying sensory dysfunction in ASD. Finally, maternal genotype may interact with embryo genotype to lead to neurodevelopmental phenotypes, a possibility that we hope to evaluate in future studies.
In conclusion, we have demonstrated a maternal SERT genotype effect on fetal development in offspring of SERT Ala56 dams. Though the patterns of fetal abnormalities indicate disruption of the placental 5-HT system in SERT Ala56 dams, there is no apparent maternal effect on tryptophan hydroxylase function. Instead, we found evidence that enhanced and dysregulated maternal SERT function causes changes that may arise from broader metabolic changes and altered immune signaling in the placenta, adding to a growing body of literature that suggests these systems play a role in the etiology of ASD. Collectively, our findings indicate that ASD susceptibility factors, such as the Ala56 variant, may increase risk for the disorder by altering the maternal prenatal environment.
FUNDING AND DISCLOSURE
This work was funded, in part, by NIH grant MH094604 (JV-V), an American Academy of Child and Adolescent Psychiatry Junior Investigator Award (JV-V), and an Autism Speaks Weatherstone Predoctoral Fellowship (CLM). Core laboratory services were supported by the NICHD Grant P30 HD15052 to the Vanderbilt Kennedy Center for Research on Human Development. Dr Veenstra-VanderWeele has consulted with Roche, Novartis, and SynapDx, and has received research funding from Roche, Novartis, SynapDx, Seaside Therapeutics, and Forest. Dr Blakely has consulted with Forest, Lundbeck, Wyeth, Pfizer, and Prexa and has received research funding from Forest, Lundbeck, Wyeth, and Prexa. The remaining authors declare no competing financial interests.
Acknowledgments
The authors would like to thank Bing Zhang, PhD, and Jing Wang, PhD, for biostatistics and bioinformatics assistance.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Amireault P, Sibon D, Cote F (2013). Life without peripheral serotonin: insights from tryptophan hydroxylase 1 knockout mice reveal the existence of paracrine/autocrine serotonergic networks. ACS Chem Neurosci 4: 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W et al (2013). Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 8: 1765–1786. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2014). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GM, Feibel FC, Cohen DJ (1987). Determination of serotonin in whole blood, platelet-rich plasma, platelet-poor plasma and plasma ultrafiltrate. Life Sci 40: 1063–1070. [DOI] [PubMed] [Google Scholar]
- Anderson GM, Jacobs-Stannard A, Chawarska K, Volkmar FR, Kliman HJ (2007). Placental trophoblast inclusions in autism spectrum disorder. Biol Psychiatry 61: 487–491. [DOI] [PubMed] [Google Scholar]
- Balsells M, Garcia-Patterson A, Gich I, Corcoy R (2012). Major congenital malformations in women with gestational diabetes mellitus: a systematic review and meta-analysis. Diabetes Metab Res Rev 28: 252–257. [DOI] [PubMed] [Google Scholar]
- Bonnin A, Goeden N, Chen K, Wilson ML, King J, Shih JC et al (2011). A transient placental source of serotonin for the fetal forebrain. Nature 472: 347–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnin A, Peng W, Hewlett W, Levitt P (2006). Expression mapping of 5-HT1 serotonin receptor subtypes during fetal and early postnatal mouse forebrain development. Neuroscience 141: 781–794. [DOI] [PubMed] [Google Scholar]
- Bonnin A, Torii M, Wang L, Rakic P, Levitt P (2007). Serotonin modulates the response of embryonic thalamocortical axons to netrin-1. Nature Neurosci 10: 588–597. [DOI] [PubMed] [Google Scholar]
- Bronson SL, Bale TL (2014). Prenatal stress-induced increases in placental inflammation and offspring hyperactivity are male-specific and ameliorated by maternal antiinflammatory treatment. Endocrinology 155: 2635–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases O, Vitalis T, Seif I, De Maeyer E, Sotelo C, Gaspar P (1996). Lack of barrels in the somatosensory cortex of monoamine oxidase A-deficient mice: role of a serotonin excess during the critical period. Neuron 16: 297–307. [DOI] [PubMed] [Google Scholar]
- Chen X, Ye R, Gargus JJ, Blakely RD, Dobrenis K, Sze JY (2015). Disruption of transient serotonin accumulation by non-serotonin-producing neurons impairs cortical map development. Cell Rep 10: 346–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote F, Fligny C, Bayard E, Launay JM, Gershon MD, Mallet J et al (2007). Maternal serotonin is crucial for murine embryonic development. Proc Natl Acad Sci USA 104: 329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cray C (2012). Acute phase proteins in animals. Prog Mol Biol Transl Sci 105: 113–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Cook EH Jr, Coon H, Dawson G, Grigorenko EL, McMahon W et al (2005). Autism and the serotonin transporter: the long and short of it. Mol Psychiatry 10: 1110–1116. [DOI] [PubMed] [Google Scholar]
- Gabriele S, Sacco R, Persico AM (2014). Blood serotonin levels in autism spectrum disorder: a systematic review and meta-analysis. Eur Neuropsychopharmacol 24: 919–929. [DOI] [PubMed] [Google Scholar]
- Gaspar P, Cases O, Maroteaux L (2003). The developmental role of serotonin: news from mouse molecular genetics. Nat Rev Neurosci 4: 1002–1012. [DOI] [PubMed] [Google Scholar]
- Gleason G, Liu B, Bruening S, Zupan B, Auerbach A, Mark W et al (2010). The serotonin1A receptor gene as a genetic and prenatal maternal environmental factor in anxiety. Proc Natl Acad Sci USA 107: 7592–7597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeden N, Bonnin A (2013). Ex vivo perfusion of mid-to-late-gestation mouse placenta for maternal-fetal interaction studies during pregnancy. Nat Protoc 8: 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeden N, Velasquez J, Arnold KA, Chan Y, Lund BT, Anderson GM et al (2016). Maternal inflammation disrupts fetal neurodevelopment via increased placental output of serotonin to the fetal brain. J Neurosci 36: 6041–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C, Tontonoz P (2014). Liver X receptors in lipid metabolism: opportunities for drug discovery. Nat Rev Drug Discov 13: 433–444. [DOI] [PubMed] [Google Scholar]
- Hsiao EY, Patterson PH (2011). Activation of the maternal immune system induces endocrine changes in the placenta via IL-6. Brain Behav Immun 25: 604–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao EY, Patterson PH (2012). Placental regulation of maternal-fetal interactions and brain development. Dev Neurobiol 72: 1317–1326. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ (1996). An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature 383: 728–731. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner-Griffin E, Brune CW, Davis LK, Sutcliffe JS, Cox NJ, Cook EH Jr (2011). Parent-of-origin effects of the serotonin transporter gene associated with autism. Am J Med Genet B Neuropsychiatr Genet 156: 139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers F, Bloks VW, Groen AK (2014). Beyond intestinal soap—bile acids in metabolic control. Nat Rev Endocrinol 10: 488–498. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al (2009a). The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Huang SJ (2009b). Innate immunity, coagulation and placenta-related adverse pregnancy outcomes. Thromb Res 124: 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hadden C, Singh P, Mercado CP, Murphy P, Dajani NK et al (2014). GDM-associated insulin deficiency hinders the dissociation of SERT from ERp44 and down-regulates placental 5-HT uptake. Proc Natl Acad Sci USA 111: E5697–E5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova NV, Yu CZ, Hsiao EY, Moore MJ, Patterson PH (2012). Maternal immune activation yields offspring displaying mouse versions of the three core symptoms of autism. Brain Behav Immun 26: 607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40: 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller CL, Anacker AM, Veenstra-VanderWeele J (2015). The serotonin system in autism spectrum disorder: From biomarker to animal models. Neuroscience 321: 24–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA et al (1999). Bile acids: natural ligands for an orphan nuclear receptor. Science 284: 1365–1368. [DOI] [PubMed] [Google Scholar]
- Prasad HC, Steiner JA, Sutcliffe JS, Blakely RD (2009). Enhanced activity of human serotonin transporter variants associated with autism. Philos Trans Roy Soc Lond B Biol Sci 364: 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radaelli T, Varastehpour A, Catalano P, Hauguel-de Mouzon S (2003). Gestational diabetes induces placental genes for chronic stress and inflammatory pathways. Diabetes 52: 2951–2958. [DOI] [PubMed] [Google Scholar]
- Robson JM, Senior JB (1964). The 5-hydroxytryptamine content of the placenta and foetus during pregnancy in mice. Br J Pharmacol Chemother 22: 380–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Reichert J, Hoffman EJ, Cai G, Jones HB, Faham M et al (2008). A large-scale screen for coding variants in SERT/SLC6A4 in autism spectrum disorders. Autism Res 1: 251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder KB, Muller CL, Veenstra-VanderWeele J, Cascio CJ (2015). Genetic variation in serotonin transporter modulates tactile hyperresponsiveness in ASD. Res Autism Spectr Disord 10: 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M et al (2006). The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JS, Delahanty RJ, Prasad HC, McCauley JL, Han Q, Jiang L et al (2005). Allelic heterogeneity at the serotonin transporter locus (SLC6A4) confers susceptibility to autism and rigid-compulsive behaviors. Am J Hum Genet 77: 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth M (2014). Mechanisms of non-genetic inheritance and psychiatric disorders. Neuropsychopharmacology 40: 129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang KM, Croen LA, Torres AR, Kharrazi M, Delorenze GN, Windham GC et al (2013). A genome-wide survey of transgenerational genetic effects in autism. PLoS One 8: e76978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra-VanderWeele J, Jessen TN, Thompson BJ, Carter M, Prasad HC, Steiner JA et al (2009). Modeling rare gene variation to gain insight into the oldest biomarker in autism: construction of the serotonin transporter Gly56Ala knock-in mouse. J Neurodev Disord 1: 158–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra-VanderWeele J, Muller CL, Iwamoto H, Sauer JE, Owens WA, Shah CR et al (2012). Autism gene variant causes hyperserotonemia, serotonin receptor hypersensitivity, social impairment and repetitive behavior. Proc Natl Acad Sci USA 109: 5469–5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaagh S, Barlow DP, Zwart R (2001). The extraneuronal monoamine transporter Slc22a3/Orct3 co-localizes with the Maoa metabolizing enzyme in mouse placenta. Mech Dev 100: 127–130. [DOI] [PubMed] [Google Scholar]
- Walker CK, Anderson KW, Milano KM, Ye S, Tancredi DJ, Pessah IN et al (2013). Trophoblast inclusions are significantly increased in the placentas of children in families at risk for autism. Biol Psychiatry 74: 204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CK, Krakowiak P, Baker A, Hansen RL, Ozonoff S, Hertz-Picciotto I (2015). Preeclampsia, placental insufficiency, and autism spectrum disorder or developmental delay. JAMA Pediatr 169: 154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Abney M, Cook EH Jr, Ober C (2005). Sex-specific genetic architecture of whole blood serotonin levels. Am J Hum Genet 76: 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HH, Choi S, Levitt P (2016). Differential patterning of genes involved in serotonin metabolism and transport in extra-embryonic tissues of the mouse. Placenta 42: 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonan AL, Alarcon M, Cheng R, Magnusson PK, Spence SJ, Palmer AA et al (2003). A genomewide screen of 345 families for autism-susceptibility loci. Am J Hum Genet 73: 886–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Dougherty JD (2014). Investigation of maternal genotype effects in autism by genome-wide association. Autism Res 7: 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.