Abstract
APOBEC3s (A3s) are single-stranded DNA cytosine deaminases that provide innate immune defences against retroviruses and mobile elements. A3s are specific to eutherian mammals because no direct homologs exist at the syntenic genomic locus in metatherian (marsupial) or prototherian (monotreme) mammals. However, the A3s in these species have the likely evolutionary precursors, the antibody gene deaminase AID and the RNA/DNA editing enzyme APOBEC1 (A1). Here, we used cell culture-based assays to determine whether opossum A1 restricts the infectivity of retroviruses including human immunodeficiency virus type 1 (HIV-1) and the mobility of LTR/non-LTR retrotransposons. Opossum A1 partially inhibited HIV-1, as well as simian immunodeficiency virus (SIV), murine leukemia virus (MLV), and the retrotransposon MusD. The mechanism of inhibition required catalytic activity, except for human LINE1 (L1) restriction, which was deamination-independent. These results indicate that opossum A1 functions as an innate barrier to infection by retroviruses such as HIV-1, and controls LTR/non-LTR retrotransposition in marsupials.
Apolipoprotein B (apoB) mRNA editing enzyme catalytic subunit 1 (APOBEC1, A1) is a cytidine deaminase that physiologically edits apoB mRNA, which encodes a key protein involved in lipid transport1,2,3. A1 is a member of the AID/APOBEC family, which catalyses the conversion of cytosines to uracils within single-stranded DNA and RNA polynucleotides4,5. In this family, the APOBEC3 members (A3s; mainly human A3F and A3G) are well-characterised innate immune effector proteins that restrict the spread of retroviruses and LTR/non-LTR retrotransposons4,6,7,8.
Cell culture experiments have demonstrated that A1 enzymes are capable of inhibiting the replication of HIV-1, regardless of the presence of the HIV-1 Vif protein9,10,11,12. Analogous to A3 enzymes, A1 is encapsidated into assembling viral particles, and deaminates cytosines to uracils in nascent single-stranded viral cDNAs during reverse transcription10,11. This activity results in hallmark genomic strand G-to-A mutations. In addition, genomic strand C-to-T mutations are readily detectable indicating that A1 enzymes also have the capacity to edit viral genomic RNA10,11,12. Further cell culture studies have shown that A1 enzymes can suppress the infectivity of several viruses such as SIV, feline immunodeficiency virus (FIV), MLV, hepatitis B virus (HBV), and herpes simplex virus 1 (HSV-1)11,13,14,15,16 and the mobility of autonomous retrotransposons17,18. Although the biological functions of A1 in controlling viral infection and mobile elements in vivo remain unclear, the fact that proviral DNAs recovered from HIV-1-infected rabbit macrophages contained hallmarks of A1-mediated deamination suggests that A1 is a natural barrier to retroviral infection19. However, studies in A1-null mice did not support the function of A1 in blocking Friend retroviral infection in vivo20.
The grey, short-tailed opossum, Monodelphis domestica, belongs to the Metatheria, which is one of the three major groups of modern mammals and the closest relative of the Eutheria (Fig. S1a). Although eutherians encode at least one A3 family protein, there is no A3-like gene in the genomes of non-eutherian mammals (Fig. S1b), which was confirmed by the opossum genomic DNA sequence21, a pan-species Z1 PCR analysis22 and a BLAST search in this study (Fig. S1b). These observations suggest that the contribution of A1 to the innate immunity pathways of the marsupials could be greater than their contribution in eutherian mammals.
The opossum A1 is a protein of 235 amino acids that shares ~70% amino acid identity with those encoded by eutherian mammals (Fig. S2). Although the ability of this protein to edit apoB mRNA has previously been characterised23, whether it inhibits retroviruses including HIV-1 and LTR/non-LTR mobile elements remains to be determined. We addressed these questions using cell culture-based assays and found that opossum A1 from the small intestine is capable of restricting the infectivity of several retroviruses and the mobility of LTR/non-LTR retrotransposons. Taken together, our data indicate that the ability of A1 enzymes to protect the host genome from the invasion of foreign nucleic acids is conserved in marsupial and eutherian mammals.
Results
Opossum A1 is mutagenic in Escherichia coli
Although the A1 protein encoded in marsupials can edit a synthetic apoB mRNA substrate23, its DNA-editing activity has not been determined. The bacterial mutator assay is widely used to evaluate the mutagenic activity of the AID/APOBEC family proteins17,18,24,25,26. In this assay, the expression of these proteins enhances the mutation of the bacterial RNA polymerase beta (rpoB) gene, which is reported as the frequency of rifampicin resistant (RifR) colonies. Therefore, we used this system to measure the mutagenic activity of opossum A1 by counting the RifR mutant colonies that emerged during growth of multiple independent cultures (Fig. 1a). A1 cDNA was isolated from opossum small intestine and cloned into an expression vector. Consistent with previous observations12,18, rabbit A1 increased the number of RifR colonies (24.1-fold relative to the vector control), whereas human A1 had only a slight effect (2.1-fold). Interestingly, the expression of wild-type (WT) opossum A1 in bacteria markedly enhanced the frequency of rpoB mutations (390-fold). However, this mutator phenotype was completely abrogated in catalytic mutant derivatives of opossum A1, E62A and E62Q (0.7-fold), even though they were expressed at levels similar to those of WT opossum A1 (Fig. 1b). These results demonstrate that opossum A1 has greater mutagenic activity than rabbit A1 in this E. coli-based mutation assay.
Figure 1. Mutagenic activity of opossum A1 in E. coli.
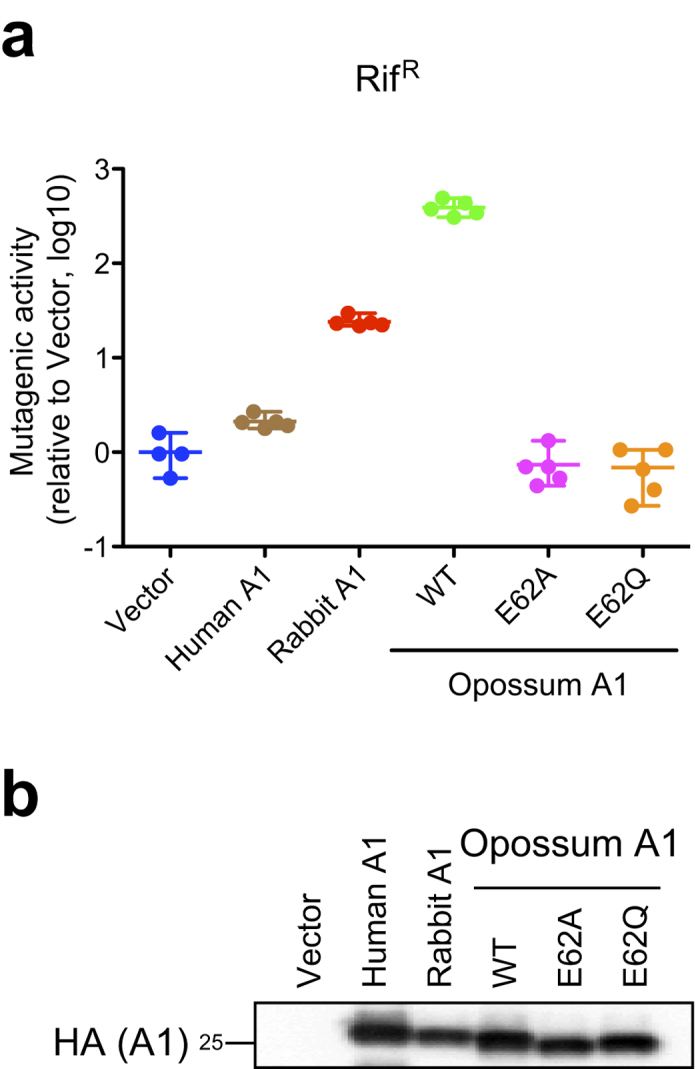
(a) Dot plots reporting the mutagenic activity of the indicated constructs (RifR colonies per 107 viable cells). Data are plotted relative to the vector control (n = 5, median with range). (b) Immunoblot of the indicated A1s expressed in bacteria.
Opossum A1 partially restricts HIV-1 infection
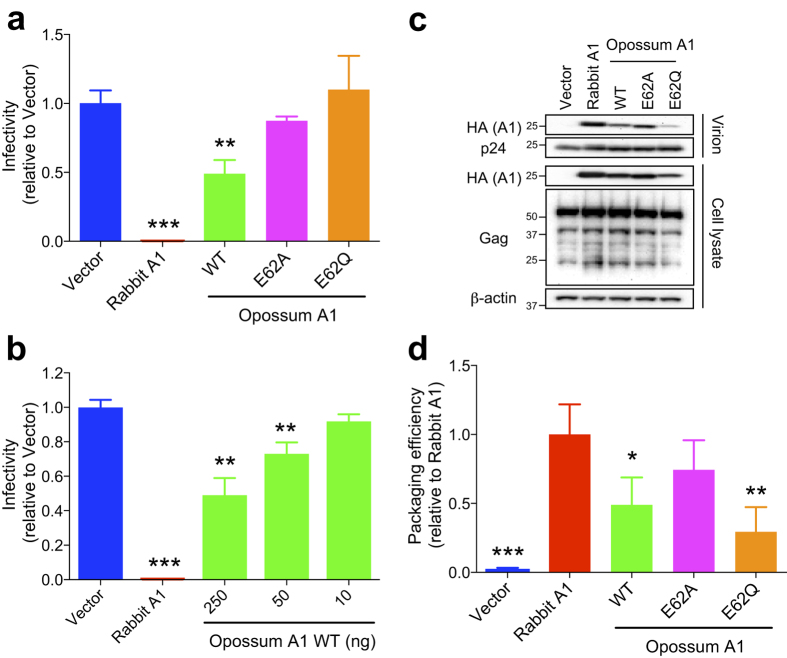
Several cell-based studies have demonstrated that the A1 proteins of rabbit and rodents inhibit HIV-1 replication independently of HIV-1 Vif expression9,10,11,12. To determine whether opossum A1 suppresses HIV-1 infectivity, we performed single-cycle HIV-1 infection assays in HEK293T cells. As expected, rabbit A1 caused a ~200-fold reduction in HIV-1 infectivity, with significant levels of A1 packaged into viral particles (Fig. 2). WT opossum A1 also inhibited HIV-1 replication compared with the vector control (~2-fold; P < 0.01), and the antiviral activity of opossum A1 was dose-dependent (Fig. 2a,b). The packaging level of the opossum protein was 2-fold lower than that of rabbit A1 (Fig. 2c,d). The antiviral activity of the WT opossum protein was abolished in the catalytic mutants, E62A and E62Q, regardless of the packaging level of the E62A mutant. These results indicate that opossum A1 partially restricts HIV-1 infectivity, and this restriction requires a catalytically active enzyme.
Figure 2. HIV-1 restriction by opossum A1.
(a) Representative single-cycle assays with WT opossum A1 or deaminase-defective mutants (E62A or E62Q). Viral infectivity is plotted relative to vector control and is proportional to luciferase activity (n = 3, average +/− SD). P values are derived by comparisons with vector control data. (b) Representative single-cycle assays with varying concentrations of WT opossum A1 (10, 50 and 250 ng). Data represent the luciferase activity relative to vector control data (n = 3, average +/− SD; P-values as above). (c) Immunoblots of A1 in viral particles and cell lysates. A1 and HIV-1 Gag were detected using anti-HA anti-p24 antibodies, respectively. β-actin expression was used a loading control. (d) A histogram showing the relative packaging efficiency of each A1 construct. Band intensity of each HA-tagged A1 was normalised to the corresponding p24 level. Packaging efficiency is shown graphically relative to the packaging of rabbit A1 (n = 3, average +/− SD). P values represent comparisons with rabbit A1 data.
Opossum A1 mainly localises to the nucleus
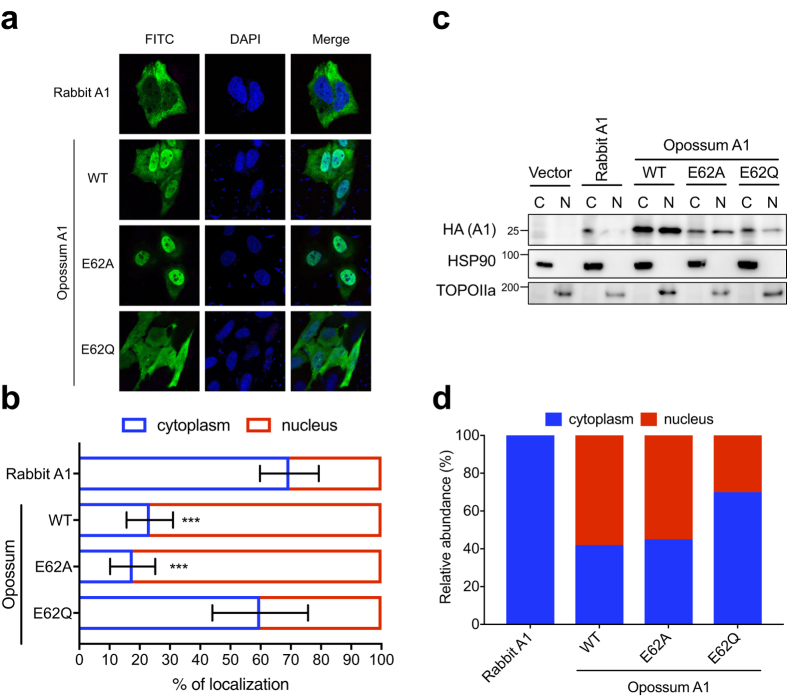
It has been demonstrated that the degree of A1 cytoplasmic localisation correlates with anti-HIV1 activity12. To assess this relationship here, we examined the subcellular localisation of opossum A1 in HeLa and HEK293T cells. The rabbit A1, which is the most active A1 protein against HIV-111, was mainly distributed in the cytoplasm of both HeLa and HEK293T cells, where ~70% of the total signal was observed (Fig. 3a,b, S3). In contrast, ~80% of the total signal for WT opossum A1 was detected in the nuclei of both HeLa and HEK293T cells. The catalytic mutant E62A displayed the same subcellular localisation in HeLa and HEK293T cells as the WT protein. However, the E62Q mutation impaired the distribution of the protein, and the mutant protein was detected in the cytoplasm, implying aberrant localisation. These findings were confirmed by immunoblotting fractionated cell extracts (Fig. 3c,d). These results suggest that the moderate antiviral activity of opossum A1 correlates, at least in part, with lower steady state cytoplasmic levels of protein in cells.
Figure 3. Subcellular localisation and expression levels of opossum A1 in HeLa cells.
(a) Representative images showing HA-tagged A1s in green (FITC) and nuclei in blue (DAPI). (b) Quantification of the subcellular distribution of the indicated A1 constructs. The percentage nuclear and cytoplasmic localisation is indicated as the graphs (n = 20, average +/− SD). P values represent comparisons with rabbit A1. (c) Nuclear (N) and cytoplasmic (C) expression levels of A1s. Each cell fraction was blotted and probed with anti-HA, anti-HSP90, and anti-TOPOIIa antibodies. (d) Relative abundance of each A1 in nuclear or cytoplasmic fractions (percentage calculated based on combined intensity of C and N bands).
Opossum A1 partially restricts SIVs and MLV
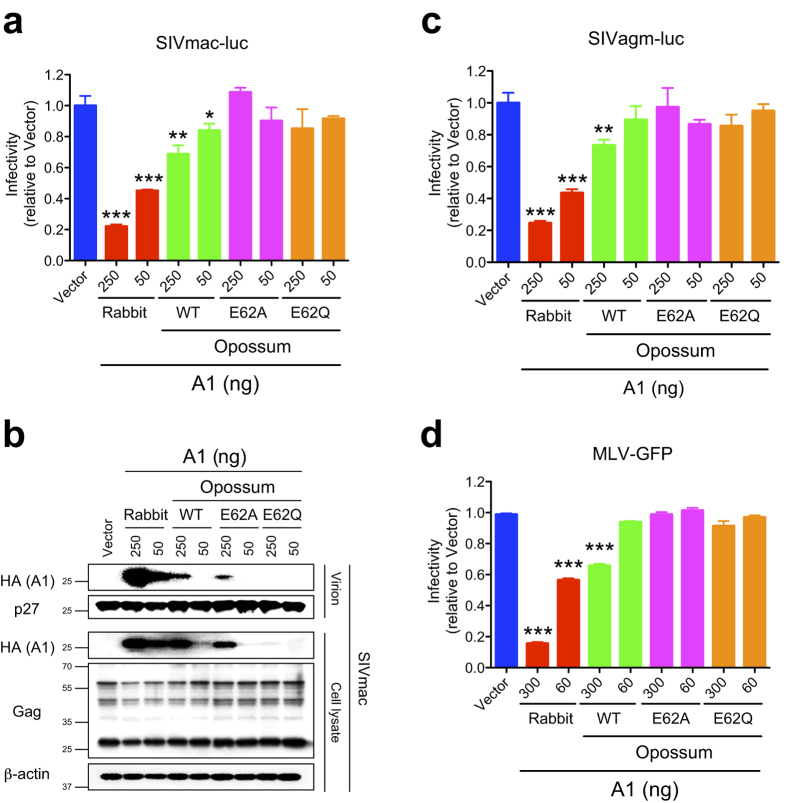
We have also demonstrated that the A1 proteins of rabbit and rodents inhibit the replication of several retroviruses including SIVs from rhesus macaque and African green monkey, and MLV11. To assess the capacity of opossum A1 to inhibit other retroviruses in addition to HIV-1, SIV and MLV pseudotyped viruses encoding a reporter gene were produced in the presence of opossum A1, and the effect of opossum A1 on retroviral replication was evaluated by infecting into HEK293T cells. Consistent with previous observation11, rabbit A1 inhibited the single cycle infection of SIVs from rhesus macaque and African green monkey (Fig. 4a to c), and MLV (Fig. 4d). A reduction in the infectivity of these viruses was also observed in the presence of opossum A1, and the degree of inhibition was similar to that of the anti-HIV-1 activity (see Fig. 2a,b versus Fig. 4a,c,d). The anti-retroviral activity against SIVs and MLV was similarly diminished by catalytic glutamate mutations (Fig. 4a,c,d), suggesting that catalytic activity is required for the restriction of these retroviruses by opossum A1. Thus, taken together with the HIV-1 data discussed above, these HEK293T-based experiments indicate that opossum A1 may have broad anti-retroviral activity.
Figure 4. Inhibition of SIV and MLV replication by opossum A1.
(a) Representative SIVmac single-cycle assays with varying concentrations (50 and 250 ng) of WT opossum A1 or deaminase-defective mutants (E62A or E62Q). Data represent the luciferase activity relative to vector control data (n = 3, average +/− SD with P values represented by asterisks). (b) Immunoblots of A1 in viral particles and cell lysates. A1 and SIVmac Gag were detected using anti-HA anti-p27 antibodies, respectively. β-actin was used a loading control. (c) Representative SIVagm single-cycle assays with varying concentrations (50 and 250 ng) of WT opossum A1 or deaminase-defective mutants. Data represent the luciferase activity relative to vector control data (n = 3, average +/− SD). P values represent comparisons with vector control data. (d) Representative MLV single-cycle assays with varying concentrations (60 and 300 ng) of WT opossum A1 or deaminase-defective mutants. Data represent the percentage of GFP+ cells relative to vector control data (n = 3, average +/− SD). P values represent comparisons with vector control data.
Opossum A1 inhibits human L1 retrotransposition using a deaminase-independent mechanism
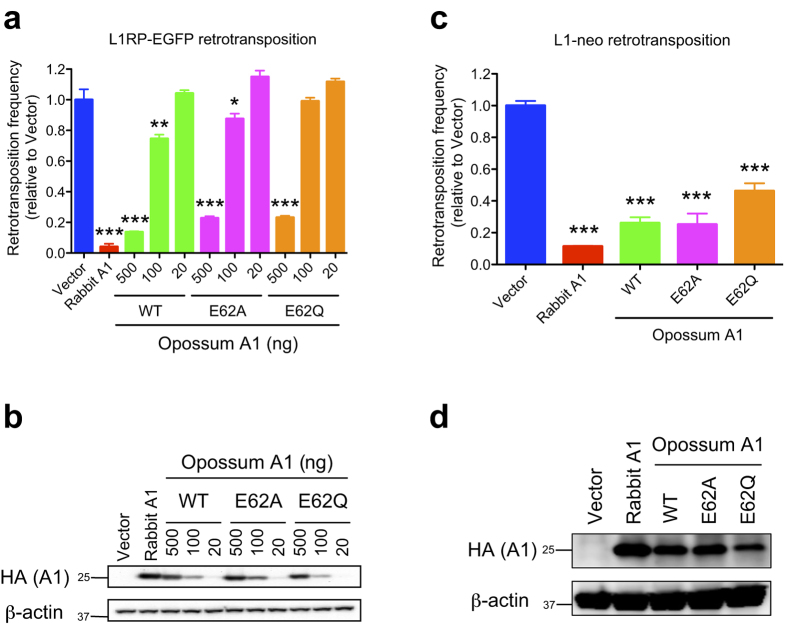
We have previously shown that the A1 homologues of eutherian mammals use a deaminase-independent mechanism to inhibit L1 retrotransposition in human cell lines18. We tested the ability of opossum A1 to inhibit human L1 retrotransposition using enhanced green fluorescent protein (EGFP)-based retrotransposition assays in HEK293T cells18,27. Consistent with our previous observation18, rabbit A1 suppressed human L1 retrotransposition (~25-fold) (Fig. 5a,b). Similarly, human L1 retrotransposition was reduced ~7.7-fold in the presence of WT opossum A1 (500 ng), and the level of suppression was proportional to the amount of enzyme expression. This anti-L1 activity was not abolished by the inactivation of the deaminase activity of opossum A1 (Fig. 5a, E62A and E62Q). The anti-L1 activity of opossum A1 was also supported by similar results with neo-marked L1 retrotransposition assays in HeLa cells (Fig. 5c,d). These data indicate that opossum A1 inhibits human L1 retrotransposition independently of its catalytic activity.
Figure 5. Restriction of L1-GFP retrotransposition by opossum A1.
(a) Representative L1RP-GFP retrotransposition assays with varying concentrations (20, 100, or 500 ng) of WT opossum A1, or its catalytic mutants (E62A or E62Q). Retrotransposition frequency is presented relative to vector control data (n = 3, average +/− SD). P values represent comparisons with vector control data. (b) Immunoblotting of the indicated HA-tagged A1s expressed in HEK293T cells, with β-actin as the loading control. (c) A representative L1-neo retrotransposition assay with opossum A1 WT, or its catalytic mutants (E62A or E62Q). Retrotransposition frequency is presented relative to vector control data (n = 3, average +/− SD). P values represent comparisons with vector control data. (d) Immunoblotting of the indicated HA-tagged A1s expressed in HEK293T cells, with β-actin as the loading control.
Opossum A1 suppresses MusD retrotransposition
It has been shown that eutherian mammal A1 proteins also inhibit the mobility of LTR retrotransposons18. To test whether this activity is present in the opossum A1, we performed a neo-marked MusD retrotransposition assay in HeLa cells. As previously observed, rabbit A1 reduced MusD retrotransposition by ~4-fold (Fig. 6a). In the presence of opossum A1, a 2-fold reduction in the MusD retrotransposition frequency was observed, and this reduction was abolished when catalytic mutants E62A and E62Q were tested (Fig. 6b). These findings indicate that opossum A1 inhibits MusD retrotransposition in a deaminase-dependent manner.
Figure 6. Suppression of MusD retrotransposition by opossum A1.
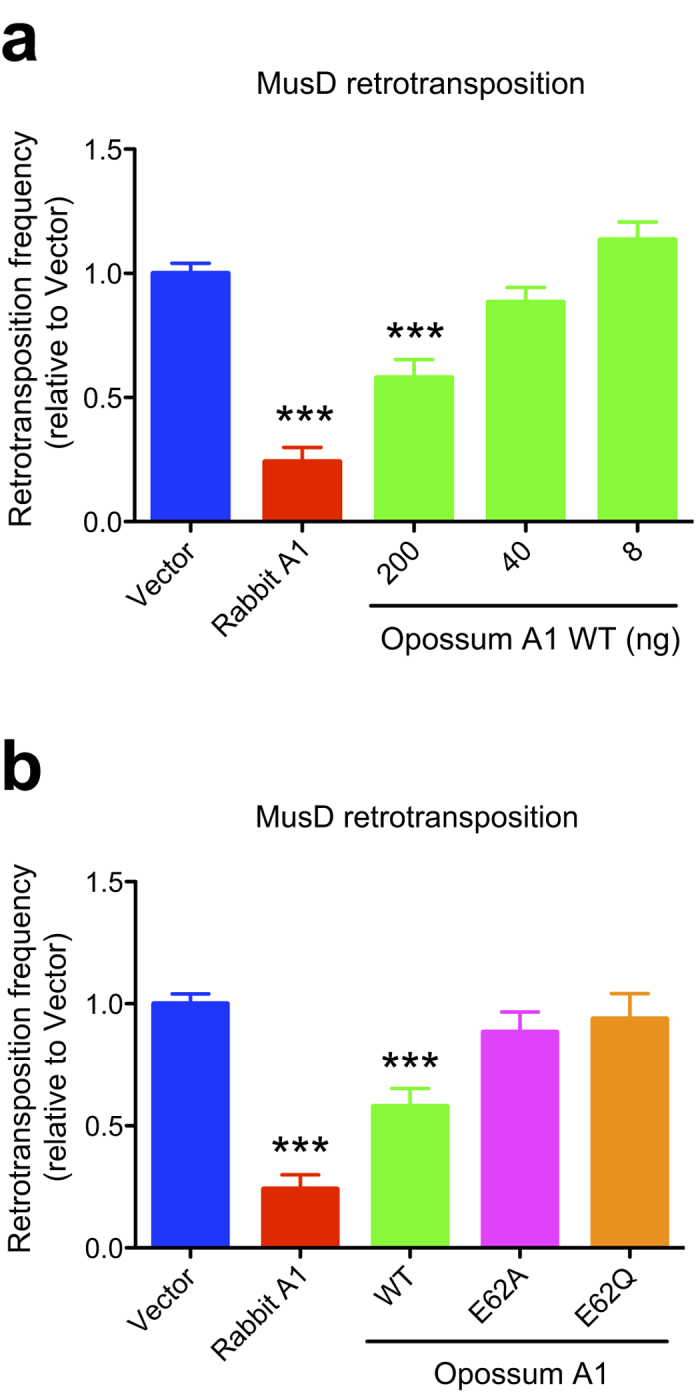
(a) Representative MusD retrotransposition results in the presence of increasing amounts of WT opossum A1 (8, 40, or 200 ng). (b) MusD retrotransposition data with WT opossum A1 or deaminase-defective mutants (E62A and E62Q). Results are presented as retrotransposition frequencies relative to vector control data (n = 3, average +/− SD).
Opossum A1 mRNA is widely expressed in primary tissues in vivo
A1 mRNAs from rabbit18, mouse18,28, and rat28,29 are broadly expressed in primary tissues such as the small intestine, spleen and gonads including both testis and ovary. In contrast, the expression of human A1 mRNA is largely limited to gastrointestinal tissues30,31. We quantified the expression level of opossum A1 mRNA in several primary opossum tissues. As anticipated, opossum A1 mRNA was abundantly expressed in the small intestine (280-fold compared with the ovary), where it appears that A1 edits its apoB mRNA (Fig. 7). Interestingly, opossum A1 mRNA was widely expressed in several tissues including the testis (22-fold) and lymphoid tissues (thymus, 4.1-fold; spleen, 3.3-fold) (Fig. 7). This expression pattern appears to be different from that of human A1, which was not reported to be expressed in the testis and lymphoid tissues30,31.
Figure 7. Expression levels of opossum A1 mRNA in primary tissues.
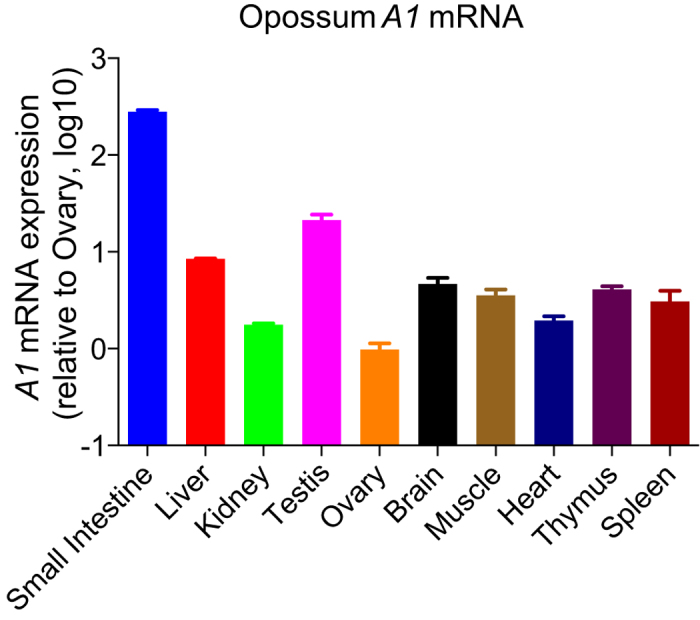
Representative opossum A1 mRNA expression levels in primary tissues by RTqPCR. Data are presented as A1 mRNA expression levels relative to those of the ovary normalized by endogenous expression levels of gapdh mRNA (n = 3, average +/− SD).
Discussion
A3 enzymes are specific proteins of eutherian mammals that are involved in the innate immunity pathways directed against retroviruses and retrotransposons4,5,6,7,8. A3 homologs do not appear to exist in non-eutherian mammals such as the opossum21,22 (Fig. S1b). In this study, we used a series of cell culture studies to examine whether opossum A1 has activity against retroviruses and LTR/non-LTR retrotransposons. Our results show that opossum A1 inhibits retrovirus replication and MusD retrotransposition in a deaminase-dependent manner (Figs 2, 4 and 6). This enzyme also suppresses human L1 retrotransposition in a deaminase-independent manner (Fig. 5). These results suggest that A1 plays role in protecting marsupial host genomes from parasitic genetic elements.
Despite potent mutagenic activity in bacteria (Fig. 1), opossum A1 showed modest HIV-1 restriction activity in HEK293T cells (Fig. 2). This relatively weak phenotype may reflect the predominantly nuclear localisation of opossum A1 in comparison to the more potent and predominantly cytoplasmic rabbit A1 enzyme (Figs 3 and S3). These results are consistent with our prior studies reporting that the degree of nuclear localisation of A1 correlates inversely with its antiviral activity12. It is likely that the nuclear distribution of opossum A1 prevents interaction with the Gag nucleocapsid protein, which in turn reduces encapsidation into virions. Alternatively, the physiological localisation of opossum A1 could differ from its localisation when overexpressed experimentally because endogenous A1 levels are below the limit of detection with currently available antibodies32. Furthermore, the nuclear localisation of A1 enzymes from different species appears to be influenced by cell-type33, suggesting that cellular factors regulate A1 localisation. A1s are expected to be more active against HIV-1 when they are in the cytoplasm. It must be noted that a chimeric human A1 fused to the rabbit homologue redistributed to the cytoplasm from the nucleus, was packaged more efficiently, and subsequently inhibited HIV-1 infectivity more effectively than the original human A1 protein12. Further studies, including the development of more sensitive antibodies and the identification of cellular factors that regulate the subcellular localisation of A1 will clarify the correlation between the subcellular localisation of A1 and its anti-HIV-1 activity.
Our cell-based assays did not use opossum cells and, to our knowledge, vectors have yet to be developed for opossum viruses and mobile elements. Nevertheless, the results presented here suggest that A1 likely functions as an innate barrier to infection by retroviruses and controls LTR/non-LTR retrotransposition in marsupials. First, opossum A1 possesses restriction activity against several different retroviruses (Figs 2 and 4) and LTR/non-LTR retroelements (Figs 5 and 6). Second, ~50% of the opossum genome is composed of repetitive elements (LINEs: ~29%, SINEs: ~10%, endogenous retroviruses: ~11% and DNA transposons: ~2%)21. Of note, the proportion of LINEs in the opossum genome is approximately 1.5 times higher than that in human and mouse genomes (~20% LINE). The greater anti-L1 activity of opossum A1 in comparison to anti-retroviral activity might be associated with the relatively higher proportion of LINEs in the opossum genome. Third, A1 mRNA is expressed in many different opossum tissues (Fig. 7), which is distinct from the gastrointestinal expression profile of A1 in human tissues30,31. This broad expression profile suggests that opossum A1 has additional functions besides editing of apoB mRNA. Future investigations should include the development of an infectious molecular clone using viruses that naturally infect opossum and assay systems using opossum cells, to determine whether opossum A1 is indeed a bona fide anti-viral restriction factor.
One of the common functions among AID/APOBEC family proteins is restriction of non-LTR retrotransposon, L1, using a mechanism that is independent of enzymatic activity17,18,34,35,36,37,38. However, it has been proposed that the mechanism by which L1 retrotransposition is suppressed by A1 is different from the deamination-independent mechanisms used by A3 enzymes17. In contrast to human A3s that are able to restrict human L1 retrotransposition even in the absence of a L1-encoded ORF1 protein39, A1s from human and lizard require the binding to L1 ORF1 protein for the inhibition of human L1 retrotransposition17. Interestingly, human and mouse ORF1 proteins form ribonucleoprotein (RNP) complexes with L1 RNA40,41, and the targeting of the L1 RNP complexes into cytoplasmic foci like stress granules is likely to control L1 retrotransposition42,43. It is noteworthy that human L1 RNA is found in A1 RNP complexes when they are cotransfected into HEK293T cells18. These observations suggest that the formation of A1 RNP complexes could sequester L1 RNA and/or ORF1 protein from its appropriate trafficking and translation, and ultimately could interfere with its retrotransposition into genome. This is also in contrast to A3s that are not necessary to form intracellular RNP complexes for L1 inhibition36. It is unclear what steps of L1 replication the A1 and A3 enzymes affect using deaminase-independent mechanisms. Elucidation of this deamination-independent repressive activity of APOBECs on L1 retrotransposition may provide important insights into the continuous arms race between viruses and their hosts.
It has been demonstrated that A1s from amniotes, including eutherian mammals, inhibit L1 retrotransposition in vitro17,18, and that these A1s also function as DNA mutators18,44. However, although A1 is expressed in the lizard, no deamination at the site corresponding to mammalian apoB mRNA editing has been observed44, suggesting that RNA editing may not be the ancestral function of A1. Because opossum A1 has RNA editing activity against apoB mRNA23, it appears that at least the A1s of mammals acquired the ability to edit apoB mRNA after the divergence of this group from the common ancestor they shared with other vertebrates (i.e., amphibians and birds). Therefore, the most likely original function of A1 was probably DNA editing and protecting cells against mobile elements, because the A1s of amniotes, including mammals, maintain anti-L1 activity in vitro17,18. Although several studies have suggested that non-human A1s (e.g., rabbit A1) may be involved in innate immune pathways11,13,14,16,18,19, it seems that the function of human A1 is limited to editing apoB mRNA in the small intestine30, and that the functions of A1 in innate immunity have been taken over by the expansion of A3s in the primate lineages such as in humans. Further studies are needed to shed light on the evolutionary history of RNA editing by the A1 deaminases and on their ancestral and physiological functions other than apoB mRNA editing.
Methods
Cells and antibodies
HEK293T, and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% bovine fetal serum (Gibco, Life Technologies). The anti-p24 capsid (CA) antibody used has been described previously45. The anti-SIVmac p27 antibody (55-2F12, #1610)46 was obtained from the NIH AIDS Reagent program. Antibodies against hemagglutinin (HA; C29F4, Cell Signaling Technologies, and HA.11, Covance), HSP90 (AC88, Stressgen), TOPOIIa (#4733, Cell Signaling Technologies), and β-actin (AC-74, Sigma) were commercially available.
DNA constructs
Vectors expressing Vif-proficient HIV-1, SIVmac and SIVagm proviral DNAs (pNL4-3 Luc E−R−, SIVmac Luc E−R− and SIVagm Luc E−R−, kindly provided by N.R. Landau, New York University)47, EGFP-based human L1 (pL1RP-EGFP, kindly provided by E.T. Luning Prak, University of Pennsylvania)27, neoR-gene-marked human L1 (pCEP4/L1mneoI/ColE1, kindly provided by N. Gilbert, Institut de Génétique Humaine, CNRS)48, and neoR-gene-marked murine MusD (pCMV L1Mus-6DneoTNF, kindly provided by T. Heidmann, Institut de Cancérologie Gustave Roussy)49 have been described elsewhere. The generation of a C-terminal HA-tagged rabbit A1 expression plasmid and bacterial expression plasmids encoding HA-tagged human and rabbit A1s has been described previously18.
Cloning of opossum A1 cDNA
Production, maintenance and experimental manipulations of the animals were approved by the Institutional Animal Care and Use Committee of the Texas Biomedical Research Institute (John L. VandeBerg; previous affiliation). All procedures were carried out in accordance with the approved guidelines. Small intestine has been removed aseptically from euthanized Monodelphis Domestica (opossum). Total RNA was isolated using TRIzol reagent (Invitrogen), and then treated with DNase I (Takara). cDNA encoding the opossum A1 gene was synthesised using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) with a primer set designed so that a single HA epitope tag was attached to the C-terminus of the protein as previously described23. (5′-NNNNNGATATCGAAGCCATGAATTCTAAGACAGGTCCA-3′ and 5′-NGCGGCCGCTCAAGCGTAATCTGGAACATCGTATGGGTATCTCCAGGTCACAAATGGCTGG-3′, Restriction enzyme sites are underlined). The amplified fragment was cloned into the pCR-Blunt vector (Invitrogen) and sequenced. The amplified product was then inserted into the EcoRV and NotI sites of the pCAGGS expression vector and sequenced. The nucleotide sequence of an open reading frame encoding opossum A1 was identical to that enrolled in GenBank (NM001032982). The catalytic mutants of the opossum A1 were constructed using the oligonucleotide primers. E62A: 5′-TCTCAACATGCTGCAATCAACTTCATG-3′ and 5′-CATGAAGTTGATTGCAGCATGTTGAGA-3′. E62Q: 5′-TCTCAACATGCTCAAATCAACTTCATGG-3′ and 5′-CCATGAAGTTGATTTGAGCATGTTGAGA-3′. The amplified products were inserted into the pCAGGS vector at the EcoRV and NotI sites and sequenced. Bacterial expression plasmids encoding C-terminal HA-tagged opossum A1 and its catalytic mutants (E62A and E62Q) were created with the following primer set: 5′-NNNNCTCGAGATGAATTCTAAGACAGGTCCATCAGTAGGAGA-3′ and 5′-NNNATGCATTCAAGCGTAATCTGGAACATCGTATGGGTA-3′. The amplified products were then cloned into the pTrcHis A vector (Invitrogen) with restriction enzymes XhoI and NsiI and sequenced.
Sequence alignment
The amino acid sequences of full-length mammalian A1s were aligned as previously described11. The GenBank accession numbers of the A1 sequences used for this comparison are: human (NM001644), rabbit (U10695), rat (MN012907), mouse (NM031159), and opossum (NM001032982).
Immunoblot analysis
Cell and virion lysates were resolved with sodium dodecyl sulphate (SDS) polyacrylamide gel electrophoresis (PAGE), transferred to a polyvinylidene difluoride membrane (Millipore), and blocked with 4% milk in phosphate-buffered saline (PBS) containing 0.1% Tween 20. The membranes were then incubated with a primary antibody, a biotin-conjugated secondary anti-mouse or anti-rabbit IgG antibody (Sigma) and streptavidin-conjugated horseradish peroxidase (Sigma). The signals were visualised with Chemi-Lumi One (Nacalai Tesque) and a VersaDoc 5000 Imager (Bio-Rad).
Subcellular localisation
An immunofluorescence analysis was performed as previously reported12,18. Briefly, 2 × 104 HEK293T or HeLa cells were plated in each well of an eight-well Lab-Tek Chamber Slide (Nalge Nunc International). The cells were transfected with the pCAGGS expression vector (500 ng) encoding either HA-tagged rabbit A1, opossum A1, or its catalytic mutants (E62A or E62Q), using FuGENE HD (Roche). After 24 h, the transfected cells were fixed with 4% formaldehyde in PBS for 30 min, and permeabilised with 0.1% Triton X-100 in PBS. The cells were then treated with 0.1 M glycine/PBS to quench the reaction and blocked with 0.3% bovine serum albumin (BSA)/PBS. To stain A1, the cells on coverslips were incubated with an anti-HA antibody (HA.11; 1:1000 dilution) in 0.3% BSA/PBS in a humid chamber at 37 °C for 1 h, and then with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG antibody (Sigma; 1:300)/0.3% BSA/PBS for 1 h. The cells were then treated with 1 μg/mL 4,6-diamidino-2-phenylindole staining (DAPI; Invitrogen) for 5 min. The coverslips were mounted with Fluorescent Mounting Medium (Dako). Fluorescence was visualised with a Zeiss LSM 700 laser-scanning confocal microscope. The images were captured with IPLab and processed with the Adobe PhotoShop 4.0 software. The fluorescence intensity of A1 in the cytoplasm was calculated as previously described12. Briefly, the total FITC signal intensity of 20 cells for each A1 was measured using Image J software. The intensity of FITC signal overlapped with the portion of DAPI staining was measured to calculate the proportion of nuclear distribution of A1. Then, the proportion of cytoplasmic distribution of each A1 was calculated by subtracting the nuclear FITC signal from the total FITC signal. The densitometric analysis of the blotting bands was quantified using an image analysis system.
Preparation of nuclear and cytoplasmic extracts
The nuclear and cytoplasmic extracts from HEK293T were prepared with the Subcellular Protein Fractionation Kit for Cultured Cells (Thermo Fisher Scientific), according to the manufacturer’s protocol.
Bacterial mutator assay
The generation of bacterial expression plasmids encoding cDNAs encoding HA-epitope-tagged opossum A1 and its derivatives is described above. The Escherichia coli–based DNA mutation assay was performed as described previously12,18. The uracil DNA glycosylase (ung)-deficient E. coli strain BW310 was transformed with the parental pTrcHisA plasmid or vectors encoding the various A1 cDNAs. Twenty colonies of the transformed bacteria, selected on plates containing ampicillin, were cultured overnight at 37 °C in 2 mL of LB medium containing ampicillin and 1 mM IPTG. The saturated cultures (100 μL) were then plated on LB agar containing 100 μg/mL rifampicin. The total number of RifR colonies per plate was counted after 24 h. The appropriate dilution of viable cells was plated onto an LB plate containing ampicillin and the mutation frequencies were calculated as RifR colonies per viable cell. To verify protein expression, 100 μL of each saturated IPTG-induced culture was lysed and subjected to immunoblotting, as described above.
Single-cycle infection assays
HEK293T cells (2.5 × 105) were cotransfected with 0.75 μg of Vif-proficient NL4-3 Luc E−R− proviral DNA construct, together with 0.5 μg of pVSV-G vector and 0.25 μg of the control vector or one of several expression vectors encoding C-terminal HA-epitope-tagged A1 protein supplemented with the appropriate concentration of the control vector, using Effectene® (Qiagen). For SIVs, 0.75 μg of Vif-proficient SIVmac or SIVagm Luc E−R− proviral DNA construct was cotransfected into HEK293T cells, with 0.5 μg of pVSV-G vector and 0.25 μg of the control vector or HA-tagged A1 expression vector. MLV-GFP virus stock was produced as previously described50,51. HEK293T cells were cotransfected with 0.3 μg of MLV genome expressing a GFP reporter M3P-GFP, together with 0.3 μg of MLV packaging construct pMD-MLV-OGP, 0.1 μg of pVSV-G and 0.3 μg of the control vector or HA-tagged A1 expression vector. After 48 h, the virus-containing supernatants were filtered with 0.45 μm filters and frozen as aliquots. The p24 or p27 content of the viruses was determined with a HIV-1 p24 or SIV p27 antigen ELISA kit (ZeptoMetrix). Fresh target HEK293T cells were infected with the equivalent amounts of the luciferase reporter viruses and after 48 h, the intracellular luciferase activity of the infected cells was measured, as previously described11,12. Infectivity of MLV-GFP virus was analysed by flow cytometery. To assess the packaging of the A1 proteins into viral particles, the virus-containing supernatants were spun through a 20% sucrose cushion and the viral pellets were solubilised in 2× SDS sample buffer. Equivalent amounts of p24 or p27 antigen from each solubilised sample were analysed with immunoblotting, as described above. Band intensities were analysed with the ImageJ software.
L1 retrotransposition assays
The EGFP-based human L1 retrotransposition assay was performed as previously described18. Briefly, 3 × 105 HEK293T cells were cotransfected with 1.5 μg of pL1RP-EGFP or the pIRESpuro vector (Clontech) and 0.5 μg of the control vector or the respective A1 expression vector with the appropriate concentration of a control vector with Effectene. At 24 h posttransfection, the cells were selected with 1.0 μg/mL puromycin and 7-9 days after selection, the EGFP expression resulting from retrotransposition was measured with flow cytometry. The neoR-gene-marked human L1 retrotransposition assay was performed by cotransfecting HeLa cells (5 × 105 cells) with 0.4 μg of the respective A1 expression plasmid and 1.2 μg of the human L1-neo reporter vector together with 0.4 μg of pIRES-EGFP (Clontech) as the transfection control, using FuGENE, as previously described18. After 72 h, 5 × 105 cells were plated onto 100 mm dishes and selected with 0.75 mg/mL G418. At 12–14 days after selection, the resultant G418R colonies were stained with crystal violet (Sigma) and counted. The retrotransposition frequency was calculated as the number of G418R colonies/transfection efficiency (percentage of GFP+ cells).
MusD retrotransposition assay
The murine MusD retrotransposition assay was performed as previously described18. Briefly, 5 × 105 HeLa cells were cotransfected with 0.2 μg of the control vector or the respective A1 expression plasmid supplemented with the appropriate concentration of the control vector, together with 0.6 μg of the neor-based murine MusD reporter vector together with 0.4 μg of pIRES-EGFP, using FuGENE. After 72 h, 5 × 105 cells were reseeded onto 100 mm dishes for G418 (1.0 mg/mL) selection, and the resultant G418R colonies were counted. The retrotransposition frequency was calculated as described above.
Quantitation of opossum A1 transcripts in vivo
cDNAs from opossum primary tissues were synthesized using the High Capacity cDNA Reverse Transcription Kit. Amplification was performed using the following primers: A1, 5′-CGCGTCCACCTGATTC-3′ and 5′-CGCGTCCACCTGATTC-3′. gapdh, 5′-CGCGTCCACCTGATTC-3′ and 5′-CGCGTCCACCTGATTC-3′. PCR amplification conditions were 95 °C for 10 min, followed by 50 cycles of 95 °C for 30 s, 53 °C for 20 s, and 68 °C for 30 s. Quantitation was performed with SYBR Green PCR master mix (Applied Biosystems) using a Realplex2 Mastercycler EPgradient S (Eppendorf). Expression levels of target cDNA were normalized to the amount of endogenous mRNA of gapdh.
Statistical analysis
The statistical analysis was performed with Student’s t test. All data are the means and standard deviations (SDs) of three experiments, unless otherwise stated. *P < 0.05, **P < 0.01, ***P < 0.001.
Additional Information
How to cite this article: Ikeda, T. et al. Opossum APOBEC1 is a DNA mutator with retrovirus and retroelement restriction activity. Sci. Rep. 7, 46719; doi: 10.1038/srep46719 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Drs N. Gilbert, T. Heidmann, N.R. Landau, and E.T. Luning Prak for providing reagents. We also thank Ms. E. Kamada for secretarial assistance, Ms. K. Fukuda for technical assistance, and M. Araki, T. Ohsugi, and A. Sarai for valuable comments. This study was supported, in part, by JSPS KAKENHI (grant number 23590546 to A.K.; 26460580 to K.M), Global Centers of Excellence (COE) Program Global Education Research Center Aiming to Control AIDS, JSPS Research Fellowship for Young Scientists and Postdoctoral Fellowship (to T.I.), Okukubo Memorial Fund for Medical Research at Kumamoto University School of Medicine (to T.I.), SENSHIN Medical Research Foundation (to K.M.), NIH NIAID R37 AI064046 (to R.S.H.), and NIH NCI CA206309 (to R.S.H.). R.S.H. in an Investigator of the Howard Hughes Medical Institute.
Footnotes
R.S.H. is a co-founder, shareholder, and consultant of ApoGen Biotechnologies Inc. The other authors declare that they have no competing financial interests.
Author Contributions T.I. and M.S. conducted the experiments. T.I., D.E., A.K. and K.M. acquired and analysed the data. A.K. and R.S.H. supervised the project. J.L.V. provided opossum total RNA. T.I., J.L.V., D.E., R.S.H. and K.M. critically evaluated and revised the final manuscript. All authors commented on the manuscript.
References
- Blanc V. & Davidson N. O. APOBEC-1-mediated RNA editing. Wiley Interdiscip Rev Syst Biol Med 2, 594–602 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prohaska K. M., Bennett R. P., Salter J. D. & Smith H. C. The multifaceted roles of RNA binding in APOBEC cytidine deaminase functions. Wiley Interdiscip Rev RNA 5, 493–508 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester A., Scott J., Anant S. & Navaratnam N. RNA editing: cytidine to uridine conversion in apolipoprotein B mRNA. Biochim Biophys Acta 1494, 1–13 (2000). [DOI] [PubMed] [Google Scholar]
- Harris R. S. & Dudley J. P. APOBECs and virus restriction. Virology 479–480, 131–145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter J. D., Bennett R. P. & Smith H. C. The APOBEC protein family: united by structure, divergent in function. Trends Biochem Sci 41, 578–594 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koito A. & Ikeda T. Apolipoprotein B mRNA-editing, catalytic polypeptide cytidine deaminases and retroviral restriction. Wiley Interdiscip Rev RNA 3, 529–541 (2012). [DOI] [PubMed] [Google Scholar]
- Koito A. & Ikeda T. Intrinsic immunity against retrotransposons by APOBEC cytidine deaminases. Front Microbiol 4, 28 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon V., Bloch N. & Landau N. R. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol 16, 546–553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop K. N. et al. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol 14, 1392–1396 (2004). [DOI] [PubMed] [Google Scholar]
- Bishop K. N., Holmes R. K., Sheehy A. M. & Malim M. H. APOBEC-mediated editing of viral RNA. Science 305, 645 (2004). [DOI] [PubMed] [Google Scholar]
- Ikeda T. et al. The antiretroviral potency of APOBEC1 deaminase from small animal species. Nucleic Acids Res 36, 6859–6871 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T. et al. Creation of chimeric human/rabbit APOBEC1 with HIV-1 restriction and DNA mutation activities. Sci Rep 6, 19035 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit V. et al. Murine APOBEC1 is a powerful mutator of retroviral and cellular RNA in vitro and in vivo. J Mol Biol 385, 65–78 (2009). [DOI] [PubMed] [Google Scholar]
- Renard M., Henry M., Guetard D. & Vartanian J. P. Wain-Hobson S. APOBEC1 and APOBEC3 cytidine deaminases as restriction factors for hepadnaviral genomes in non-humans in vivo. J Mol Biol 400, 323–334 (2010). [DOI] [PubMed] [Google Scholar]
- Gee P. et al. APOBEC1-mediated editing and attenuation of herpes simplex virus 1 DNA indicate that neurons have an antiviral role during herpes simplex encephalitis. J Virol 85, 9726–9736 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M. C. et al. Human APOBEC1 cytidine deaminase edits HBV DNA. Retrovirology 6, 96 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindic N. et al. Differential inhibition of LINE1 and LINE2 retrotransposition by vertebrate AID/APOBEC proteins. Retrovirology 10, 156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T. et al. Intrinsic restriction activity by apolipoprotein B mRNA editing enzyme APOBEC1 against the mobility of autonomous retrotransposons. Nucleic Acids Res 39, 5538–5554 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tervo H. M. & Keppler O. T. High natural permissivity of primary rabbit cells for HIV-1, with a virion infectivity defect in macrophages as the final replication barrier. J Virol 84, 12300–12314 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett B. S. et al. Reassessment of murine APOBEC1 as a retrovirus restriction factor in vivo. Virology 468–470, 601–608 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T. S. et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447, 167–177 (2007). [DOI] [PubMed] [Google Scholar]
- LaRue R. S. et al. The, artiodactyl APOBEC3 innateimmune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol Biol 9, 104 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino T., Navaratnam N., Jarmuz A., von Haeseler A. & Scott J. C–>U editing of apolipoprotein B mRNA in marsupials: identification and characterisation of APOBEC-1 from the American opossum Monodelphus domestica. Nucleic Acids Res 27, 2662–2671 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. S., Petersen-Mahrt S. K. & Neuberger M. S. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell 10, 1247–1253 (2002). [DOI] [PubMed] [Google Scholar]
- Petersen-Mahrt S. K., Harris R. S. & Neuberger M. S. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418, 99–103 (2002). [DOI] [PubMed] [Google Scholar]
- Jonsson S. R. et al. Evolutionarily conserved and non-conserved retrovirus restriction activities of artiodactyl APOBEC3F proteins. Nucleic Acids Res 34, 5683–5694 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prak E. T., Dodson A. W., Farkash E. A. & Kazazian H. H. Jr. Tracking an embryonic L1 retrotransposition event. Proc Natl Acad Sci USA 100, 1832–1837 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamuta M. et al. Alternative mRNA splicing and differential promoter utilization determine tissue-specific expression of the apolipoprotein B mRNA-editing protein (Apobec1) gene in mice. Structure and evolution of Apobec1 and related nucleoside/nucleotide deaminases. J Biol Chem 270, 13042–13056 (1995). [DOI] [PubMed] [Google Scholar]
- Funahashi T., Giannoni F., DePaoli A. M., Skarosi S. F. & Davidson N. O. Tissue-specific, developmental and nutritional regulation of the gene encoding the catalytic subunit of the rat apolipoprotein B mRNA editing enzyme: functional role in the modulation of apoB mRNA editing. J Lipid Res 36, 414–428 (1995). [PubMed] [Google Scholar]
- Hadjiagapiou C., Giannoni F., Funahashi T., Skarosi S. F. & Davidson N. O. Molecular cloning of a human small intestinal apolipoprotein B mRNA editing protein. Nucleic Acids Res 22, 1874–1879 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns M. B. et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossat N. et al. C to U RNA editing mediated by APOBEC1 requires RNA-binding protein RBM47. EMBO Rep 15, 903–910 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Yang Y. & Smith H. C. Multiple protein domains determine the cell type-specific nuclear distribution of the catalytic subunit required for apolipoprotein B mRNA editing. Proc Natl Acad Sci USA 94, 13075–13080 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDuff D. A., Demorest Z. L. & Harris R. S. AID can restrict L1 retrotransposition suggesting a dual role in innate and adaptive immunity. Nucleic Acids Res 37, 1854–1867 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinomoto M. et al. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res 35, 2955–2964 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomska A. M. et al. Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J Virol 81, 9577–9583 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd H. P. et al. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci USA 103, 8780–8785 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein M. D. & Harris R. S. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem 281, 16837–16841 (2006). [DOI] [PubMed] [Google Scholar]
- Lovsin N. & Peterlin B. M. APOBEC3 proteins inhibit LINE-1 retrotransposition in the absence of ORF1p binding. Ann N Y Acad Sci 1178, 268–275 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. L. & Branciforte D. Synchronous expression of LINE-1 RNA and protein in mouse embryonal carcinoma cells. Mol Cell Biol 13, 5383–5392 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohjoh H. & Singer M. F. Ribonuclease and high salt sensitivity of the ribonucleoprotein complex formed by the human LINE-1 retrotransposon. J Mol Biol 271, 7–12 (1997). [DOI] [PubMed] [Google Scholar]
- Goodier J. L., Zhang L., Vetter M. R. & Kazazian H. H. Jr. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol Cell Biol 27, 6469–6483 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier J. L., Ostertag E. M., Engleka K. A., Seleme M. C. & Kazazian H. H. Jr. A potential role for the nucleolus in L1 retrotransposition. Hum Mol Genet 13, 1041–1048 (2004). [DOI] [PubMed] [Google Scholar]
- Severi F., Chicca A. & Conticello S. G. Analysis of reptilian APOBEC1 suggests that RNA editing may not be its ancestral function. Mol Biol Evol 28, 1125–1129 (2011). [DOI] [PubMed] [Google Scholar]
- Koito A. et al. Conserved immunogenic region of a major core protein (p24) of human and simian immunodeficiency viruses. AIDS Res Hum Retroviruses 4, 409–417 (1988). [DOI] [PubMed] [Google Scholar]
- Higgins J. R., Sutjipto S., Marx P. A. & Pedersen N. C. Shared antigenic epitopes of the major core proteins of human and simian immunodeficiency virus isolates. J Med Primatol 21, 265–269 (1992). [PubMed] [Google Scholar]
- Mariani R. et al. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114, 21–31 (2003). [DOI] [PubMed] [Google Scholar]
- Gilbert N., Lutz-Prigge S. & Moran J. V. Genomic deletions created upon LINE-1 retrotransposition. Cell 110, 315–325 (2002). [DOI] [PubMed] [Google Scholar]
- Esnault C., Casella J. F. & Heidmann T. A Tetrahymena thermophila ribozyme-based indicator gene to detect transposition of marked retroelements in mammalian cells. Nucleic Acids Res 30, e49 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. S. et al. DNA deamination mediates innate immunity to retroviral infection. Cell 113, 803–809 (2003). [DOI] [PubMed] [Google Scholar]
- Liddament M. T., Brown W. L., Schumacher A. J. & Harris R. S. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol 14, 1385–1391 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.