Abstract
Context:
A single microRNA gene may give rise to several mature products that differ in length, called isomiRs. IsomiRs are known to be tissue specific and functionally relevant. The microRNA sequence heterogeneity of the thyroid gland has yet to be determined.
Objective:
The objective of the study was to provide a comprehensive view of the microRNA transcriptome in normal thyroid and papillary thyroid carcinoma (PTC).
Design:
We used next-generation deep sequencing to analyze microRNA length heterogeneity and expression profiles of PTC tumors (n = 14), unaffected tissue adjacent to tumors (n = 14), and control, noncancerous thyroid tissue (n = 14). The results were validated with a microarray on an additional set of 9 PTC tumor/normal tissue pairs.
Results:
Eighty-nine microRNAs were significantly deregulated in PTC compared with normal thyroid tissue (false discovery rate < 0.05, fold change 0.13–20.7). Top deregulated miRNAs included miR-146b-5p, miR-221-3p, miR-7-3p, miR-551b-3p, miR-486-3p, and miR-144-3p, confirming previous microarray profiling. The expression of miRNAs did not depend on the BRAF mutation status. Interestingly, 85% of the most abundant microRNAs consisted of isoforms that differed from the standard reference sequence deposited in miRBase. Moreover, the reference microRNAs were completely absent in 42.4% and 35.9% of the microRNAs expressed in normal thyroid and PTC tumors, respectively. Numerous isomiRs had altered seed sequences, which led to a different set of target genes. For highly deregulated miR-146b-5p, we detected 6 isoforms (tumor/normal fold change 14.4–28.7, false discovery rate < 0.002) that varied at their 5′ ends with a 1-nt difference that created 2 alternative seeds. The target genes for those 2 seeds overlapped in only 13.1% of genes.
Conclusions:
Almost all microRNAs exhibit isoforms of variable length and potentially distinct function in thyroid tumorigenesis.
Papillary thyroid carcinoma (PTC) accounts for approximately 85% of all thyroid carcinomas, with 60 200 new cases estimated in the United States in 2013 (http://www.cancer.gov/cancertopics/types/thyroid). Unlike many other cancers, its incidence is increasing (1). PTC displays a strong genetic predisposition as evidenced by case-control population studies. In first-degree relatives of a PTC patient, the risk of acquiring this kind of cancer is increased 8–12 times compared with the general population, which ranks PTC with one of the highest scores among all cancers in terms of heritability (2, 3). Nevertheless, no predisposing protein-encoding gene has been convincingly described, even though several markers have been identified by linkage analyses and genome-wide association studies (4–7). We reasoned therefore that the mechanisms underlying thyroid carcinogenesis may require the interaction of 2 or more genes of low penetrance and thus most probably involve regulatory genes, such as microRNAs, rather than protein-encoding genes.
MicroRNAs (miRNAs or miRs) are short, noncoding RNAs that regulate the expression of protein-coding genes binding to complementary sequences in the 3′-untranslated regions of their transcripts. The sequence that determines the binding of a miRNA to its target mRNA comprises nucleotides 2–8 of a miRNA and is called a seed region (8–10). Sequences of newly discovered miRNAs are deposited in a specifically designated public repository; miRBase (11). Recent deep-sequencing studies revealed significant miRNA length heterogeneity, showing that individual miRNA genes may give rise to several mature miRNA products that differ in length. These products, called isomiRs, originate most likely from imperfect specificity of both Drosha and Dicer cleavage of miRNA precursors, in the latter case due to asymmetrical structural motifs present in precursor hairpins (12). IsomiRs proved to be tissue specific and functionally relevant (13, 14) and were shown to be functionally cooperative partners of canonical miRNAs (14), even when a fraction of isomiRs underwent some 5′-end nucleotide biases at the AGO2 loading step (15).
miRNAs are implicated in the development of several types of human cancers. Their role in carcinogenesis consists mainly in abnormal levels of expression of mature miRNA transcripts in tumors compared with the corresponding unaffected tissues (16), resulting in aberrant expression of their target mRNAs. Because a single miRNA can regulate the expression of dozens of genes, abnormal levels of miRNAs lead to the deregulation of whole signaling pathways within the tumor cell (17). MicroRNA expression profiling of human tumors has identified signatures associated with diagnosis, staging, prognosis, and response to treatment (18, 19).
Our recent data strongly support the role of miRNAs in carcinogenesis because a polymorphism in one microRNA (miR-146a) has been identified as predisposing to thyroid carcinoma, whereas numerous other miRNAs, deregulated in PTC, have been shown to be involved in the regulation of signaling (mainly phosphatase and tensin homolog, phosphatidylinositol 3-kinase, AKT) that is central to thyroid carcinogenesis (20–23). Thus, both expression and sequence alterations of miRNAs can contribute to neoplastic transformation. However, comprehensive information on the thyroid miRNome is still lacking. To elucidate the role of miRNAs in the predisposition to thyroid carcinoma, it is necessary to identify the sequences and expression levels of all miRNAs expressed in normal thyroid gland and in tumor tissue. The observed differences in expression levels of miRNAs, as well as alterations in their nucleotide sequences, will allow for identification of miRNA-related predisposition to PTC, for a better understanding of the biology of the disease process, and for the design of miRNA-based experiments aiming at the inhibition of specific genes. Therefore, previous experiments based on microarray or real-time PCR profiling and thus studying only limited numbers of known miRNAs (24, 25) need to be updated with the wealth of currently available data on novel miRNAs.
Materials and Methods
Thyroid tissue samples
For the sequencing experiment, samples from PTC tumors (PTC-T, n = 14), unaffected tissue adjacent to but not infiltrated by tumor from the same patient (PTC-N, n = 14), and control, noncancerous thyroid specimens (NN, n = 14) were collected at the Medical University of Warsaw (Warsaw, Poland). The clinical characteristics of patients are summarized in Supplemental Table 1, published on The Endocrine Society's Journals Online web site at http://jcem.endojournals.org. For the microarray experiment, an additional validation set of PTC-T (n = 9) and PTC-N (n = 9) samples was collected at the same institution. After approval of the institutional review board and patient consent, fresh tissue samples were obtained from patients undergoing surgical resection; the samples were snap frozen on dry ice and stored at −80°C. Total RNA was extracted with TRIzol solution (Invitrogen, Carlsbad, California), and the integrity of RNA was assessed using an Agilent BioAnalyzer 2100 (Agilent Technologies, Palo Alto, California).
Next-generation sequencing
The RNAs were gel fractionated and the small RNA fraction (with length not exceeding 40 nt) was subjected to hybridization and ligation with Adaptor Mix (Applied Biosystems, Foster City, California). Subsequently, RNAs were reverse transcribed and sequenced using whole-transcriptome sequencing on the SOLiD platform (Applied Biosystems). On this basis, a sequence and expression database of all small RNAs was created.
miRNA microarrays
Biotin-labeled miRNA was hybridized on miRNA chips as described elsewhere (26). Briefly, 5 μg of total RNA from each sample was reverse transcribed using biotin end-labeled random octamers. Hybridization was carried out on a custom miRNA microarray chip (OSU-CCC version 2.0, Gene Expression Omnibus accession number GPL4700). The hybridized chips were washed and processed to detect biotin-containing transcripts by streptavidin-Alexa647 conjugate and scanned on the Axon 4000B (Axon Instruments Inc, Foster City, California). Scanned images were quantified by GENEPIX PRO 6.0 (Axon Instruments).
BRAF mutation and RET/PTC rearrangement analysis
DNA from thyroid samples that were subject to next-generation sequencing (NGS) analysis was extracted using standard TRIzol solution (Invitrogen) extraction protocol. For BRAF, the samples were genotyped using high-resolution melting BRAF V600E assay (27) with a modified protocol in LightCycler¨ 480 instrument (Roche Molecular Diagnostics, Basel, Switzerland). RET/PTC1 and RET/PTC3 rearrangements were analyzed using RT-PCR amplification according to protocol reported by Nikiforov et al (28).
Data analysis
To detect all human isomiRs, a library of reference sequences was prepared by identifying the sequences of mature miRNAs, together with 5 flanking nucleotides, within the hairpins deposited in miRBase, version 19 (29). Because the SOLiD platform requires a read length of 35 nucleotides, all the small RNAs were extended with specific adapters and annealed to their 3′ ends during library preparation. Thus, all sequences studied here included adapters, and their removal was performed on the raw, 35-nt-long SOLiD sequence reads using Cutadapt software (30). Obtained sequences with the length of 15–28 nucleotides were subject to further analysis as potential miRNAs. The sequences were mapped on the prepared reference library using Bowtie version 0.12.7 (31), with the requirement of perfect matching. The numbers of mapped reads were subsequently calculated for each miRNA and provided in 2 ways: as a number of each of unique reads mapped to each reference sequence, and as a number of all reads mapped to each reference sequence (see example in Supplemental Table 2). The data obtained for each sample was normalized using the reads per million (RPM) normalization according to the formula: RPM = (Nref/Nall) × 106, where Nref is the number of reads mapped to the miRNA reference and Nall is the total number of reads mapped in the sample. Microarray raw data of 9 pairs PTC-T/PTC-N were preprocessed by the subtraction of background intensity, followed by quantile normalization. Sequences of 44 miRNAs identified in NGS analysis as deregulated in PTC-T compared with PTC-N were matched to the sequences of microarrays probes. The sequences for 30 genes overlapped in NGS and microarray analysis thus could serve as a basis for further correlation analysis.
Statistical analysis
The ratios of the number of isomiRs per miRNA among different sample types were computed using a Pearson's χ2 test. Selection of miRNAs and isomiRs deregulated between the analyzed groups, analysis of differences between PTC samples from patients with and without BRAF V600E mutation, and the comparison of PTC-T and PTC-N samples in microarray analyses were performed using a Welch t test (unpaired for comparisons between PTC-T and NN, PTC-N and NN, and BRAF and BRAF V600E in the NGS study, paired for comparison between PTC-T and PTC-N in both the NGS and microarray studies). The false discovery rate (FDR) was used to assess the multiple testing errors. Association between log2-transformed fold changes obtained in the NGS and microarray analyses was tested using a Pearson's product moment correlation coefficient. Identification of target genes for each miRNA seed sequence whose expression was significantly deregulated between PTC-T and PTC-N samples was performed by TargetRank software (8).
Results
Significant miRNAs
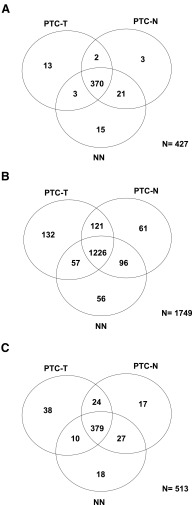
We detected 427 miRNAs significantly expressed in thyroid tissue (median RPM ≥ 5 in at least 50% of samples within any of the 3 studied groups), including 388 miRNAs expressed in PTC-T, 396 expressed in PTC-N, and 409 expressed in NN (Supplemental Table 3 and Figure 1A).
Figure 1.
Venn diagrams of significantly expressed miRNAs (A), isomiRs (B), and seeds (C) in PTC-T, PTC-N, and NN. Significant miRNAs are defined as those with a median RPM of 5 or greater in at least 50% of the samples within any of the study groups. Significant isomiRs are defined as having an expression of greater than 1% of the total expression of a particular miRNA, in at least 80% of the samples within any of the study groups.
Deregulated miRNAs
Among 427 miRNAs significantly expressed in normal or tumorous thyroid tissue, 124 were deregulated in PTC tumors compared with control, noncancerous thyroid (PTC-T vs NN, FDR < 0.05), including 24 strongly up-regulated (fold change > 2, maximum 20.7), and 65 strongly down-regulated miRNAs (fold change < 0.5, minimum 0.13, Supplemental Table 3). The list of top deregulated miRNAs includes miR-146b-5p, miR-221-3p, miR-7-3p, miR-551b-3p, miR-486-3p, and miR-144-3p (fold change 20.7, 8.1, 0.13, 7.0, 0.17, and 0.18, respectively, Supplemental Table 3). In addition, top deregulated miRNAs include several miRNAs derived from the passenger strand of miRNA precursors, eg, miR-146b-3p, miR-7-5p, miR-221-5p, and miR-144-5p (fold change 17.5, 0.14, 5.0, and 0.26, respectively, Supplemental Table 3).
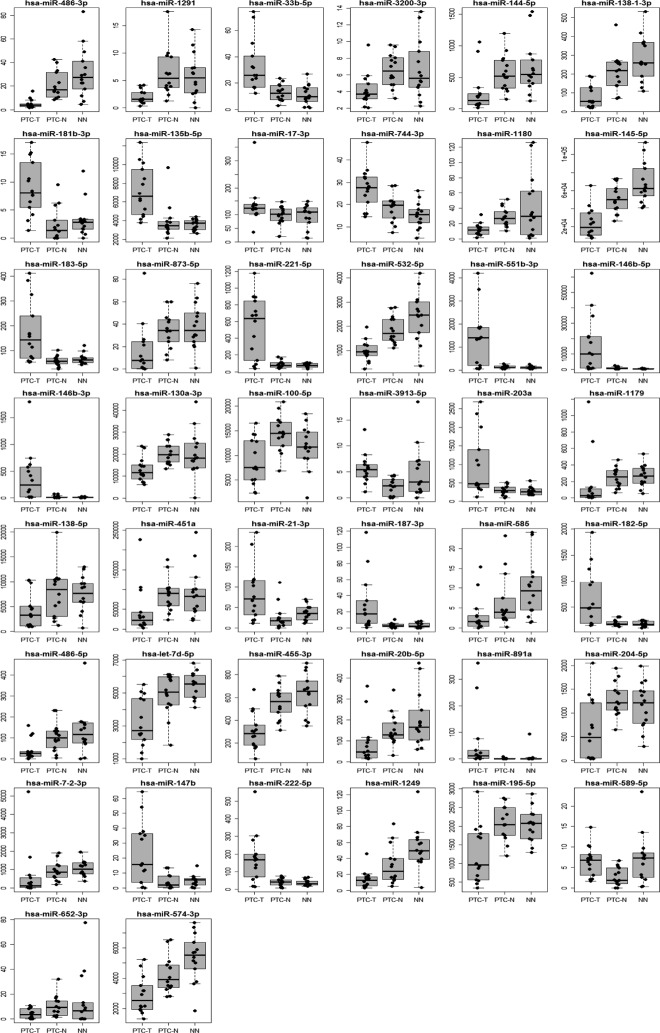
The paired t test analysis revealed that 44 miRNAs were expressed differentially between PTC tumors and adjacent tissue, not infiltrated by tumor (PTC-T vs PTC-N) at a significance level of FDR less than 0.05 (Table 1 and Figure 2). We identified 16 miRNAs that were strongly overexpressed in PTC-T vs PTC-N (fold change > 2, maximum 10.8), including miR-146b-5p and miR-146b-3p at the top [tumor/normal (T/N) fold change 10.81 and 10.51, respectively, Table 1 and Figure 2]. The list of the most down-regulated genes (fold change < 0.5, minimum 0.14) included 19 miRNAs, with the most deregulated miR-1179 and miR-7-2-3p (T/N fold change 0.14 and 0.17, respectively, Table 1 and Figure 2). Importantly, 33 of 44 miRNAs were among the miRNAs significantly deregulated in the PTC-T compared with the NN thyroid samples (PTC-T vs NN, FDR < 0.05, Table 1).
Table 1.
Significantly expressed miRNAs deregulated in PTC compared with adjacent normal tissue (PTC-T vs PTC-N, FDR < 0.05) and with control, noncancerous thyroid (PTC-T vs NN, FDR < 0.05)
| miRNA | Mean Expression in PTC-T | Mean Expression in PTC-N | Mean Expression in NN | Paired P Value, T vs N | Paired FDR, T vs N | Fold Change, T vs N | P Value, T vs NN | FDR T, vs NN | fold change T vs. NN |
|---|---|---|---|---|---|---|---|---|---|
| hsa-miR-146b-3p | 119 | 11 | 6.8 | .00142 | 0.031 | 10.8 | .00002 | 0.001 | 17.5 |
| hsa-miR-146b-5p | 5346 | 507 | 258 | .00116 | 0.027 | 10.5 | .00001 | 0.001 | 20.7 |
| hsa-miR-891a | 12 | 1.6 | 2.0 | .00214 | 0.031 | 7.68 | .00705 | 0.027 | 5.92 |
| hsa-miR-551b-3p | 713 | 109 | 102 | .00101 | 0.025 | 6.53 | .00011 | 0.002 | 7 |
| hsa-miR-187–3p | 14 | 2.8 | 2.6 | .00176 | 0.031 | 5.10 | .00036 | 0.004 | 5.58 |
| hsa-miR-221–5p | 356 | 78 | 71 | .0008 | 0.024 | 4.57 | .00003 | 0.001 | 5.03 |
| hsa-miR-147b | 11 | 2.5 | 3.8 | .0032 | 0.036 | 4.53 | .0254 | 0.073 | 3.01 |
| hsa-miR-21–3p | 62 | 15 | 32 | .00188 | 0.031 | 4.04 | .02784 | 0.078 | 1.95 |
| hsa-miR-181b-3p | 7.5 | 1.9 | 2.5 | .00034 | 0.02 | 3.91 | .00048 | 0.005 | 2.94 |
| hsa-miR-222–5p | 123 | 34 | 35 | .00359 | 0.039 | 3.63 | .00022 | 0.003 | 3.51 |
| hsa-miR-182–5p | 487 | 185 | 168 | .00235 | 0.031 | 2.63 | .0002 | 0.003 | 2.9 |
| hsa-miR-33b-5p | 28 | 11 | 8.3 | .00005 | 0.01 | 2.55 | .00023 | 0.003 | 3.44 |
| hsa-miR-203a | 678 | 272 | 258 | .00223 | 0.031 | 2.49 | .00093 | 0.007 | 2.63 |
| hsa-miR-183–5p | 135 | 54 | 63 | .00065 | 0.024 | 2.48 | .00113 | 0.008 | 2.14 |
| hsa-miR-589–5p | 5.4 | 2.3 | 5.2 | .00484 | 0.049 | 2.39 | .87155 | 0.917 | 1.05 |
| hsa-miR-3913–5p | 4.9 | 2.1 | 3.3 | .00244 | 0.031 | 2.29 | .20154 | 0.334 | 1.48 |
| hsa-miR-135b-5p | 6699 | 3644 | 3512 | .00079 | 0.024 | 1.84 | 0 | 0 | 1.91 |
| hsa-miR-744–3p | 26 | 18 | 14 | .00073 | 0.024 | 1.49 | .00027 | 0.003 | 1.85 |
| hsa-miR-17–3p | 122 | 90 | 75 | .00072 | 0.024 | 1.35 | .06266 | 0.142 | 1.62 |
| hsa-miR-574–3p | 2649 | 4086 | 5085 | .0052 | 0.05 | 0.65 | .00016 | 0.003 | 0.52 |
| hsa-miR-3200–3p | 3.9 | 6.3 | 5.9 | .00012 | 0.012 | 0.62 | .02797 | 0.078 | 0.66 |
| hsa-let-7d-5p | 2872 | 4688 | 5415 | .00174 | 0.031 | 0.61 | .00017 | 0.003 | 0.53 |
| hsa-miR-130a-3p | 11926 | 19878 | 14234 | .00253 | 0.031 | 0.6 | .65199 | 0.757 | 0.84 |
| hsa-miR-100–5p | 7424 | 13920 | 10393 | .00211 | 0.031 | 0.53 | .17437 | 0.301 | 0.71 |
| hsa-miR-195–5p | 1031 | 2019 | 1973 | .00495 | 0.049 | 0.51 | .00164 | 0.01 | 0.52 |
| hsa-miR-532–5p | 869 | 1777 | 2116 | .00088 | 0.024 | 0.49 | .0003 | 0.003 | 0.41 |
| hsa-miR-585 | 2.2 | 4.4 | 7.1 | .00179 | 0.031 | 0.49 | .00164 | 0.01 | 0.3 |
| hsa-miR-455–3p | 258 | 537 | 608 | .00235 | 0.031 | 0.48 | .00007 | 0.001 | 0.42 |
| hsa-miR-138–5p | 2831 | 6088 | 6265 | .00244 | 0.031 | 0.47 | .01952 | 0.062 | 0.45 |
| hsa-miR-1249 | 10 | 24 | 44 | .00399 | 0.043 | 0.43 | .00004 | 0.001 | 0.23 |
| hsa-miR-145–5p | 19539 | 48752 | 65418 | .00055 | 0.024 | 0.4 | 0 | 0 | 0.3 |
| hsa-miR-20b-5p | 54 | 133 | 168 | .00198 | 0.031 | 0.4 | .00151 | 0.009 | 0.32 |
| hsa-miR-1180 | 10 | 26 | 21 | .00061 | 0.024 | 0.39 | .10622 | 0.203 | 0.48 |
| hsa-miR-652–3p | 2.9 | 7.4 | 5.0 | .00471 | 0.049 | 0.38 | .27762 | 0.407 | 0.57 |
| hsa-miR-1291 | 1.8 | 5.2 | 4.5 | .00007 | 0.01 | 0.35 | .00157 | 0.01 | 0.4 |
| hsa-miR-138–1-3p | 60 | 189 | 260 | .00024 | 0.017 | 0.31 | 0 | 0 | 0.23 |
| hsa-miR-451a | 23566 | 76540 | 73460 | .0015 | 0.031 | 0.31 | .00441 | 0.019 | 0.32 |
| hsa-miR-486–5p | 24 | 77 | 75 | .00254 | 0.031 | 0.31 | .04174 | 0.105 | 0.31 |
| hsa-miR-144–5p | 138 | 499 | 535 | .00023 | 0.017 | 0.28 | .00073 | 0.006 | 0.26 |
| hsa-miR-486–3p | 4.2 | 18 | 24 | .00001 | 0.002 | 0.23 | 0 | 0 | 0.17 |
| hsa-miR-873–5p | 6.6 | 30 | 29 | .0009 | 0.024 | 0.22 | .00678 | 0.027 | 0.23 |
| hsa-miR-204–5p | 254 | 1241 | 1041 | .00275 | 0.033 | 0.2 | .00423 | 0.019 | 0.24 |
| hsa-miR-7–2-3p | 128 | 773 | 981 | .00292 | 0.034 | 0.17 | .00065 | 0.006 | 0.13 |
| hsa-miR-1179 | 29 | 214 | 229 | .00214 | 0.031 | 0.14 | .00102 | 0.008 | 0.13 |
Figure 2.
Expression levels of 44 miRNAs deregulated in PTC tumors compared with adjacent normal tissue (FDR < 0.05) shown in PTC-T, PTC-N, and NN.
Validation of deregulated miRNAs with microarray
To confirm the observed deregulation of 44 miRNAs in PTC-T compared with PTC-N, we analyzed miRNA expression in a miRNA microarray experiment on an additional set of 9 PTC T/N tissue pairs. It has to be underlined, however, that due to cross-hybridization, the signal on the microarray platform is captured from both canonical miRNAs and isomiRs, even though the probes are designed for canonical miRNAs only. The probes for 30 of 44 miRNAs were present in the miRNA microarray platform, and we found a significant correlation between the fold change PTC-T/PTC-N detected by the microarray and NGS study (r = 0.64, P = .0001, Supplemental Table 4).
microRNA expression and status of BRAF V600E mutation
We examined the BRAF (V600E) mutation in DNA from PTC-T and PTC-N samples and found 64.3% of tumors (9 of 14 samples) to be indeed mutation positive. None of the PTC-N samples were positive, which confirms their control status. We compared the miRNA expression profiles between the BRAF and BRAF V600E samples and found that the expression of miRNAs did not depend on the BRAF mutation status (Supplemental Table 5). Neither of the analyzed patients had a RET/PTC1 or RET/PTC3 rearrangement.
Significant isomiRs
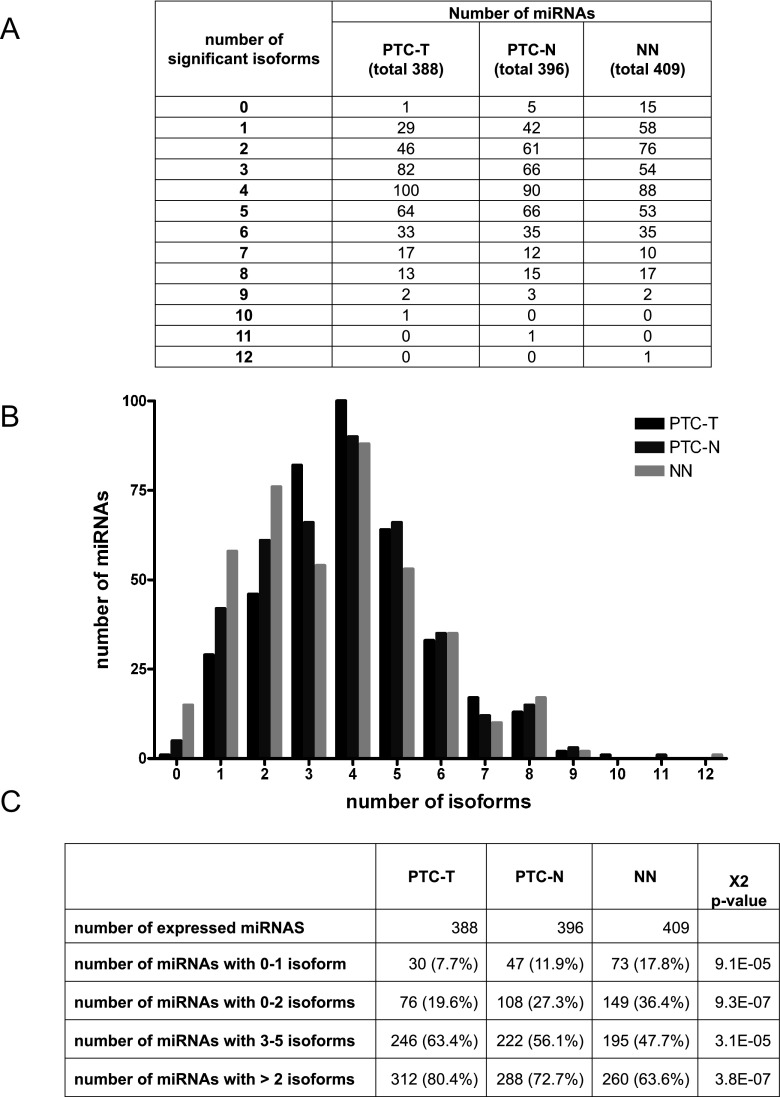
Almost all individual miRNA genes produced several mature miRNA products that differed in length, called isomiRs (Figure 3A). We detected 1749 significantly expressed isomiRs (expression > 1% of the total expression of a particular miRNA, in at least 80% of samples within any of the studied groups, Supplemental Table 6), including 132, 61, and 56 isoforms specific for PTC-T, PTC-N, and NN, respectively (Figure 1B). The average number of isoforms per miRNA was 3.96 ± 1.75, 3.8 ± 1.9, and 3.51 ± 2 for PTC-T, PTC-N, and NN, respectively (Figure 3B). The distribution of expressed isomiRs per miRNA differed between tumor tissue and normal tissues, with more isoforms expressed in tumors: 2 or more isoforms produced by 80.4%, 72.7%, and 63.6% of miRNAs in PTC-T, PTC-N, and NN, respectively (P = 3.8 × 10−7, Figure 3, B and C).
Figure 3.
Distribution of isomiRs per miRNA. Table (A) and graph (B) show the numbers of significant miRNAs producing the definite numbers of significant isoforms, and P values (C) of a χ2 test of the distribution of isoforms per miRNA in PTC-T, PTC-N, and NN.
Correlation between the expression of the reference molecules deposited in the miRbase (canonical miRNAs) and their corresponding isoforms in most cases was high [Pearson's correlation coefficient median (r) = 0.68, median FDR = 1.6e-6; Supplemental Figure 1]. However, the sequence analysis of expressed miRNAs revealed that canonical miRNAs were present in only 57.6%–64.1% of miRNAs (depending on tissue type); moreover, the reference sequences were dominant (of highest expression) in only 14.1%–15.8% of miRNA genes (Supplemental Table 7).
Significant seeds
Because recognition of a target gene by a miRNA is mainly dependent on its seed sequence and all miRNAs sharing the same seed sequence potentially regulate the same target genes, we have calculated the tissue-specific seed power in terms of the expression of all isomiRs carrying the same seed sequence. The analysis has shown that miRNAs expressed in thyroid tissue can be grouped based on the seed sequence that is shared by a number of isomiRs, often belonging to different miRNA families. As a result, we received a list of 513 seeds that are present in identified isomiRs and can serve as a basis for the genome-wide prediction of genes targeted by the thyroid-expressed miRNAs (Supplemental Table 8). Of the identified seeds, 38, 17, and 18 were specific for PTC-T, PTC-N, and NN, respectively (Figure 1C). Such specific seeds reside in a group of miRNAs that regulate genes, depending specifically on the disease status of the thyroid gland.
Although it was shown that individual miRNA genes produce several isomiRs, most of such isoforms share the same seed region (61.5%–66.5%, depending on the tissue type, Table 2), so their functionality should be similar. Nevertheless, depending on the tissue type, 28.4%–32.8% of miRNAs produce isomiRs with 2 alternative seed regions, and 4.8%–5.7% of miRNAs produce isomiRs with 3 alternative seed regions. Two miRNAs were shown to have isoforms with 4 different seeds, each targeting a unique set of genes.
Table 2.
Distribution of the Seed Sequences
| Number of Seeds per miRNA | Number of miRNAs |
||
|---|---|---|---|
| PTC-T (n = 388) | PTC-N (n = 396) | NN (n = 409) | |
| 1 | 238 (61.5%) | 244 (62.4%) | 262 (66.5%) |
| 2 | 127 (32.8%) | 124 (31.7%) | 112 (28.4%) |
| 3 | 22 (5.7%) | 22 (5.6%) | 19 (4.8%) |
| 4 | 0 | 1 (0.3%) | 1 (0.3%) |
The numbers of significant miRNAs producing the definite numbers of significant isoforms of unique seeds are shown.
Target genes
In further steps, we used in silico analysis to identify target genes for 44 miRNAs whose expression was significantly deregulated between PTC-T and PTC-N tissue samples (FDR < 0.05, Table 1). This group potentially includes miRNAs whose deregulation is directly linked with development of PTC. Among identified miRNAs, we found a number of isomiRs produced from a single precursor that differed in their seed region. In 15 miRNAs, the isoforms contained 2 alternative seed regions, and 1 miRNA (miR-455-3p) produced isomiRs of 4 seeds. No miRNA was found to produce isoforms with 3 alternative seeds (Table 3). In case of a miRNA, which is expressed in isoforms exhibiting 4 different seeds, the number of potential target genes is increased 3.2 times compared with a list of genes regulated solely by the canonical form. More than 80% of all target genes were unique for each seed, even if the sequences of isoforms differed by only 1 nucleotide (Table 3).
Table 3.
Number of Putative Target Genes for miRNAs Deregulated in PTC Tumor
| Number of Seeds per miRNA | Number of miRNAs (n = 44) | Number of Target Genes per miRNA (Mean) |
||||
|---|---|---|---|---|---|---|
| Total | Unique | Shared by 2 Seeds | Shared by 3 Seeds | Shared by 4 Seeds | ||
| 1 | 28 | 423.8 ± 370.7 | n/a | n/a | n/a | n/a |
| 2 | 15 | 1303.9 ± 712.5 | 1047 ± 454.8 (80.3%) | 256.9 ± 293.1 (19.7%) | n/a | n/a |
| 3 | 0 | n/a | n/a | n/a | n/a | n/a |
| 4 | 1 | 1467 | 1196 (81.5%) | 229 (15.6%) | 31 (2.1%) | 11 (0.7%) |
Abbreviation: n/a, not applicable. The table shows how many miRNAs deregulated in PTC tumors produce significant isoforms of unique seeds and the number of their unique and shared target genes.
Discussion
The standard method of miRNA identification used up to now relied on in silico prediction and laborious small RNA cloning. Newly introduced NGS techniques significantly facilitated miRNA studies, providing robust tools for comprehensive, simultaneous analysis of both nucleotide sequence and expression levels of all the miRNAs detected in the analyzed tissue (32, 33). Recently published studies revealed the full miRNA profiles of human tissues including the brain, heart, kidney, liver, lung, ovary, placenta, spleen, testes, and thymus as well as the human embryonic stem cells (14). A number of performed studies focused on the elucidation of aberrances of the miRNome in cancers, such as acute myeloid leukemia (34) and breast (35), prostate (36), and bladder (37) cancers. However, no in-depth study of the miRNome of the thyroid gland has been performed so far. Therefore, we have used the SOLiD massive parallel sequencing to obtain comprehensive information on the sequences and expression levels of all the microRNAs present in the normal thyroid gland tissue (control, noncancerous thyroid specimens) as well as on their changes seen in thyroid tumorigenesis (PTC tumor and unaffected tissues adjacent to tumors). Our results disclosed a complete miRNome of the thyroid gland and PTC, showing severe deregulation of several miRNAs and profound length variation of microRNAs leading to an even more complicated network of gene expression regulation. Interestingly, many of the miRNAs expressed in the thyroid gland are produced from the passenger strand of a pre-miRNA. Their deregulation in cancer additionally indicates that these molecules are of no lesser importance than their counterparts derived from the leading strand.
The sequence variations of many of the identified isomiRs consist of the addition or deletion of nucleotides at their 5′ end, which results in a change of their seed region. The existence of isomiRs is therefore highly important due to a pool of seed sequences they introduce to the cell's regulatory pathways. Because all miRNAs sharing the same seed potentially bind and regulate the same target genes, we analyzed the abundance of seed sequences among identified isomiRs. The analysis has shown that 1749 isomiRs expressed in the thyroid tissue can be grouped to produce 513 seeds (Figure 1 and Supplemental Table 8). IsomiRs sharing the same seed sequence very often belong to distinct miRNA families but will potentially regulate the same target genes. Thus, even though expression of a single isomiR might not be very high, the concert action of all the isoforms sharing a seed region (so-called seed power) will induce strong biological responses. Interestingly, many of the novel isomiRs are expressed in significantly higher levels than their canonical counterparts previously identified by other teams and deposited in the miRBase, indicating that cleavage and processing of a pre-miRNA is most probably highly tissue specific.
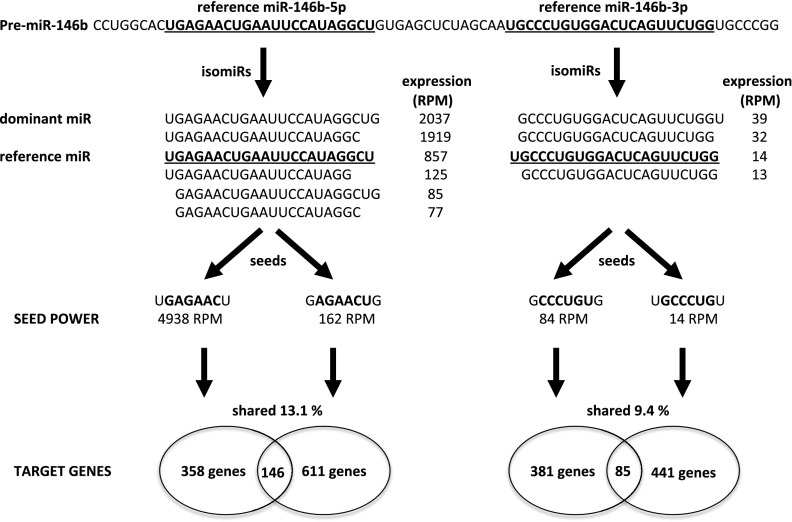
Among the identified miRNAs, both mature products of mir-146b are of particular interest due to their high deregulation in tumors and a large number of isoforms (Figure 4). The leading strand of the pre-miRNA produces 6 isoforms whose expression in PTC samples is 14.4–28.7 times higher than in control tissue. Notably, the reference sequence from the miRbase is not the dominant, most highly expressed isoform (Figure 4). The isoforms of miR-146b-5p vary at their 5′ ends with a 1-nt difference, creating 2 alternative seeds. Additionally, the passenger strand of the pre-miRNA produces 4 significantly expressed isoforms, whose expression is 16–39 times higher in PTC tumors compared with control tissue. These isoforms also differ at their 5′ end, and this alteration leads to the production of isomiRs with 2 distinct seeds. The TargetRank prediction revealed that each seed binds a unique set of target genes with only 13.1% and 9.4% of target genes concertedly regulated by both significant seeds of miR-146b-5p and miR-146b-3p, respectively. These shared targets seem to be the most interesting, first because the increased number of miRNAs that bind a transcript of a gene will result in a higher impact on its expression, and second because this specific kind of regulation depends solely on the products of a single miRNA precursor and, moreover, is immune to the biases of Dicer activity. Even if the Dicer cleavage site within the pre-miRNA sequence is changed and an alternative isoform produced, gene regulation will still occur because all the existing isoforms bind the same transcript. Although the miRNA's inhibitory effects on individual mRNAs are generally modest, the combined effects of alterations of numerous miRNAs on their multiple target mRNAs, in fact whole pathways, can induce strong biological responses.
Figure 4.
Functional pathway of pre-miR-146b. Pre-miR-146b produces 2 mature miRNAs existing in several isoforms, which harbor 4 alternative binding seeds, each targeting a unique set of genes. Shared target genes cooperatively regulate 2 important pathways of tumorigenesis. RPM are in PTC-T samples (mean).
The obtained data are of great importance for understanding gene regulation networks within the thyroid gland because it provides information on the miRNAs that can be important players in this process. As our study has shown, not all miRNAs deposited in the miRbase are expressed in the thyroid; thus, even though in silico analyses reveal potential binding of such miRNAs to thyroid-expressed genes, such predicted interaction will have no biological relevance. Simultaneously, newly discovered isoforms of miRNAs constitute a group of previously unknown molecules that regulate a large number of target genes. The striking differences in the miRNA expression levels between control and cancer tissues indicate that the role of miRNAs in thyroid tumorigenesis is profound and complex.
Comprehensive information on the miRNAs expressed in the thyroid gland provided in this paper is a unique tool for researchers working on gene regulation pathways in the thyroid gland. Understanding of the miRNAs and their isoforms specifically expressed in the thyroid allows for proper design of experiments focused on miRNAs, whereas relying solely on data deposited in the miRbase and related miRNA repositories can be misleading because the expression of miRNAs is highly tissue specific. For the same reason, the identified isomiRs may serve as diagnostic and prognostic tools for thyroid cancers as well as for identification of thyroid metastases to other organs.
Acknowledgments
This work was supported by Polish Ministry of Science and Higher Education Grant NN401584838 and the Foundation for Polish Science, Programme FOCUS, and Programme TEAM, cofinanced by the European Union European Regional Development Fund. The Laboratory of Genomic Medicine of the Medical University of Warsaw participates in Bastion, a program financed by the European Union (Grant FP7-REGPOT-2012-CT2012-316254-BASTION).
Disclosure Summary: The authors have nothing to disclose.
Funding Statement
This work was supported by Polish Ministry of Science and Higher Education Grant NN401584838 and the Foundation for Polish Science, Programme FOCUS, and Programme TEAM, cofinanced by the European Union European Regional Development Fund. The Laboratory of Genomic Medicine of the Medical University of Warsaw participates in Bastion, a program financed by the European Union (Grant FP7-REGPOT-2012-CT2012-316254-BASTION).
Footnotes
- FDR
- false discovery rate
- isomiR
- single microRNA gene giving rise to a mature products that differ in length
- miRNA
- microRNA
- NGS
- next-generation sequencing
- NN
- control, noncancerous thyroid specimens
- PTC
- papillary thyroid carcinoma
- PTC-N
- unaffected tissue adjacent to but not infiltrated by tumor from the same patient
- PTC-T
- PTC tumors
- RPM
- reads per million
- T/N
- tumor/normal.
References
- 1. Chen AY, Jemal A, Ward EM. Increasing incidence of differentiated thyroid cancer in the United States, 1988–2005. Cancer. 2009;115:3801–3807. [DOI] [PubMed] [Google Scholar]
- 2. Risch N. The genetic epidemiology of cancer: interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol Biomarkers Prev. 2001;10:733–741. [PubMed] [Google Scholar]
- 3. Frich L, Glattre E, Akslen LA. Familial occurrence of nonmedullary thyroid cancer: a population-based study of 5673 first-degree relatives of thyroid cancer patients from Norway. Cancer Epidemiol Biomarkers Prev. 2001;10:113–117. [PubMed] [Google Scholar]
- 4. Gudmundsson J, Sulem P, Gudbjartsson DF, et al. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat Genet. 2009;41:460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gudmundsson J, Sulem P, Gudbjartsson DF, et al. Discovery of common variants associated with low TSH levels and thyroid cancer risk. Nat Genet. 2012;44:319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jendrzejewski J, He H, Radomska HS, et al. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci USA. 2012;109:8646–8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. He H, Nagy R, Liyanarachchi S, et al. A susceptibility locus for papillary thyroid carcinoma on chromosome 8q24. Cancer Res. 2009;69:625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. [DOI] [PubMed] [Google Scholar]
- 10. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–138. [DOI] [PubMed] [Google Scholar]
- 12. Starega-Roslan J, Krol J, Koscianska E, et al. Structural basis of microRNA length variety. Nucleic Acids Res. 2011;39:257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chiang HR, Schoenfeld LW, Ruby JG, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cloonan N, Wani S, Xu Q, et al. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 2012;12:R126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. [DOI] [PubMed] [Google Scholar]
- 16. Barbarotto E, Schmittgen TD, Calin GA. MicroRNAs and cancer: profile, profile, profile. Int J Cancer. 2008;122:969–977. [DOI] [PubMed] [Google Scholar]
- 17. Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769. [DOI] [PubMed] [Google Scholar]
- 18. Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. [DOI] [PubMed] [Google Scholar]
- 19. Marcucci G, Radmacher MD, Maharry K, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1919–1928. [DOI] [PubMed] [Google Scholar]
- 20. Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2008;105:7269–7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jazdzewski K, Liyanarachchi S, Swierniak M, et al. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci USA. 2009;106:1502–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jazdzewski K, de la Chapelle A. Genomic sequence matters: a SNP in microRNA-146a can turn anti-apoptotic. Cell Cycle. 2009;8:1642–1643. [PMC free article] [PubMed] [Google Scholar]
- 23. de la Chapelle A, Jazdzewski K. MicroRNAs in thyroid cancer. J Clin Endocrinol Metab. 2011;96:3326–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. He H, Jazdzewski K, Liyanarachchi S, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–19080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pallante P, Visone R, Ferracin M, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13:497–508. [DOI] [PubMed] [Google Scholar]
- 26. Liu CG, Calin GA, Meloon B, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101:9740–9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pichler M, Balic M, Stadelmeyer E, et al. Evaluation of high-resolution melting analysis as a diagnostic tool to detect the BRAF V600E mutation in colorectal tumors. J Mol Diagn. 2009;11:140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nikiforov YE, Rowland JM, Bove KE, Monforte-Munoz H, Fagin JA. Distinct pattern of ret oncogene rearrangements in morphological variants of radiation-induced and sporadic thyroid papillary carcinomas in children. Cancer Res. 1997;57:1690–1694. [PubMed] [Google Scholar]
- 29. Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martin MM. Cutadapt removes adaptor sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:1. [Google Scholar]
- 31. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cloonan N, Grimmond SM. Transcriptome content and dynamics at single-nucleotide resolution. Genome Biol. 2008;9:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morin RD, O'Connor MD, Griffith M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kuchenbauer F, Morin RD, Argiropoulos B, et al. In-depth characterization of the microRNA transcriptome in a leukemia progression model. Genome Res. 2008;18:1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Farazi TA, Horlings HM, Ten Hoeve JJ, et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71:4443–4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Szczyrba J, Loprich E, Wach S, et al. The microRNA profile of prostate carcinoma obtained by deep sequencing. Mol Cancer Res. 2010;8:529–538. [DOI] [PubMed] [Google Scholar]
- 37. Han Y, Chen J, Zhao X, et al. MicroRNA expression signatures of bladder cancer revealed by deep sequencing. PLoS One. 2011;6:e18286. [DOI] [PMC free article] [PubMed] [Google Scholar]