Key Points
We demonstrate frequent plasma detection of multiple double-stranded DNA viruses after allogeneic hematopoietic cell transplantation.
There was a dose-response relationship of the cumulative burden of virus exposure with early (days 0-100) and late (days 101-365) mortality.
Abstract
Strategies to prevent active infection with certain double-stranded DNA (dsDNA) viruses after allogeneic hematopoietic cell transplantation (HCT) are limited by incomplete understanding of their epidemiology and clinical impact. We retrospectively tested weekly plasma samples from allogeneic HCT recipients at our center from 2007 to 2014. We used quantitative PCR to test for cytomegalovirus, BK polyomavirus, human herpesvirus 6B, HHV-6A, adenovirus, and Epstein-Barr virus between days 0 and 100 post-HCT. We evaluated risk factors for detection of multiple viruses and association of viruses with mortality through day 365 post-HCT with Cox models. Among 404 allogeneic HCT recipients, including 125 cord blood, 125 HLA-mismatched, and 154 HLA-matched HCTs, detection of multiple viruses was common through day 100: 90% had ≥1, 62% had ≥2, 28% had ≥3, and 5% had 4 or 5 viruses. Risk factors for detection of multiple viruses included cord blood or HLA-mismatched HCT, myeloablative conditioning, and acute graft-versus-host disease (P values < .01). Absolute lymphocyte count of <200 cells/mm3 was associated with greater virus exposure on the basis of the maximum cumulative viral load area under the curve (AUC) (P = .054). The maximum cumulative viral load AUC was the best predictor of early (days 0-100) and late (days 101-365) overall mortality (adjusted hazard ratio [aHR] = 1.36, 95% confidence interval [CI] [1.25, 1.49], and aHR = 1.04, 95% CI [1.0, 1.08], respectively) after accounting for immune reconstitution and graft-versus-host disease. In conclusion, detection of multiple dsDNA viruses was frequent after allogeneic HCT and had a dose-dependent association with increased mortality. These data suggest opportunities to improve outcomes with better antiviral strategies.
Introduction
Double-stranded DNA (dsDNA) viruses contribute to substantial morbidity after allogeneic hematopoietic cell transplantation (HCT) despite major advances in treatment and prevention. Clinically relevant dsDNA viruses include the herpesviruses, BK polyomavirus (BKV), and adenovirus (AdV). Preventive strategies reduce disease due to herpes simplex virus (HSV), varicella zoster virus (VZV), and human cytomegalovirus (CMV) after HCT,1 but other dsDNA viral infections lack definitive treatment and continue to contribute to morbidity and mortality.2-6 Use of alternative donor sources and graft manipulation (eg, cord blood cells and haploidentical donors, T-cell depletion) allows for greater access to HCT but may result in delayed engraftment and adaptive immune reconstitution with a consequent increased burden of dsDNA virus-associated complications.7-10
The relative lack of approved antiviral therapies for dsDNA viruses, and the toxicities of currently available medications, are key factors limiting broader antiviral prevention strategies. However, novel therapeutics with broad activity are in varying stages of development and may improve HCT outcomes.11-13 Although many studies have reported the epidemiology of individual dsDNA virus infections after HCT, few of them comprehensively describe the clinical significance of multiple dsDNA viruses. Additionally, the attributable impact of viral reactivation on outcomes in the context of poor immune reconstitution is not well studied. To inform the application of new therapeutic approaches, we performed a retrospective observational study of plasma detection of CMV, BKV, human herpesvirus 6B (HHV-6B), HHV-6A, AdV, and Epstein-Barr virus (EBV) in a large cohort of allogeneic HCT recipients.
Materials and methods
Patients and samples
We identified patients of any age who received a first allogeneic HCT at our center. We prespecified a target cohort enriched for higher-risk patients that consisted of human leukocyte antigen–matched (HLA-matched) HCT recipients (ie, 10/10 allele or antigen match), HLA-mismatched HCT recipients, and cord blood HCT recipients with 125 to 150 patients per group. These 3 groupings were specified a priori on the basis of expected differential risks for viral infections and to establish cohorts of sufficient size for informative analyses. We selected consecutive patients who had availability of ≥60% of possible weekly plasma samples while alive between days 0 and 100 post-HCT. Specimens were left over from clinical CMV testing and preserved when available in an unbiased fashion. To establish the prespecified cohort, we included patients through February 2007. Our center performed 1926 allogeneic HCTs during this period. Of these patients, we included 125 (54%) of 232 cord blood HCT recipients, 125 (41%) of 302 HLA-mismatched HCT recipients (including 47 haploidentical donors), and 154 (11%) of 1392 HLA-matched HCT recipients. Additional details of donor relatedness and matching are in Table 1.
Table 1.
Demographic and clinical characteristics of the study cohort compared with all other HCT recipients during the study period
| Characteristic | All HCTs during study period, n (%); N = 1926 | Selected cohort; N = 404 | Unselected cohort; N = 1522* |
|---|---|---|---|
| Age >21 y | 1591 (83) | 358 (89) | 1233 (81) |
| Female sex | 809 (42) | 179 (44) | 630 (41) |
| White | 1430 (74) | 277 (69) | 1153 (76) |
| CMV donor negative, recipient negative | 563 (29) | 57 (14) | 506 (33) |
| HCT comorbidity index† | |||
| Low (0) | 223 (18) | 70 (17) | 153 (18) |
| Intermediate (1-2) | 391 (31) | 133 (33) | 258 (30) |
| High (≥3) | 658 (52) | 201 (50) | 457 (53) |
| Underlying disease | |||
| Acute leukemia | 928 (48) | 195 (48) | 733 (48) |
| Chronic leukemia | 186 (10) | 58 (14) | 128 (8) |
| Lymphoma | 232 (12) | 41 (10) | 191 (13) |
| Nonmalignant | 158 (8) | 24 (6) | 134 (9) |
| Other | 422 (22) | 86 (21) | 336 (22) |
| Higher-risk disease‡ | 394 (20) | 75 (19) | 319 (21) |
| Myeloablative conditioning regimen§ | 683 (35) | 155 (38) | 528 (35) |
| HCT type‖ | |||
| HLA matched | 1392 (72) | 154 (38) | 1,238 (81) |
| HLA mismatched | 302 (16) | 125 (31) | 177 (12) |
| Cord blood | 232 (12) | 125 (31) | 107 (7) |
| Donor related | 703 (37) | 131 (32) | 572 (38) |
| Acute GVHD, grade 3-4¶ | 246 (13) | 46 (11) | 200 (13) |
Unknown data for N patients in the following categories: 89, race; 54, CMV serostatus; 654, HCT comorbidity index.
Based on the HCT comorbidity index.40
Higher-risk disease refers to diagnoses other than acute myeloid leukemia, acute lymphoblastic leukemia, or lymphoma in first remission, chronic myeloid leukemia in chronic phase, and refractory anemia without excess blasts.
Myeloablative regimens included any regimen containing ≥800 cGY total body irradiation, any regimen containing carmustine/etoposide/cytarabine/melphalan (BEAM), or any regimen containing busulfan/cyclophosphamide with or without antithymocyte globulin.
Categories included related and unrelated donors. HLA-matched HCTs included 76 related donors and 78 unrelated donors. HLA-mismatched HCTs included 55 related donors and 70 unrelated donors; this group consisted of 47 haploidentical donors, 76 9/10-matched donors, and 2 8/10-matched donors. All cord blood HCTs were mismatched, and 12 were single unit.
Acute graft-versus-host disease grades were categorized as previously described.41
We excluded patients who received experimental therapies for dsDNA virus prophylaxis as part of a clinical trial. Two patients with likely inherited chromosomally integrated (ci)HHV-6 were excluded.14
All patients provided written consent, and the protocol was approved by the Fred Hutchinson Cancer Research Center institutional review board.
Antiviral prophylaxis and treatment per clinical standards of care
All patients seropositive for HSV or VZV received acyclovir prophylaxis through ≥100 or 365 days post-HCT, respectively.1 CMV seropositive cord blood HCT recipients received an intensified strategy using ganciclovir pre-HCT and high-dose valacyclovir post-HCT after June 2008.15 CMV reactivation was monitored by polymerase chain reaction (PCR) and treated per clinical standards of care.15,16 Ganciclovir was the first-line antiviral postengraftment, and foscarnet was the second. Clinical testing and treatment of non-CMV viruses was at the discretion of treating providers and HCT protocols.
Virologic testing
Samples were retrospectively tested for BKV, HHV-6B, HHV-6A, AdV, and EBV. CMV results were obtained from routine clinical testing. Real-time quantitative PCR with a lower limit of detection of 1 copy of virus DNA/reaction (50 copies/mL of plasma) was performed for each virus as previously described.17-21 These cutoffs were optimized to minimize the proportion of false positives or false negatives. CMV results reported as international units were converted to copies per milliliter to allow for comparisons on the same scale. Technicians were blinded to patients’ clinical status. Patients with HHV-6 detection in >50% of samples were tested for inherited ciHHV-6 by droplet digital PCR as previously described.14
Immune reconstitution
For the entire cohort, we captured neutrophil engraftment, absolute lymphocyte count (ALC) of <200 cells/mm3, and absolute monocyte count (AMC) of <300 cells/mm3 at day 30 ±2 days post-HCT as biomarkers for immune reconstitution.22-24 Neutrophil engraftment was defined as the first of 3 consecutive days of an absolute neutrophil count of >500 cells/mm3. Additionally, we captured the dose and days of steroid use during the first 100 days post-HCT. Steroid use was categorized as 0, <1 mg/kg, 1 to <2 mg/kg, and ≥2 mg/kg prednisone equivalent per day.
Among the cord blood HCT group, we also captured absolute CD3 count of <50 cells/mm3, absolute CD4 count of <50 cells/mm3, absolute CD8 count of <50 cells/mm3, and absolute CD56 count of <50 cells/mm3 at day 28 post-HCT as additional biomarkers of immune reconstitution.22-24 Data were available for 87 (70%) of the 125 cord blood HCT recipients included in this study.
Definitions
Overall mortality was defined as mortality occurring for any reason. Nonrelapse mortality (NRM) was defined as mortality occurring for reasons other than relapse in patients receiving myeloablative HCTs or for reasons other than relapse or progression of underlying disease in patients receiving nonmyeloablative HCTs. Cause of death was classified by using an established hierarchical schema.25 Definitions of other covariates and secondary endpoints are in Table 1 and the supplemental Methods (available on the Blood Web site), respectively.
Statistical analyses
We abstracted demographic and clinical information from medical records and databases. Cumulative incidence curves for the number of viruses detected between days 0 and 100 post-HCT were generated, treating death or second HCT as competing risk events. We used Cox proportional hazards regression models to evaluate univariate and multivariate hazard ratios (HRs) and their 95% confidence intervals (CIs) for characteristics associated with detection of multiple viruses.
Cox models were used to analyze the association of virus detection between days 0 and 100 post-HCT, the primary predictor of interest, and an endpoint of overall mortality through day 365. Viral detection was incorporated as a time-dependent variable during the testing period through day 100 and in separate models as a time-invariant variable between days 101 and 365, using the last recorded time-dependent value. We tested 4 primary models, each using a different definition of virus detection. Virus detection was analyzed as a (1) categorical variable (0-1, 2-3, ≥4 viruses) determined from the cumulative number of different viruses detected at any time, (2) discrete variable (0-5) for the cumulative number of different viruses detected at any time, and (3) discrete variable for the cumulative maximum number of different viruses detected at the same time. Any single positive test counted as detection of a given virus. In a fourth model, we accounted for the cumulative burden of viral exposure by calculating the average area under the curve (AUC) (log10 copies/mL) for each virus. Individual virus AUCs were summated to calculate a cumulative viral load AUC per patient. We also performed these analyses using a weighted viral load AUC to account for differential viral loads per virus. Similar models were used for an endpoint of NRM. We also generated a model among day-30 survivors to incorporate biomarkers of immune reconstitution. Kaplan-Meier plots of time to overall survival and cumulative incidence curves of time to NRM were generated to display the data.
Secondary endpoints that were explored in additional analyses included National Institutes of Health late acute or chronic graft-versus-host disease (GVHD), chronic kidney disease, acute kidney or liver injury, lower respiratory tract disease, and respiratory failure (see the supplemental Methods section).
We performed an a priori power calculation to inform sample size on the basis of estimated minimum detectable HRs for the association of multiple viruses with overall mortality (see the supplemental Methods section). For adjusted analyses, variables with P ≤ .2 in univariable analyses and those considered to have important biological relevance were selected. Variables were retained in final models if P < .1 or if their inclusion modified the effect of the primary predictor of interest by >10%. To account for multiple comparisons, P values between .05 and .01 should be considered suggestive and those that are ≤.01 considered significant. SAS version 9.3 (SAS Institute, Cary, NC) was used for analyses.
Statistical considerations relating to weighted analyses to adjust for HCT type sampling, variable selection, interactions, and methods for secondary endpoint and exploratory analyses are in the supplemental Methods section.
Results
We included 404 pediatric and adult first allogeneic HCT recipients from 2007 to 2014. Patient demographics and clinical characteristics for the study cohort and the overall cohort from which they were selected are shown in Table 1. The distribution of characteristics was similar between selected and unselected patients aside from HCT type (as prespecified) and CMV serostatus. Supplemental Table 1 demonstrates characteristics stratified by the number of viruses detected.
Detection of dsDNA viruses
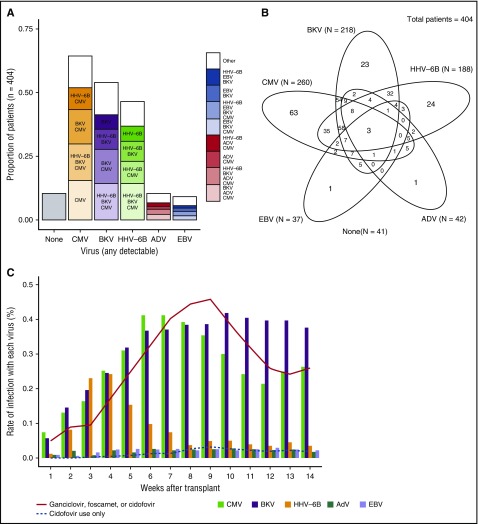
We retrospectively tested a total of 4996 plasma samples obtained between days 0 and 100 post-HCT. The majority of patients had weekly samples available for each time point, with a median of 13 samples per patient (interquartile range [IQR], 12-14) and 7 days between samples (IQR, 7-7). CMV was the most frequently detected virus (65% of patients), followed by BKV (54%), HHV-6B (46%), AdV (10%), and EBV (9%) (Figure 1A). HHV-6A was detected in 1 (0.2%) patient who did not have inherited ciHHV-6A; this patient is included in the HHV-6B category for analyses. Median viral loads were 191 copies/mL (IQR, 81-617) for CMV, 3090 (617-16 596) for BKV, 741 (229-3236) for HHV-6B, 1023 (91-16 982) for AdV, and 195 (91-589) for EBV.
Figure 1.
Proportion of patients with dsDNA virus detection. (A) Proportion of patients with individual viruses detected through day 100 post-HCT, as well as the proportion with additional viruses detected. (B) Venn diagram depicting the number of patients with detection of dsDNA viruses alone and in combination; ovals are not to scale. (C) Proportion of patients with detection of each virus by week post-HCT.
CMV, BKV, and HHV-6B were the viruses most frequently seen in combination. CMV was the most frequently detected in isolation (24%), whereas AdV and EBV were detected as the sole virus in only 2% and 3% of patients, respectively (Figure 1B). HHV-6B detection peaked the earliest, followed by CMV and then BKV; AdV and EBV were detected at relatively stable rates throughout this period (Figure 1C).
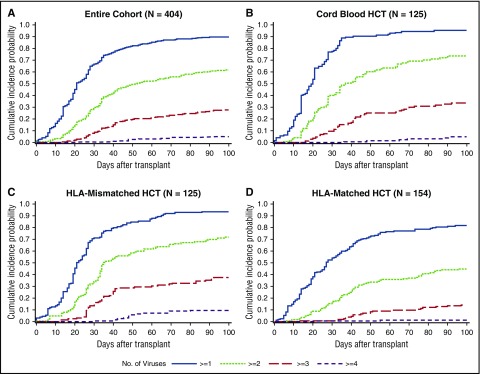
The cumulative incidence of detection of multiple viruses based on any single positive PCR result at any time within 100 days post-HCT was frequent; 90% of patients had ≥1, 62% had ≥2, 28% had ≥3, and 5% had 4 or 5 viruses detected (Figure 2A). Median time to first virus detection was day 20 (IQR, 13-32), and median time to the maximum number of viruses detected per patient was day 31 (IQR, 21-48). Cumulative incidences were higher in each category after cord blood and HLA-mismatched HCT in comparison with HLA-matched HCT (Figure 2B-D).
Figure 2.
Cumulative incidence plot of time to any dsDNA virus detection by day 100 post-HCT. (A) The cumulative incidence of each category of the number of viruses detected is demonstrated in the entire cohort. The cumulative incidences are higher in each category among cord blood HCT recipients (B) and HLA-mismatched HCT recipients (C) than among HLA-matched HCT recipients (D).
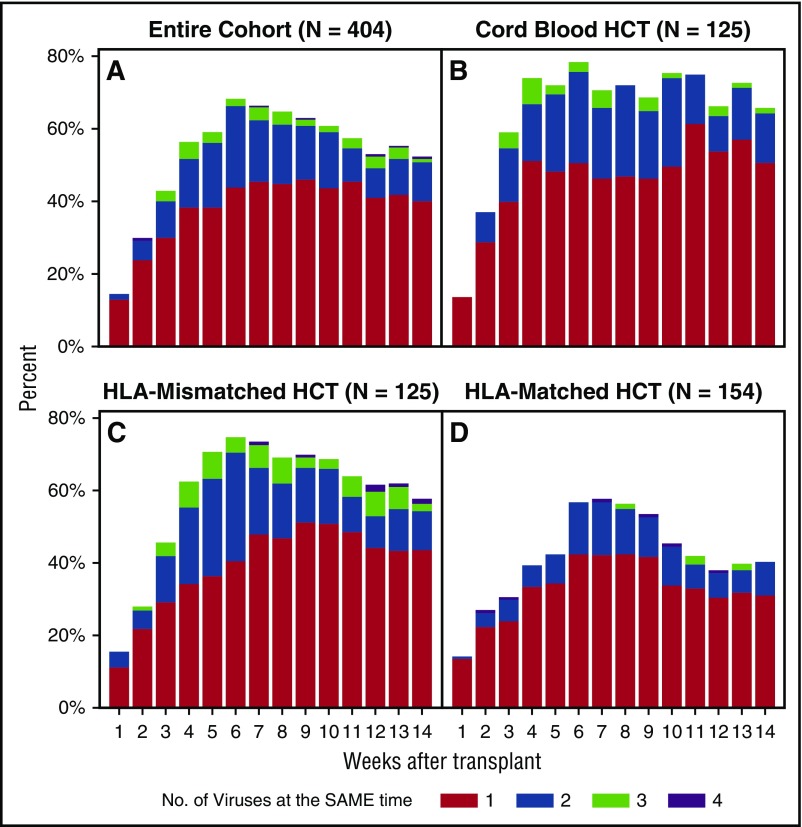
When assessed by the maximum number of viruses detected at the same time in the overall cohort, 37% of patients had 2 viruses and 12% had 3 or 4 viruses in the blood concurrently. The proportion of patients with multiple viruses detected per week is depicted in Figure 3A. Single and multiple virus detection peaked by week 6 and remained relatively stable through week 14. Patients receiving cord blood HCTs had the earliest, most frequent, and most persistent detection of viruses (Figure 3B). HLA-mismatched HCT recipients had a similar pattern (Figure 3C), whereas HLA-matched HCT recipients had less viral detection overall (Figure 3D). The subgroup of haploidentical HCT recipients (n = 47) had higher rates of detection than did other patients in the HLA-mismatched group and the cord blood group (data not shown).
Figure 3.
Histogram of the weekly proportion of patients with concurrent detection of multiple dsDNA viruses. The proportion of patients with multiple viruses detected per week in the entire cohort (A), cord blood HCT recipients (B), HLA-mismatched related or unrelated HCT recipients (C), and HLA-matched related or unrelated HCT recipients (D).
Clinically guided testing for dsDNA viruses (except CMV) detected ∼75% fewer patients with any given virus (see the supplemental Results section). Antiviral treatment was administered to 247 patients (61%) within the first 100 days (Figure 1C). Ganciclovir, foscarnet, and cidofovir were administered in 128 (32%), 108 (27%), and 20 (5%) patients at any time during this period, respectively (not mutually exclusive).
Risk factors for detection of dsDNA viruses
In adjusted models, patients receiving a cord blood or HLA-mismatched HCT had a significantly higher risk for detection of ≥2, ≥3, and ≥ viruses with increasing adjusted hazard ratios (aHRs) ranging from 2.03 to 7.36 (Table 2). Acute GVHD grade 3 to 4 was associated with detection of ≥2 viruses (aHR, 2.14).
Table 2.
Multivariable Cox models of risk factors for detection of multiple viruses
| Covariate | ≥2 viruses | ≥3 viruses | ≥4 viruses* |
|---|---|---|---|
| aHR (95% CI) | aHR (95% CI) | aHR (95% CI) | |
| Age ≤21 y | 2.65 (1.46-4.81)† | ||
| Male | 1.18 (1.04-1.33)‡ | ||
| White | |||
| CMV donor or recipient positive | 2.47 (1.91-3.19)† | 3.10 (1.95-4.94)† | |
| Donor related | 0.93 (0.81-1.07) | 1.00 (0.80-1.25) | |
| HCT comorbidity index | |||
| Low | Ref | Ref | |
| Intermediate | 1.03 (0.86-1.24) | 1.64 (1.22-2.22)† | |
| High | 1.07 (0.90-1.28) | 1.13 (0.83-1.53) | |
| Myeloablative conditioning | 1.36 (1.11-1.67)† | ||
| HCT type | |||
| HLA-matched | Ref | Ref | Ref |
| HLA-mismatched | 2.03 (1.75-2.37)† | 3.32 (2.64-4.18)† | 7.36 (4.13-13.1)† |
| Cord blood | 2.60 (2.16-3.14)† | 3.29 (2.46-4.40)† | 3.45 (1.62-7.36)† |
| Acute GVHD grades 3-4 | 2.14 (1.54-2.97)† |
Ref, reference.
This model supported only 2 variables because of limited events. Myeloablative conditioning was also significant after adjustment for HCT type: aHR, 4.64; 95% CI, <0.001.
P ≤ .001; ‡P ≤ .01.
Myeloablative conditioning was associated with a significantly higher risk for ≥3 (aHR, 1.36) and ≥4 viruses (aHR, 4.64; see Table 2 footnote). Age of ≤21 years was associated with detection of ≥4 viruses (aHR, 2.65).
In a linear regression model among day-30 survivors to incorporate biomarkers of immune reconstitution, day 30 ALC < 200 cells/mm3 was associated with a higher maximum cumulative viral load AUC (P = .054); no association was seen with day 30 AMC < 300 cells/mm3, neutrophil engraftment, or cumulative steroid dose AUC. In a subgroup analysis of 87 cord blood HCT recipients with CD3, CD4, CD8, and CD56 cell counts tested at day 28 post-HCT, none of these parameters were significantly associated with the maximum cumulative viral load AUC in an adjusted model (data not shown).
Association of dsDNA virus detection with overall mortality and NRM
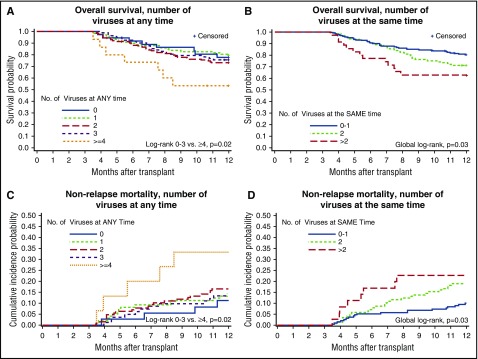
There were 125 deaths (32%) by day 365 post-HCT; 36 patients (9%) died in days 0 to 100, and 89 patients (23%) died in days 101 to 365 (supplemental Table 2). End-organ disease and death clinically attributed to dsDNA viruses was infrequent (see the supplemental Results section). Patients with more viruses detected in their plasma had significantly increased risk for overall mortality after day 100 post-HCT, as demonstrated in Kaplan-Meier plots (Figure 4). This effect appeared to be driven by patients with >3 viruses at any time or ≥2 viruses at the same time. Analyses that focused on higher viral load thresholds resulted in similar patterns of association with overall mortality (see the supplemental Results and supplemental Figure 1).
Figure 4.
Plots of time to overall mortality and NRM through day 365 post-HCT among day-100 survivors (n = 358). Kaplan-Meier plots of overall mortality stratified by the cumulative number of different viruses detected at any time by day 100 (A) and cumulative maximum number of viruses detected at the same time by day 100 (B). Cumulative incidence curves of NRM stratified by the cumulative number of different viruses detected at any time by day 100 (C) and cumulative maximum number of viruses detected at the same time by day 100 (D).
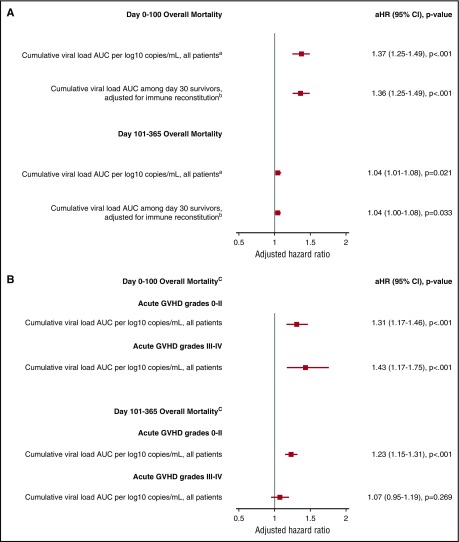
The independent association of additional virus detection (any positive PCR test) with overall mortality as a time-dependent variable through day 100 and a time-invariant variable between days 101 and 365 post-HCT was recapitulated in adjusted Cox models. The cumulative viral load AUC definition of virus detection was most consistently associated with overall mortality and NRM and will be the focus of these results. For each log10 copies/mL increase in the cumulative viral load AUC per patient, there was a 1.37 increased hazard for overall mortality through day 100 and a 1.04 increased hazard between days 101 and 365 (Figure 5A) in a model adjusted for age, underlying disease risk, HCT comorbidity index, conditioning regimen, acute GVHD grade (time dependent), and cumulative steroid dose AUC (time dependent). Other significant risk factors included age, acute GVHD grade 3 to 4, high-HCT comorbidity index score, high-risk disease, and the cumulative steroid dose AUC (data not shown). HCT type (ie, HLA matched, HLA mismatched, or cord blood) and antiviral use were not associated with overall mortality after accounting for other variables. Analyses using a weighted viral load AUC to account for differential viral loads per virus showed similar findings but were less interpretable in a clinical context (data not shown). Similar findings were demonstrated for NRM (data not shown).
Figure 5.
Forest plot demonstrating the association of the cumulative viral load AUC with overall mortality from adjusted Cox models. (A) Models of the association of the cumulative viral load AUC with overall mortality in the entire cohort and among day-30 survivors with adjustment for immune reconstitution parameters. (B) Models that considered the effect of the cumulative viral load AUC within strata of acute GVHD to account for effect modification. In all models, viral detection was included as a time-dependent variable for overall mortality by day 100 and a time-invariant variable for overall mortality between day 101 and day 365 (because virus testing stopped at day 100). aAdjusted for age, HCT comorbidity index, underlying disease risk, conditioning regimen intensity, and acute GVHD (time dependent), and cumulative steroid dose AUC. bAdjusted for age, HCT comorbidity index, underlying disease risk, conditioning regimen intensity, and acute GVHD (time dependent), day 30 neutrophil engraftment, day 30 ALC < 200 cells/mm3, day 30 AMC < 300 cells/mm3, and cumulative steroid dose AUC. cAdjusted for age, HCT comorbidity index, conditioning regimen intensity, and underlying disease risk.
Models incorporating alternative definitions of virus detection, that is, as the number of viruses detected at any time or the same time, also demonstrated that a higher number of viruses was associated with overall mortality and NRM (supplemental Figure 2). Comparison of specific viruses and virus combinations lacked power to detect an association with mortality (see the supplemental Results section and supplemental Figure 3).
Association of dsDNA virus detection and overall mortality with adjustment for immune reconstitution
We developed additional models that considered biomarkers of early immune reconstitution at day 30 ±2 days. ALC < 200 cells/mm3, AMC < 300 cells/mm3, and neutrophil engraftment were associated with overall mortality in a univariate model (data not shown). In a multivariable model, neutrophil engraftment was protective (aHR, 0.48; P < .001) and AMC < 300 cells/mm3 increased risk (aHR, 1.23; P = .001) for overall mortality. The cumulative viral load AUC remained a significant predictor of early and late overall mortality when added to this model (Figure 5B). Supplemental Figure 2 presents the association of the number of viruses detected with overall mortality after adjusting for immune reconstitution. Similar patterns were seen for NRM (data not shown).
In a subgroup analysis of 87 cord blood HCT recipients with CD3, CD4, CD8, and CD56 cell counts tested at day 28 post-HCT, CD56 < 50 cells/mm3 (aHR, 1.45; P = .04) and the number of viruses detected at the same time (aHR, 2.44 per additional virus; 95% CI [1.5, 3.98]; P < .001) were both associated with overall mortality when included in the same model.
Association of dsDNA viruses and mortality stratified by GVHD
We identified an interaction between acute GVHD and the number of viruses detected. Accordingly, we modeled the impact of the cumulative viral load AUC in different strata of acute GVHD severity. This demonstrated that the association between cumulative viral load AUC and overall mortality by day 100 was maintained, regardless of acute GVHD severity (Figure 5B). Increased virus detection through day 100 was not associated with death after day 100 among patients who had grades 3 to 4 acute GVHD.
Association of dsDNA virus detection with other outcomes
Models exploring the association of virus detection with other clinically relevant endpoints, including respiratory failure, lower respiratory tract disease, late-onset acute GVHD or chronic GVHD, acute kidney or liver injury within 100 days post-HCT, and chronic kidney disease are presented in the supplemental Results section.
Discussion
We demonstrated frequent and persistent detection of single and multiple dsDNA viruses in the first 100 days after allogeneic HCT. Important risk factors included cord blood or mismatched HCT, myeloablative conditioning, younger age, and day 30 ALC < 200 cells/mm3. We found that the net burden of virus exposure, or cumulative viral load AUC in the first 100 days post-HCT, was consistently associated with increased risk for early (by day 100) and late (between days 101 and 365) overall mortality and NRM in multivariable models adjusted for important confounders. Increased virus detection remained an important predictor of overall mortality after accounting for biomarkers of delayed immune reconstitution, acute GVHD severity, and steroid use.
Three or more dsDNA viruses were detected in the plasma in 28% of this patient population. The additive effects of multiple virus reactivations or infections in allogeneic HCT recipients are not well studied. One observational study of CMV, HHV-6, EBV, and AdV detection in 124 pediatric patients reported infection with ≥1 herpesvirus in 74% of patients and ≥2 in 35% of patients.26 Patients with ≥2 herpesvirus infections had increased risk for chronic GVHD and allograft failure. Similar findings of high viral burden and association with chronic GVHD were reported in an observational study of 215 pediatric patients.9 Patients with detection of multiple viruses also have increased health care utilization.27 Smaller studies of HCT recipients have explored multiple viral reactivation events but were limited by intermittent testing, heterogeneous test methods, or analyses restricted to individual viruses.28-30
Cord blood and HLA-mismatched HCTs, myeloablative conditioning, acute GVHD, age of ≤21 years, and low ALC at day 30 increased risk for virus detection in this study. Discerning the attributable impact of dsDNA viremia on mortality in the context of poor immune function is challenging, because delayed immune reconstitution by day 30 was also associated with higher overall mortality in this study and others.22-24 However, the association of greater virus detection with overall mortality was maintained after adjusting for multiple biomarkers of immune reconstitution, acute GVHD severity, and cumulative steroid dose AUC. Defined T-cell subsets did not predict viral reactivation in a subgroup of cord blood HCT recipients, although future studies should consider incorporating T-cell subsets in addition to ALC, AMC, and neutrophils.
There were only 34 patients with end-organ disease and 8 deaths attributed to dsDNA viruses in this cohort (see the supplemental Results section). Whether dsDNA viremia without apparent end-organ disease modulates morbidity and mortality remains poorly understood. Direct virus-mediated tissue injury may be underrecognized or misattributed to other causes in the HCT setting, which in turn would predispose patients to greater toxicity from other insults. Viremia with dsDNA viruses may have indirect effects due to increased production of proinflammatory and immunomodulatory cytokines that contribute to the pathogenesis of important complications after HCT.31 This phenomenon is well described in the setting of CMV and HHV-6B,32-34 and viremia has been associated with increased risk for bacterial and fungal infections, prolonged hospitalization, and death.6,16,33,35 Immunomodulation is therefore a plausible, albeit unproven, mechanistic link between increased burden of viruses and worse outcomes through independent or synergistic pathways.
Our analyses demonstrated novel associations between greater virus detection and respiratory failure, which may account for some of the association with mortality (see the supplemental Results section). Clinically guided testing for dsDNA viruses (except CMV) missed an average of ∼75% of patients with any given virus in comparison with weekly screening (see the supplemental Results section), suggesting opportunities for intervention through strategies such as multivirus-specific T cells11 and novel small molecules.12,13 Methods to further improve immune reconstitution, such as adoptive transfer of immune cell subsets or progenitor cells,24,36 and to reduce the incidence of acute GVHD, such as post-HCT cyclophosphamide,37 have great potential to mitigate virus reactivation and other complications leading to poor outcomes.
Strengths of our study include the use of a large cohort of diverse HCT recipients and donor types with weekly plasma samples available for dsDNA virus testing by sensitive quantitative PCR assays. Use of plasma samples limited confounding introduced by detection of latent virus in whole blood samples. We used any positive test for virus detection in these analyses rather than specific thresholds, which are poorly established (except for CMV) and dictated by treatment toxicities, to allow for an objective approach to any virus detection. This also allowed us to analyze the cumulative viral load AUC as a novel predictor of mortality. Detection of herpesviruses in plasma may underestimate tissue-level reactivation,38 and safer interventions may allow for prophylactic or earlier preemptive treatment in the future. The study design allowed for comprehensive analyses of virus detection in time-dependent models adjusted for important risk factors, including biomarkers of immune reconstitution and cumulative steroid dose exposure.
This study also had limitations. Although we did not separate the type of HCT by degree of mismatch due to limited numbers of patients available for inclusion in each group, the increased cumulative incidence and weekly proportion of multivirus detection in HLA-mismatched and cord blood groups in comparison with the HLA-matched group supports our patient-grouping approach. Whether past infection (ie, seropositivity) with these viruses can modulate the immune system such that patients have worse outcomes independent of viral reactivation, as has been shown in the setting of CMV seropositivity,39 is not well understood. Although our models demonstrate a dose-response relationship between dsDNA virus burden and mortality, these variables are less predictive at the individual patient level. It remains unclear whether detection of multiple dsDNA viruses is simply a useful biomarker for poor outcomes or whether it lies on the causal pathway for mortality in the absence of prospective interventional studies.
In conclusion, we demonstrate a high burden of dsDNA virus detection in a large and diverse cohort of allogeneic HCT recipients. Detection of a greater cumulative burden of dsDNA viruses was associated with increased mortality independent of poor immune reconstitution and acute GVHD. These data provide the rationale and inform clinical and virologic endpoints for the development and implementation of randomized prevention and treatment trials using novel antiviral strategies to determine whether prevention of single or multiple viral infections will improve outcomes.
Acknowledgments
The authors thank E. Lisa Chung, Laurel Joncas-Schronce, Laura Sissons-Ross, Emily Cox, and Zach Stednick for help with data collection and management, and Heather Andrew, Lizanne Ngo, Jo Tono, Jessica Yi, Tracy Santo Hayes, and Susan McArdle for preparing and testing samples. Samples were obtained from the Infectious Disease Sciences Biorepository at Fred Hutch.
This was an investigator-initiated study funded in part by Chimerix, Inc., to J.A.H. and M.B. J.A.H. and M.B. received support from the National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases grant 5K23AI119133-02 and NIH, National Heart, Lung, and Blood Institute grant K24HL093294, respectively. Additional resources were provided by NIH, National Heart, Lung, and Blood Institute grants HL088021 and HL122173, and NIH, National Cancer Institute grants CA78902 and CA18029.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: J.A.H., W.M.L., G.N., B.T.M., J.T.S., D.M.Z., and M.B. designed the study and interpreted the data; J.A.H., W.M.L., H.X., and B.T.M. analyzed the data and created the figures; J.A.H., M.L.S., B.M.S., F.M., and C.D. collected the data; M.-L.H., T.S.-A., and K.R.J. carried out or supervised the laboratory work; all authors contributed to the writing and revision of the manuscript and approved the final version.
Conflict-of-interest disclosure: J.A.H. reports grants from Chimerix, Inc. W.M.L., H.X., B.T.M., and J.T.S. received grants from Chimerix, Inc., for the conduct of the study. G.N. is an employee of, and owns stock in, Chimerix, Inc. M.L.S. reports personal fees from Jazz Pharmaceuticals, outside of the submitted work. D.M.Z. reports grant support from Chimerix, Inc., outside of the submitted work. J.T.S. receives research funding from Chimerix, Inc. M.B. reports grants and personal fees from Merck and Co.; grants and personal fees from Astellas, Shire, Roche/Genentech, Gilead, and Chimerix; and personal fees from Clinigen and Microbiotix, outside of the submitted work. The remaining authors declare no competing financial interests.
Correspondence: Joshua A. Hill, Fred Hutchinson Cancer Research Center, University of Washington, 1100 Fairview Ave N, Mail Stop E-400, Seattle, WA 98109; e-mail: jahill3@fredhutch.org.
References
- 1.Tomblyn M, Chiller T, Einsele H, et al. ; Center for International Blood and Marrow Research; National Marrow Donor program; European Blood and Marrow Transplant Group; American Society of Blood and Marrow Transplantation; Canadian Blood and Marrow Transplant Group; Infectious Diseases Society of America; Society for Healthcare Epidemiology of America; Association of Medical Microbiology and Infectious Disease Canada; Centers for Disease Control and Prevention. Guidelines for preventing infectious complications among hematopoietic cell transplantation recipients: a global perspective. Biol Blood Marrow Transplant. 2009;15(10):1143-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kontoyiannis DP, Lewis RE, Marr K. The burden of bacterial and viral infections in hematopoietic stem cell transplant. Biol Blood Marrow Transplant. 2009;15(1 Suppl):128-133. [DOI] [PubMed] [Google Scholar]
- 3.Mynarek M, Ganzenmueller T, Mueller-Heine A, et al. Patient, virus, and treatment-related risk factors in pediatric adenovirus infection after stem cell transplantation: results of a routine monitoring program. Biol Blood Marrow Transplant. 2014;20(2):250-256. [DOI] [PubMed] [Google Scholar]
- 4.Rasche L, Kapp M, Einsele H, Mielke S. EBV-induced post transplant lymphoproliferative disorders: a persisting challenge in allogeneic hematopoetic SCT. Bone Marrow Transplant. 2014;49(2):163-167. [DOI] [PubMed] [Google Scholar]
- 5.Rorije NMG, Shea MM, Satyanarayana G, et al. BK virus disease after allogeneic stem cell transplantation: a cohort analysis. Biol Blood Marrow Transplant. 2014;20(4):564-570. [DOI] [PubMed] [Google Scholar]
- 6.Zerr DM, Boeckh M, Delaney C, et al. HHV-6 reactivation and associated sequelae after hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2012;18(11):1700-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGoldrick SM, Bleakley ME, Guerrero A, et al. Cytomegalovirus-specific T cells are primed early after cord blood transplant but fail to control virus in vivo. Blood. 2013;121(14):2796-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunstein CG, Gutman JA, Weisdorf DJ, et al. Allogeneic hematopoietic cell transplantation for hematologic malignancy: relative risks and benefits of double umbilical cord blood. Blood. 2010;116(22):4693-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verdeguer A, de Heredia CD, González M, et al. ; GETMON: Spanish Working Party for Blood and Marrow Transplantation in Children. Observational prospective study of viral infections in children undergoing allogeneic hematopoietic cell transplantation: a 3-year GETMON experience. Bone Marrow Transplant. 2011;46(1):119-124. [DOI] [PubMed] [Google Scholar]
- 10.Wagner JE, Thompson JS, Carter SL, Kernan NA; Unrelated Donor Marrow Transplantation Trial. Effect of graft-versus-host disease prophylaxis on 3-year disease-free survival in recipients of unrelated donor bone marrow (T-cell depletion trial): a multi-centre, randomised phase II-III trial. Lancet. 2005;366(9487):733-741. [DOI] [PubMed] [Google Scholar]
- 11.Papadopoulou A, Gerdemann U, Katari UL, et al. Activity of broad-spectrum T cells as treatment for AdV, EBV, CMV, BKV, and HHV6 infections after HSCT. Sci Transl Med. 2014;6(242):242ra83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Florescu DF, Keck MA. Development of CMX001 (Brincidofovir) for the treatment of serious diseases or conditions caused by dsDNA viruses. Expert Rev Anti Infect Ther. 2014;12(10):1171-1178. [DOI] [PubMed] [Google Scholar]
- 13.Prichard MN, Williams JD, Komazin-Meredith G, et al. Synthesis and antiviral activities of methylenecyclopropane analogs with 6-alkoxy and 6-alkylthio substitutions that exhibit broad-spectrum antiviral activity against human herpesviruses. Antimicrob Agents Chemother. 2013;57(8):3518-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sedlak RH, Cook L, Huang M-L, et al. Identification of chromosomally integrated human herpesvirus 6 by droplet digital PCR. Clin Chem. 2014;60(5):765-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milano F, Pergam SA, Xie H, et al. Intensive strategy to prevent CMV disease in seropositive umbilical cord blood transplant recipients. Blood. 2011;118(20):5689-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green ML, Leisenring W, Xie H, et al. Cytomegalovirus viral load and mortality after haemopoietic stem cell transplantation in the era of pre-emptive therapy: a retrospective cohort study. Lancet Haematol. 2016;3(3):e119-e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zerr DM, Gupta D, Huang M-L, Carter R, Corey L. Effect of antivirals on human herpesvirus 6 replication in hematopoietic stem cell transplant recipients. Clin Infect Dis. 2002;34(3):309-317. [DOI] [PubMed] [Google Scholar]
- 18.Boeckh M, Huang M, Ferrenberg J, et al. Optimization of quantitative detection of cytomegalovirus DNA in plasma by real-time PCR. J Clin Microbiol. 2004;42(3):1142-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heugel J, Boeckh M, Huang M-L, et al. Detection of adeno-associated virus viremia in hematopoietic cell transplant recipients. J Infect Dis. 2011;204(11):1746-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erard V, Storer B, Corey L, et al. BK virus infection in hematopoietic stem cell transplant recipients: frequency, risk factors, and association with postengraftment hemorrhagic cystitis. Clin Infect Dis. 2004;39(12):1861-1865. [DOI] [PubMed] [Google Scholar]
- 21.Kimura H, Morita M, Yabuta Y, et al. Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. J Clin Microbiol. 1999;37(1):132-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Storek J, Dawson MA, Storer B, et al. Immune reconstitution after allogeneic marrow transplantation compared with blood stem cell transplantation. Blood. 2001;97(11):3380-3389. [DOI] [PubMed] [Google Scholar]
- 23.Forcina A, Noviello M, Carbone MR, Bonini C, Bondanza A. Predicting the clinical outcome of allogeneic hematopoietic stem cell transplantation: the long and winding road toward validated immune biomarkers. Front Immunol. 2013;4:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang YJ, Zhao XY, Huang XJ. Immune reconstitution after haploidentical hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2014;20(4):440-449. [DOI] [PubMed] [Google Scholar]
- 25.Copelan E, Casper JT, Carter SL, et al. A scheme for defining cause of death and its application in the T cell depletion trial. Biol Blood Marrow Transplant. 2007;13(12):1469-1476. [DOI] [PubMed] [Google Scholar]
- 26.Olkinuora HA, Taskinen MH, Saarinen-Pihkala UM, Vettenranta KK. Multiple viral infections post-hematopoietic stem cell transplantation are linked to the appearance of chronic GVHD among pediatric recipients of allogeneic grafts. Pediatr Transplant. 2010;14(2):242-248. [DOI] [PubMed] [Google Scholar]
- 27.Rustia E, Violago L, Jin Z, et al. Risk factors and utility of a risk-based algorithm for monitoring cytomegalovirus, Epstein-Barr virus, and adenovirus infections in pediatric recipients after allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2016;22(9):1646-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Debaugnies F, Busson L, Ferster A, et al. Detection of Herpesviridae in whole blood by multiplex PCR DNA-based microarray analysis after hematopoietic stem cell transplantation. J Clin Microbiol. 2014;52(7):2552-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamin DK Jr, Miller WC, Bayliff S, Martel L, Alexander KA, Martin PL. Infections diagnosed in the first year after pediatric stem cell transplantation. Pediatr Infect Dis J. 2002;21(3):227-234. [DOI] [PubMed] [Google Scholar]
- 30.Inazawa N, Hori T, Hatakeyama N, et al. Large-scale multiplex polymerase chain reaction assay for diagnosis of viral reactivations after allogeneic hematopoietic stem cell transplantation. J Med Virol. 2015;87(8):1427-1435. [DOI] [PubMed] [Google Scholar]
- 31.Schots R, Kaufman L, Van Riet I, et al. Proinflammatory cytokines and their role in the development of major transplant-related complications in the early phase after allogeneic bone marrow transplantation. Leukemia. 2003;17(6):1150-1156. [DOI] [PubMed] [Google Scholar]
- 32.Kawamura Y, Yamazaki Y, Ohashi M, Ihira M, Yoshikawa T. Cytokine and chemokine responses in the blood and cerebrospinal fluid of patients with human herpesvirus 6B-associated acute encephalopathy with biphasic seizures and late reduced diffusion. J Med Virol. 2014;86(3):512-518. [DOI] [PubMed] [Google Scholar]
- 33.Nichols WG, Corey L, Gooley T, Davis C, Boeckh M. High risk of death due to bacterial and fungal infection among cytomegalovirus (CMV)-seronegative recipients of stem cell transplants from seropositive donors: evidence for indirect effects of primary CMV infection. J Infect Dis. 2002;185(3):273-282. [DOI] [PubMed] [Google Scholar]
- 34.Boeckh M, Nichols WG. Immunosuppressive effects of beta-herpesviruses. Herpes. 2003;10(1):12-16. [PubMed] [Google Scholar]
- 35.Lopez Roa P, Hill JA, Kirby KA, et al. Coreactivation of human herpesvirus 6 and cytomegalovirus is associated with worse clinical outcome in critically ill adults. Crit Care Med. 2015;43(7):1415-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delaney C, Milano F, Nicoud I, et al. Dose dependent enhancement of neutrophil recovery by infusion of notch ligand ex vivo expanded cord blood progenitors: results of a multi-center phase I trial. Blood. 2013;122(21):297.23847190 [Google Scholar]
- 37.Ciurea SO, Zhang M-J, Bacigalupo AA, et al. Haploidentical transplant with posttransplant cyclophosphamide vs matched unrelated donor transplant for acute myeloid leukemia. Blood. 2015;126(8):1033-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fotheringham J, Akhyani N, Vortmeyer A, et al. Detection of active human herpesvirus-6 infection in the brain: correlation with polymerase chain reaction detection in cerebrospinal fluid. J Infect Dis. 2007;195(3):450-454. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt-Hieber M, Labopin M, Beelen D, et al. CMV serostatus still has an important prognostic impact in de novo acute leukemia patients after allogeneic stem cell transplantation: a report from the Acute Leukemia Working Party of EBMT. Blood. 2013;122(19):3359-3364. [DOI] [PubMed] [Google Scholar]
- 40.Sorror ML. Hematopoietic cell transplantation (hct)-specific comorbidity index: a new tool for risk assessment before allogeneic hct. Blood. 2005;106(8):2912-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leisenring WM, Martin PJ, Petersdorf EW, et al. An acute graft-versus-host disease activity index to predict survival after hematopoietic cell transplantation with myeloablative conditioning regimens. Blood. 2006;108:749-755. [DOI] [PMC free article] [PubMed] [Google Scholar]