Abstract
PDB_TM is a database for transmembrane proteins with known structures. It aims to collect all transmembrane proteins that are deposited in the protein structure database (PDB) and to determine their membrane-spanning regions. These assignments are based on the TMDET algorithm, which uses only structural information to locate the most likely position of the lipid bilayer and to distinguish between transmembrane and globular proteins. This algorithm was applied to all PDB entries and the results were collected in the PDB_TM database. By using TMDET algorithm, the PDB_TM database can be automatically updated every week, keeping it synchronized with the latest PDB updates. The PDB_TM database is available at http://www.enzim.hu/PDB_TM.
INTRODUCTION
Integral membrane proteins represent about 20–30% of the total proteins of various organisms (1–3). Despite their important physiological roles, they are highly underrepresented in the protein structure database (PDB) (4), due to the difficulties in crystallizing them in an aqueous environment. Among the >25 000 structures that are deposited in the PDB so far, the number of membrane protein structures hardly exceeds 400, which represents around 30–40 different folds only. A complete list of transmembrane proteins (TMPs) is difficult to extract from the PDB that is dominated by water-soluble proteins, mainly because the annotation of PDB files is not reliable in terms of the transmembrane character of the deposited protein structures. For example, on entering the keyword ‘transmembrane’ into the search field on the PDB home page, the search resulted in 1508 hits (in July, 2004). There are only 310 of these proteins, which are really TMPs, whereas many others contain only the water-soluble domain of TMPs. At the same time, 114 actual TMPs were missed by the keyword search. Therefore, to determine the type of a protein in the PDB, the actual coordinates of the proteins have to be analyzed.
A separate issue that is specific to TMP structures is the identification of membrane-spanning regions. This information is not included in the PDB files, as the lipid molecules are removed before the process of structure determination. Therefore, no direct experimental data is available about how the protein spans the membrane (5). Although the transmembrane character and the likely position of the membrane can be guessed by inspecting individual cases to ensure completeness and consistency, an automated approach is required to identify membrane-spanning regions. The structures of these proteins are often of low resolution, or correspond to only very small fragments, in which cases automatic assignment can be quite challenging.
We have recently developed an algorithm (6), called TMDET, to find the most likely position of the membrane, and to distinguish between transmembrane and non-transmembrane proteins or protein segments by using their coordinates only. The TMDET algorithm allows the appropriate classification of low-resolution structures, as well as cases when only a fragment or a partial structure of a multichain protein complex is available. The discrimination power of TMDET algorithm is >98%; therefore, it can be used to create and maintain a database that contains the TMPs of known 3D structures, as well as to calculate the possible membrane localization of these proteins.
Here we present the details of the resulting database, PDB_TM, and its usage on the world wide web. The aim of this database is 2-fold. First, it assigns a transmembrane character for each entry in the PDB, which allows the construction of a comprehensive and up-to-date list of all membrane proteins and segments with known structures. Second, it identifies the location of the lipid bilayer that is relative to the coordinates system of the molecule. This database would be useful to anyone who seeks to study any particular TMP. For example, it can help to assign whether a binding site is located in the lipid or in the aqueous phase, which can be important to design a drug that binds to a certain part of a receptor. Bioinformatists who compile statistical analysis on TMPs are also potential users. What is more, since PDB_TM identifies all TMPs in the PDB, the rest of the PDB is a pure database of non-transmembrane proteins, therefore such information might be useful for scientists who work on databases of water-soluble proteins.
DATABASE CREATION AND MAINTENANCE
The technical details of the TMDET algorithm, as well as the initial version of PDB_TM database can be found in our recent article (6). Here we briefly summarize the creation of the database: TMDET algorithm was applied to all files in the PDB. For the sake of simplicity, virus and pilus proteins as well as entries that contained nucleotides (RNA or DNA sequences) and peptides <15 residues in length were omitted. The TMDET algorithm is based on an empirical measure, the Q-value (6), which describes the manner in which a given slice of a protein fits into a membranous environment. For each protein, an exhaustive search is carried out by varying the position and orientation of the slice, in order to find the maximum of the Q-values with the corresponding most likely location of the membrane. If the TMDET algorithm resulted in a maximum Q-value below a predefined threshold, the PDB entry was classified as a globular protein. If the value was above an upper selection limit, the PDB entry was classified as a TMP. In a small number of cases, for which the Q-values fell between the lower and upper selection limit, the transmembrane character could not be assigned confidently. In theses cases, the decision was made manually based on the annotation of the PDB file, the annotation of the corresponding swiss-prot entries (7), and when in doubt by checking the literature. For all identified TMPs, our algorithm also determined the number and type of transmembrane segments, localization of membrane that is relative to the protein coordinates and localization of each sequence part that is relative to the membrane. The number of transmembrane segments does not include non-canonical elements within the membrane region. Some alpha helical proteins contain segments that do not cross the membrane, but turn back at the middle of the membrane [e.g. aquaporin, 1fqy, (8)], whereas the inside of the pore that is formed by beta-barrel proteins are basically shielded from the membrane, and can accommodate alpha helical segments as well [e.g. the translocator domain of a bacterial autotransporter, 1uyo, (9)]. The algorithm is able to identify the correct topology even when these elements are present in the structure.
The database is updated every week when the new PDB entries are released, by running the TMDET algorithm on each new PDB file. A typical run on an average PDB file takes a few seconds on a Pentium 4, 2.4 GHz personal computer, thus the update of the database is around one or two hours for the computer plus some minutes for checking the results by a human expert.
DATABASE ORGANIZATION, FILE FORMATS AND ANNOTATIONS
The PDB_TM database can be regarded as an extension of the PDB database, because it contains additional information for each PDB entry. Therefore, the database is organized similarly to the PDB, the entries are identified by their PDB code and are grouped in subdirectories according to the middle two characters of their codes. The properties of proteins determined by the TMDET algorithm are described in xml format, and the corresponding Document Type Definition can be found on the PDB_TM homepage.
For non-transmembrane proteins, the xml files contain only the information that the protein is not of the transmembrane type, whereas for transmembrane proteins, the xml files contain all the information that were gathered and calculated by the TMDET algorithm. These are the Q-value on which the selection was made; the type of the protein according to the secondary structure of its membrane-spanning residues that were determined by the DSSP algorithm (alpha-helical, beta-barrel or unstructured) (10); and some additional information on keyword search in the corresponding swiss-prot and PDB files. The next part of the files contains the membrane plane definition. It is given by the transformation matrix, which rotates the molecule such that the normal of the membrane plane is parallel with the z-axis and translates it, placing the new origin along this axis at the membrane half-width. The start and end position of the normal vector of the membrane plane is also specified. The length of the normal vector is equal to the half-width of the membrane. The last part of the files contains the localization of each sequence parts that is relative to the membrane for each chain that builds the biological active molecule. Because the inside and outside orientation of proteins cannot be determined from its coordinates, we use side-one and side-two notation to distinguish between the two sides of the membrane. The file contains information for all protein chains included in the biological oligomer structure; therefore, the number of chains can be different in a PDB_TM entry and in the corresponding PDB file. The definition and the calculation of biological active molecule are described in our previous article.
DATA ACCESS AND VISUALIZATION
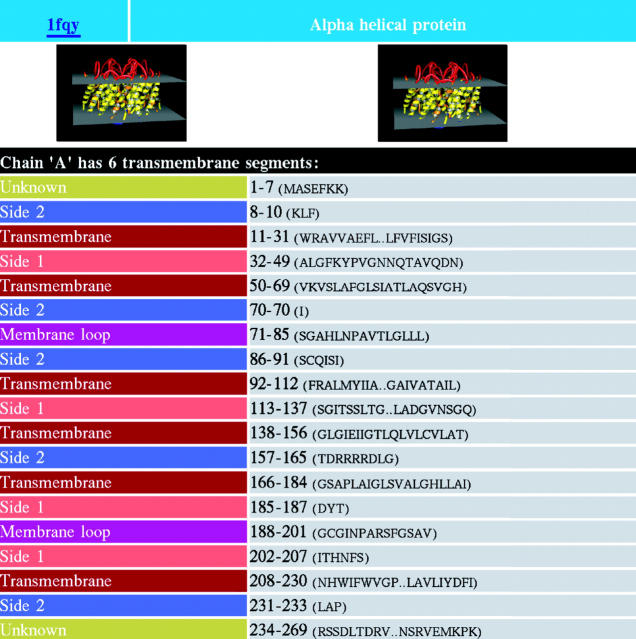
The PDB_TM database is available at http://www.enzim.hu/PDB_TM. There are various ways to find information on a protein in the PDB_TM database: the user can find it by entering the protein PDB code, by searching with keywords in the PDB or by entering some properties of the TMP, such as its type or the number of transmembrane segments. If the search resulted in multiple hits, the list of hits is displayed, and the wanted molecule can be easily selected from the list. Figure 1 shows the explore window of the aquaporin molecule (PDB code: 1fqy), which is a graphical representation of the corresponding xml file. The database is downloadable, either the full database in one big file, or its subsets containing the alpha helical or the beta-barrel TMPs. There is a possibility to browse all the files as well.
Figure 1.
The explore window of PDB_TM database containing part of the results for aquaporin protein (1fqy).
For each TMP, two typical pictures can be viewed on the visualization window, which is made by the Pymol molecular graphics software (11). The molecule in this picture is colored according to the identified localization of the segments, which discriminates between transmembrane regions, membrane loops, inner membrane segments as well as the two sides of the water-soluble regions.
FUTURE DIRECTION
Currently the PDB_TM database is updated semi-automatically every week right after the weekly update of the PDB. There is a family classification on the home page representing the situation as of early 2004. Work is in progress that will allow the automated update of the family classification as well. We also plan to include the biological complex definition in the xml files. While currently TMPs can be viewed on pictures made by the Pymol molecular graphic software, we plan to extend the range of visualization tools to provide rasmol script, pymol script or VRML, similar to the PDB site. Naturally, we are open to any user's advice as well.
Acknowledgments
ACKNOWLEDGEMENTS
This work was sponsored by grants BIO-0005/2001, OTKA T34131, D42207 and F043609. Z.D. and G.E.T. were supported by the Bolyai Janos Scholarship.
REFERENCES
- 1.Jones D.T. (1998) Do transmembrane protein superfolds exist? FEBS Lett., 423, 281–285. [DOI] [PubMed] [Google Scholar]
- 2.Krogh A., Larsson,B., von Heijne,G. and Sonnhammer,E.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol., 305, 567–580. [DOI] [PubMed] [Google Scholar]
- 3.Wallin E. and von Heijne,G. (1998) Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci., 7, 1029–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tusnady G.E. and Simon,I. (2001) Topology of membrane proteins. J. Chem. Inf. Comput. Sci., 41, 364–368. [DOI] [PubMed] [Google Scholar]
- 6.Tusnady G.E., Dosztanyi,Z. and Simon,I. (2004) Transmembrane proteins in protein data bank: identification and classification. Bioinformatics, in press; doi:10.1093/bioinformatics/bth340. [DOI] [PubMed] [Google Scholar]
- 7.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O'Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murata K., Mitsuoka,K., Hirai,T., Walz,T., Agre,P., Heymann,J.B., Engel,A. and Fujiyoshi,Y. (2000) Structural determinants of water permeation through aquaporin-1. Nature, 407, 599–605. [DOI] [PubMed] [Google Scholar]
- 9.Oomen C.J., Van Ulsen,P., Van Gelder,P., Feijen,M., Tommassen,J. and Gros,P. (2004) Structure of the translocator domain of a bacterial autotransporter. EMBO J, 23, 1257–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- 11.DeLano W.L. (1998–2003) The PyMOL Molecular Graphics System. DeLano Scientific LLC. San Carlos, CA, USA. http://pymol.sourceforge.net. [Google Scholar]