ABSTRACT
Research using animal model systems has been instrumental in delivering improved therapies for breast cancer, as well as in generating new insights into the mechanisms that underpin development of the disease. A large number of different models are now available, reflecting different types and stages of the disease; choosing which one to use depends on the specific research question(s) to be investigated. Based on presentations and discussions from leading experts who attended a recent workshop focused on in vivo models of breast cancer, this article provides a perspective on the many varied uses of these models in breast cancer research, their strengths, associated challenges and future directions. Among the questions discussed were: how well do models represent the different stages of human disease; how can we model the involvement of the human immune system and microenvironment in breast cancer; what are the appropriate models of metastatic disease; can we use models to carry out preclinical drug trials and identify pathways responsible for drug resistance; and what are the limitations of patient-derived xenograft models? We briefly outline the areas where the existing breast cancer models require improvement in light of the increased understanding of the disease process, reflecting the drive towards more personalised therapies and identification of mechanisms of drug resistance.
KEY WORDS: Breast cancer, Mouse models, GEMM, PDX, CDX, SEARCHBreast, EurOPDX
Summary: This Review provides a summary of the many varied uses of mouse models in breast cancer research focusing on their strengths, challenges and future directions.
Introduction
Clinical management of breast cancer has improved significantly over the past 30 years, with almost 87% of women surviving their diagnosis for at least 5 years compared with only 53% of those diagnosed in the early 1970s (cancerresearchuk.org; accessed December 2016). Nonetheless, breast cancer remains the leading cause of cancer-related female death worldwide with more than half a million women succumbing to the disease annually (Torre et al., 2016) including around 11,500 in the UK (breastcancernow.org; accessed December 2016). One of the reasons for this is that breast cancer is not a single disease entity (Curtis et al., 2012) and ‘one size’ does not ‘fit all’ for clinical management, treatment nor, as discussed here, modelling of the disease. Breast cancer is treated based on the receptor status of the tumour, specifically oestrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor-2 (HER2), and the main molecular subtypes are termed Luminal A (ER/PR-positive); Luminal B (ER/PR-positive, higher histological grade than Luminal A); HER2-positive; and triple-negative (ER/PR/HER2-negative) (Cardiff and Kenney, 2011). Tailored therapies have led to considerable success in treating some breast cancers, such as hormone therapies (e.g. tamoxifen, and inhibitors of the enzyme aromatase, involved in oestrogen synthesis) for ER-positive disease, and trastuzumab (Herceptin) for HER2-positive breast cancer; however, drug resistance to these regimes is common (Osborne and Schiff, 2011; Palmieri et al., 2014; Luque-Cabal et al., 2016). Furthermore, there is still no good targeted therapy for triple-negative breast cancer, which is one of the more aggressive subtypes of the disease (Kalimutho et al., 2015; Gu et al., 2016). In addition, whilst primary breast cancer is highly treatable [80-99% of women diagnosed with stage I/II breast cancer survive to 5 years; (cancerresearchuk.org; wcrf.org)], there is no cure currently available for metastatic breast cancer, which affects an estimated 40% of UK patients (breastcancernow.org) and likely accounts for the decline in survival rate to 65% at 20 years post-diagnosis. Genetic sequencing endeavours have identified many of the mutations implicated in breast cancer (Nik-Zainal et al., 2016), which may lead to the development of new therapeutic options; however, the functional role of these alterations in the different subtypes has still to be confirmed, and their distinct roles during disease progression, tumour heterogeneity and dormancy, clarified (Eccles et al., 2013).
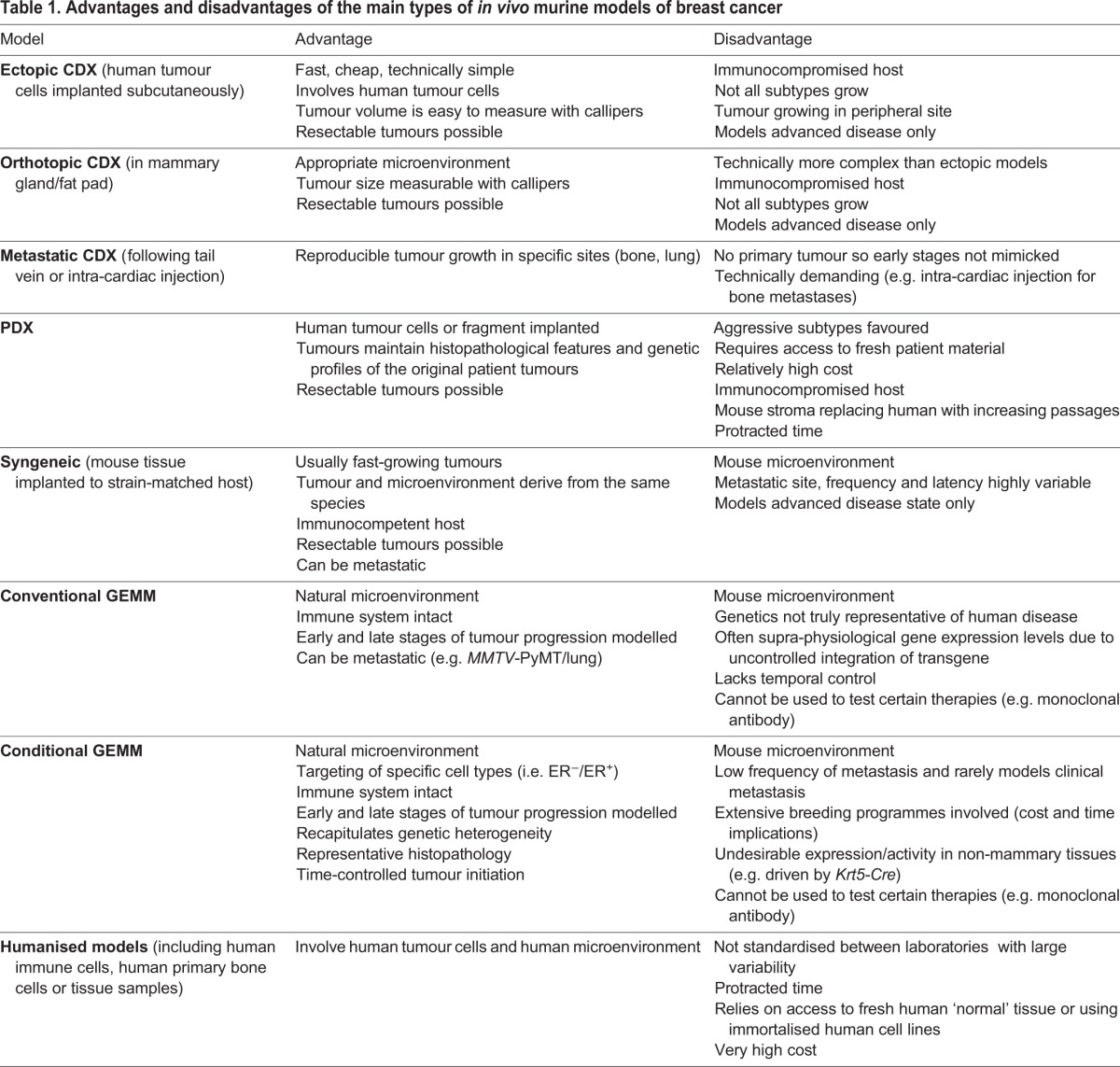
Scientists have capitalised on in vivo models as important research tools to study the pertinent questions in breast cancer research (Fig. 1). By ‘in vivo’ we refer to a living organism and we will restrict our discussions to the mouse, a physiologically relevant system in which to explore cancer initiation, invasion and metastasis, and which represents an essential step between in vitro systems and clinical studies. Researchers now have access to a broad range of mouse models, each with its own strengths and limitations (see overview in Table 1). A clear understanding of these parameters is paramount in order to choose the model best suited to address the specific research questions posed. SEARCHBreast, an organisation dedicated to sharing animal resources (see Box 1), hosted a workshop (searchbreast.org/workshop3.html) to showcase the contribution these complex models have made to breast cancer research highlighting recent developments and discussions on how shortfalls in existing models could be addressed in the foreseeable future. The workshop, open to all UK researchers with an interest in breast cancer, involved presentations and discussions from leading groups with broad expertise in utilising breast cancer mouse models in a variety of areas, from disease mechanisms to preclinical trials. In this workshop-inspired perspective, we highlight recent progress in mouse models of breast cancer, and discuss some of the outstanding issues researchers are grappling with in their pursuit of the best in vivo model for their research question. The article is not intended to give an exhaustive review of the vast literature but will give readers a current overview of the advantages, limitations and challenges that lie ahead for breast cancer researchers and especially those utilising, or wishing to adopt, mouse models to study this heterogeneous disease.
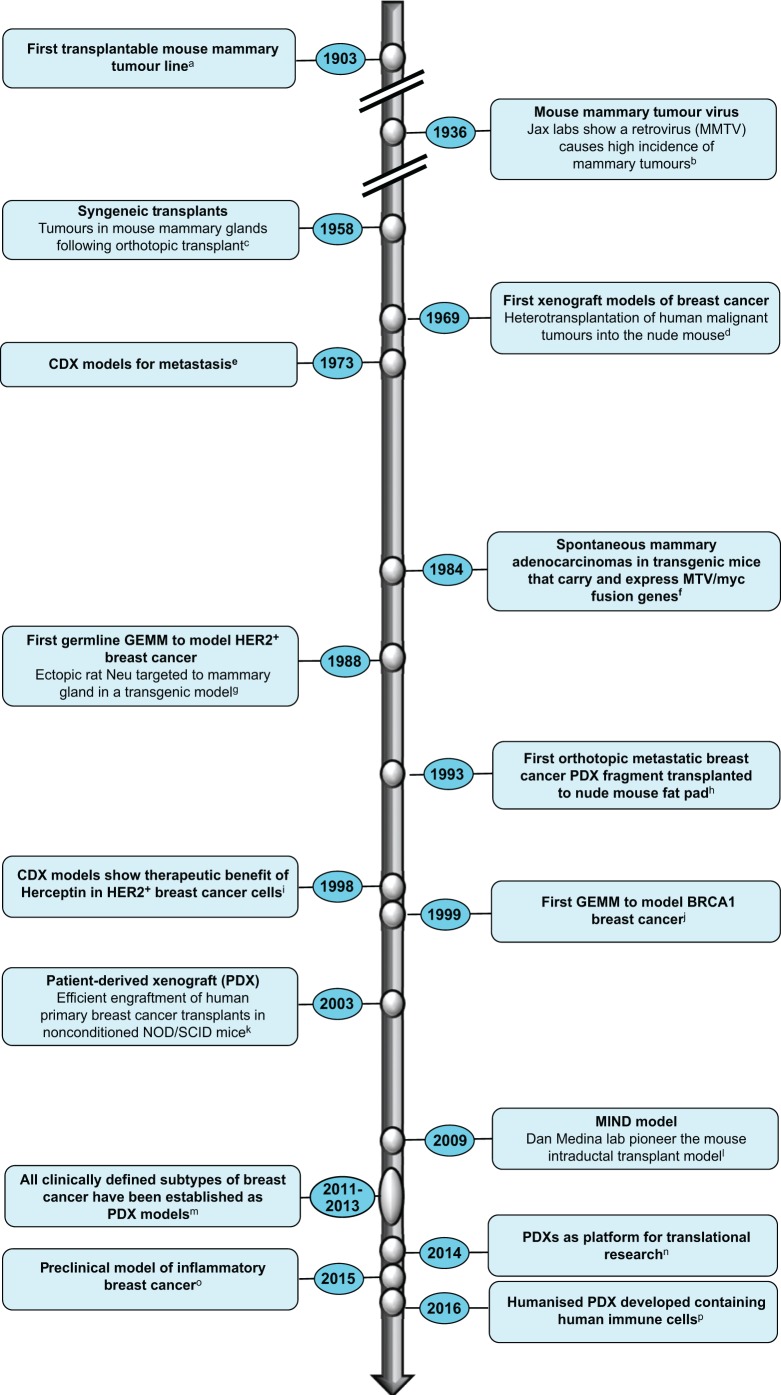
Fig. 1.
Modelling breast cancer through the ages. The figure depicts a timeline of key events or developments in the evolution of mouse models of breast cancer over the years. Lowercase letters denote the following references, to which the reader is referred for further reading on specific milestones: aCardiff and Kenney, 2011; bBittner, 1936; cDeOme et al., 1959; dRygaard and Povsen, 2007; eFidler, 1973; fStewart et al., 1984; gMuller et al., 1988; hFu et al., 1993; iBaselga et al., 1998; jXu et al., 1999; kBeckhove et al., 2003; lBehbod et al., 2009; mDeRose et al., 2011; nWhittle et al., 2015; oWurth et al., 2015; phttps://www.jax.org/news-and-insights/jax-blog/2015/april/the-next-big-thing-in-cancer-modeling-patient-derived-xenografts-in-humaniz.
Table 1.
Advantages and disadvantages of the main types of in vivo murine models of breast cancer
Box 1. SEARCHBreast and EurOPDX.
The workshop was hosted by SEARCHBreast (sharing experimental animal resources: coordinating holdings – Breast, www.searchbreast.org); a novel resource funded by the National Centre for the Replacement, Refinement and Reduction of Animals in Research (NC3Rs) to create a secure, searchable database of archived material derived from in vivo breast cancer models, which is available to share between researchers (Blyth et al., 2016). Use of this resource could allow new animal experiments to be avoided, or refined, ultimately reducing the number of animals used for research, as well as saving the time and money required to run in vivo experiments from scratch (Morrissey et al., 2016). At the heart of SEARCHBreast is the fact that many labs have surplus tissues from in vivo experiments left over at the end of a study, while other researchers would benefit from access to such material. SEARCHBreast is a no-cost mediator that brings together these collaborations. The web-based platform contains information on thousands of tissue samples that are available for immediate use following a simple online request. Samples are available free of charge from over 85 different mouse models, including GEMM, xenograft and patient-derived xenografts. With almost 250 members across the UK and internationally, SEARCHBreast is a large network of breast cancer researchers with collective experience with in vivo, in vitro and in silico models of breast cancer.
The EurOPDX consortium (www.europdx.eu), established from partners in 10 different European countries covering 16 academic institutions, is a network of cancer scientists and clinicians who have pioneered the standardisation of PDX models as clinically relevant models of human cancer. EurOPDX has created an extensive virtual collection of PDX models (>1500) encompassing many cancer types that have been genomically and histologically characterised in well-established laboratories. The aim of the consortia is to unify and harmonise the use of PDX models, which could lead to better resources for investigating predictive biomarkers and testing novel therapeutic approaches, and ultimately, personalised cancer treatment.
Modelling breast cancer development, therapeutic targeting and drug resistance
One of the main challenges in developing in vivo models has been the increasing understanding of the many different subtypes of breast cancer (Perou et al., 2000; Sorlie et al., 2003; Curtis et al., 2012; Cancer Genome Atlas Network, 2012). Ideally, this complexity should be faithfully reflected in model systems. Although the field has access to a wide variety of mouse models that have evolved over the years (Fig. 1), certain models are much more commonly used than others and not all subtypes are represented. The main model systems and their application to date are briefly outlined below.
Cell-line-derived xenografts
One of the simplest and therefore most commonly used model systems is based on engraftment of human cell lines (Neve et al., 2006) to immunocompromised animals [cell-derived xenografts (CDX)]. These have proven to be extremely useful for assessment of breast cancer genetics, biological processes, and to some degree, metastatic potential; but are limited by their reduced intra-tumoural heterogeneity and their poor record of predicting clinically effective therapies (Whittle et al., 2015 and references therein). In addition, the lines used are frequently derived from highly aggressive malignant tumours or plural effusions (fluids drained from lung metastasis) such as the frequently studied MDA-MB-231 line, making these less useful for modelling early events in the evolution of the primary tumour. Although well-characterised cell lines representing the common clinical subtypes – luminal A (e.g. MCF-7, T47D), luminal B (e.g. BT474, MDA-MB-361), HER2+ (e.g. SKBR3, HCC202) and triple negative (e.g. BT20, MDA-MB-231, MDA-MB-468) – have been extensively studied, not all can be established in vivo (Holliday and Speirs, 2011) and in particular there is a dearth of tumourigenic HER2+ lines. Furthermore, long-term growth in vitro can result in aberrant selection pressures.
CDX models represent a relatively homogenous mass of transformed breast epithelial cells and as such do not capture the heterogeneity of human breast tumours, which arise in a niche of cell types that have symbiotically evolved together (Hanahan and Weinberg, 2011). Indeed the use of CDX to model the native tumour microenvironment is difficult, particularly because of the necessity to use immunocompromised recipients lacking an effective immune system. The site of transplantation should also be considered. This can be straightforwardly achieved via subcutaneous injection (ectopic), or by implanting cells in the mouse mammary gland (orthotopic), which is more complicated. The diverse microenvironment of these sites, particularly the tumour vasculature, significantly impacts tumour growth rate (Fleming et al., 2010), drug delivery and therapeutic efficacy – something that should be considered when choosing a cell-line-based model for preclinical testing of novel agents and/or treatment schedules (Talmadge et al., 2007; Fung et al., 2015).
Finally, spontaneous metastasis from CDX models is rare, hampering studies of breast cancer metastasis using this type of model. However, a few characterised murine cell lines (e.g. 4T1) do metastasize in syngeneic models (involving transplant of mouse-derived cells to an independent strain-matched mouse), although these are currently a limited resource (Johnstone et al., 2015; Erler et al., 2009; Paschall and Liu, 2016). Otherwise, direct injection of breast cancer cells into ectopic sites (e.g. bloodstream, femur) can be used as an experimental metastasis assay.
Patient-derived xenografts
Patient-derived xenografts (PDXs) – involving the transplantation of primary human cancer cells or tumour pieces into host mice – were developed to address the shortcomings of CDX, heralding hope for models with improved clinical relevance. Although transplantation of human tumour fragments into immunocompromised mice has a long history (reviewed in Hoffman, 2015), there is renewed interest in PDX models because of the preservation of many relevant features of the primary human tumour, including growth kinetics, histological features, behavioural characteristics (such as invasiveness and metastatic capacity) and most importantly, response to therapy (Marangoni et al., 2007; DeRose et al., 2011; Tentler et al., 2012; Hidalgo et al., 2014). Indeed, Ledford (2016) reported that the US National Cancer Institute (NCI) are planning to replace their NCI-60 cell line resource with PDX samples, which underscores the importance and acceptance of this resource.
Use of PDX models to model disease subtypes and metastasis
Modelling luminal ER+ subtypes, which are found in over 70% of diagnosed individuals, making these the commonest form of human breast cancer, has been particularly challenging. This is partly because successful xenotransplantation selects for the most aggressive subtypes, hence biasing both CDX and PDX towards the triple-negative subtype (reviewed by Tentler et al., 2012; Cottu et al., 2012). The recent demonstration, however, that transplanting ER+ cells (derived from cell lines and primary patient material) directly into the mouse ductal epithelium rather than the fat pad (Behbod et al., 2009) can preserve the luminal/ER+ phenotype of these cells through avoiding activation of the TGFβ signalling pathway, is encouraging (Richard et al., 2016; Sflomos et al., 2016). Not only does this facilitate establishment of a less-aggressive breast cancer subtype, but more importantly maintains faithful representation of the naïve human disease and has been described as a ‘potential game-changer’ for preclinical modelling of ER+ breast cancer (Haricharan et al., 2016). Indeed, PDXs derived from ER+, HER2+ and triple-negative disease, representing the main clinical subtypes, have been described (Dobrolecki et al., 2016).
Importantly, encouraging results have been obtained in which PDX models effectively model metastasis, with recent protocols showing preservation of patterns of metastatic spread representative of the clinical situation (DeRose et al., 2013). However, as PDXs are most commonly generated at the point of breast cancer surgery, it is difficult to assess to what extent their capacity to form metastases recapitulate that of the patient without a follow-up period of at least 5 years. A number of studies have reported apparent discrepancies between the metastatic patterns of PDX compared with those of the patient from which they originate. The most common metastatic site in the PDX models are lungs and lymph nodes, whereas brain and bone metastases are rarely reported, despite occurring frequently in patients (Eyre et al., 2016; Whittle et al., 2015).
PDX models for preclinical therapy testing
Evidence supporting the usefulness of PDX models in identifying clinically relevant treatment resistance mechanisms is amassing (ter Brugge et al., 2016). This has even been demonstrated in hard-to-model ER+ breast cancer, in which phosphoinositide 3-kinase (PI3K) pathway activation and outgrowth of stem cell populations, have been found during acquired resistance to anti-oestrogen therapies (Cottu et al., 2014; Simões et al., 2015). Key challenges that still need to be addressed, however, include the overall under-representation of ER+ tumours, as well as the aforementioned bias towards more aggressive tumours. Furthermore, such bias towards aggressive tumours may falsely infer therapeutic benefit (or indeed not) considering that less-aggressive cancers may not respond in the same way to therapy as those which are more proliferative and/or have different molecular alterations. Finally, the study of the tumour microenvironment is compromised both by intrinsic species differences (e.g. mouse fat pad) and the lack of immune cells in tolerant hosts. This latter point is notable considering the impact of immune-based therapies in the clinic (Varn et al., 2016) and is discussed below.
Some have mooted the possibility of PDX models as mouse ‘avatars’ for personalising treatment to individual patients; however, clinical decision-making is quick (weeks), compared with the time taken to establish a PDX and using it to test the response to different therapeutic regimens (months to years), so currently the potential of these models is limited to cohort-based preclinical studies. Regardless of these limitations, the wide acceptance of PDX models in the pharmaceutical industry is worthy of mention. There has been an explosion in the commercial sector in the use and availability of PDXs as the model of choice for translational research. Companies such as Champions Oncology and Crown Biosciences offer partnerships to run preclinical drug trials, while certain suppliers such as Horizon Discovery and The Jackson Laboratories make these models available to academic institutions. This is a fast-moving field and efforts are now being honed into producing humanised systems (see below).
Genetically engineered mouse models
For addressing early events in the tumour process, genetically engineered mouse models (GEMMs) come into their own. In these models, spontaneous tumour initiation occurs within the correct microenvironment from an otherwise normal mammary cell. These may be simple oncogenic-driven transgenic mice, referred to here as conventional GEMM (e.g. MMTV-PyMT). One limitation to the conventional GEMM models is that not only are the regulatory sequences used to drive transgene expression not well-defined in terms of specific lineage/expression domains, but also, the specific oncogenes may not necessarily reflect those observed in human tumours. Nonetheless, these models continue to serve a purpose in breast cancer research (Blaas et al., 2016; Arun et al., 2016).
With these limitations in mind, the field has turned to more specific models emulating the genetics of human disease with spatial and temporal activation of oncogenes and deletion of tumour suppressors targeted to the mouse mammary gland (e.g. Blg-Cre;Brca1fl/fl;p53fl/fl). These conditional GEMMs use the Cre/loxP system in which a tissue-specific promoter drives expression of the bacterial enzyme Cre recombinase (e.g. Blg-Cre) within the mammary gland to elicit recombination of DNA between loxP sites (e.g. introduced into the coding region of tumour suppressors such as p53 and Brca1). Lists of the many different breast cancer GEMMs (conventional and conditional) can be found in other recent reviews (Pfefferle et al., 2013; Borowsky, 2011; Menezes et al., 2014; Dabydeen and Furth, 2014; Greenow and Smalley, 2015; Ben-David et al., 2016).
Recreating tumour pathology in GEMMs
Pathological profiling has been essential in disease classification, prognosis and disease management and remains the clinical basis of stratification, so it is important that as well as genetic complexity, the tumour pathology is faithfully recapitulated in genetic models (Blyth et al., 2012). This has worked well for some epithelial cancers (e.g. pancreas, colon); however, there is a concern that models of breast cancer do not always reflect the pathology of the human disease. Historical models of spontaneous and mouse mammary tumour virus (MMTV)-infected tumours (Fig. 1) do not share histological features of human tumours (Cardiff et al., 2004) while specific oncogenic models in the form of conventional GEMMs (e.g. MMTV-Wnt1 and MMTV-PyMT) can demonstrate phenotypic similarities as well as disparities (Cardiff and Wellings, 1999). A consensus of medical and veterinary pathologists voiced caution over the interpretation of tumours derived from breast cancer GEMMs and advocated the inclusion of pathology expertise in any research team using these models (Cardiff et al., 2000). With an exponential rise in the number of breast cancer models now available, there has been a push to revisit these guidelines and a meeting of in vivo biologists and pathologists is planned for the near future. Meanwhile, publications (Munn et al., 1995; Cardiff et al., 2000; Ponzo et al., 2009; Cardiff, 2010; McCarthy et al., 2007; Fathers et al., 2010; Meyer et al., 2013; Melchor et al., 2014; Bao et al., 2015) and online tools (tvmouse.ucdavis.edu/pathology/) are available to aid in the comparative assessment of GEMM tumour pathology.
Modelling breast cancer subtypes in genetic models
The mature mammary gland is principally composed of two epithelial lineages comprising the luminal and basal cells. At the simplest level, breast cancer heterogeneity arises from origins in these different cell types with unique and associated gene signatures in distinct breast cancer subtypes (Sorlie et al., 2001; Prat and Perou, 2011). Recreating these subtypes in GEMMs has been attempted by targeting a variety of oncogenic drivers to the different mammary lineages. To this end, keratin 14 promoter (Krt14)-Cre or keratin 5 promoter (Krt5)-Cre have been used to direct oncogenic events to the basal lineage; whey acidic protein promoter (Wap)-Cre and keratin 8 (Krt8)-Cre to target luminal cells, and β-lactoglobulin (Blg)-Cre and Cited1-Cre to specifically target ER− and ER+ luminal cells, respectively. However, despite the use of lineage-specific promoters, a complete characterisation in the expression pattern of these Cre recombinases has not been done, and so ‘off-target’ and/or unexpected expression can confound studies. For example, Krt14-Cre can be active in the luminal as well as basal lineages (Jonkers et al., 2001; Regan et al., 2012), hampering complete separation of events in these populations. Furthermore, there is a lack of specificity in some Cre recombinases, such as with Krt14-Cre, which is expressed in skin and dental epithelium (Dassule et al., 2000), resulting in issues with skin tumours (Liu et al., 2007) or normal feeding capability, depending on the genetic changes introduced by that promoter. Caveats aside, genetic profiling has aligned GEMMs with specific molecular subtypes (Herschkowitz et al., 2007; Pfefferle et al., 2013; Hollern and Andrechek, 2014), highlighting the utility of genetic models in modelling the human disease.
Usefulness of GEMMs in determining the cell of origin
The use of lineage-restricted GEMMs has been instrumental in studies to identify the cell of tumour origin (Lindeman and Visvader, 2010; Beck and Blanpain, 2013; Brooks et al., 2015; Koren and Bentires-Alj, 2015). A prime example of this was the demonstration that breast cancer 1 susceptibility gene (Brca1)-deficient breast cancers originated from luminal progenitors, contrary to the expectation that these were of basal stem cell origin (Molyneux et al., 2010; Lim et al., 2009). However, breast cancer is not just one disease and whether there are one or more different types of tumour-initiating cell remains to be determined. The origin of the normal post-natal mammary tissue itself (from either a multi-lineage basal mammary stem cell, or a lineage-restricted progenitor) has been a source of contention, and extensively explored using in vivo lineage tracing and transplantation models (Visvader and Stingl, 2014; Wuidart et al., 2016). Studying the tumour-initiating cell in genetic models is also hindered by the variable efficiencies of different reporters that are used to ‘tag’ cell populations and the specificity/off-target expression of Cre recombinase, as described above. Furthermore, the effect of the driver mutation on the targeted cell needs to be considered. PIK3CA (phosphatidylinositol 3-kinase, catalytic, alpha polypeptide), a common genetic driver of breast cancer, invokes mixed-lineage tumours even when expressed in unipotent progenitor cells, demonstrating plasticity (Van Keymeulen et al., 2015; Koren et al., 2015) as well as lineage switching (i.e. expression of PIK3CA in a basal-progenitor cell giving rise to luminal tumours).
Preclinical testing in genetic models
Whilst targeted therapies have been a resounding success in breast cancer, relapse due to drug resistance remains a problem. GEMMs have been used to define the molecular mechanisms of drug resistance in vivo and have the advantage that pathway inactivation can be achieved either genetically and/or pharmacologically. For example, deregulation of cell cycle control (Goel et al., 2016) or loss of phosphatase and tensin homologue (PTEN) (Creedon et al., 2016) were shown to be important in conferring resistance to anti-HER2 therapies (e.g. trastuzumab). Of course, these mechanisms have to be shown to be relevant to the human disease; hence validation in primary patient material is essential. Towards this goal, c-Myc amplification in a genetic model of PIK3CA-related breast cancer was discovered to circumvent PI3K-targeted treatment, in agreement with the observation that high MYC levels aligned with mutation of PIK3CA in patient samples (Liu et al., 2011). Also, when searching for mechanisms of resistance to poly(ADP ribose) polymerase (PARP) inhibitors in BRCA1-associated disease, downregulation of the tumour suppressor p53-binding protein 1 (53BP1) and its downstream effector MAD2L2 (MAD2 mitotic arrest deficient-like 2; previously known as REV7 ), led to rescue of homologous recombination in BRCA1-deficient cells through reinstatement of double-strand-break signalling (Jaspers et al., 2013; Xu et al., 2015). These genes are also downregulated in the human disease, showing their credibility for clinical relevance, while there is some doubt over whether upregulated expression of ATP-binding cassette 1 (ABC1) transporters, also seen in the same mouse models, is a bone fide resistance mechanism in patients (Rottenberg and Borst, 2012; Borst, 2012).
The use of GEMMs in ‘preclinical breast cancer trials’ for new drug combinations is gaining momentum, but as highlighted in the SEARCHBreast workshop, the successful design and execution of appropriate preclinical in vivo trials requires close collaboration between clinicians and scientists from the outset. As a proof of concept, a BRCA1 breast cancer model shown to respond to a combination of cisplatin with PARP inhibition (Rottenberg et al., 2008) with further benefit provided by long-term PARP inhibition (Jaspers et al., 2013), led to the US Food and Drug Administration (FDA) approving such a regime for treatment of platinum-sensitive relapsed ovarian cancer (Ledermann et al., 2012; Matulonis et al., 2016). Disease-specific genetic and syngeneic models (in particular involving transplant of GEMM tissue to recipient strain-matched mice) together with PDX models, hold great potential for the evaluation of patient-relevant regimes. For instance, GEMM-derived and PDX tumours could be used as platforms for preclinical testing of monotherapies versus dual/combination treatment for direct comparison; or testing the merits of neo-adjuvant (first-line) therapy against surgical resection plus adjuvant treatment for efficacy.
Modelling clinically relevant metastasis in GEMMs
A few key conventional GEMM models (e.g. MMTV-PyMT and MMTV-Erbb2) show manifestation of spontaneously arising metastatic disease in lymph nodes and lungs. These models have been, and continue to be, used to study metastatic disease (Kabeer et al., 2016). So for example by genetically deleting their gene of interest, investigators showed how transforming growth factor beta (Tgfb1) and β1-integrin (Itgb1) are important in metastasis (Bierie et al., 2008; Huck et al., 2010). Furthermore, conventional GEMMs were instrumental in demonstrating that metastatic spread was an early step in breast cancer progression (Hüsemann et al., 2008). However, as with CDXs and some PDXs described above, an impediment of GEMMs is that relatively few mimic clinical metastasis (i.e. to the brain and bone), which is responsible for the majority of breast-cancer-associated deaths.
A drawback of using conditional GEMMs to study metastatic disease is low penetrance of metastasis. Therefore, large cohorts of animals have to be followed for a prolonged period of time in order for metastatic disease, and effects of therapies thereon, to be accurately measured. While individual GEMM cohorts develop tumours with highly variable latency and inconsistent metastatic penetrance, the syngeneic system used by the Jonkers lab of implanting orthotopic GEMM tumour fragments (Rottenberg et al., 2007) allows a more homogeneous cohort at the study outset. Surgical resection of the mammary tumours in the Jonkers' syngeneic model then permits the development and subsequent evaluation of metastasis, although the authors still report considerable heterogeneity in latency and number of organs affected (Coffelt et al., 2015). This approach could prove useful if it can be extended to other genetic models, although one must consider that in vivo transplant/passage of GEMM tissue requires the use of an inbred mouse strain to benefit from immune competency. Propagation of tumours originating from a mixed strain can of course still be performed using immunocompromised recipients to avoid host-graft rejection but this is more costly and limited by a non-physiological microenvironment lacking a functional immune component.
The impact of technological advances on breast cancer models
The sophisticated models now at our disposal have only been made possible through ongoing technological advancement. It thus seems fitting to chart some of the relevant history as a preface to how we could improve the toolbox to assist in addressing the topical issues of the here and now.
The imaging revolution
Precise imaging modalities, which are now at the disposal of researchers, have permitted imaging at the single-cell and whole-organism levels. A new era of cancer imaging was heralded with the discovery of green fluorescent protein (GFP) and the realisation that cells could be genetically engineered to express this without detrimental effects (Misteli and Spector, 1997), enabling researchers to locate and track cancer cells and tumour colonies without the need for specific markers or antibodies. This has facilitated mapping of breast tumour development and metastatic progression, as well as evaluation of responses to therapy (Hoffman, 2002). GFP continues to be a useful tool in most breast cancer research laboratories, despite the development of the next generation of imaging probes, allowing easy identification and/or separation of GFP+ tumour cells from a mixed population (Lizier et al., 2016; Holen et al., 2015).
Subsequently, the use of bioluminescence allowed sensitive and rapid in vivo imaging following injection of the substrate luciferin in animals bearing tumour cells genetically engineered to express firefly luciferase (Edinger et al., 1999). Tumour growth kinetics, as well as therapeutic responses, can now be monitored in the same animal over time (Gross and Piwnica-Worms, 2005; Henriquez et al., 2007). Researchers have embraced this new capability and used it to explore the role of specific molecules in breast cancer development, progression and response to treatment (examples in Serganova et al., 2009; Lu et al., 2010).
Technological advances, such as the use of mammary imaging windows pioneered by the Condeelis and van Rheenen labs (Kedrin et al., 2008), in parallel with the increasing sophistication of molecular probes and other tools have enabled visualisation of tumours in vivo. A new generation of confocal and multi-photon microscopes allows visualisation of the interactions between individual breast tumour cells and the microenvironment (e.g. the microvasculature) (Malladi et al., 2016). Initially, this was only possible ex vivo, but increased capability for intra-vital microscopy has provided new information regarding how cancer cells move and respond to stimuli in the live animal (Wyckoff et al., 2011; Patsialou et al., 2013; Nakasone et al., 2013). For example, the early stages of local breast cancer invasion (Giampieri et al., 2009) and stromal cell dynamics (Lohela and Werb, 2010; Ewald et al., 2011) have been elucidated using this technology. Furthermore, elegant in vivo imaging studies have been used to decipher the behaviour of metastatic cells in real time (Zomer et al., 2015; Harney et al., 2015; Beerling et al., 2016).
Although not widely available, a range of quantitative functional imaging capabilities, e.g. PET (positron emission tomography)/CT, SPECT (single photon emission computed tomography)/CT and PET/MRI (magnetic resonance imaging) are being adapted for preclinical research, leading to increased understanding of tumour development and effects of therapy (Consolino et al., 2016). Furthermore, the advancements in imaging capability have facilitated sophisticated lineage-tracing studies in GEMMs, enabling insights into different cell populations, and in particular the tracking of the mammary stem cell (Rios et al., 2014; van Amerongen, 2015; Sale and Pavelic, 2015) and cancer stem cell (Zomer et al., 2013).
Improved molecular engineering and the development of targeted genetic models
The application of gene targeting to manipulate the mouse genome (Thomas and Capecchi, 1987; Capecchi, 2005) revolutionised the use of GEMMs to model cancer. Another key advance was the ability to direct these changes to the tissue of interest using site-specific recombinases (e.g. the Cre/loxP system, described above) circumventing lethal and off-target effects elicited by germline deletion of fundamental genes (Sauer and Henderson, 1988). This has permitted targeting of breast cancer mutations, such as deletion of key tumour suppressors BRCA1, BRCA2, p53 and PTEN, specifically to the mouse mammary epithelium to provide models for the human disease (Jonkers et al., 2001; Diaz-Cruz et al., 2010; Melchor et al., 2014). Whilst conditional GEMMs are a key resource, one limitation is the inability to model natural tumour evolution (early versus late events), because of the simultaneous rather than temporal activation of multiple genetic events. The principle of different site-specific recombinases (Olorunniji et al., 2016) to achieve sequential mutations in a time-dependent way is possible, although have not yet been widely applied.
Another major impediment to using GEMMs for dissecting multistage tumourigenesis is the extensive, and time-consuming, breeding programmes required to achieve relevant compound genetic alleles (sometimes requiring in excess of five alleles). Some groups have expedited this process by targeting new alleles in embryonic stem cells derived from pre-existing GEMMs (GEMM-ESCs), an approach that shows excellent potential for streamlined and high-throughput studies (Huijbers et al., 2014, 2015; Henneman et al., 2015). Others meanwhile have applied inducible in vivo short hairpin RNA (shRNA)-based knockdown as a means to investigate loss of specific tumour suppressor genes without the need for protracted breeding programmes (Ebbesen et al., 2016).
Within the last decade, increasingly fast, cheap and reproducible genetic sequencing, coupled with improved bioinformatics and pathway analysis tools, has resulted in the identification of mutations that are potentially involved in breast cancer development and progression (Banerji et al., 2012; Ellis et al., 2012; Curtis et al., 2012; Nik-Zainal et al., 2016; Ferrari et al., 2016). Whether these genetic alterations are indeed functional can only be established through experimental validation using in vivo tumourigenesis models. High-throughput approaches – such as those using GEMM-ESCs and shRNA – will be necessary if biological verifications of new genes are to match the speed of these discoveries. A recent report, describing how the CRISPR/Cas9 gene-editing system could be combined with GEMM to provide in vivo validation of a candidate tumour suppressor in invasive lobular breast cancer demonstrates a powerful new system for the elucidation of functional relevance in the context of mammary carcinomas (Annunziato et al., 2016). This is a perfect example of how advances in one field (gene editing) can be utilised to drive continued development of preclinical modelling in breast cancer. Furthermore, applying high-throughput ‘omics’ and forward genetic screens (e.g. retrovirus or transposons) (McIntyre et al., 2012) to murine-derived tumours provides a platform for discovery of additional cancer-associated mutations (Francis et al., 2015). Of course, these also need to be cross-referenced for relevance in the human disease, but have demonstrable potential to open up new therapeutic avenues; for example, where profiling of mouse tumours highlighted the druggable target Met as a secondary mutation in p53-deficient tumours, pointing to a putative new target for treating triple-negative breast cancer (Francis et al., 2015; Pfefferle et al., 2016).
The expertise in cultivating PDXs provides more clinically relevant models
As discussed above, PDX models have been welcomed by the community as possibly the most promising for precision medicine (Hidalgo et al., 2014; Whittle et al., 2015). Thus, refinement in the protocols to manipulate and propagate primary patient material over recent years (Zhang and Lewis, 2013; DeRose et al., 2013) has been a key advancement. The choice of recipient strains, use of hormone supplementation and site of implant (i.e. intraductal delivery) have all contributed to the success of creating a renewable resource in the form of stably transplantable PDX models that faithfully maintain the characteristics of the original tumour (DeRose et al., 2011). Access to patient samples via clinical interactions or engagement with specialist biobanks such as the Breast Cancer Now Tissue Bank (breastcancertissuebank.org/) is a necessity and the relevant expertise is currently limited to a relatively small number of labs. However, as increasing numbers of breast cancer PDX models are made available through commercial entities (as discussed above) and consortia such as EurOPDX (see Box 1) and the International Breast Cancer Patient-derived Xenograft Consortium (Dobrolecki et al., 2016), enhanced use of these models is expected. In addition, short-term cultures have been generated from some of these and used as models for high-throughput drug screening (Bruna et al., 2016) and to determine mammosphere-forming efficiency in vitro (Eyre et al., 2016). These studies illustrate the power of these models, which are likely to be used increasingly in the future. For a comprehensive review of breast cancer PDXs, including their limitations, the reader is referred to Dobrolecki et al. (2016).
Building a toolkit fit for the future
Although models are continually evolving to incorporate novel technological and biological advances to improve their utility and clinical relevance, there are still refinements to be made. These require tailoring to address the pertinent questions in the field. In this section, we forecast what the future landscape in development of optimised models and tools might be.
Using GEMMs to model the microenvironment
The importance of the microenvironment in tumour evolution is undisputed (Hanahan and Weinberg, 2011). For example, researchers have demonstrated how macrophages and a highly interactive stroma with cross-talk between the microenvironmental cell types and tumour cells are part of tumour evolution (Qian and Pollard, 2010; Dovas et al., 2013; Noy and Pollard, 2014; Lewis et al., 2016). Furthermore, the immune system can be hijacked into fuelling breast cancer growth and metastasis (Coffelt et al., 2015). Whilst co-injection of stromal and cancer cells to generate CDXs has its place (Orimo et al., 2005), it is time to refine these models. GEMM and syngeneic models are superior for studying the microenvironment as they have an intact immune system, which needs to be given due consideration when testing therapeutic strategies (Coffelt and de Visser, 2015). One caveat of all models is that the microenvironment is murine-derived; even in PDX tumours, the stroma of the mouse host gradually replaces that of the human within 3-4 passages. Regardless, it is noteworthy that studies using GEMMs have elegantly demonstrated just how important the (murine) stroma is for tumour evolution. For example, specific deletion of Pten in stromal fibroblasts (Trimboli et al., 2009) or E2f3 in macrophages (Trikha et al., 2016) modifies tumour growth and metastasis, respectively. Limitations arise, however, because the micro-environmental gene alteration tends to be under Cre/loxP control (e.g. LysCre:E2f3fl/fl), which prohibits application in Cre/loxP-driven tumour models. Therefore, syngeneic tumour implants or conventional transgenic tumour models (e.g. MMTV-Erbb2; MMTV-PyMT) are used rather than more physiologically relevant GEMMs. As stromal influences are likely to impact differently on subtypes of breast cancer (Wallace et al., 2011), it is important to consider the use of alternative site-specific recombinases (Olorunniji et al., 2016) to direct the stromal gene modification (e.g. Flp/Frt), which would permit investigation with the many available Cre/loxP GEMMs.
Humanisation of models in CDX and PDX models
For many situations – such as preclinical studies using monoclonal antibody therapy, which do not act on the murine gene homologue – there is a desire to use human cells in transplantation models. In order to make in vivo models more clinically relevant, researchers have incorporated cells and tissues to increase the human component of the tumour microenvironment i.e. to humanise models. Examples are the implantation of human bone pieces (Kuperwasser et al., 2005; Holen et al., 2015) or tissue-engineered scaffolds seeded with human osteoblasts in immunocompromised animals for studies of bone metastasis (Thibaudeau et al., 2014). The bone discs were either pre-seeded with human tumour cells or tumour cells were injected via the intra-cardiac route once the humanised bone scaffolds were established in the host. These models allow investigation of the interactions between human tumour cells and components of the human bone microenvironment, but they are not yet widely available and standardisation between laboratories is limited by the intrinsic variability in donor bone specimens (Holen et al., 2015).
Furthermore, with the increasing interest in immuno-oncology, efforts have been made to create PDX models that incorporate human immune cells, such as the MiXeno platform from Crown Biosciences. One approach to achieve this is by incorporation of immune cells through co-transplantation of human CD34+ hematopoietic stem cells and breast cancer cells in immunodeficient mice (Wege et al., 2011, 2014).
Recently, the Jackson Laboratories described humanised PDX models as the next ‘big thing’ in cancer modelling (www.jax.org/news-and-insights). Indeed, humanised mice in which triple-negative breast cancer PDX responds to immunotherapy are now commercially available. The high cost of these animals is likely to present a barrier to their general uptake, but they represent an important step forward in terms of our ability to investigate the potential utility of immunotherapies in breast cancer.
Optimising the timelines
A particular challenge for researchers is that breast tumour growth in vivo is most often quite rapid (progressing within weeks), in contrast to the longer latency of human disease (involving years and even decades). In this respect, model systems do not represent the clinical reality; however, it is notoriously difficult to work with slow-growing models because results need to be delivered within the normal funding cycle or within the duration of a standard PhD project (3-5 years). As discussed in the workshop, the realisation that ‘slow is better’ is often balanced against the need to know whether a specific genetic or therapeutic manipulation works in a model whilst sufficient time is still left on a particular grant. It was agreed that science funding bodies generally support the use of short-term (and therefore less representative) models, rather than accepting a more limited (but more clinically relevant) output from models where it may take 1-2 years before tumours develop. Many of the workshop participants are members of scientific committees that make decisions on which projects to fund, so there are opportunities to influence thinking in this area with the aim of providing the longer-term funding required to implement the use of more clinically relevant model systems.
Increasing access to models and material
Working with in vivo model systems is expensive, requires expert staff and facilities, and can be time-consuming, presenting a barrier for many researchers. In addition, translation of basic findings into patient benefit requires evidence to be obtained from several model systems. Paradoxically, those who routinely use in vivo models are almost always generating more material than they need for the purpose of testing the original hypothesis. The SEARCHBreast initiative (see Box 1) was established in order to bridge this gap by allowing researchers to register the availability of surplus archival material from breast cancer models, providing resources that can be accessed on a collaborative basis, saving time and money (Blyth et al., 2016). Although SEARCHBreast contains material from a range of models, including PDXs, a much larger PDX collection is available through the EurOPDX consortium (europdx.eu/, see Box 1). A rapidly expanding resource, this currently contains 54 luminal, 89 triple-negative and 18 HER2+ PDX models, many with associated transcriptomic characterisation; researchers can access these models instead of establishing them de novo. With the increasing cost of carrying out large-scale animal studies, we envisage that the sharing of models and material will become the norm, supporting collaborations with laboratories with extensive expertise, with SEARCHBreast as the trailblazer for this type of thinking.
Developing the models for the future
Breast cancer progression is characterised by the extended period – usually several years – between successful treatment of the primary disease and subsequent relapse (most likely due to the reactivation of dormant disseminated tumour cells in the bone marrow) (Zhang et al., 2013). Model systems that incorporate a dormant phase have been lacking, but some recent studies presented at the workshop have demonstrated that intra-cardiac injection of human tumour cells in mature animals (>12 weeks) shows promise in recapitulating the latency of breast cancer bone metastasis (Ottewell et al., 2014).
As for other models, it was interesting to see one described recently for inflammatory breast cancer, a rare but very aggressive type of breast cancer (Wurth et al., 2015), but there is still a need for a generation of models of male breast cancer and of systems that allow separation of pre- versus post-menopausal disease. We highlight the importance of this point regards disease before and after menopause because recent clinical trials demonstrated therapeutic benefit of bone-targeted agents only in post-menopausal women [Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) et al., 2015]. In addition, new models will be needed as molecular profiling of human breast cancers further stratifies this heterogeneous and complex disease. Better consideration must be given to the specific gene mutations too; for example, genetic modelling of BRCA1 disease has been achieved predominantly through allelic deletions, but these are not truly representative of the patient-derived pathogenic mutant alleles. The importance of this was recently demonstrated: different Brca1 mutations elicited similar disease patterns but showed differences in therapeutic response and resistance mechanisms (Drost et al., 2011, 2016). Modelling metastatic disease remains a challenge due to the need to resect the primary tumour to allow sufficient time for metastatic disease to develop, as well as the highly variable number, sites and growth kinetics of resulting metastases. This means that experimental groups need to be large in order to produce meaningful/significant data, which increases the number of animals used in research, puts a burden on resources and can be costly. Furthermore, a flexible study design, such as treatment starting at different time points in different animals, is required for such studies. Finally, the use of models in experimental designs that mimic clinical trials (including in the context of adjuvant therapy) may become more common, suggesting that the clinical relevance of experimental systems will have greater significance in the future.
Summary and conclusions
In vivo models of breast cancer have proven their usefulness in many different contexts and will continue to contribute to our understanding of disease progression, therapeutic response and resistance mechanisms. The field has progressed incredibly in the last 20-30 years and is likely to evolve further with technological advances that enhance the growing arsenal with which researchers can probe unanswered questions. Nonetheless, scientists need to consider the limitations of each model and choose the one that best represents the process they aim to model and addresses their specific research question. New models being developed using patient-derived tumour material have shown promise, but sharing of these resources is very important to allow comparison between studies and make them widely available to the research community. Meanwhile, new and improved models for metastasis and study of the microenvironment are urgently required. In sum, although no perfect in vivo model of human breast cancer will ever exist, such models remain valuable research tools complemented by clinical material, ex vivo and in vitro systems, and we are equipped with the knowledge and technologies to continue improving them.
Acknowledgements
We would like to thank all the speakers at the workshop, and in particular the plenary speakers Dr Mohamed Bentires-Alj, Dr Matthew Smalley, Prof Valerie Brunton, Dr Robert Clarke and Prof Jos Jonkers, who together with all the participants at the In vivo Models of Breast Cancer workshop contributed to the discussions this manuscript is based on. Only a small selection of the vast literature is cited as examples to illustrate the points discussed in this brief overview. Numerous excellent publications are not included, and we ask for the understanding of those authors whose work has not been cited. Thanks also to Ewan Cameron and Catherine Winchester for critical reading of the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
All authors contributed to the drafting of the manuscript.
Funding
SEARCHBreast was funded by an Infrastructure for Impact grant from the National Centre for the Replacement, Refinement and Reduction of Animals in Research (NC3Rs), UK (NC/L001004/1). Work in the corresponding author's lab is funded by Cancer Research UK (C596/A17196).
References
- Annunziato S., Kas S. M., Nethe M., Yücel H., Del Bravo J., Pritchard C., Bin Ali R., van Gerwen B., Siteur B., Drenth A. P. et al. (2016). Modeling invasive lobular breast carcinoma by CRISPR/Cas9-mediated somatic genome editing of the mammary gland. Genes Dev. 30, 1470-1480. 10.1101/gad.279190.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun G., Diermeier S., Akerman M., Chang K.-C., Wilkinson J. E., Hearn S., Kim Y., MacLeod A. R., Krainer A. R., Norton L. et al. (2016). Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 30, 34-51. 10.1101/gad.270959.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji S., Cibulskis K., Rangel-Escareno C., Brown K. K., Carter S. L., Frederick A. M., Lawrence M. S., Sivachenko A. Y., Sougnez C., Zou L. et al. (2012). Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 486, 405-409. 10.1038/nature11154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Cardiff R. D., Steinbach P., Messer K. S. and Ellies L. G. (2015). Multipotent luminal mammary cancer stem cells model tumor heterogeneity. Breast Cancer Res. 17, 137 10.1186/s13058-015-0615-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselga J., Norton L., Albanell J., Kim Y. M. and Mendelsohn J. (1998). Recombinant humanized anti-HER2 antibody (Herceptin) enhances the antitumor activity of paclitaxel and doxorubicin against HER2/neu overexpressing human breast cancer xenografts. Cancer Res. 58, 2825-2831. [PubMed] [Google Scholar]
- Beck B. and Blanpain C. (2013). Unravelling cancer stem cell potential. Nat. Rev. Cancer 13, 727-738. 10.1038/nrc3597 [DOI] [PubMed] [Google Scholar]
- Beckhove P., Schütz F., Diel I. J., Solomayer E.-F., Bastert G., Foerster J., Feuerer M., Bai L., Sinn H.-P., Umansky V. et al. (2003). Efficient engraftment of human primary breast cancer transplants in nonconditioned NOD/Scid mice. Int. J. Cancer 105, 444-453. 10.1002/ijc.11125 [DOI] [PubMed] [Google Scholar]
- Beerling E., Seinstra D., de Wit E., Kester L., van der Velden D., Maynard C., Schäfer R., van Diest P., Voest E., van Oudenaarden A. et al. (2016). Plasticity between epithelial and mesenchymal states unlinks EMT from metastasis-enhancing stem cell capacity. Cell Rep. 14, 2281-2288. 10.1016/j.celrep.2016.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behbod F., Kittrell F. S., LaMarca H., Edwards D., Kerbawy S., Heestand J. C., Young E., Mukhopadhyay P., Yeh H.-W., Allred D. C. et al. (2009). An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast Cancer Res. 11, R66 10.1186/bcr2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U., Ha G., Khadka P., Jin X., Wong B., Franke L. and Golub T. R. (2016). The landscape of chromosomal aberrations in breast cancer mouse models reveals driver-specific routes to tumorigenesis. Nat. Commun. 7, 12160 10.1038/ncomms12160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierie B., Stover D. G., Abel T. W., Chytil A., Gorska A. E., Aakre M., Forrester E., Yang L., Wagner K.-U. and Moses H. L. (2008). Transforming growth factor-beta regulates mammary carcinoma cell survival and interaction with the adjacent microenvironment. Cancer Res. 68, 1809-1819. 10.1158/0008-5472.CAN-07-5597 [DOI] [PubMed] [Google Scholar]
- Bittner J. J. (1936). Some possible effects of nursing on the mammary gland tumor incidence in mice. Science 84, 162 10.1126/science.84.2172.162 [DOI] [PubMed] [Google Scholar]
- Blaas L., Pucci F., Messal H. A., Andersson A. B., Josue Ruiz E., Gerling M., Douagi I., Spencer-Dene B., Musch A., Mitter R. et al. (2016). Lgr6 labels a rare population of mammary gland progenitor cells that are able to originate luminal mammary tumours. Nat. Cell Biol. 18, 1346-1356. 10.1038/ncb3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyth K., Morton J. P. and Sansom O. J. (2012). The right time. the right place: will targeting human cancer-associated mutations to the mouse provide the perfect preclinical model? Curr. Opin. Genet. Dev. 22, 28-35. 10.1016/j.gde.2012.02.009 [DOI] [PubMed] [Google Scholar]
- Blyth K., Carter P., Morrissey B., Chelala C., Jones L., Holen I. and Speirs V. (2016). SEARCHBreast: a new resource to locate and share surplus archival material from breast cancer animal models to help address the 3Rs. Breast Cancer Res. Treat. 156, 447-452. 10.1007/s10549-016-3785-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowsky A. D. (2011). Choosing a mouse model: experimental biology in context--the utility and limitations of mouse models of breast cancer. Cold Spring Harb. Perspect. Biol. 3, a009670 10.1101/cshperspect.a009670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst P. (2012). Cancer drug pan-resistance: pumps. cancer stem cells, quiescence, epithelial to mesenchymal transition, blocked cell death pathways, persisters or what? Open Biol. 2, 120066 10.1098/rsob.120066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks M. D., Burness M. L. and Wicha M. S. (2015). Therapeutic implications of cellular heterogeneity and plasticity in breast cancer. Cell Stem Cell 17, 260-271. 10.1016/j.stem.2015.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruna A., Rueda O. M., Greenwood W., Batra A. S., Callari M., Batra R. N., Pogrebniak K., Sandoval J., Cassidy J. W., Tufegdzic-Vidakovic A. et al. (2016). A biobank of breast cancer explants with preserved intra-tumor heterogeneity to screen anticancer compounds. Cell 167, 260-274.e22. 10.1016/j.cell.2016.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. (2012). Comprehensive molecular portraits of human breast tumours. Nature 490, 61-70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi M. R. (2005). Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 6, 507-512. 10.1038/nrg1619 [DOI] [PubMed] [Google Scholar]
- Cardiff R. D. (2010). The pathology of EMT in mouse mammary tumorigenesis. J. Mammary Gland Biol. Neoplasia 15, 225-233. 10.1007/s10911-010-9184-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardiff R. D. and Kenney N. (2011). A compendium of the mouse mammary tumor biologist: from the initial observations in the house mouse to the development of genetically engineered mice. Cold Spring Harb. Perspect. Biol. 3, a003111 10.1101/cshperspect.a003111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardiff R. D. and Wellings S. R. (1999). The comparative pathology of human and mouse mammary glands. J. Mammary Gland Biol. Neoplasia 4, 105-122. 10.1023/A:1018712905244 [DOI] [PubMed] [Google Scholar]
- Cardiff R. D., Anver M. R., Gusterson B. A., Hennighausen L., Jensen R. A., Merino M. J., Rehm S., Russo J., Tavassoli F. A., Wakefield L. M. et al. (2000). The mammary pathology of genetically engineered mice: the consensus report and recommendations from the Annapolis meeting. Oncogene 19, 968-988. 10.1038/sj.onc.1203277 [DOI] [PubMed] [Google Scholar]
- Cardiff R. D., Rosner A., Hogarth M. A., Galvez J. J., Borowsky A. D. and Gregg J. P. (2004). Validation: the new challenge for pathology. Toxicol. Pathol. 32, 31-39. 10.1080/01926230490424662 [DOI] [PubMed] [Google Scholar]
- Coffelt S. B. and de Visser K. E. (2015). Immune-mediated mechanisms influencing the efficacy of anticancer therapies. Trends Immunol. 36, 198-216. 10.1016/j.it.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Coffelt S. B., Kersten K., Doornebal C. W., Weiden J., Vrijland K., Hau C.-S., Verstegen N. J. M., Ciampricotti M., Hawinkels L. J. A. C., Jonkers J. et al. (2015). IL-17-producing gammadelta T cells and neutrophils conspire to promote breast cancer metastasis. Nature 522, 345-348. 10.1038/nature14282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consolino L., Longo D. L., Dastrù W., Cutrin J. C., Dettori D., Lanzardo S., Oliviero S., Cavallo F. and Aime S. (2016). Functional imaging of the angiogenic switch in a transgenic mouse model of human breast cancer by dynamic contrast enhanced magnetic resonance imaging. Int. J. Cancer 139, 404-413. 10.1002/ijc.30073 [DOI] [PubMed] [Google Scholar]
- Cottu P., Marangoni E., Assayag F., de Cremoux P., Vincent-Salomon A., Guyader C., de Plater L., Elbaz C., Karboul N., Fontaine J. J. et al. (2012). Modeling of response to endocrine therapy in a panel of human luminal breast cancer xenografts. Breast Cancer Res. Treat. 133, 595-606. 10.1007/s10549-011-1815-5 [DOI] [PubMed] [Google Scholar]
- Cottu P., Bieche I., Assayag F., El Botty R., Chateau-Joubert S., Thuleau A., Bagarre T., Albaud B., Rapinat A., Gentien D. et al. (2014). Acquired resistance to endocrine treatments is associated with tumor-specific molecular changes in patient-derived luminal breast cancer xenografts. Clin. Cancer Res. 20, 4314-4325. 10.1158/1078-0432.CCR-13-3230 [DOI] [PubMed] [Google Scholar]
- Creedon H., Balderstone L. A., Muir M., Balla J., Gomez-Cuadrado L., Tracey N., Loane J., Klinowska T., Muller W. J. and Brunton V. G. (2016). Use of a genetically engineered mouse model as a preclinical tool for HER2 breast cancer. Dis. Model. Mech. 9, 131-140. 10.1242/dmm.023143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C., Shah S. P., Chin S.-F., Turashvili G., Rueda O. M., Dunning M. J., Speed D., Lynch A. G., Samarajiwa S., Yuan Y. et al. (2012). The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346-352. 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabydeen S. A. and Furth P. A. (2014). Genetically engineered ERalpha-positive breast cancer mouse models. Endocr Relat. Cancer 21, R195-R208. 10.1530/ERC-13-0512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassule H. R., Lewis P., Bei M., Maas R. and Mcmahon A. P. (2000). Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 127, 4775-4785. [DOI] [PubMed] [Google Scholar]
- Deome K. B., Faulkin L. J. Jr, Bern H. A. and Blair P. B. (1959). Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 19, 515-520. [PubMed] [Google Scholar]
- DeRose Y. S., Wang G., Lin Y.-C., Bernard P. S., Buys S. S., Ebbert M. T. W., Factor R., Matsen C., Milash B. A., Nelson E. et al. (2011). Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat. Med. 17, 1514-1520. 10.1038/nm.2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRose Y. S., Gligorich K. M., Wang G., Georgelas A., Bowman P., Courdy S. J., Welm A. L. and Welm B. E. (2013). Patient-derived models of human breast cancer: protocols for in vitro and in vivo applications in tumor biology and translational medicine. Curr. Protoc. Pharmacol. 14, Unit 14.23 10.1002/0471141755.ph1423s60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Cruz E. S., Cabrera M. C., Nakles R., Rutstein B. H. and Furth P. A. (2010). BRCA1 deficient mouse models to study pathogenesis and therapy of triple negative breast cancer. Breast Dis. 32, 85-97. 10.3233/BD-2010-0308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrolecki L. E., Airhart S. D., Alferez D. G., Aparicio S., Behbod F., Bentires-Alj M., Brisken C., Bult C. J., Cai S., Clarke R. B. et al. (2016). Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev. 35, 547-573. 10.1007/s10555-016-9653-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovas A., Patsialou A., Harney A. S., Condeelis J. and Cox D. (2013). Imaging interactions between macrophages and tumour cells that are involved in metastasis in vivo and in vitro. J. Microsc. 251, 261-269. 10.1111/j.1365-2818.2012.03667.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost R., Bouwman P., Rottenberg S., Boon U., Schut E., Klarenbeek S., Klijn C., van der Heijden I., van der Gulden H., Wientjens E. et al. (2011). BRCA1 RING function is essential for tumor suppression but dispensable for therapy resistance. Cancer Cell 20, 797-809. 10.1016/j.ccr.2011.11.014 [DOI] [PubMed] [Google Scholar]
- Drost R., Dhillon K. K., van der Gulden H., van der Heijden I., Brandsma I., Cruz C., Chondronasiou D., Castroviejo-Bermejo M., Boon U., Schut E. et al. (2016). BRCA1185delAG tumors may acquire therapy resistance through expression of RING-less BRCA1. J. Clin. Invest. 126, 2903-2918. 10.1172/JCI70196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG), Coleman R., Powles T., Paterson A., Gnant M., Anderson S., Diel I., Gralow J., Von Minckwitz G., Moebus V., Bergh J. et al. (2015). Adjuvant bisphosphonate treatment in early breast cancer: meta-analyses of individual patient data from randomised trials. Lancet 386, 1353-1361. 10.1016/S0140-6736(15)60908-4 [DOI] [PubMed] [Google Scholar]
- Ebbesen S. H., Scaltriti M., Bialucha C. U., Morse N., Kastenhuber E. R., Wen H. Y., Dow L. E., Baselga J. and Lowe S. W. (2016). Pten loss promotes MAPK pathway dependency in HER2/neu breast carcinomas. Proc. Natl. Acad. Sci. USA 113, 3030-3035. 10.1073/pnas.1523693113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles S. A., Aboagye E. O., Ali S., Anderson A. S., Armes J., Berditchevski F., Blaydes J. P., Brennan K., Brown N. J., Bryant H. E. et al. (2013). Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res. 15, R92 10.1186/bcr3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger M., Sweeney T. J., Tucker A. A., Olomu A. B., Negrin R. S. and Contag C. H. (1999). Noninvasive assessment of tumor cell proliferation in animal models. Neoplasia 1, 303-310. 10.1038/sj.neo.7900048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M. J., Ding L., Shen D., Luo J., Suman V. J., Wallis J. W., Van Tine B. A., Hoog J., Goiffon R. J., Goldstein T. C. et al. (2012). Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486, 353-360. 10.1038/nature11143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erler J. T., Bennewith K. L., Cox T. R., Lang G., Bird D., Koong A., Le Q.-T. and Giaccia A. J. (2009). Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell 15, 35-44. 10.1016/j.ccr.2008.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald A. J., Werb Z. and Egeblad M. (2011). Dynamic, long-term in vivo imaging of tumor-stroma interactions in mouse models of breast cancer using spinning-disk confocal microscopy. Cold Spring Harb. Protoc. 2011, pdb.top97 10.1101/pdb.top97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre R., Alférez D. G., Spence K., Kamal M., Shaw F. L., Simões B. M., Santiago-Gómez A., Sarmiento-Castro A., Bramley M., Absar M. et al. (2016). Patient-derived Mammosphere and Xenograft Tumour Initiation Correlates with Progression to Metastasis. J. Mammary Gland Biol. Neoplasia 21, 99-109. 10.1007/s10911-016-9361-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathers K. E., Rodrigues S., Zuo D., Murthy I. V., Hallett M., Cardiff R. and Park M. (2010). CrkII transgene induces atypical mammary gland development and tumorigenesis. Am. J. Pathol. 176, 446-460. 10.2353/ajpath.2010.090383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari A., Vincent-Salomon A., Pivot X., Sertier A. S., Thomas E., Tonon L., Boyault S., Mulugeta E., Treilleux I., MacGrogan G. et al. (2016). A whole-genome sequence and transcriptome perspective on HER2-positive breast cancers. Nat. Commun. 7, 12222 10.1038/ncomms12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler I. J. (1973). Selection of successive tumour lines for metastasis. Nat. New Biol. 242, 148-149. 10.1038/newbio242148a0 [DOI] [PubMed] [Google Scholar]
- Fleming J. M., Miller T. C., Meyer M. J., Ginsburg E. and Vonderhaar B. K. (2010). Local regulation of human breast xenograft models. J. Cell. Physiol. 224, 795-806. 10.1002/jcp.22190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis J. C., Melchor L., Campbell J., Kendrick H., Wei W., Armisen-Garrido J., Assiotis I., Chen L., Kozarewa I., Fenwick K. et al. (2015). Whole-exome DNA sequence analysis of Brca2- and Trp53-deficient mouse mammary gland tumours. J. Pathol. 236, 186-200. 10.1002/path.4517 [DOI] [PubMed] [Google Scholar]
- Fu X., Le P. and Hoffman R. M. (1993). A metastatic orthotopic-transplant nude-mouse model of human patient breast cancer. Anticancer Res. 13, 901-904. [PubMed] [Google Scholar]
- Fung A. S., Lee C., Yu M. and Tannock I. F. (2015). The effect of chemotherapeutic agents on tumor vasculature in subcutaneous and orthotopic human tumor xenografts. BMC Cancer 15, 112 10.1186/s12885-015-1091-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giampieri S., Manning C., Hooper S., Jones L., Hill C. S. and Sahai E. (2009). Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat. Cell Biol. 11, 1287-1296. 10.1038/ncb1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel S., Wang Q., Watt A. C., Tolaney S. M., Dillon D. A., Li W., Ramm S., Palmer A. C., Yuzugullu H., Varadan V. et al. (2016). Overcoming therapeutic resistance in HER2-positive breast cancers with CDK4/6 inhibitors. Cancer Cell 29, 255-269. 10.1016/j.ccell.2016.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenow K. R. and Smalley M. J. (2015). Overview of genetically engineered mouse models of breast cancer used in translational biology and drug development. Curr. Protoc. Pharmacol. 70, 1-14. 10.1002/0471141755.ph1436s70 [DOI] [PubMed] [Google Scholar]
- Gross S. and Piwnica-Worms D. (2005). Spying on cancer: molecular imaging in vivo with genetically encoded reporters. Cancer Cell 7, 5-15. 10.1016/j.ccr.2004.12.011 [DOI] [PubMed] [Google Scholar]
- Gu G., Dustin D. and Fuqua S. A. W. (2016). Targeted therapy for breast cancer and molecular mechanisms of resistance to treatment. Curr. Opin. Pharmacol. 31, 97-103. 10.1016/j.coph.2016.11.005 [DOI] [PubMed] [Google Scholar]
- Hanahan D. and Weinberg R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646-674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Haricharan S., Lei J. and Ellis M. (2016). Mammary ductal environment is necessary for faithful maintenance of estrogen signaling in ER(+) breast cancer. Cancer Cell 29, 249-250. 10.1016/j.ccell.2016.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harney A. S., Arwert E. N., Entenberg D., Wang Y., Guo P., Qian B.-Z., Oktay M. H., Pollard J. W., Jones J. G. and Condeelis J. S. (2015). Real-time imaging reveals local, transient vascular permeability, and tumor cell intravasation stimulated by TIE2hi macrophage-derived VEGFA. Cancer Discov. 5, 932-943. 10.1158/2159-8290.CD-15-0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneman L., van Miltenburg M. H., Michalak E. M., Braumuller T. M., Jaspers J. E., Drenth A. P., de Korte-Grimmerink R., Gogola E., Szuhai K., Schlicker A. et al. (2015). Selective resistance to the PARP inhibitor olaparib in a mouse model for BRCA1-deficient metaplastic breast cancer. Proc. Natl. Acad. Sci. USA 112, 8409-8414. 10.1073/pnas.1500223112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriquez N. V., van Overveld P. G. M., Que I., Buijs J. T., Bachelier R., Kaijzel E. L., Löwik C. W. G. M., Clezardin P. and van der Pluijm G. (2007). Advances in optical imaging and novel model systems for cancer metastasis research. Clin. Exp. Metastasis 24, 699-705. 10.1007/s10585-007-9115-5 [DOI] [PubMed] [Google Scholar]
- Herschkowitz J. I., Simin K., Weigman V. J., Mikaelian I., Usary J., Hu Z., Rasmussen K. E., Jones L. P., Assefnia S., Chandrasekharan S. et al. (2007). Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 8, R76 10.1186/gb-2007-8-5-r76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo M., Amant F., Biankin A. V., Budinska E., Byrne A. T., Caldas C., Clarke R. B., de Jong S., Jonkers J., Maelandsmo G. M. et al. (2014). Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 4, 998-1013. 10.1158/2159-8290.CD-14-0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman R. M. (2002). Green fluorescent protein imaging of tumour growth, metastasis, and angiogenesis in mouse models. Lancet Oncol. 3, 546-556. 10.1016/S1470-2045(02)00848-3 [DOI] [PubMed] [Google Scholar]
- Hoffman R. M. (2015). Patient-derived orthotopic xenografts: better mimic of metastasis than subcutaneous xenografts. Nat. Rev. Cancer 15, 451-452. 10.1038/nrc3972 [DOI] [PubMed] [Google Scholar]
- Holen I., Nutter F., Wilkinson J. M., Evans C. A., Avgoustou P. and Ottewell P. D. (2015). Human breast cancer bone metastasis in vitro and in vivo: a novel 3D model system for studies of tumour cell-bone cell interactions. Clin. Exp. Metastasis 32, 689-702. 10.1007/s10585-015-9737-y [DOI] [PubMed] [Google Scholar]
- Hollern D. P. and Andrechek E. R. (2014). A genomic analysis of mouse models of breast cancer reveals molecular features of mouse models and relationships to human breast cancer. Breast Cancer Res. 16, R59 10.1186/bcr3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday D. L. and Speirs V. (2011). Choosing the right cell line for breast cancer research. Breast Cancer Res. 13, 215. 10.1186/bcr2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck L., Pontier S. M., Zuo D. M. and Muller W. J. (2010). beta1-integrin is dispensable for the induction of ErbB2 mammary tumors but plays a critical role in the metastatic phase of tumor progression. Proc. Natl. Acad. Sci. U. S. A. 107, 15559-15564. 10.1073/pnas.1003034107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers I. J., Bin Ali R., Pritchard C., Cozijnsen M., Kwon M.-C., Proost N., Song J.-Y., de Vries H., Badhai J., Sutherland K. et al. (2014). Rapid target gene validation in complex cancer mouse models using re-derived embryonic stem cells. EMBO Mol. Med. 6, 212-225. 10.1002/emmm.201303297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers I. J., Del Bravo J., Bin Ali R., Pritchard C., Braumuller T. M., van Miltenburg M. H., Henneman L., Michalak E. M., Berns A. and Jonkers J. (2015). Using the GEMM-ESC strategy to study gene function in mouse models. Nat. Protoc. 10, 1755-1785. 10.1038/nprot.2015.114 [DOI] [PubMed] [Google Scholar]
- Hüsemann Y., Geigl J. B., Schubert F., Musiani P., Meyer M., Burghart E., Forni G., Eils R., Fehm T., Riethmüller G. et al. (2008). Systemic spread is an early step in breast cancer. Cancer Cell 13, 58-68. 10.1016/j.ccr.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Jaspers J. E., Kersbergen A., Boon U., Sol W., van Deemter L., Zander S. A., Drost R., Wientjens E., Ji J., Aly A. et al. (2013). Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer Discov. 3, 68-81. 10.1158/2159-8290.CD-12-0049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone C. N., Smith Y. E., Cao Y., Burrows A. D., Cross R. S. N., Ling X., Redvers R. P., Doherty J. P., Eckhardt B. L., Natoli A. L. et al. (2015). Functional and molecular characterisation of EO771.LMB tumours, a new C57BL/6-mouse-derived model of spontaneously metastatic mammary cancer. Dis. Model. Mech. 8, 237-251. 10.1242/dmm.017830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers J., Meuwissen R., van der Gulden H., Peterse H., van der Valk M. and Berns A. (2001). Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29, 418-425. 10.1038/ng747 [DOI] [PubMed] [Google Scholar]
- Kabeer F., Beverly L. J., Darrasse-Jeze G. and Podsypanina K. (2016). Methods to study metastasis in genetically modified mice. Cold Spring Harb. Protoc. 2016, pdb.top069948 10.1101/pdb.top069948 [DOI] [PubMed] [Google Scholar]
- Kalimutho M., Parsons K., Mittal D., López J. A., Srihari S. and Khanna K. K. (2015). Targeted therapies for triple-negative breast cancer: combating a stubborn disease. Trends Pharmacol. Sci. 36, 822-846. 10.1016/j.tips.2015.08.009 [DOI] [PubMed] [Google Scholar]
- Kedrin D., Gligorijevic B., Wyckoff J., Verkhusha V. V., Condeelis J., Segall J. E. and van Rheenen J. (2008). Intravital imaging of metastatic behavior through a mammary imaging window. Nat. Methods 5, 1019-1021. 10.1038/nmeth.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S. and Bentires-Alj M. (2015). Breast tumor heterogeneity: source of fitness, hurdle for therapy. Mol. Cell 60, 537-546. 10.1016/j.molcel.2015.10.031 [DOI] [PubMed] [Google Scholar]
- Koren S., Reavie L., Couto J. P., De Silva D., Stadler M. B., Roloff T., Britschgi A., Eichlisberger T., Kohler H., Aina O. et al. (2015). PIK3CA(H1047R) induces multipotency and multi-lineage mammary tumours. Nature 525, 114-118. 10.1038/nature14669 [DOI] [PubMed] [Google Scholar]
- Kuperwasser C., Dessain S., Bierbaum B. E., Garnet D., Sperandio K., Gauvin G. P., Naber S. P., Weinberg R. A. and Rosenblatt M. (2005). A mouse model of human breast cancer metastasis to human bone. Cancer Res. 65, 6130-6138. 10.1158/0008-5472.CAN-04-1408 [DOI] [PubMed] [Google Scholar]
- Ledermann J., Harter P., Gourley C., Friedlander M., Vergote I., Rustin G., Scott C., Meier W., Shapira-Frommer R., Safra T. et al. (2012). Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N. Engl. J. Med. 366, 1382-1392. 10.1056/NEJMoa1105535 [DOI] [PubMed] [Google Scholar]
- Ledford H. (2016). US cancer institute to overhaul tumour cell lines. Nature 530, 391 10.1038/nature.2016.19364 [DOI] [PubMed] [Google Scholar]
- Lewis C. E., Harney A. S. and Pollard J. W. (2016). The multifaceted role of perivascular macrophages in tumors. Cancer Cell 30, 365 10.1016/j.ccell.2016.07.009 [DOI] [PubMed] [Google Scholar]
- Lim E., Vaillant F., Wu D., Forrest N. C., Pal B., Hart A. H., Asselin-Labat M.-L., Gyorki D. E., Ward T., Partanen A. et al. (2009). Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 15, 907-913. 10.1038/nm.2000 [DOI] [PubMed] [Google Scholar]
- Lindeman G. J. and Visvader J. E. (2010). Insights into the cell of origin in breast cancer and breast cancer stem cells. Asia Pac. J. Clin. Oncol. 6, 89-97. 10.1111/j.1743-7563.2010.01279.x [DOI] [PubMed] [Google Scholar]
- Liu X., Holstege H., van der Gulden H., Treur-Mulder M., Zevenhoven J., Velds A., Kerkhoven R. M., van Vliet M. H., Wessels L. F. A., Peterse J. L. et al. (2007). Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc. Natl. Acad. Sci. USA 104, 12111-12116. 10.1073/pnas.0702969104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Cheng H., Santiago S., Raeder M., Zhang F., Isabella A., Yang J., Semaan D. J., Chen C., Fox E. A. et al. (2011). Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat. Med. 17, 1116-1120. 10.1038/nm.2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizier M., Anselmo A., Mantero S., Ficara F., Paulis M., Vezzoni P., Lucchini F. and Pacchiana G. (2016). Fusion between cancer cells and macrophages occurs in a murine model of spontaneous neu+ breast cancer without increasing its metastatic potential. Oncotarget 7, 60793-60806. 10.18632/oncotarget.11508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohela M. and Werb Z. (2010). Intravital imaging of stromal cell dynamics in tumors. Curr. Opin. Genet. Dev. 20, 72-78. 10.1016/j.gde.2009.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Yan C. H., Yuan M., Wei Y., Hu G. and Kang Y. (2010). In vivo dynamics and distinct functions of hypoxia in primary tumor growth and organotropic metastasis of breast cancer. Cancer Res. 70, 3905-3914. 10.1158/0008-5472.CAN-09-3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque-Cabal M., Garcia-Teijido P., Fernandez-Perez Y., Sanchez-Lorenzo L. and Palacio-Vazquez I. (2016). Mechanisms behind the resistance to trastuzumab in HER2-amplified breast cancer and strategies to overcome it. Clin. Med. Insights Oncol. 10, 21-30. 10.4137/CMO.S34537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malladi S., Macalinao D. G., Jin X., He L., Basnet H., Zou Y., de Stanchina E. and Massagué J. (2016). Metastatic latency and immune evasion through autocrine inhibition of WNT. Cell 165, 45-60. 10.1016/j.cell.2016.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marangoni E., Vincent-Salomon A., Auger N., Degeorges A., Assayag F., de Cremoux P., de Plater L., Guyader C., De Pinieux G., Judde J.-G. et al. (2007). A new model of patient tumor-derived breast cancer xenografts for preclinical assays. Clin. Cancer Res. 13, 3989-3998. 10.1158/1078-0432.CCR-07-0078 [DOI] [PubMed] [Google Scholar]
- Matulonis U. A., Harter P., Gourley C., Friedlander M., Vergote I., Rustin G., Scott C., Meier W., Shapira-Frommer R., Safra T. et al. (2016). Olaparib maintenance therapy in patients with platinum-sensitive, relapsed serous ovarian cancer and a BRCA mutation: Overall survival adjusted for postprogression poly(adenosine diphosphate ribose) polymerase inhibitor therapy. Cancer 122, 1844-1852. 10.1002/cncr.29995 [DOI] [PubMed] [Google Scholar]
- McCarthy A., Savage K., Gabriel A., Naceur C., Reis-Filho J. S. and Ashworth A. (2007). A mouse model of basal-like breast carcinoma with metaplastic elements. J. Pathol. 211, 389-398. 10.1002/path.2124 [DOI] [PubMed] [Google Scholar]
- McIntyre R. E., van der Weyden L. and Adams D. J. (2012). Cancer gene discovery in the mouse. Curr. Opin. Genet. Dev. 22, 14-20. 10.1016/j.gde.2011.12.003 [DOI] [PubMed] [Google Scholar]
- Melchor L., Molyneux G., Mackay A., Magnay F.-A., Atienza M., Kendrick H., Nava-Rodrigues D., López-García M. A., Milanezi F., Greenow K. et al. (2014). Identification of cellular and genetic drivers of breast cancer heterogeneity in genetically engineered mouse tumour models. J. Pathol. 233, 124-137. 10.1002/path.4345 [DOI] [PubMed] [Google Scholar]
- Menezes M. E., Das S. K., Emdad L., Windle J. J., Wang X.-Y., Sarkar D. and Fisher P. B. (2014). Genetically engineered mice as experimental tools to dissect the critical events in breast cancer. Adv. Cancer Res. 121, 331-382. 10.1016/B978-0-12-800249-0.00008-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D. S., Koren S., Leroy C., Brinkhaus H., Muller U., Klebba I., Muller M., Cardiff R. D. and Bentires-Alj M. (2013). Expression of PIK3CA mutant E545K in the mammary gland induces heterogeneous tumors but is less potent than mutant H1047R. Oncogenesis 2, e74 10.1038/oncsis.2013.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. and Spector D. L. (1997). Applications of the green fluorescent protein in cell biology and biotechnology. Nat. Biotechnol. 15, 961-964. 10.1038/nbt1097-961 [DOI] [PubMed] [Google Scholar]
- Molyneux G., Geyer F. C., Magnay F.-A., McCarthy A., Kendrick H., Natrajan R., MacKay A., Grigoriadis A., Tutt A., Ashworth A. et al. (2010). BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell 7, 403-417. 10.1016/j.stem.2010.07.010 [DOI] [PubMed] [Google Scholar]
- Morrissey B., Blyth K., Carter P., Chelala C., Jones L., Holen I. and Speirs V. (2016). SEARCHBreast: a new online resource to make surplus material from in vivo models of breast cancer visible and accessible to researchers. Breast Cancer Res. 18, 59 10.1186/s13058-016-0716-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller W. J., Sinn E., Pattengale P. K., Wallace R. and Leder P. (1988). Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell 54, 105-115. 10.1016/0092-8674(88)90184-5 [DOI] [PubMed] [Google Scholar]
- Munn R. J., Webster M., Muller W. J. and Cardiff R. D. (1995). Histopathology of transgenic mouse mammary tumors (a short atlas). Semin. Cancer Biol. 6, 153-158. 10.1006/scbi.1995.0020 [DOI] [PubMed] [Google Scholar]
- Nakasone E. S., Askautrud H. A. and Egeblad M. (2013). Live imaging of drug responses in the tumor microenvironment in mouse models of breast cancer. J. Vis. Exp. 24, 50088 10.3791/50088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve R. M., Chin K., Fridlyand J., Yeh J., Baehner F. L., Fevr T., Clark L., Bayani N., Coppe J.-P., Tong F. et al. (2006). A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10, 515-527. 10.1016/j.ccr.2006.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S., Davies H., Staaf J., Ramakrishna M., Glodzik D., Zou X., Martincorena I., Alexandrov L. B., Martin S., Wedge D. C. et al. (2016). Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47-54. 10.1038/nature17676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy R. and Pollard J. W. (2014). Tumor-associated macrophages: from mechanisms to therapy. Immunity 41, 49-61. 10.1016/j.immuni.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olorunniji F. J., Rosser S. J. and Stark W. M. (2016). Site-specific recombinases: molecular machines for the Genetic Revolution. Biochem. J. 473, 673-684. 10.1042/BJ20151112 [DOI] [PubMed] [Google Scholar]
- Orimo A., Gupta P. B., Sgroi D. C., Arenzana-Seisdedos F., Delaunay T., Naeem R., Carey V. J., Richardson A. L. and Weinberg R. A. (2005). Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 121, 335-348. 10.1016/j.cell.2005.02.034 [DOI] [PubMed] [Google Scholar]
- Osborne C. K. and Schiff R. (2011). Mechanisms of endocrine resistance in breast cancer. Annu. Rev. Med. 62, 233-247. 10.1146/annurev-med-070909-182917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottewell P. D., Wang N., Brown H. K., Reeves K. J., Fowles C. A., Croucher P. I., Eaton C. L. and Holen I. (2014). Zoledronic acid has differential antitumor activity in the pre- and postmenopausal bone microenvironment in vivo. Clin. Cancer Res. 20, 2922-2932. 10.1158/1078-0432.CCR-13-1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri C., Patten D. K., Januszewski A., Zucchini G. and Howell S. J. (2014). Breast cancer: current and future endocrine therapies. Mol. Cell. Endocrinol. 382, 695-723. 10.1016/j.mce.2013.08.001 [DOI] [PubMed] [Google Scholar]
- Paschall A. V. and Liu K. (2016). An orthotopic mouse model of spontaneous breast cancer metastasis. J. Vis. Exp. 14, 54040 10.3791/54040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsialou A., Bravo-Cordero J. J., Wang Y., Entenberg D., Liu H., Clarke M. and Condeelis J. S. (2013). Intravital multiphoton imaging reveals multicellular streaming as a crucial component of in vivo cell migration in human breast tumors. Intravital 2, e25294 10.4161/intv.25294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou C. M., Sørlie T., Eisen M. B., van de Rijn M., Jeffrey S. S., Rees C. A., Pollack J. R., Ross D. T., Johnsen H., Akslen L. A. et al. (2000). Molecular portraits of human breast tumours. Nature 406, 747-752. 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- Pfefferle A. D., Herschkowitz J. I., Usary J., Harrell J. C., Spike B. T., Adams J. R., Torres-Arzayus M. I., Brown M., Egan S. E., Wahl G. M. et al. (2013). Transcriptomic classification of genetically engineered mouse models of breast cancer identifies human subtype counterparts. Genome Biol. 14, R125 10.1186/gb-2013-14-11-r125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferle A. D., Agrawal Y. N., Koboldt D. C., Kanchi K. L., Herschkowitz J. I., Mardis E. R., Rosen J. M. and Perou C. M. (2016). Genomic profiling of murine mammary tumors identifies potential personalized drug targets for p53-deficient mammary cancers. Dis. Model. Mech. 9, 749-757. 10.1242/dmm.025239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzo M. G., Lesurf R., Petkiewicz S., O'Malley F. P., Pinnaduwage D., Andrulis I. L., Bull S. B., Chughtai N., Zuo D., Souleimanova M. et al. (2009). Met induces mammary tumors with diverse histologies and is associated with poor outcome and human basal breast cancer. Proc. Natl. Acad. Sci. USA 106, 12903-12908. 10.1073/pnas.0810402106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A. and Perou C. M. (2011). Deconstructing the molecular portraits of breast cancer. Mol. Oncol. 5, 5-23. 10.1016/j.molonc.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian B.-Z. and Pollard J. W. (2010). Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39-51. 10.1016/j.cell.2010.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan J. L., Kendrick H., Magnay F.-A., Vafaizadeh V., Groner B. and Smalley M. J. (2012). c-Kit is required for growth and survival of the cells of origin of Brca1-mutation-associated breast cancer. Oncogene 31, 869-883. 10.1038/onc.2011.289 [DOI] [PubMed] [Google Scholar]
- Richard E., Grellety T., Velasco V., MacGrogan G., Bonnefoi H. and Iggo R. (2016). The mammary ducts create a favourable microenvironment for xenografting of luminal and molecular apocrine breast tumours. J. Pathol. 22, 256-261. 10.1002/path.4772 [DOI] [PubMed] [Google Scholar]
- Rios A. C., Fu N. Y., Lindeman G. J. and Visvader J. E. (2014). In situ identification of bipotent stem cells in the mammary gland. Nature 506, 322-327. 10.1038/nature12948 [DOI] [PubMed] [Google Scholar]
- Rottenberg S. and Borst P. (2012). Drug resistance in the mouse cancer clinic. Drug Resist. Update 15, 81-89. 10.1016/j.drup.2012.01.001 [DOI] [PubMed] [Google Scholar]
- Rottenberg S., Nygren A. O. H., Pajic M., van Leeuwen F. W. B., van der Heijden I., van de Wetering K., Liu X., de Visser K. E., Gilhuijs K. G., van Tellingen O. et al. (2007). Selective induction of chemotherapy resistance of mammary tumors in a conditional mouse model for hereditary breast cancer. Proc. Natl. Acad. Sci. USA 104, 12117-12122. 10.1073/pnas.0702955104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottenberg S., Jaspers J. E., Kersbergen A., van der Burg E., Nygren A. O. H., Zander S. A. L., Derksen P. W. B., de Bruin M., Zevenhoven J., Lau A. et al. (2008). High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc. Natl. Acad. Sci. USA 105, 17079-17084. 10.1073/pnas.0806092105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rygaard J. and Povsen C. O. (2007). Heterotransplantation of a human malignant tumour to “nude” mice. 1969. Apmis 115, 604-606. 10.1111/j.1600-0463.2007.apm_689a.x [DOI] [PubMed] [Google Scholar]
- Sale S. and Pavelic K. (2015). Mammary lineage tracing: the coming of age. Cell. Mol. Life Sci. 72, 1577-1583. 10.1007/s00018-014-1817-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B. and Henderson N. (1988). Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. USA 85, 5166-5170. 10.1073/pnas.85.14.5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganova I., Moroz E., Vider J., Gogiberidze G., Moroz M., Pillarsetty N., Doubrovin M., Minn A., Thaler H. T., Massague J. et al. (2009). Multimodality imaging of TGFbeta signaling in breast cancer metastases. FASEB J. 23, 2662-2672. 10.1096/fj.08-126920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sflomos G., Dormoy V., Metsalu T., Jeitziner R., Battista L., Scabia V., Raffoul W., Delaloye J.-F., Treboux A., Fiche M. et al. (2016). A preclinical model for ERalpha-positive breast cancer points to the epithelial microenvironment as determinant of luminal phenotype and hormone response. Cancer Cell 29, 407-422. 10.1016/j.ccell.2016.02.002 [DOI] [PubMed] [Google Scholar]
- Simões B. M., O'Brien C. S., Eyre R., Silva A., Yu L., Sarmiento-Castro A., Alférez D. G., Spence K., Santiago-Gómez A., Chemi F. et al. (2015). Anti-estrogen resistance in human breast tumors is driven by JAG1-NOTCH4-dependent cancer stem cell activity. Cell Rep. 12, 1968-1977. 10.1016/j.celrep.2015.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T., Perou C. M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M. B., van de Rijn M., Jeffrey S. S. et al. (2001). Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 98, 10869-10874. 10.1073/pnas.191367098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T., Tibshirani R., Parker J., Hastie T., Marron J. S., Nobel A., Deng S., Johnsen H., Pesich R., Geisler S. et al. (2003). Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. USA 100, 8418-8423. 10.1073/pnas.0932692100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart T. A., Pattengale P. K. and Leder P. (1984). Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell 38, 627-637. 10.1016/0092-8674(84)90257-5 [DOI] [PubMed] [Google Scholar]
- Talmadge J. E., Singh R. K., Fidler I. J. and Raz A. (2007). Murine models to evaluate novel and conventional therapeutic strategies for cancer. Am. J. Pathol. 170, 793-804. 10.2353/ajpath.2007.060929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tentler J. J., Tan A. C., Weekes C. D., Jimeno A., Leong S., Pitts T. M., Arcaroli J. J., Messersmith W. A. and Eckhardt S. G. (2012). Patient-derived tumour xenografts as models for oncology drug development. Nat. Rev. Clin. Oncol. 9, 338-350. 10.1038/nrclinonc.2012.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Brugge P., Kristel P., van der Burg E., Boon U., de Maaker M., Lips E., Mulder L., de Ruiter J., Moutinho C., Gevensleben H. et al. (2016). Mechanisms of therapy resistance in patient-derived xenograft models of BRCA1-deficient breast cancer. J. Natl. Cancer Inst. 108, djw148 10.1093/jnci/djw148 [DOI] [PubMed] [Google Scholar]
- Thibaudeau L., Taubenberger A. V., Holzapfel B. M., Quent V. M., Fuehrmann T., Hesami P., Brown T. D., Dalton P. D., Power C. A., Hollier B. G. et al. (2014). A tissue-engineered humanized xenograft model of human breast cancer metastasis to bone. Dis. Model. Mech. 7, 299-309. 10.1242/dmm.014076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas K. R. and Capecchi M. R. (1987). Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51, 503-512. 10.1016/0092-8674(87)90646-5 [DOI] [PubMed] [Google Scholar]
- Torre L. A., Siegel R. L., Ward E. M. and Jemal A. (2016). Global cancer incidence and mortality rates and trends--an update. Cancer Epidemiol. Biomarkers Prev. 25, 16-27. 10.1158/1055-9965.EPI-15-0578 [DOI] [PubMed] [Google Scholar]
- Trikha P., Sharma N., Pena C., Reyes A., Pécot T., Khurshid S., Rawahneh M., Moffitt J., Stephens J. A., Fernandez S. A. et al. (2016). E2f3 in tumor macrophages promotes lung metastasis. Oncogene 35, 3636-3646. 10.1038/onc.2015.429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimboli A. J., Cantemir-Stone C. Z., Li F., Wallace J. A., Merchant A., Creasap N., Thompson J. C., Caserta E., Wang H., Chong J.-L. et al. (2009). Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature 461, 1084-1091. 10.1038/nature08486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R. (2015). Lineage tracing in the mammary gland using Cre/lox technology and fluorescent reporter alleles. Methods Mol. Biol. 1293, 187-211. 10.1007/978-1-4939-2519-3_11 [DOI] [PubMed] [Google Scholar]
- van Keymeulen A., Lee M. Y., Ousset M., Brohée S., Rorive S., Giraddi R. R., Wuidart A., Bouvencourt G., Dubois C., Salmon I. et al. (2015). Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature 525, 119-123. 10.1038/nature14665 [DOI] [PubMed] [Google Scholar]
- Varn F. S., Mullins D. W., Arias-Pulido H., Fiering S. and Cheng C. (2016). Adaptive immunity programs in breast cancer. Immunology 26, 12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader J. E. and Stingl J. (2014). Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 28, 1143-1158. 10.1101/gad.242511.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace J. A., Li F., Leone G. and Ostrowski M. C. (2011). Pten in the breast tumor microenvironment: modeling tumor-stroma coevolution. Cancer Res. 71, 1203-1207. 10.1158/0008-5472.CAN-10-3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wege A. K., Ernst W., Eckl J., Frankenberger B., Vollmann-Zwerenz A., Männel D. N., Ortmann O., Kroemer A. and Brockhoff G. (2011). Humanized tumor mice--a new model to study and manipulate the immune response in advanced cancer therapy. Int. J. Cancer 129, 2194-2206. 10.1002/ijc.26159 [DOI] [PubMed] [Google Scholar]
- Wege A. K., Schmidt M., Ueberham E., Ponnath M., Ortmann O., Brockhoff G. and Lehmann J. (2014). Co-transplantation of human hematopoietic stem cells and human breast cancer cells in NSG mice: a novel approach to generate tumor cell specific human antibodies. MAbs 6, 968-977. 10.4161/mabs.29111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle J. R., Lewis M. T., Lindeman G. J. and Visvader J. E. (2015). Patient-derived xenograft models of breast cancer and their predictive power. Breast Cancer Res. 17, 17 10.1186/s13058-015-0523-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuidart A., Ousset M., Rulands S., Simons B. D., Van Keymeulen A. and Blanpain C. (2016). Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells. Genes Dev. 30, 1261-1277. 10.1101/gad.280057.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurth R., Tarn K., Jernigan D., Fernandez S. V., Cristofanilli M., Fatatis A. and Meucci O. (2015). A preclinical model of inflammatory breast cancer to study the involvement of CXCR4 and ACKR3 in the metastatic process. Transl. Oncol. 8, 358-367. 10.1016/j.tranon.2015.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff J., Gligorijevic B., Entenberg D., Segall J. and Condeelis J. (2011). High-resolution multiphoton imaging of tumors in vivo. Cold Spring Harb. Protoc. 1, 1167-1184. 10.1101/pdb.top065904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Wagner K.-U., Larson D., Weaver Z., Li C., Ried T., Hennighausen L., Wynshaw-Boris A. and Deng C. X. (1999). Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat. Genet. 22, 37-43. 10.1038/8743 [DOI] [PubMed] [Google Scholar]
- Xu G., Chapman J. R., Brandsma I., Yuan J., Mistrik M., Bouwman P., Bartkova J., Gogola E., Warmerdam D., Barazas M. et al. (2015). REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 521, 541-544. 10.1038/nature14328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. and Lewis M. T. (2013). Establishment of Patient-Derived Xenograft (PDX) Models of human breast cancer. Curr. Protoc. Mouse Biol. 3, 21-29. 10.1002/9780470942390.mo120140 [DOI] [PubMed] [Google Scholar]
- Zhang X. H.-F., Giuliano M., Trivedi M. V., Schiff R. and Osborne C. K. (2013). Metastasis dormancy in estrogen receptor-positive breast cancer. Clin. Cancer Res. 19, 6389-6397. 10.1158/1078-0432.CCR-13-0838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer A., Ellenbroek S. I. J., Ritsma L., Beerling E., Vrisekoop N. and Van Rheenen J. (2013). Intravital imaging of cancer stem cell plasticity in mammary tumors. Stem Cells 31, 602-606. 10.1002/stem.1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer A., Maynard C., Verweij F. J., Kamermans A., Schäfer R., Beerling E., Schiffelers R. M., de Wit E., Berenguer J., Ellenbroek S. I. J. et al. (2015). In Vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 161, 1046-1057. 10.1016/j.cell.2015.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]