Abstract
A full description of a protein's function requires knowledge of all partner proteins with which it specifically associates. From a functional perspective, ‘association’ can mean direct physical binding, but can also mean indirect interaction such as participation in the same metabolic pathway or cellular process. Currently, information about protein association is scattered over a wide variety of resources and model organisms. STRING aims to simplify access to this information by providing a comprehensive, yet quality-controlled collection of protein–protein associations for a large number of organisms. The associations are derived from high-throughput experimental data, from the mining of databases and literature, and from predictions based on genomic context analysis. STRING integrates and ranks these associations by benchmarking them against a common reference set, and presents evidence in a consistent and intuitive web interface. Importantly, the associations are extended beyond the organism in which they were originally described, by automatic transfer to orthologous protein pairs in other organisms, where applicable. STRING currently holds 730 000 proteins in 180 fully sequenced organisms, and is available at http://string.embl.de/.
INTRODUCTION
Several databases exist, whose main purpose is to collect and curate direct experimental evidence about protein–protein interactions (1–4). Other databases take a more generalized perspective on proteins and their associations, by functionally grouping proteins into metabolic, signaling or transcriptional pathways (5–8). Finally, a third class of resources attempts to fill gaps in both datasets, by predicting protein–protein associations de novo, using a variety of computational techniques (9–13).
The database STRING (‘Search Tool for the Retrieval of Interacting Genes/Proteins’) represents an ongoing effort to provide these three types of protein–protein association evidence under one common framework. Such an integrated approach offers several unique advantages: (i) various types of evidence are mapped onto a single, stable set of proteins, thereby facilitating comparative analysis; (ii) known and predicted interactions often partially complement each other, leading to increased coverage; (iii) an integrated scoring scheme can provide higher confidence when independent evidence types agree; and (iv) mapping and transferring interactions onto a large number of organisms facilitates evolutionary studies.
Because STRING is fully pre-computed, all information can be quickly accessed—both at the high-level network view and at the level of the individual interaction record. The various evidence types can be enabled or disabled separately, which allows the searches to be customized at run-time, and dedicated viewers allow the inspection of all the evidence underlying an association (Figure 1). The database is an exploratory resource: it contains a much larger number of associations than primary interaction databases—albeit with varying confidence scores. It is thus best used for getting a quick initial overview of the functional partners of a query protein, especially for proteins that are still poorly characterized.
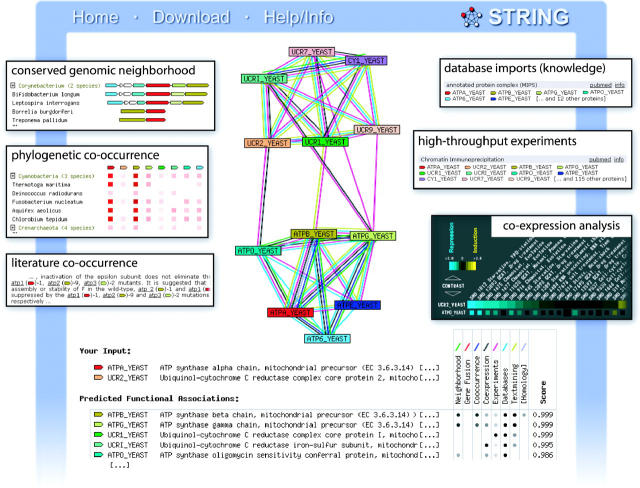
Figure 1.
Results from a STRING search. Inserts show partial screen shots from evidence pages, which are accessible from the main result page. Two proteins were used as inputs to the query—one is a subunit from the yeast ATP synthase complex, the other a subunit from the ubiquinol–cytochrome C reductase complex. The number of requested partners was limited to 10 (default settings). STRING reports both proteins to be members of functional modules, which are in turn connected as part of a larger unit. The diversity of evidence types supporting the modules is noted.
DATA SOURCES AND SCORING
Many of the protein–protein associations in STRING are imported from other databases (see below), but STRING also contains a large body of predicted associations that are produced de novo. These predictions are based on systematic genome comparisons [‘genomic context’, (14,15)]. We periodically import completely sequenced genomes [metazoan genomes from Ensembl, all others from SwissProt, (16)], and search them for three types of genomic context associations: conserved genomic neighborhood, gene fusion events, and co-occurrence of genes across genomes. All three searches aim to identify pairs of genes which appear to be under common selective pressures during evolution (more so than expected by chance), and which are therefore thought to be functionally associated.
As for all other types of associations in STRING, we assign a confidence score to each predicted association. The scores are derived by benchmarking the performance of the predictions against a common reference set of trusted, true associations. We chose the functional grouping of proteins maintained at KEGG [Kyoto Encyclopedia of Genes and Genomes, (5)] as the reference. Any predicted association for which both proteins are assigned to the same ‘KEGG pathway’ is counted as a true positive. KEGG pathways are particularly suitable as a reference because they are based on manual curation, are available for a number of organisms, and cover several functional areas. The benchmarked confidence scores in STRING generally correspond to the probability of finding the linked proteins within the same KEGG pathway. STRING performs a similar benchmark for high-throughput experimental interaction data, separately for each dataset. Scores vary within one dataset because they include additional, intrinsic information from the data itself, such as the frequency or reciprocality of the detection (see Figure 2 for a typical benchmark). In contrast to high-throughput data, validated small-scale interactions, protein complexes, and annotated pathways are directly imported from databases (2,5,17), and given a uniform confidence score per dataset.
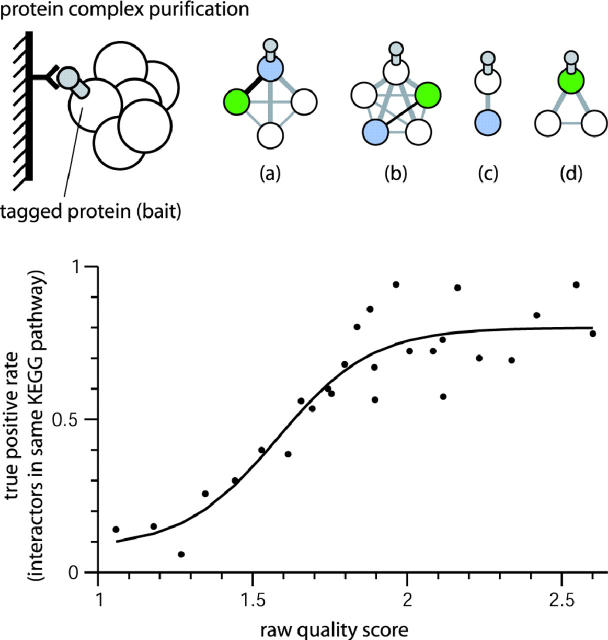
Figure 2.
Deriving confidence scores for high-throughput interaction data [exemplified here for a dataset of protein complex purifications (22)]. In this case, the relative confidence depends on how often two proteins are pulled down together (a and b), versus how often they are pulled down alone (c and d). A purification is counted twice when one of the partners is the bait (a and d). Raw quality is: Q = log{(Ntogether · Ntotal)/[(Nalone1 + 1) · (Nalone2 + 1)]}.
Another important source of protein association information is the published literature (18,19). We systematically extract associations from PubMed, by searching for recurrent co-mentioning of gene names in abstracts. This search relies on gene names and synonyms parsed from SwissProt as well as from organism-specific databases, and we utilize a benchmarked scoring system based on the frequencies and distributions of gene names in abstracts (not shown).
Finally, we also derive protein–protein associations from functional genomics data: co-regulation of genes across diverse experimental conditions, as measured by using microarray analysis, can be a predictor of functional associations (20). We import these associations from the ArrayProspector server (12), which is based on the same benchmarks and genomes as STRING itself.
TRANSFER OF ASSOCIATIONS ACROSS ORGANISMS
STRING employs two different strategies for transferring known and predicted associations between organisms (Figure 3): the first (‘COG-mode’) relies on externally provided orthology assignments and transfers interactions in an all-or-none fashion, whereas the second (‘protein-mode’) uses quantitative sequence similarity searches and often distributes a given interaction fractionally among several protein pairs of the target organism. Both approaches have strengths and weaknesses, and users can choose either one of them before starting their query (a color change helps them to distinguish the modus throughout the user interface).
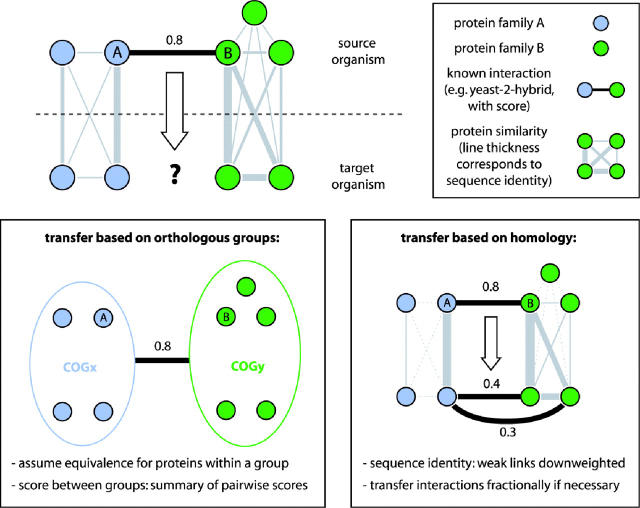
Figure 3.
Transferring association scores between organisms. Initial situation (top): a scored association between two proteins in a source organism—how confidently can it be transferred to a target organism by a postulated association among homologous proteins? Bottom left: in ‘COG-mode’, all proteins in an orthologous group (COG) are considered equivalent. The highest association score between any two proteins in the two COGs is assumed to be valid for all pairs. Bottom right: in ‘protein-mode’, all sequence similarity relations between the two organisms are considered. Associations are transferred fractionally, such that the pair with the highest similarity receives the bulk of the score. The relation is not linear: empiric analysis (not shown) suggests that competing similarity links should be down weighted, relative to the best link, as follows: (i) express similarities as values between zero and one, i.e. normalize by self-hit; (ii) transform similarities using s′ = exp(−k1/s), thereby amplifying their ‘spread’; (iii) re-normalize so that, between the two species, all similarities for a protein family add up to one; (iv) each pair of proteins, A and B in the target species now receives a share of the association score: Starget = Ssource · k2 · s′A · s′B. (optimal values for k1 and k2 were empirically found to be 0.7 for both).
The COG mode requires an assignment of proteins into orthologous groups; all proteins within such a group are assumed to be functionally equivalent across genomes. This orthology information is imported from the COGs database [(21), we extend the groups to cover all organisms in STRING]. Any association score observed between a pair of proteins from two different COGs is assumed to be valid for all protein pairs spanning these two COGs. Repeated observations of links, e.g. occurrence of genes in the same operon, increase the association score—but only when they are observed in phylogenetically distant organisms.
In the newly developed protein mode, there is no preassigned orthology information. Instead, the transfer relies on a precomputed all-against-all similarity search of the 730 000 proteins in STRING (using the sensitive Smith-Waterman algorithm). For each association to be transferred, the algorithm searches for potential orthologs of the interacting partners in other genomes. Orthology is assumed if proteins form reciprocal best matches in the searches, in the absence of any close, second-best hits (paralogs) in either species. In such an ideal situation, the interactions can be transferred in toto. However, in reality there will often be additional paralogs in one or both of the genomes, which complicates the transfer. We have devised and benchmarked an empirical scheme that is based on the relative sequence similarity of competing paralogous proteins (Figure 3). Essentially, the pair of proteins exhibiting the highest sequence similarity to the source pair receives the highest ‘share’ of the transferred interaction.
INTEGRATION
After assignment of association scores and transfer between species, we compute a final ‘combined score’ between any pair of proteins (or pair of COGs). This score is often higher than the individual sub-scores, expressing increased confidence when an association is supported by several types of evidence (Table 1). It is computed under the assumption of independence for the various sources, in a naïve Bayesian fashion. It is thus a simple expression of the individual scores:
Table 1. The number of associations stored in STRING, shown separately for each data source and confidence range (low confidence: scores <0.4; medium: 0.4 to 0.7; high: >0.7).
| Association evidence type | Low confidence | Medium confidence | High confidence |
|---|---|---|---|
| Conserved neighborhood | 441 385 | 111 785 | 32 401 |
| Gene fusions | 33 578 | 3765 | 4393 |
| Phylogenetic co-occurrence | 10 634 199 | 1 091 997 | 175 774 |
| Co-expression | 4 556 189 | 615,825 | 29 879 |
| Database imports | 38 638 | 11 193 | 7,695 |
| Large-scale experiments | 207 829 | 26 226 | 3,401 |
| Literature co-occurrence | 413 521 | 154 594 | 55 387 |
| Combined score | 15 498 524 | 1 937 091 | 368 669 |
The confidence increases when methods are combined (e.g. there are more high-confidence links in the last row than the simple sum). Data from version 5.1 of STRING.
![]()
The assumption of independence is valid here because datasets that are based on similar technologies (e.g. different yeast two-hybrid datasets) have been joined previously and are benchmarked as a single information source. Along with the combined score, the individual sub-scores are always displayed as well, because they provide valuable information about the nature of a particular association.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by grants from the Bundesministerium für Forschung und Bildung, Germany, from the Netherlands Organization of Scientific Research (NOW), and from The Knut and Alice Wallenberg Foundation (to S.D.H.).
REFERENCES
- 1.Salwinski L., Miller,C.S., Smith,A.J., Pettit,F.K., Bowie,J.U. and Eisenberg,D. (2004) The database of interacting proteins: 2004 update. Nucleic Acids Res., 32, D449–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bader G.D., Betel,D. and Hogue,C.W. (2003) BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res., 31, 248–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hermjakob H., Montecchi-Palazzi,L., Lewington,C., Mudali,S., Kerrien,S., Orchard,S., Vingron,M., Roechert,B., Roepstorff,P., Valencia,A. et al. (2004) IntAct: an open source molecular interaction database. Nucleic Acids Res., 32, D452–D455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zanzoni A., Montecchi-Palazzi,L., Quondam,M., Ausiello,G., Helmer-Citterich,M. and Cesareni,G. (2002) MINT: a Molecular INTeraction database. FEBS Lett., 513, 135–140. [DOI] [PubMed] [Google Scholar]
- 5.Kanehisa M., Goto,S., Kawashima,S., Okuno,Y. and Hattori,M. (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res., 32, D277–D280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krieger C.J., Zhang,P., Mueller,L.A., Wang,A., Paley,S., Arnaud,M., Pick,J., Rhee,S.Y. and Karp,P.D. (2004) MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res., 32, D438–D442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joshi-Tope G., Vastrik,I., Gopinath,G.R., Matthews,L., Schmidt,E., Gillespie,M., D'Eustachio,P., Jassal,B., Lewis,S., Wu,G. et al. (2003) The Genome Knowledgebase: a resource for biologists and bioinformaticists. Cold Spring Harb. Symp. Quant. Biol., 68, 237–243. [DOI] [PubMed] [Google Scholar]
- 8.Salgado H., Gama-Castro,S., Martinez-Antonio,A., Diaz-Peredo,E., Sanchez-Solano,F., Peralta-Gil,M., Garcia-Alonso,D., Jimenez-Jacinto,V., Santos-Zavaleta,A., Bonavides-Martinez,C. et al. (2004) RegulonDB (version 4.0): transcriptional regulation, operon organization and growth conditions in Escherichia coli K-12. Nucleic Acids Res., 32, D303–D306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enault F., Suhre,K., Poirot,O., Abergel,C. and Claverie,J.M. (2004) Phydbac2: improved inference of gene function using interactive phylogenomic profiling and chromosomal location analysis. Nucleic Acids Res., 32, W336–W339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mellor J.C., Yanai,I., Clodfelter,K.H., Mintseris,J. and DeLisi,C. (2002) Predictome: a database of putative functional links between proteins. Nucleic Acids Res., 30, 306–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Mering C., Huynen,M., Jaeggi,D., Schmidt,S., Bork,P. and Snel,B. (2003) STRING: a database of predicted functional associations between proteins. Nucleic Acids Res., 31, 258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen L.J., Lagarde,J., von Mering,C. and Bork,P. (2004) ArrayProspector: a web resource of functional associations inferred from microarray expression data. Nucleic Acids Res., 32, W445–W448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowers P.M., Pellegrini,M., Thompson,M.J., Fierro,J., Yeates,T.O. and Eisenberg,D. (2004) Prolinks: a database of protein functional linkages derived from coevolution. Genome Biol., 5, R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valencia A. and Pazos,F. (2003) Prediction of protein-protein interactions from evolutionary information. Methods Biochem Anal., 44, 411–426. [PubMed] [Google Scholar]
- 15.Huynen M.A., Snel,B., von Mering,C. and Bork,P. (2003) Function prediction and protein networks. Curr. Opin. Cell Biol., 15, 191–198. [DOI] [PubMed] [Google Scholar]
- 16.Brooksbank C., Camon,E., Harris,M.A., Magrane,M., Martin,M.J., Mulder,N., O'Donovan,C., Parkinson,H., Tuli,M.A., Apweiler,R. et al. (2003) The European Bioinformatics Institute's data resources. Nucleic Acids Res., 31, 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mewes H.W., Amid,C., Arnold,R., Frishman,D., Guldener,U., Mannhaupt,G., Munsterkotter,M., Pagel,P., Strack,N., Stumpflen,V. et al. (2004) MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res., 32, D41–D44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donaldson I., Martin,J., de Bruijn,B., Wolting,C., Lay,V., Tuekam,B., Zhang,S., Baskin,B., Bader,G.D., Michalickova,K. et al. (2003) PreBIND and Textomy—mining the biomedical literature for protein–protein interactions using a support vector machine. BMC Bioinformatics, 4, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcotte E.M., Xenarios,I. and Eisenberg,D. (2001) Mining literature for protein-protein interactions. Bioinformatics, 17, 359–363. [DOI] [PubMed] [Google Scholar]
- 20.Stuart J.M., Segal,E., Koller,D. and Kim,S.K. (2003) A gene-coexpression network for global discovery of conserved genetic modules. Science, 302, 249–255. [DOI] [PubMed] [Google Scholar]
- 21.Tatusov R.L., Fedorova,N.D., Jackson,J.D., Jacobs,A.R., Kiryutin,B., Koonin,E.V., Krylov,D.M., Mazumder,R., Mekhedov,S.L., Nikolskaya,A.N. et al. (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics, 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gavin A.C., Bosche,M., Krause,R., Grandi,P., Marzioch,M., Bauer,A., Schultz,J., Rick,J.M., Michon,A.M., Cruciat,C.M. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]