Abstract
The Stanford Microarray Database (SMD) (http://smd.stanford.edu) is a research tool for hundreds of Stanford researchers and their collaborators. In addition, SMD functions as a resource for the entire biological research community by providing unrestricted access to microarray data published by SMD users and by disseminating its source code. In addition to storing GenePix (Axon Instruments) and ScanAlyze output from spotted microarrays, SMD has recently added the ability to store, retrieve, display and analyze the complete raw data produced by several additional microarray platforms and image analysis software packages, so that we can also now accept data from Affymetrix GeneChips (MAS5/GCOS or dChip), Agilent Catalog or Custom arrays (using Agilent's Feature Extraction software) or data created by SpotReader (Niles Scientific). We have implemented software that allows us to accept MAGE-ML documents from array manufacturers and to submit MIAME-compliant data in MAGE-ML format directly to ArrayExpress and GEO, greatly increasing the ease with which data from SMD can be published adhering to accepted standards and also increasing the accessibility of published microarray data to the general public. We have introduced a new tool to facilitate data sharing among our users, so that datasets can be shared during, before or after the completion of data analysis. The latest version of the source code for the complete database package was released in November 2004 (http://smd.stanford.edu/download/), allowing researchers around the world to deploy their own installations of SMD.
INTRODUCTION
The Stanford Microarray Database (SMD) (1) (http://smd.stanford.edu) was initially developed in 1999 to serve a small team of researchers using spotted DNA microarrays for human and yeast research at Stanford University. Since then, it has become a research tool for a much larger scientific community using multiple microarray platforms to study a myriad of biomedical research problems. SMD now supports the research of more than 1000 users in over 260 laboratories at Stanford and around the world. These users have entered data generated from more than 50 000 microarrays used to study the biology of 34 organisms, published more than 190 papers referring to data in SMD and have made the complete raw data from more than 7000 microarrays freely available via the SMD website. The public data can be selected, viewed, downloaded and analyzed by the public using most of the tools that are available to registered SMD users. The source code for SMD has been downloaded and installed at several academic and private locations.
Here, we discuss some of the recent developments at SMD that have enabled us to accept microarray data from additional platforms and image analysis software, export data in MAGE-ML format and permit greater data sharing between researchers. In addition, we present information about the latest release of the SMD source code.
RESULTS
Microarray platforms and software
Until 2003, all data in SMD were obtained from two-channel cDNA microarrays extracted from scanned images using either GenePix (www.axon.com) or ScanAlyze (http://rana.lbl.gov/EisenSoftware.htm). The increased interest among SMD users in other microarray platforms and other data acquisition software provided impetus to accommodate new data formats in SMD. Specifically, we have added the ability to accept data from Agilent arrays acquired by Agilent's Feature Extraction software, gene expression data from Affymetrix arrays acquired by the Affymetrix Microarray Analysis Suite v5.0 (MAS5, Affymetrix), Gene Chip Operating System (GCOS, Affymetrix) or DNA-Chip Analyzer (dChip) (2) and two-color data acquired using SpotReader (http://www.nilesscientific.com/) software.
In order to accept data from additional microarray platforms and software packages, we were faced with several hurdles. First, the data models used to describe both the microarray designs and the associated results had to be re-designed andre-implemented. We decided to store all fields available from every software package that we supported, which can be up to several dozen, rather than store only the data fields common to many different software packages. Second, we modified the software for entry and retrieval of array designs and for microarray data entry, retrieval, display and analysis. The design and implementation are object-oriented and relatively easy to extend, so that additional platforms and data formats can be accommodated in the future. These features are available in the 11/04 software release.
Data export to public repositories
In close collaboration with the staff of ArrayExpress, a public repository for microarray data (3) (http://www.ebi.ac.uk/arrayexpress/), we have constructed and implemented a pipeline that converts sets of microarray data within SMD into MAGE-ML files that support the MIAME standards for information content (4,5) that can be directly deposited to ArrayExpress, and more recently GEO (6; http://www.ncbi.nlm.nih.gov/geo). We took advantage of software developed by members of the microarray informatics community (MAGE-stk; http://mged.sourceforge.net/), to create this pipeline, which translates data from SMD into the MAGE-OM structure, export it as MAGE-ML, and then test its formatting with the ArrayExpress MAGE-ML validator (ftp://ftp.ebi.ac.uk/pub/databases/arrayexpress/MAGEvalidator-DISTRIB/). The MAGE-ML files are then transferred via ftp to ArrayExpress and GEO where they can be immediately entered.
Several new tables and user interfaces were designed and added to allow SMD users to provide the annotation of their experiments and biological samples that are required for MIAME compliance. These include tools to annotate protocols, experimental factors and experimental designs. Using MGED Ontology terms (http://mged.sourceforge.net/ontologies/MGEDontology.php), SMD users can describe the overall experimental design and how each member of a set of microarrays fits into that design, such as time points in a time series experiment or tumor samples from many individuals in a molecular survey of a type of cancer. Users can also describe the biological properties and parameters, including HIPAA-compliant clinical data, of each sample and the procedures and protocols used to treat the sample, extract the RNA or DNA, amplify and label the nucleic acids, hybridize the arrays, and scan and acquire data from the resulting microarray image. In this way, each set of microarrays associated with a publication in SMD can be adequately annotated and easily submitted to a public data repository.
Tools for collaboration
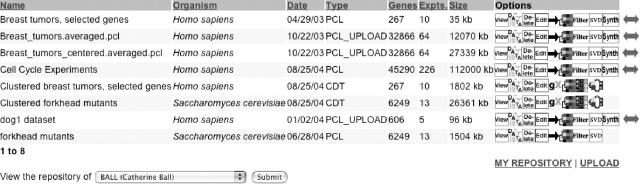
The large community of researchers using SMD participates in widespread and active collaborations. SMD's tools for sharing data facilitate collaboration and accurate communication of experimental details. To complement existing tools for organizing and annotating shared data, we have implemented a ‘data repository’ that allows users to save and share the results of data analysis (Figure 1). Users are able to save the results of data selection, filtering, transformation and analysis at each of these different steps and then specify other users or groups who should be able to view the datasets. In addition to providing a means to help users save their work and facilitate collaboration, the data repository provides a jumping-off point for various analysis tools, such as hierarchical clustering and singular value decomposition.
Figure 1.
Screenshot of SMD's Microarray Data Repository. The repository allows the users to store their data sets created within SMD, upload datasets from other sources, share them with colleagues and collaborators and jump off to a variety of data retrieval, selection, transformation and analysis options. The interface provides the option to view any other repository for which a user has permission to view data. SMD users can store datasets before and after filtering and centering in pre-clustering (.pcl) files. Using repository options, a user can view, download, cluster and filter data, delete data from the repository, edit descriptions and permissions for a data set, perform singular value decomposition and collapse the data based on human genes into UniGene clusters, sections of chromosome or other ‘synthetic genes’ constructed by the researcher. Clustered data can also be stored, and users have additional options to view a heat-map cluster using GeneXplorer (8) to view spot-image clusters displaying or to view both heat-map and spot-image clusters side by side.
Availability
An updated version of the source code for SMD, including all the functions described in this paper, was made publicly available in November 2004 (http://smd.stanford.edu/download/). The source code is freely available under the liberal, open-source MIT license (http://www.opensource.org/licenses/mit-license.php).
Future directions
In the future, we will update and release our software with greater frequency and with better support for users who are upgrading their SMD installations and must therefore migrate data from one database schema to another. In this way, other installations of SMD will be able to benefit from improvements and new tools in a timely manner that will require less work to deploy. We also plan to provide web services through BioMOBY (7) to allow for more creative and flexible use of SMD by researchers or data miners who wish to be able to automate complex queries of the available data. Finally, techniques for determining data quality in an automated fashion remain an active area of research at SMD. Automatically recognizing problematic data will allow researchers to reject it from analysis, correct errors and, ideally, identify and prevent potential sources of poor-quality data. The large body of data in SMD will provide excellent training and test sets for quality assurance studies.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the US National Institutes of Health to P.O.B. (5R01 CA77097) and to G.S. (1R01 HG002732).
REFERENCES
- 1.Gollub J., Ball,C.A., Binkley,G., Demeter,J., Finkelstein,D.B., Hebert,J.M., Hernandez-Boussard,T., Jin,H., Kaloper,M., Matese,J.C. et al. (2003) The Stanford Microarray Database: data access and quality assessment tools. Nucleic Acids Res., 31, 94–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li C. and Hung Wong,W. (2001) Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol., 2, RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brazma A., Parkinson,H., Sarkans,U., Shojatalab,M., Vilo,J., Abeygunawardena,N., Holloway,E., Kapushesky,M., Kemmeren,P., Lara,G.G. et al. (2003) ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res., 31, 68–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spellman P.T., Miller,M., Stewart,J., Troup,C., Sarkans,U., Chervitz,S., Bernhart,D., Sherlock,G., Ball,C., Lepage,M. et al. (2002) Design and implementation of microarray gene expression markup language (MAGE-ML). Genome Biol., 3, RESEARCH0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brazma A., Hingamp,P., Quackenbush,J., Sherlock,G., Spellman,P., Stoeckert,C., Aach,J., Ansorge,W., Ball,C.A., Causton,H.C. et al. (2001) Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature Genet., 29, 365–371. [DOI] [PubMed] [Google Scholar]
- 6.Edgar R. Domrachev,M. and Lash,A.E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res., 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkinson M.D. and Links,M. (2002) BioMOBY: an open source biological web services proposal. Brief. Bioinform, 3, 331–341. [DOI] [PubMed] [Google Scholar]
- 8.Rees C.A., Demeter,J., Matese,J.M., Botstein,D. and Sherlock,G. (2004) GeneXplorer: an interactive web application for microarray data visualization and analysis. BMC Bioinformatics, 5, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]