Abstract
Few genome-wide association studies (GWAS) of type 2 diabetes (T2D) have been conducted in U.S. Hispanics/Latinos of diverse backgrounds who are disproportionately affected by diabetes. We conducted a GWAS in 2,499 T2D case subjects and 5,247 control subjects from six Hispanic/Latino background groups in the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). Our GWAS identified two known loci (TCF7L2 and KCNQ1) reaching genome-wide significance levels. Conditional analysis on known index single nucleotide polymorphisms (SNPs) indicated an additional independent signal at KCNQ1, represented by an African ancestry–specific variant, rs1049549 (odds ratio 1.49 [95% CI 1.27–1.75]). This association was consistent across Hispanic/Latino background groups and replicated in the MEta-analysis of type 2 DIabetes in African Americans (MEDIA) Consortium. Among 80 previously known index SNPs at T2D loci, 66 SNPs showed consistency with the reported direction of associations and 14 SNPs significantly generalized to the HCHS/SOL. A genetic risk score based on these 80 index SNPs was significantly associated with T2D (odds ratio 1.07 [1.06–1.09] per risk allele), with a stronger effect observed in nonobese than in obese individuals. Our study identified a novel independent signal suggesting an African ancestry–specific allele at KCNQ1 for T2D. Associations between previously identified loci and T2D were generally shown in a large cohort of U.S. Hispanics/Latinos.
Introduction
U.S. Hispanics/Latinos, who now make up the nation’s largest minority group, are disproportionately affected by diabetes (1). Our recent data from the Hispanic Community Health Study/Study of Latinos (HCHS/SOL) showed a diabetes prevalence of ∼17% in Hispanic adults (2). Previous genome-wide association studies (GWAS) have identified a number of type 2 diabetes (T2D) susceptibility loci primarily in populations of European ancestry (3). Few GWAS have been conducted in Hispanic/Latino populations (mostly comprising individuals of Mexican origin) and identified novel T2D loci (e.g., SLC16A11) (4,5). To further understand the genetic basis of T2D in U.S. Hispanics/Latinos of diverse backgrounds, we conducted a GWAS including 2,499 T2D case subjects and 5,247 control subjects from the HCHS/SOL with the purpose of 1) searching for novel T2D loci and 2) examining previously known T2D loci in U.S. Hispanics/Latinos.
Research Design and Methods
Participants
The HCHS/SOL is a population-based study of 16,415 Hispanic/Latino adults, 18–74 years old, living in four U.S. metropolitan areas (Bronx, NY; Chicago, IL; Miami, FL; and San Diego, CA) (6,7). A comprehensive battery of interviews relating to personal and family characteristics and health status and behaviors, as well as a clinical assessment with blood draw, were conducted at an in-person clinic baseline visit during 2008–2011. The study was approved by the institutional review boards at all participating institutions, and all participants gave written informed consent. The analyses described here included 7,746 individuals comprising 2,499 T2D case subjects and 5,247 control subjects who consented to participation in genetic studies (Supplementary Table 1).
Ascertainment of T2D
Individuals with T2D were defined as those with fasting time >8 h and fasting glucose levels ≥126 mg/dL, fasting ≤8 h and fasting glucose ≥200 mg/dL, post–oral glucose tolerance test glucose ≥200 mg/dL, HbA1c ≥6.5% (48 mmol/mol), or on current treatment with antihyperglycemia medications. Control subjects were defined as those with fasting time >8 h and fasting glucose levels <100 mg/dL, post–oral glucose tolerance test glucose <140 mg/dL, and HbA1c <5.6% (38 mmol/mol). Individuals with intermediate phenotypes (prediabetes) were excluded from this analysis.
Genotyping and Imputation
Genotyping was performed with an Illumina custom array (15041502 B3), which consists of the Illumina Omni 2.5M array (HumanOmni2.5-8 v1.1) plus ∼150,000 custom single nucleotide polymorphisms (SNPs), with the quality control performed at HCHS/SOL Genetic Analysis Center (8). Genome-wide imputation was carried out using the 1000 Genomes Project phase 1 reference panel SHAPEIT2 and IMPUTE2 software, as described previously (8). An iterative procedure was used to simultaneously estimate principal components (PCs) reflecting population structure and kinship coefficients measuring familial relatedness (8). Genetic analysis groups (Hispanic/Latino background groups: Cuban, Dominican, Puerto Rican, Mexican, Central American, or South American) were constructed based on a combination of self-identified Hispanic/Latino background and genetic similarity (8).
Statistical Analysis
GWAS
We used the generalized linear mixed model association test (GMMAT) (9) to test SNP–T2D genome-wide associations using the score test applied on a logistic model. Our study had a power of 80% to detect a genetic variant that could explain 0.64% variance of T2D on a liability scale, at the genome-wide significance level (P < 5.0 × 10−8). Correlations between participants were accounted for by incorporating covariance matrices corresponding to genetic relatedness (kinship), household, and census block group as random effects. The model also included center, age, sex, the first five PCs to adjust for ancestry (8), and sampling weights (10).
Conditional Analysis and Haplotype Analysis
Conditional analyses were performed at the KCNQ1 locus using GMMAT, with known index SNP and lead SNPs as covariates. We identified haplotypes of KCNQ1 based on four SNPs using the phased imputation data. Haplotypes that appeared at least 50 times in both case and control subjects were tested for associations with T2D using GMMAT, with the most common haplotype as a reference.
Known Index SNP Analysis
For 80 known index SNPs identified and/or summarized in previous studies (4,11–14), we used GMMAT to estimate the odds ratios (ORs) and 95% CI per risk allele of each SNP on T2D. We calculated post hoc power for these 80 SNPs using a previously reported method (15), under the risk allele frequencies and effect sizes of SNPs observed in the HCHS/SOL, at a P of 6.25 × 10−4 (Bonferroni correction for 80 SNPs). To study whether these associations vary by Hispanic/Latino groups, we included SNP × group interaction terms (as fixed effects) in the models. Binomial tests were conducted to examine whether the observed consistency of 80 known T2D index SNPs with the previously reported associations is due to chance. We tested these 80 known index SNPs for generalization in the HCHS/SOL by calculating false discovery rate–controlling r values, to account for multiple testing of the generalization null hypothesis (16).
Genetic Risk Score
An unweighted genetic risk score (GRS) was constructed for each participant by summing all risk allele dosages of 80 known T2D index SNPs. To test potential differences between two sets of T2D SNPs according to their possible biological categories (17), two sub-GRSs were calculated based on 36 β-cell function–related SNPs and 18 insulin resistance–related SNPs, respectively. Similar GMMAT models were used to examine the ORs and 95% CI per unit of the GRSs on T2D in all samples, and subgroups were stratified by age-group (dichotomized at median: 45 years old), sex, BMI status (dichotomized at the cutoff for obesity: 30 kg/m2), and Hispanic/Latino background.
Results
In a GWAS of T2D including all individuals, there was no evidence of genomic inflation due to population stratification (λ = 0.99; Supplementary Fig. 1). There were two genome-wide significant T2D loci (P < 5.0 × 10−8), and both are known: TCF7L2 (lead SNP rs7903146, P = 1.3 × 10−11) and KCNQ1 (lead SNP rs2283228, P = 8.4 × 10−13) (Supplementary Fig. 2). Further adjustment for BMI did not change the results materially.
We then evaluated association results at 80 known T2D loci to examine whether lead SNPs in the HCHS/SOL are different from the reported index SNPs. There were eight loci with lead SNPs of P < 6.25 × 10−4 (Bonferroni adjustment for 80 tested loci), four of which (TCF7L2, INS-IGF2, KCNQ1, and MTNR1B) reached significance after correction for the number of independent SNPs calculated using the simpleM methods (18) at the respective locus (Supplementary Fig. 3). The observed lead SNPs in the HCHS/SOL were identical or in moderate-to-high linkage disequilibrium (LD) (r2 > 0.5) with known index SNPs at these loci, except SNP rs1049549 at INS-IGF2 and SNP rs2283228 at KCNQ1 (r2 < 0.2 with known index SNPs). Of note, SNP rs1049549 is located between the INS-IGF2 and KCNQ1 genes and relatively closer to KCNQ1 than to INS-IGF2. Thus, rs1049549 was considered as a SNP at the KCNQ1 locus in our following analyses.
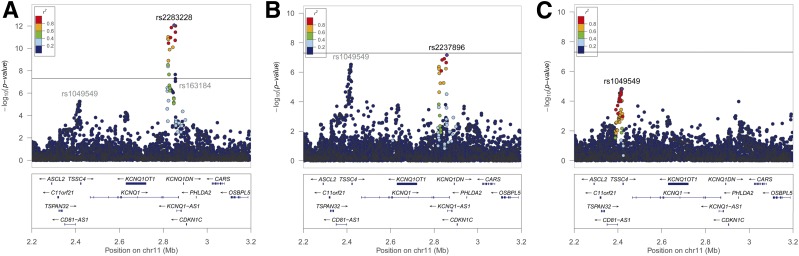
Conditional analyses at the KCNQ1 locus also yielded two additional independent signals represented by SNP rs2283228/rs2237896 (in high LD with r2 = 0.83) and SNP rs1049549, respectively (Fig. 1 and Supplementary Table 2). A potentially novel signal represented by SNP rs1049549 showed a significant association with T2D (OR 1.49 [95% CI 1.27–1.75]; P = 5.5 × 10−6) in primary analysis (Fig. 1A), and the result did not change materially after conditioning on index SNP rs163184 (Fig. 1B) or after conditioning on both index SNP rs163184 and lead SNP rs2237896 (Fig. 1C). In addition, there was another signal represented by SNP rs3888647 showing varied associations with T2D in primary analysis and conditional analyses (Supplementary Table 2). This might be due to the correlations between this SNP and SNPs rs2283228/rs2237896 (Supplementary Table 3), indicating that this might not be an independent signal.
Figure 1.
Regional association plots for the KCNQ1 locus in the HCHS/SOL. A: Primary analysis. B: Conditional analysis on the index SNP rs163184. C: Second conditional analysis on the index SNP rs163184 and the conditional lead SNP rs2237896. Values of r2 for LD were estimated in the HCHS/SOL.
Haplotype analyses of the KCNQ1 locus based on four SNPs, rs1049549, rs163184, rs2237896, and rs3888647, further supported the independence of SNP rs1049549 in association with T2D (Supplementary Table 4). The most common haplotype T-G-G-G in the HCHS/SOL (frequency 0.30) has nonrisk allele (C allele) of rs1049549 and risk alleles (G alleles) of other three SNPs for T2D. The haplotype C-G-G-G, with the risk allele (C allele) of rs1049549, showed a significant association with increased T2D risk compared with the haplotype T-G-G-G (P = 9.6 ×10−4).
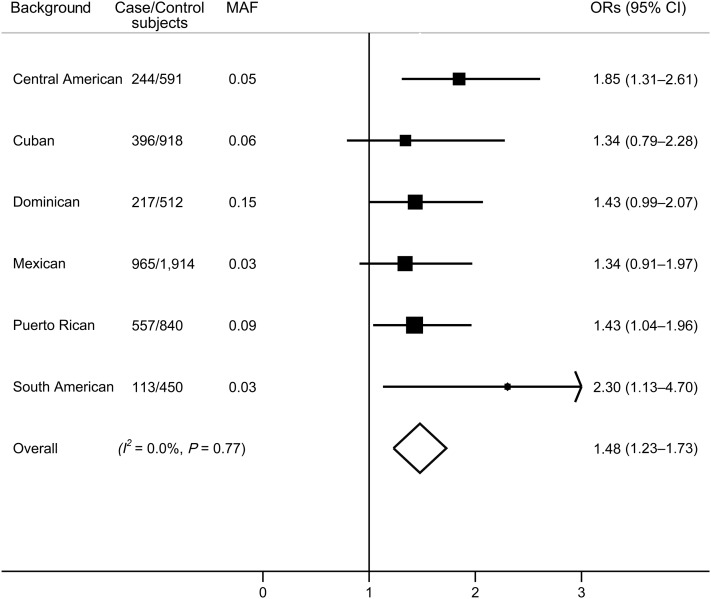
Associations between SNP rs1049549 and T2D were highly consistent across Hispanic/Latino background groups (I2 = 0; Pheterogeneity = 0.77) (Fig. 2). Previously inferred local ancestry estimates in the HCHS/SOL (19) indicated that this C allele is an African ancestry–specific allele, with relative high frequency in African ancestry (minor allele frequency [MAF] 0.32) and very low frequency in European ancestry and Native American ancestry (MAF <0.02). We replicated this association in the MEta-analysis of type 2 DIabetes in African Americans (MEDIA) Consortium (11). A proxy SNP rs7124991 (r2 = 1 with rs1049549 in both Hispanics and Africans; MAF 0.23 in African Americans) was significantly associated with T2D (P = 0.006).
Figure 2.
Associations between SNP rs1049549 at KCNQ1 and T2D across Hispanic/Latino background groups. Data are ORs (95% CI) for each minor allele of rs1049549 on T2D, adjusted for age, sex, center, sampling weights, relatedness, and population structure (kinship coefficients and eigenvectors). Overall results were combined by fixed-effects meta-analysis and did not account for low-level relatedness between groups.
Table 1 provides information on consistency of 80 index SNPs at known T2D loci between the HCHS/SOL and previous studies. A total of 69 SNPs showed consistency with the previously reported direction of associations (binomial test P = 8.67 × 10−12). Characteristics of 80 known index SNPs, SNP–T2D associations, and post hoc power calculation in the HCHS/SOL are shown in Supplementary Table 5. There were no significant interactions between these SNPs and Hispanic/Latino backgrounds on T2D (all Pinteraction > 0.05). In the generalization analysis, 14 index SNPs were generalized to the HCHS/SOL.
Table 1.
Consistency of the direction of effects of 80 known index SNPs on T2D between the HCHS/SOL and previously reported studies
| Results in the HCHS/SOL | Consistent SNPs/total SNPs (%) | Binomial test P |
|---|---|---|
| SNPs showed consistency of direction | 69/80 (86.3) | 8.67 × 10−12 |
| SNPs showed P < 0.05 | 27/80 (33.8) | 7.56 × 10−16 |
| SNPs showed P < 6.25 × 10−4* | 10/80 (12.5) | 1.43 × 10−20 |
*Bonferroni corrected significance in the current analysis (0.05/80 SNPs).
The total GRS based on 80 known index SNPs showed a highly significant association with T2D (OR 1.07 [95% CI 1.06–1.08] per risk allele; P = 2.31 × 10−39) (Table 2). Association between GRS and T2D was stronger in nonobese (BMI <30 kg/m2) compared with obese (BMI ≥30 kg/m2) individuals (OR 1.10 [95% CI 1.08–1.11] vs. 1.06 [1.04–1.08]; Pinteraction = 2.84 × 10−3). Obesity status showed significant interactions with the β-cell function GRS (Pinteraction = 8.55 × 10−5) but not the insulin resistance GRS (Pinteraction=0.34). We did not find significant interactions between the GRSs and age, sex, or Hispanic/Latino background on T2D (all Pinteraction > 0.05). Another GRS based on 66 SNPs that did not pass generalization testing also showed a significant association with T2D (P = 6.12 × 10−14).
Table 2.
Associations between the GRSs and T2D according to age, sex, BMI status, and Hispanic background in the HCHS/SOL
| T2D case/control subjects |
Total GRS | β-Cell function GRS | Insulin resistance GRS | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI)* |
P* | P for interaction | OR (95% CI)* | P* | P for interaction | OR (95% CI)* | P* | P for interaction | ||
| Overall |
2,499/5,247 |
1.07 (1.06–1.08) |
2.31 × 10−39 |
1.10 (1.09–1.12) |
1.89 × 10−36 |
1.06 (1.06–1.08) |
1.53 × 10−7 |
|||
| Age, years |
0.12 |
0.07 |
0.51 |
|||||||
| <45 |
320/3,284 |
1.06 (1.04–1.08) |
1.05 × 10−8 |
1.08 (1.05–1.11) |
1.76 × 10−7 |
1.05 (1.04–1.08) |
0.03 |
|||
| ≥45 |
2,179/1,936 |
1.08 (1.07–1.09) |
1.01 × 10−31 |
1.11 (1.09–1.14) |
1.40 × 10−30 |
1.06 (1.07–1.09) |
2.59 × 10−6 |
|||
| Sex |
0.65 |
0.44 |
0.93 |
|||||||
| Male |
995/2,076 |
1.07 (1.05–1.09) |
5.92 × 10−16 |
1.10 (1.07–1.12) |
9.58 × 10−14 |
1.06 (1.05–1.09) |
6.63 × 10−4 |
|||
| Female |
1,504/3,197 |
1.08 (1.06–1.09) |
7.33 × 10−26 |
1.11 (1.09–1.13) |
2.49 × 10−25 |
1.06 (1.06–1.09) |
5.48 × 10−5 |
|||
| BMI, kg/m2 |
2.84 × 10−3 |
8.55 × 10−5 |
0.34 |
|||||||
| <30 |
1,050/3,639 |
1.10 (1.08–1.11) |
1.08 × 10−31 |
1.15 (1.13–1.18) |
3.50 × 10−34 |
1.05 (1.08–1.11) |
1.73 × 10−3 |
|||
| ≥30 |
1,449/1,608 |
1.06 (1.04–1.08) |
8.90 × 10−14 |
1.08 (1.06–1.11) |
1.15 × 10−11 |
1.07 (1.04–1.08) |
2.05 × 10−5 |
|||
| Hispanic/Latino background |
0.23 |
0.22 |
0.80 |
|||||||
| Central American |
244/591 |
1.08 (1.05–1.13) |
2.52 × 10−7 |
1.11 (1.06–1.17) |
7.51 × 10−6 |
1.09 (1.01–1.17) |
0.02 |
|||
| Cuban |
396/918 |
1.06 (1.04–1.09) |
3.40 × 10−6 |
1.09 (1.05–1.13) |
5.49 × 10−6 |
1.07 (1.01–1.12) |
0.01 |
|||
| Dominican |
217/523 |
1.08 (1.04–1.12) |
1.66 × 10−5 |
1.08 (1.03–1.13) |
3.54 × 10−3 |
1.09 (1.01–1.17) |
0.02 |
|||
| Mexican |
965/1,914 |
1.08 (1.07–1.10) |
3.60 × 10−20 |
1.12 (1.09–1.15) |
1.32 × 10−19 |
1.06 (1.02–1.10) |
1.29 × 10−3 |
|||
| Puerto Rican |
557/840 |
1.05 (1.03–1.07) |
4.87 × 10−5 |
1.08 (1.04–1.11) |
4.14 × 10−5 |
1.03 (0.98–1.08) |
0.22 |
|||
| South American | 113/450 | 1.09 (1.04–1.14) | 1.27 × 10−4 | 1.16 (1.08–1.23) | 1.56 × 10−5 | 1.07 (0.97–1.18) | 0.16 | |||
*ORs (95% CI) per unit of the GRSs and P values were estimated using GMMAT, incorporating covariance matrices corresponding to genetic relatedness (kinship), household, and census block group as random effects and adjusting for center, age, sex, the first five PCs, and sampling weights.
Discussion
In this GWAS of T2D in U.S. Hispanics/Latinos, we identified an independent association signal represented by an African ancestry–specific variant, SNP rs1049549, in addition to two known association signals at the KCNQ1 locus (20–22). Consistent results across different Hispanic/Latino background groups and successful replication in African Americans from the MEDIA Consortium (11) strengthened the reliability of this observed association. SNP rs1049549 is located at the 3′ untranslated region of CD81, ∼3.3 kb upstream of TSSC4 and ∼4.8 kb upstream of KCNQ1, which often contains regulatory elements (ENCODE databases can be found in the University of California, Santa Cruz, Genome Browser at https://genome.ucsc.edu/). Further examination indicated that SNP rs1049549 is in a high LD with a set of SNPs (r2 > 0.8) clustered with regulatory elements, such as the transcription factor binding regions, DNase I hypersensitivity sites, and H3K27Ac mark.
It is not surprising that only 14 known index SNPs formally generalized to the HCHS/SOL, due to limited power of our study. Top SNPs, TCF7L2 rs7903146 and KCNQ1 rs2283228, in our study explained 0.28% and 0.33% variance of T2D, respectively, indicating that our GWAS was underpowered. However, we demonstrated that both the GRS based on all 80 known index SNPs and the GRS that excluded generalized SNPs showed highly significant associations with T2D. The observed effect of the GRS in U.S. Hispanics/Latinos (∼7% increased risk of T2D per allele) is very similar to those observed in two large studies of Europeans (23) and Chinese (17). Consistent with previous studies (17,23), we found a stronger genetic association with T2D in leaner individuals, which might be driven by β-cell function–related variants (17).
One unique feature of our study is the diversity of Hispanic/Latino backgrounds, which have been rarely investigated in previous T2D GWAS (4,5). This provides both challenges and advantages for genetic association studies. Heterogeneity among diverse Hispanic/Latino groups may increase genomic inflation, confounding effects, or other artifacts and decrease statistical power (24), although appropriate statistical approaches have been applied to account for heterogeneous variances among groups in our genetic association analyses (8). On the other hand, the diversity of Hispanic/Latino groups with different ancestries provides opportunities to identify potential ancestry-specific alleles (e.g., the newly identified African ancestry–specific allele at KCNQ1 in this study) and/or group-specific genetic effects in relation to human diseases (25). Our study, the largest study of U.S. Hispanics/Latinos of diverse backgrounds to date, is still limited by inadequate sample size. Additional studies of U.S. Hispanics/Latinos are clearly needed to fill this gap.
In summary, our study identified a potentially novel independent signal suggesting an African ancestry–specific allele at KCNQ1 for T2D and provides evidence for the similarity of known genetic predisposition to T2D between U.S. Hispanics/Latinos and other populations. Larger meta-analyses of T2D GWAS efforts are needed to confirm our findings and identify more T2D loci among this understudied population.
Supplementary Material
Article Information
Funding. The baseline examination of HCHS/SOL was carried out as a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (N01-HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC65236), and San Diego State University (N01-HC65237). The following institutes/centers/offices contributed to the first phase of HCHS/SOL through a transfer of funds to the NHLBI: the National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research, National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, and National Institutes of Health (NIH) Office of Dietary Supplements. The Genetic Analysis Center at the University of Washington was supported by NHLBI and National Institute of Dental and Craniofacial Research contracts (HHSN268201300005C AM03 and MOD03, respectively). Genotyping efforts were supported by NHLBI grant HSN 26220/20054C, National Center for Advancing Translational Sciences Clinical and Translational Science Institute grant UL1TR000123, and National Institute of Diabetes and Digestive and Kidney Diseases Diabetes Research Center grant DK063491. Q.Q. is supported by an NHLBI Scientist Development Award (K01HL129892). This manuscript has been reviewed by the HCHS/SOL Publications Committee for scientific content and consistency of data interpretation with previous HCHS/SOL publications. The work for the MEDIA Consortium is partly supported by NIH grant R01 DK066358.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. Q.Q. designed the study, researched data, and wrote the manuscript. A.M.S., T.S., J.-Y.M., B.H., A.A.S., T.W., M.C.Y.N., and X.G. researched and reviewed data and edited and reviewed the manuscript. Y.-D.I.C., K.D.T., M.L.A.-S., G.P., and J.S.P. contributed to the discussion and edited and reviewed the manuscript. N.S., C.C.L., J.I.R., and R.C.K. designed the study, contributed to the discussion, and edited and reviewed the manuscript. Q.Q. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db16-1150/-/DC1.
The opinions of the authors do not represent the opinions of the National Heart, Lung, and Blood Institute, the National Institutes of Health, or the federal government.
References
- 1.Menke A, Casagrande S, Geiss L, Cowie CC. Prevalence of and trends in diabetes among adults in the United States, 1988-2012. JAMA 2015;314:1021–1029 [DOI] [PubMed] [Google Scholar]
- 2.Daviglus ML, Talavera GA, Avilés-Santa ML, et al. . Prevalence of major cardiovascular risk factors and cardiovascular diseases among Hispanic/Latino individuals of diverse backgrounds in the United States. JAMA 2012;308:1775–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Strizich G, Hu Y, Wang T, Kaplan RC, Qi Q. Genetic markers of type 2 diabetes: progress in genome-wide association studies and clinical application for risk prediction. J Diabetes 2016;8:24–35 [DOI] [PubMed] [Google Scholar]
- 4.Williams AL, Jacobs SB, Moreno-Macías H, et al.; SIGMA Type 2 Diabetes Consortium . Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature 2014;506:97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parra EJ, Below JE, Krithika S, et al.; Diabetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium . Genome-wide association study of type 2 diabetes in a sample from Mexico City and a meta-analysis of a Mexican-American sample from Starr County, Texas. Diabetologia 2011;54:2038–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lavange LM, Kalsbeek WD, Sorlie PD, et al. . Sample design and cohort selection in the Hispanic Community Health Study/Study of Latinos. Ann Epidemiol 2010;20:642–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorlie PD, Avilés-Santa LM, Wassertheil-Smoller S, et al. . Design and implementation of the Hispanic Community Health Study/Study of Latinos. Ann Epidemiol 2010;20:629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conomos MP, Laurie CA, Stilp AM, et al. . Genetic diversity and association studies in US Hispanic/Latino populations: applications in the Hispanic Community Health Study/Study of Latinos. Am J Hum Genet 2016;98:165–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H, Wang C, Conomos MP, et al. . Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am J Hum Genet 2016;98:653–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfefferman D. The role of sampling weights when modeling survey data. Int Stat Rev 1993;61:317–337 [Google Scholar]
- 11.Ng MC, Shriner D, Chen BH, et al.; FIND Consortium; eMERGE Consortium; DIAGRAM Consortium; MuTHER Consortium; MEta-analysis of type 2 DIabetes in African Americans Consortium . Meta-analysis of genome-wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. PLoS Genet 2014;10:e1004517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahajan A, Go MJ, Zhang W, et al.; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; Mexican American Type 2 Diabetes (MAT2D) Consortium; Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium . Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet 2014;46:234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saxena R, Elbers CC, Guo Y, et al.; Look AHEAD Research Group; DIAGRAM consortium . Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci [published correction appears in Am J Hum Genet. 2012;90:753]. Am J Hum Genet 2012;90:410–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara K, Fujita H, Johnson TA, et al.; DIAGRAM consortium . Genome-wide association study identifies three novel loci for type 2 diabetes. Hum Mol Genet 2014;23:239–246 [DOI] [PubMed] [Google Scholar]
- 15.Gordon D, Finch SJ, Nothnagel M, Ott J. Power and sample size calculations for case-control genetic association tests when errors are present: application to single nucleotide polymorphisms. Hum Hered 2002;54:22–33 [DOI] [PubMed] [Google Scholar]
- 16.Sofer T, Heller R, Bogomolov M, et al. . A powerful statistical framework for generalization testing in GWAS, with application to the HCHS/SOL. Genet Epidemiol 2017;41:251–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gan W, Walters RG, Holmes MV, et al.; China Kadoorie Biobank Collaborative Group . Evaluation of type 2 diabetes genetic risk variants in Chinese adults: findings from 93,000 individuals from the China Kadoorie Biobank. Diabetologia 2016;59:1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao X, Becker LC, Becker DM, Starmer JD, Province MA. Avoiding the high Bonferroni penalty in genome-wide association studies. Genet Epidemiol 2010;34:100–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Browning SR, Grinde K, Plantinga A, et al. . Local ancestry inference in a large US-based Hispanic/Latino study: Hispanic Community Health Study/Study of Latinos (HCHS/SOL). G3 (Bethesda) 2016;6:1525–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yasuda K, Miyake K, Horikawa Y, et al. . Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet 2008;40:1092–1097 [DOI] [PubMed] [Google Scholar]
- 21.Unoki H, Takahashi A, Kawaguchi T, et al. . SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet 2008;40:1098–1102 [DOI] [PubMed] [Google Scholar]
- 22.Morris AP, Voight BF, Teslovich TM, et al.; Wellcome Trust Case Control Consortium; Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012;44:981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langenberg C, Sharp SJ, Franks PW, et al. . Gene-lifestyle interaction and type 2 diabetes: the EPIC interact case-cohort study. PLoS Med 2014;11:e1001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet 2010;11:459–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schick UM, Jain D, Hodonsky CJ, et al. . Genome-wide association study of platelet count identifies ancestry-specific loci in Hispanic/Latino Americans. Am J Hum Genet 2016;98:229–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.