Abstract
Zebrafish innately regenerate amputated fins by mechanisms that expand and precisely position injury-induced progenitor cells to re-form tissue of the original size and pattern. For example, cell signaling networks direct osteoblast progenitors (pObs) to rebuild thin cylindrical bony rays with a stereotypical branched morphology. Hedgehog/Smoothened (Hh/Smo) signaling has been variably proposed to stimulate overall fin regenerative outgrowth or promote ray branching. Using a photoconvertible patched2 reporter, we resolve active Hh/Smo output to a narrow distal regenerate zone comprising pObs and adjacent motile basal epidermal cells. This Hh/Smo activity is driven by epidermal Sonic hedgehog a (Shha) rather than Ob-derived Indian hedgehog a (Ihha), which nevertheless functions atypically to support bone maturation. Using BMS-833923, a uniquely effective Smo inhibitor, and high-resolution imaging, we show that Shha/Smo is functionally dedicated to ray branching during fin regeneration. Hh/Smo activation enables transiently divided clusters of Shha-expressing epidermis to escort pObs into similarly split groups. This co-movement likely depends on epidermal cellular protrusions that directly contact pObs only where an otherwise occluding basement membrane remains incompletely assembled. Progressively separated pObs pools then continue regenerating independently to collectively re-form a now branched skeletal structure.
KEY WORDS: Zebrafish, Caudal fins, Regeneration, Ray branching, Bone patterning, Hedgehog signaling, Sonic hedgehog, Indian hedgehog, Osteoblasts, Calcification, Basal epidermis, Basement membrane, Smoothened inhibitor, Cyclopamine, BMS-833923
Highlighted article: In the regenerating zebrafish fin, local Hedgehog signalling between basal epidermal cells and osteoblast progenitors controls ray branching and hence skeletal patterning.
INTRODUCTION
Unlike mammalian appendages, the adult zebrafish caudal fin perfectly restores tissue organization, size and shape in response to injury or resection (Tornini and Poss, 2014). Regeneration of the fin skeleton, which comprises bony rays (lepidotrichia) that extend along the proximal-distal axis, depends on the coordinated growth, differentiation and positioning of osteoblasts (Obs). Individual rays are formed by two semi-cylindrical bones, or hemi-rays, that are covered with Obs and segmented by regularly spaced joints. Furthermore, each lepidotrichia, excluding the most dorsal and ventral rays, bifurcates in a highly stereotypical manner. A near identical skeletal pattern is efficiently restored within 2 weeks of fin amputation. Therefore, the zebrafish fin provides a tractable and simple model system with which to decipher mechanisms of regenerative skeletal patterning.
Bone regeneration is initiated after fin resection by Ob dedifferentiation that generates osteoblast progenitors (pObs) at the amputation site (Knopf et al., 2011; Singh et al., 2012; Sousa et al., 2011; Stewart and Stankunas, 2012; Stewart et al., 2014; Tu and Johnson, 2011). These pObs populate the lateral edges of the regenerative blastema that forms above each bony ray stump and surround a largely mesenchymal core cell population. The entire tissue is encased by a stratified epidermis. The Ob lineage remains highly organized for the duration of regeneration with distally located Runx2-expressing pObs and more proximal maturing Obs defined by sp7 (osterix) expression (Stewart et al., 2014). This arrangement is maintained by spatially segregated Wnt and bone morphogenetic protein (BMP) signaling that promotes the opposing activities of Ob growth and differentiation, respectively (Stewart et al., 2014; Wehner et al., 2014). This balanced signaling network, however, does not explain how regenerated bones become bifurcated in the same pattern as the lost fin. Earlier studies show that ray branching requires a neighboring ray (Marí-Beffa et al., 1999) and transplantation of a non-branching dorsal (or ventral) ray to a medial position results in branching of the transplant (Murciano et al., 2002). These observations suggest that cell-cell interactions between adjacent tissues are essential for ray morphogenesis.
Hedgehog (Hh) signaling has been proposed to mediate ray branching during fin regeneration (Laforest et al., 1998; Quint et al., 2002; Zhang et al., 2012). The Hh ligand family, Sonic (Shh), Indian (Ihh) and Desert (Dhh) Hedgehogs, bind to their receptor, Patched (Ptch) on target cells (Fuse et al., 1999; Marigo et al., 1996; Stone et al., 1996). Binding of Hh to Ptch relieves Smoothened (Smo) inhibition, leading to transcriptional changes, including activation of ptch genes to form a negative-feedback loop (Briscoe and Thérond, 2013; Chen and Struhl, 1996; Ingham et al., 1991). During fin regeneration, shha transcripts are expressed in basal epidermal cells on each side of the distal regenerate, a pattern recapitulated by a shha:GFP reporter transgene (Laforest et al., 1998; Lee et al., 2009; Quint et al., 2002; Zhang et al., 2012). Preceding ray bifurcation, each shha-expressing cluster bisects into two discrete domains, presaging the splitting of underlying Obs and consequently ray branching (Laforest et al., 1998; Zhang et al., 2012). Laser ablation of shha-expressing basal epidermal cells delays branching, underscoring that epidermal-Ob interactions, possibly directing localized Ob proliferation, underlie regenerative bone patterning (Zhang et al., 2012). Shha is a candidate mediator of this signaling, as ptch2 (previously called ptc1) is expressed in Obs adjacent to shha-expressing epidermal cells (Laforest et al., 1998; Murciano et al., 2007; Quint et al., 2002) and ectopic Shh promotes ray fusion and promiscuous bone formation (Quint et al., 2002). However, a role for Shha in ray branching has been questioned based on the suggestion that shha-expressing epidermal domains are constitutively split and that shha induction kinetics are inconsistent with Shha being the instructive ray bifurcation signal (Azevedo et al., 2012).
Additional and alternative roles for Hh/Smo signaling during fin regeneration are also possible. The Smo small-molecule inhibitor cyclopamine arrests proliferation of multiple cell types in the regenerate (Blum and Begemann, 2015; Lee et al., 2009; Quint et al., 2002; Wehner et al., 2014), suggesting Hh/Smo signaling contributes to general regenerative outgrowth. Furthermore, ihha is robustly expressed in blastemal Obs during regeneration (Avaron et al., 2006). Therefore, Ihha rather than Shha could account for ptch2 induction in regenerating Obs and in the direct control of osteoblast growth and/or differentiation in a manner recapitulating the proposed developmental roles of Ihh (Abzhanov et al., 2007; Huycke et al., 2012; Lenton et al., 2011; Long, 2012).
We sought to resolve the role(s) of Hh/Smo signaling during fin bone regeneration. A dynamic ptch2 transgenic reporter shows that Hh/Smo output is tightly restricted to a narrow band of distally extending basal epidermal cells and underlying pObs. These epidermal cells transiently split into two Shha-positive clusters on each side of the ray. We use viable ihha-null zebrafish to demonstrate that the Hh/Smo output in both cell types is Shha-driven and that Ob-expressed Ihha instead supports bone maturation, likely via an atypical signaling mechanism. We show that the small molecule Smo inhibitor, BMS-833923, avoids widespread off-target effects of the classic Smo inhibitor cyclopamine in zebrafish. Most strikingly, BMS-833923 unambiguously demonstrates that Shha/Smo signaling is required for ray branching morphogenesis and not regenerative outgrowth. Mechanistically, cellular protrusions from shha-expressing epidermal cells directly contact neighboring ptch2-expressing pObs at distal sites of incompletely assembled basal lamina. Rather than promoting local proliferation, the split clusters of motile Shha-positive basal epidermis progressively escort pObs into two separated pools that then independently continue regenerating to form a now bifurcated ray.
RESULTS
Hedgehog/Smoothened signaling output at the time of ray bifurcation is spatially and temporally restricted to osteoblast progenitors and basal epidermis
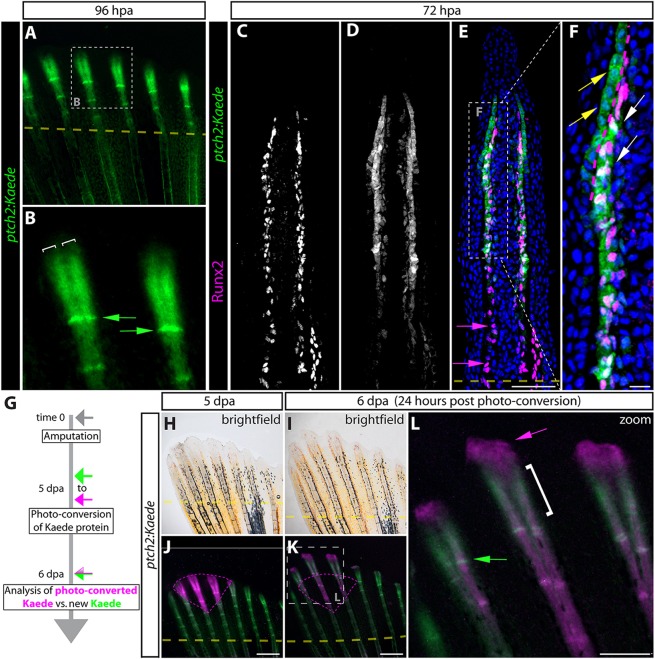
Zebrafish patched2 (ptch2; previously named ptc1) is induced by Hedgehog/Smoothened (Hh/Smo) signaling (Concordet et al., 1996; Koudijs et al., 2008) and therefore its expression serves as a reporter of pathway activity. We used the TgBAC(ptch2:Kaede)a4596 transgenic line (hereafter denoted as ptch2:Kaede), which recapitulates endogenous ptch2 expression (Huang et al., 2012), to monitor Hh/Smo signaling during caudal fin regeneration. At 96 h post-amputation (hpa), when regenerating rays begin to branch, ptch2:Kaede was expressed in all reforming bony rays and was excluded from inter-ray regions. Kaede levels were highest towards the distal regenerate, where, at each ray, it split into two domains on both sides of the fin (Fig. 1A,B), reproducing the ptch2 transcript pattern (Laforest et al., 1998; Murciano et al., 2007; Quint et al., 2002). We immunostained sections from ptch2:Kaede fins using Kaede and Runx2 antibodies to visualize sites of Hh/Smo activity relative to the position of Obs. At 72 hpa, ptch2:Kaede reporter activity was observed in distal and medial Runx2+ Obs, and was absent in more proximal osteoblasts extending new bone (Fig. 1C-F; earlier time points preceding branching are shown in Fig. S1). Therefore, pObs activate Hh/Smo signaling as they are generated upon self-renewal and then lose this Hh/Smo response when they mature into quiescent and re-epithelialized Obs. Concurrently, ptch2:Kaede was expressed in distal basal epidermal cells neighboring and extending beyond Runx2+ pObs. At the onset of ray bifurcation, canonical Hh signaling therefore is spatially restricted to two cell types: pObs and basal epidermal cells.
Fig. 1.
ptch2:Kaede expression and photoconversion reveals transient Hedgehog/Smoothened signaling restricted to distal osteoblast progenitors and basal epidermis during fin regeneration. (A,B) Whole-mount images showing Kaede expression (green) in a 96 hpa fin from a ptch2:Kaede fish. (B) A high-magnification image of the boxed region in A. White brackets mark the split domains of Kaede at the distal end of the regenerate. Green arrows indicate Kaede+ cells at newly re-forming joints. (C-F) Antibody-stained longitudinal fin sections from a 72 hpa ptch2:Kaede fish. Individual Runx2 (C) and Kaede (D) channels are shown in gray scale. Overlay images with Runx2 in magenta and Kaede in green are shown in E and F (a magnification of the boxed region in E). Nuclei are in blue. Yellow arrows indicate Kaede+ basal epidermis; white arrows mark Kaede+/Runx2+ Obs; magenta arrows indicate proximal Runx2+ Obs lacking Kaede. The dotted magenta line indicates the boundary between Kaede+ basal epidermis and pObs. (G) ptch2:Kaede photo-conversion experiment overview. (H-L) Whole-mount bright-field (H,I) and fluorescent (J-L) images of the regenerating caudal fin from a ptch2:Kaede photo-conversion experiment fish. (J) The fin is shown at 5 dpa, immediately after photoconverting Kaede protein in a distal field (tissue marked by the magenta dashed line). (K,L) The same fin 24 h post-conversion (6 dpa) imaged for Kaede expression. The dashed box in K marks the enlarged region in L. Unconverted and new Kaede is green; photo-converted Kaede is magenta. The green arrow marks cells in a forming joint that expressed Kaede within the previous 24 h. The white bracket indicates the narrow distal domain of new Kaede production since photoconversion. The magenta arrow indicates epidermis near the tip of the fin that had displaced distally while retaining photo-converted Kaede. Dashed yellow lines in A,E,J,K show amputation planes. Scale bars: 50 µm in E; 10 µm in F; 500 µm in J,K; 250 µm in L.
Osteoblast progenitors and basal epidermis transiently encounter a distal field of active Hedgehog/Smoothened signaling
The ability to stably photoconvert the Kaede protein from green to red (Ando et al., 2002) allowed us to determine whether Hh/Smo signaling in pObs and basal epidermis was transient or continuous, and to follow the fate of Hh/Smo-responsive cells during regeneration. We performed Kaede photoconversion (the ‘pulse’) by illuminating a field containing distal regions of several rays of a 96 hpa ptch2:Kaede fish with 405 nm light (Fig. 1H,J). We then monitored both the perdurance of converted red Kaede and the appearance of new green Kaede during a 24 h ‘chase’ period. Active Kaede production was found in a narrow region distal to the photoconverted field corresponding to new bone segments, marked by intervening nascent joints, that re-formed during the chase period (Fig. 1I,K,L). By contrast, minimal new Kaede was produced in more proximal Obs that retained photoconverted red Kaede. We also detected a cell population at the extreme distal end of the regenerate that expressed only photoconverted Kaede (Fig. 1L). By observing confocal z-stacks, we determined these cells were previously Hh/Smo-responsive epidermal cells (Fig. S2A-H) that had migrated distally and possibly are then shed (Fig. S2I-K). We conclude that during the outgrowth phase of fin regeneration, Hh/Smo signaling is actively transmitted only in pObs and immediately adjacent basal epidermis. However, the fate of the Hh/Smo-responding cells in each lineage are distinct. After terminating Hh/Smo signaling, differentiating pObs remain relatively stationary where they progressively extend replacement bone. By contrast, the continuous distal displacement of basal epidermal cells directs their brief passage through the same ‘signaling zone’ before moving further distally as they downregulate Hh/Smo signaling.
shha and ihha are transiently expressed in parallel bands of adjacent epidermis and osteoblast progenitors during fin regeneration
The dynamic ptch2:Kaede-marked Hh/Smo activity seen in the basal epidermis and Obs of regenerating fins could be driven by one or more of the Hh family members. By qRT-PCR on 96 hpa fins, we observed robust and comparable shha and ihha transcript levels. By contrast, ihhb was expressed at low levels (one-ninth that of ihha) and shhb mRNA was undetectable (Fig. 2A). dhh also is not expressed in regenerating fins (Avaron et al., 2006). We conclude that Shha and/or Ihha, which are transcribed in basal epidermis and Obs, respectively (Avaron et al., 2006; Laforest et al., 1998; Quint et al., 2002), are the only clear candidates to stimulate the ptch2-marked Hh/Smo activity seen in both cell types.
Fig. 2.
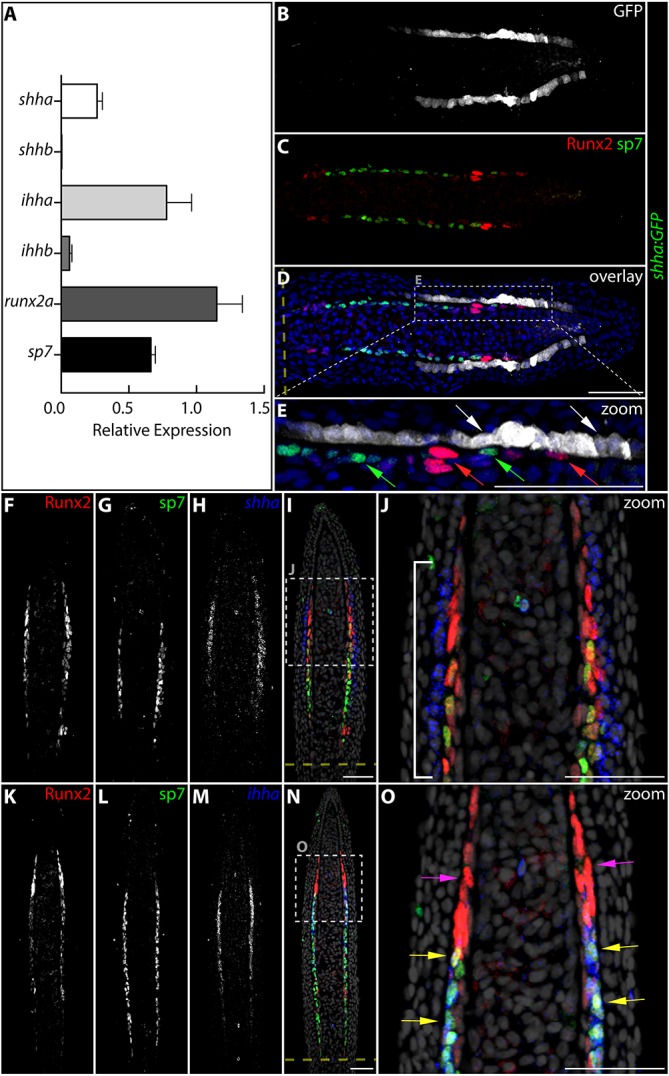
shha is briefly transcribed by distal migrating epidermal cells, whereas ihha is restricted to re-differentiating progenitor osteoblasts. (A) qRT-PCR analysis of the relative expression levels of shha, shhb, ihha, ihhb, runx2a and sp7 in 96 hpa fin tissue. The relative levels of the indicated transcripts are means of four fins normalized to rpl8 expression. Error bars represent 1 s.d. (B-E) An immunostained fin section from a 72 hpa shha:GFP fish showing GFP (white), Runx2 (red) and sp7 (green) expression. Nuclei are in blue. E is a high-magnification view of the dashed box in D. (F-O) Fin sections from a 72 hpa fish stained by RNA in situ hybridization for shha (F-J) or ihha (K-O) transcripts (blue) and with Runx2 (red) and sp7 (green) antibodies. Single channels are shown in gray scale (F-I,K-N). Nuclei are gray in the overlay images (I,J,N,O). (J,O) Enlarged regions marked in I,N, respectively. The white bracket indicates the extent of epidermal cells expressing shha relative to pObs. Yellow arrows show Runx2+/sp7+ Obs that express ihha; magenta arrows mark distal Runx2+ pObs that lack ihha mRNA. The dashed yellow lines indicate amputation sites. Scale bars: 25 μm in J,O; 50 µm in E; 50 μm in B-D,F-N.
We used the Tg(-2.4shha:GFP:ABC)sb15 (abbreviated shha:GFP) transgenic line (Ertzer et al., 2007), which recapitulates endogenous shha expression during fin regeneration (Lee et al., 2009; Zhang et al., 2012), to further resolve the spatial-temporal dynamics of shha transcription during fin regeneration. Using whole-mount analysis, we first observed faint GFP fluorescence in the distal regenerate at 48 hpa. This patch of shha:GFP expression split into two clusters on each side of the regenerating ray around 72 hpa (Fig. S3), as previously reported (Zhang et al., 2012). We used antibody staining of sectioned fins to observe shha-expressing epidermal cells relative to sp7- and Runx2-expressing Obs at various times post-fin amputation (24-48 hpa are shown in Fig. S4A-I). At 72 hpa, 1 day ahead of the initiation of ray branching, shha:GFP expression was isolated to the basal epidermis adjacent and at least five cells distal to Runx2+ pObs (Fig. 2B-E). However, using fluorescent in situ hybridization (FISH) coupled with Runx2 and sp7 immunostaining, shha transcripts were actively produced only by basal epidermal cells directly neighboring distal Runx2+ pObs (Fig. 2F-J). We conclude that the extreme distal shha:GFP expression likely represents migrating epidermal cells with residual GFP protein, rather than ongoing shha transcription. Overlapping GFP and Kaede protein expression in adherens junction-marked distal basal epidermal cells in fins of 72 hpa shha:GFP;ptch2:Kaede fish further shows the basal epidermis transduces Hh signals only while neighboring pObs and ceases responding upon its continued distal displacement (Fig. S4J-M). A summary schematic showing the migratory and Hh/Smo pathway dynamics of both epidermal and Ob cells is shown in Fig. S5.
We also used FISH to resolve ihha transcription within hierarchically arranged regenerating Obs defined by Runx2 and/or sp7 expression. The distal and proximal extents of ihha expression precisely corresponded with the distal-most sp7 expressing and proximal-most Runx2-expressing Obs, respectively. Therefore, active ihha transcription is restricted to re-differentiating Runx2+/sp7+ Obs (Fig. 2K-O) and not to Runx2+ pObs nor the sp7+ Obs that actively produce replacement bone.
Ihha is required to promote mineralization of regenerated bone
We used the ihhahu2131 null allele (Hammond and Schulte-Merker, 2009) to determine whether Ihha is responsible for the ptch2-marked Hh/Smo activity in the epidermis and/or Obs of regenerating fins. A subset of ihhahu2131/hu2131 (ihha−/−) homozygotes survived craniofacial, swim bladder and cloaca development defects (Hammond and Schulte-Merker, 2009; Huycke et al., 2012; Korzh et al., 2011; Parkin et al., 2009), allowing adult regeneration studies. The pattern of ptch2:Kaede fin expression in 5 dpa ihha-null fish was normal and, unexpectedly, levels were modestly increased rather than decreased (Fig. 3A-F). shha, shhb and ihhb transcript levels were not altered in regenerating fins of ihha-deficient fish, arguing against compensatory expression of related Hh genes (Fig. S6A). By contrast, ihha transcripts were reduced, likely due to nonsense-mediated mRNA decay (Fig. S6A). We deduce that the epidermal and Ob Hh/Smo activity during fin regeneration is Ihha independent and therefore Shha driven. Furthermore, any essential Ihha signaling is not driven by Smo-mediated transcriptional changes canonically represented by ptch2:Kaede expression.
Fig. 3.
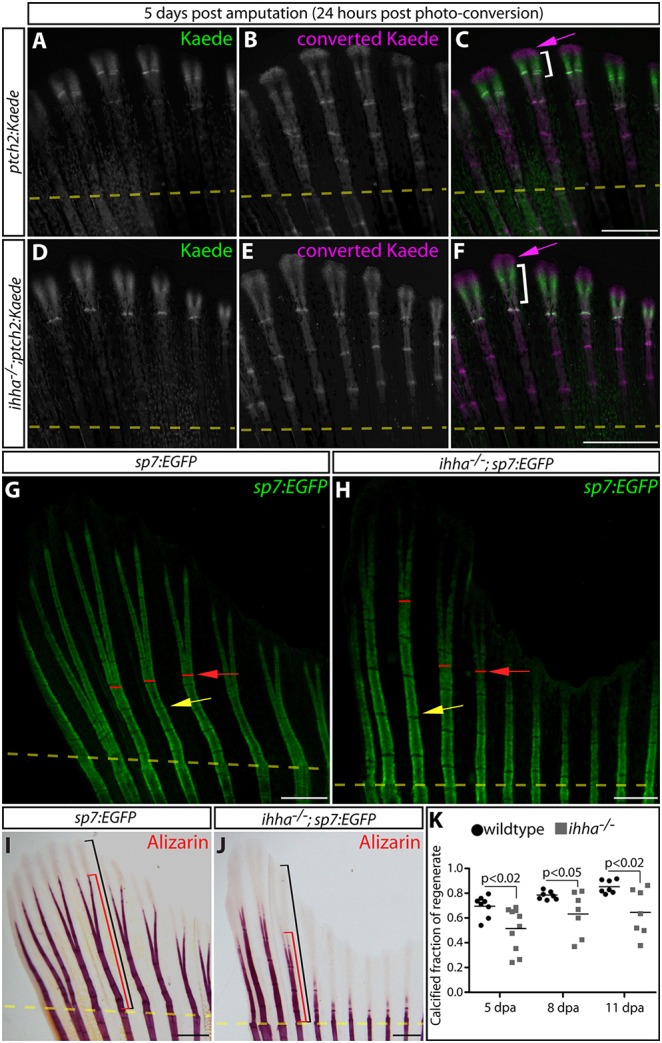
Ihha promotes the efficient calcification of regenerated bone by non-canonical Hedgehog signaling. (A-F) Whole-mount Kaede fluorescence images of 5 dpa caudal fins from ptch2:Kaede (A-C) and ihha−/−;ptch2:Kaede (D-F) fish 24 h after photoconversion. Photoconverted pre-existing Kaede is magenta; Kaede produced after photoconversion is green. Magenta arrows indicate extreme distal epidermal tissue exclusively expressing converted Kaede. The white brackets indicate the domain of newly expressed Kaede protein. (G,H) Whole-mount images showing GFP-expressing osteoblasts in fins from sp7:EGFP and ihha−/−;sp7:EGFP fish at 11 dpa. Yellow arrows mark newly formed joints. Red lines and arrows denote points of ray bifurcation. (I,J) Bright-field images of Alizarin Red-stained sp7:EGFP(I) and ihha−/−;sp7:EGFP (J) fins at 11 dpa. Black and red brackets show the total length of the regenerate and the extent of mineralization from the site of amputation, respectively. Yellow dashed lines in all panels show amputation positions. (K) Quantification of the relative extent of calcified regenerated bone in sp7:EGFP versus ihha−/−;sp7:EGFP fish at 5, 8 and 11 dpa. Means and data points representing individual fish are shown. Significant differences between control and ihha-null fish (P<0.05) were determined by two-tailed Student's t-tests. Scale bars: 500 μm.
Fins from ihha−/− fish regenerated grossly normally following resection, including restoring organized bones with branched rays as shown by Tg(sp7:EGFP)b1212 (hereafter referred to as sp7:EGFP) expression (Fig. 3G,H). However, using whole-mount Alizarin Red and von Kossa staining of tissue sections, the majority of ihha-deficient fish still displayed qualitative and quantitative calcification defects 6 weeks after amputation (Fig. 3I-K, Fig. S6B-J). Fin ray calcification was largely normal in unamputated ihha−/− adult fish, indicating that Ihha non-redundantly promotes regenerative but not developmental bone maturation (Fig. S6K-N). Consistent with a late role for Ihha in bone regeneration, ihha−/− zebrafish had no change in Ob numbers, proliferation rate or Runx2/sp7 expression during the fin regenerative response (Fig. S6O-Q). Furthermore, BMP signaling, as monitored by pSmad1/5/8 staining, was intact in the absence of ihha (Fig. S7). We conclude that Ihha expressed in re-differentiating Obs acts in parallel with the core differentiation regulatory network to support maturation of subsequently formed fully differentiated Obs.
Smoothened inhibitor BMS-833923 blocks Hedgehog signaling in zebrafish and avoids non-specific anti-proliferative effects of cyclopamine
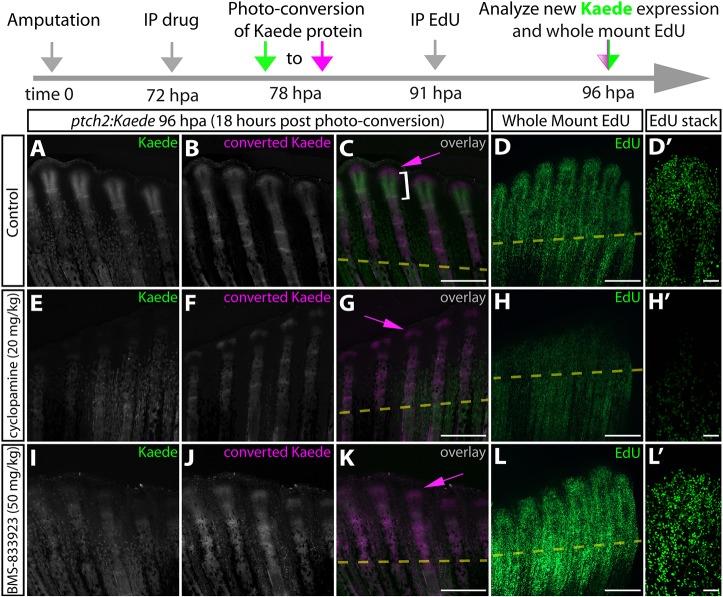
The Shha-driven Hh/Smo signaling we observed in a narrow distal band of neighboring basal epidermis and Obs does not adequately explain the widespread proliferation block observed when exposing regenerating zebrafish to the Smo-inhibitor cyclopamine (Blum and Begemann, 2015; Lee et al., 2009; Quint et al., 2002; Wehner et al., 2014). We surmise that this phenotype, which is the primary evidence supporting the hypothesis that Hh promotes regenerative outgrowth, is an off-target effect similar to that reported when using cyclopamine to study zebrafish germ cell development (Mich et al., 2009). Therefore, we performed a screen of seven additional Smo small-molecule inhibitors to determine whether any would reduce ptch2:Kaede expression and recapitulate developmental defects seen in smo-null embryos without blocking global cell proliferation.
We amputated ptch2:Kaede fins and intraperitoneally injected cyclopamine (20 mg/kg), BMS-833923 (50 mg/kg), Vismodegib (75 mg/kg), Erismodegib (50 mg/kg), SANT-1 (50 mg/kg), Taladegib (8.25 mg/kg), glasdegib (50 mg/kg) or PF-05274857 (100 mg/kg) at 72 hpa. We photoconverted the Kaede protein 6 h later (78 hpa) and analyzed fins at 96 hpa (24 h after drug delivery) for new Kaede protein expressed during the drug exposure period (n≥4 for each group). As expected, cyclopamine-treated fins produced no new Kaede expression (Fig. 4A-C,E-G). Among the other compounds tested, only BMS-833923 prevented new Kaede expression (Fig. 4I-K, Fig. S8). Whole-mount EdU analysis indicated that cyclopamine treatment rapidly arrested DNA synthesis in cells throughout the regenerating fin and correspondingly halted regenerative outgrowth (Fig. 4D,H). Importantly, this proliferation arrest was not observed in BMS-833923-treated fins (Fig. 4L). We could not identify a cyclopamine dose that separated its effects on ptch2:Kaede expression and cell proliferation. By contrast, even doses of BMS-833923 exceeding those necessary to block ptch2:Kaede had no appreciable effect on proliferation (Fig. S9). Furthermore, BMS-833923 treatment of early zebrafish embryos more closely recapitulated the gross developmental defects seen in smo-null zebrafish than did cyclopamine exposure (Fig. S10) (Aanstad et al., 2009; Chen et al., 2001; Lewis and Eisen, 2001; Loucks et al., 2007; Varga et al., 2001; Wolff et al., 2003). Notably, BMS-833923 phenocopied smo mutants in preventing the development of Engrailed-expressing muscle pioneer cells (Aanstad et al., 2009; Barresi et al., 2000; Wolff et al., 2003). Therefore, BMS-833923 exhibits preferred characteristics over cyclopamine as a small-molecule inhibitor for Hh/Smo signaling studies in zebrafish.
Fig. 4.
BMS-833923, a newly identified potent zebrafish Smoothened inhibitor, shows that Hedgehog/Smo signaling does not impact fin regenerative outgrowth. (A-L′) Whole-mount fluorescence images of 96 hpa fins from ptch2:Kaede fish 18 h after Kaede photoconversion and 5 h after EdU injection. Regenerating fish were treated for 24 h with DMSO (A-D′), cyclopamine (E-H′) or BMS-833923 (I-L′). Newly produced Kaede and converted Kaede are green and magenta, respectively, in overlay images (C,G,K). Magenta arrows indicate cells with converted Kaede. White brackets mark the domain of new Kaede expression in the 18 h since photoconversion. (D,H,L) Fins from the same fish shown for Kaede fluorescence processed to reveal EdU-incorporating nuclei in green. (D′,H′,L′) Confocal stack images showing EdU incorporation at the distal aspect of single re-forming rays. The timeline at the top of the figure details the experimental design. Scale bars: 500 μm.
Hedgehog/Smoothened signaling is dedicated to ray bifurcation during the outgrowth phase of fin regeneration
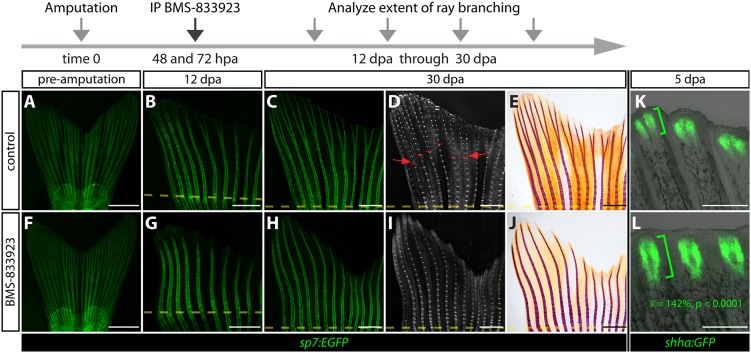
To test potential Shha/Smo signaling contributions to ray branching, we treated sp7:EGFP zebrafish with BMS-833923 at 48 and 72 hpa, and monitored the course of regeneration. At 12 dpa, when all regenerated fins from control treated fish had bifurcated lepidotrichia, BMS-833923-treated fish dramatically lacked branched rays (Fig. 5A-B,F-G). By contrast, BMS-833923 exposure did not disrupt regenerate outgrowth (control mean=3.30 mm; BMS-833923 mean=3.07 mm; n=5, P>0.212). This specific ray branching defect persisted through 30 dpa when fin regeneration, including bone growth, overt differentiation and joint formation, was otherwise completely normal (n=12; Fig. 5C-D,H-I). The permanent blockage of ray branching may reflect prolonged Hh/Smo inhibition by BMS-833923; ptch2:Kaede zebrafish treated at 48 and 72 hpa failed to express new Kaede protein through 9 dpa (Fig. S11). Bone mineralization, which was disrupted in ihha−/− zebrafish, was unaffected by Hh/Smo inhibition initiated after 48 hpa (Fig. 5E,J), reinforcing the possibility that Ihha does not function through Smo-dependent signaling to promote regenerative bone maturation. Furthermore, we conclude that Hh/Smo signaling driven by basal epidermal Shha is primarily dedicated to directing ray branching after the initiating steps of fin regeneration are complete.
Fig. 5.
BMS-833923 demonstrates that Hedgehog/Smoothened signaling is dedicated to bony ray branching during the outgrowth phase of fin regeneration. (A-J) Whole-mount images of fins from the same two sp7:EGFP fish acquired prior to amputation and at 12 and 30 days post-amputation (dpa). The fish are treated with either control DMSO or BMS-833923 at 48 and 72 hpa. (A-C,F-H) Fluorescence images showing osteoblast GFP expression in green. (D,I) Rotterman contrast images. Periodic high-contrast patches along the rays are joints. Red lines and arrows mark ray bifurcation points. (E,J) Bright-field images of Alizarin Red-stained fins collected from the same fish to visualize mineralized bone. Amputation planes are indicated with dashed yellow lines. (K,L) Wide-field whole-mount GFP fluorescence and bright-field overlay images of 5 dpa fins from shha:GFP fish following injections with DMSO or BMS-833923 at 48 and 72 hpa. Green brackets indicate the extent of epidermal shha:GFP expression along the proximal-distal axis. Student's t-test shows the 142% mean increase in the length of the shha:GFP domain is significant [n=5 control and 10 BMS-833923-treated fish (60 scored rays), P<0.0001]. Scale bars: 2 mm in A,F; 1 mm in B-E,G-J; 500 µm in K,L.
BMS-833923-exposed shha:GFP zebrafish (treated at 48 and 72 hpa) retained two distinct GFP-positive epidermal patches adjacent to each hemiray at 4 and 5 dpa. However, the shha:GFP-expressing domain was expanded along the proximal-distal axis (mean length=142%, n=5 control and 10 BMS-833923-treated fish, P<0.0001; Fig. 5K,L), reminiscent of a previous report using cyclopamine (Quint et al., 2002). Collectively, we conclude that: (1) Shha/Smo signaling is not required to generate split clusters of shha-expressing epidermis; (2) in the absence of Smo activity, shha-expressing epidermal splitting is not sufficient to promote ray branching; and (3) negative feedback restricts shha expression to a short stretch of distal basal epidermis.
The onset of shha expression splitting precedes observable ray branching and the split pattern persists for several days while branching unfolds (Laforest et al., 1998; Zhang et al., 2012). To assess whether Shh/Smo signaling is continuously required for ray bifurcation, we inhibited Hh/Smo signaling at 5 dpa, 2 days post-splitting of shha:GFP-expressing epidermis (Fig. S3; Zhang et al., 2012). Fins from these BMS-833923-exposed fish had normal regenerative outgrowth with limited and delayed ray bifurcation in the longest rays (rays 3-6, both dorsal and ventral sides) at 40 dpa (n=10; Fig. S12). Therefore, the process of Shh/Smo-induced ray branching transpires over several days, suggesting a progressive rather than switch-like mechanism.
Shh promotes ray bifurcation by directing pObs to migrate into split pools
Shh/Smo signaling has been speculated to direct branching morphogenesis by inducing local proliferation of pObs underlying the split shha-expressing basal epidermis (Zhang et al., 2012). Alternatively, Shh/Smo could direct pObs to migrate into split pools that then continue regenerating independently to form branched rays. To distinguish between these possibilities, we treated shha:GFP zebrafish with BMS-833923 at 48 and 72 hpa, and then analyzed the proliferation (by EdU incorporation) and arrangement of Runx2+ pObs at 96 hpa in multiple transverse sections along the proximal-distal axis. We tracked individual rays in control and BMS-833923-treated animals from a relatively proximal position, where shha:GFP initiated in a single basal epidermal field, through the region of shha:GFP domain splitting, to far distal sections that lacked Runx2+ pObs (Fig. 6A-F). Given our ptch2:Kaede cell tracing showed basal epidermal cells migrate distally, we conclude that shha transcriptional initiation precedes the physical assortment of shha-expressing epidermal cells into distinct clusters. Furthermore, as anticipated from our whole-mount studies, both shha induction and epidermal movements are Smo independent. Finally, shha-expressing epidermal cells only transiently split as the distal-most basal epidermal cells that retained GFP protein but no longer actively produce shha re-merged into a single population.
Fig. 6.
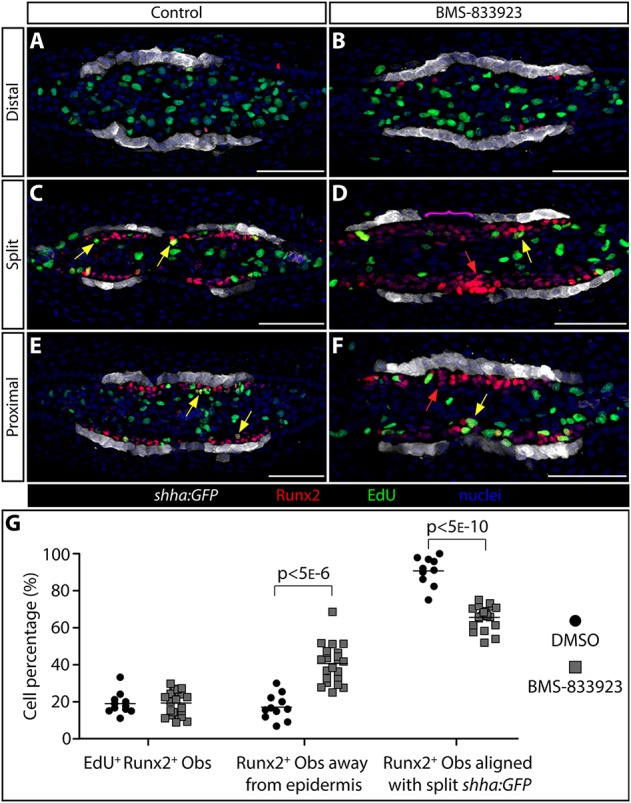
Shha-driven Smoothened signaling directs progenitor osteoblasts to migrate in step with transiently split basal epidermal clusters at the onset of ray branching. (A-F) Runx2 and EGFP immunostaining (red and white, respectively) and EdU incorporation (2 h treatment, green) on transverse 96 hpa fin sections from individual shha:GFP fish treated at 48 and 72 hpa with DMSO (A,C,E) or BMS-833923 (B,D,F). Nuclei are blue. (A,B) Far distal sections beyond the distal-most pObs. (C,D) Sections from positions where shha-expressing epidermal cells have split into two clusters on each side of the fin. (E,F) Further proximal sections where shha is first induced by distal migrating epidermal cells. Yellow arrows indicate Runx2+/EdU+ cells located more than one cell layer from shha:GFP-expressing basal epidermis. Red arrows indicate Runx2+ pObs two or more cell layers distant from shha:GFP-positive epidermal cells. The magenta bracket in D highlights Runx2+ pObs that span the junction between split shha:GFP domains in BMS-833923-treated fish. (G) Quantification of pOb proliferation and the relative position of pObs to epidermal cells in regenerating fins from the above and similar DMSO versus BMS-833923 treated shha:GFP fish. Only images at ‘split’ positions are scored. Left plots: the fraction of EdU incorporating Runx2+ pObs. Middle plots: the fraction of pObs not directly adjacent to GFP+ epidermal cells. Right plots: the fraction of pObs aligned with split clusters of shha:GFP-expressing epidermis. Each data point represents a scored independent section (11 individual rays from five DMSO-treated fish and 23 rays from seven BMS-833923-treated fish). Two-tailed Student's t-tests were used to determine statistically significant differences (P<0.05) between the means of control versus small-molecule-treated samples. Scale bars: 50 μm.
At sites of transiently split shha:GFP-expressing epidermis, most Runx2+ pObs in control fins were arranged in a single layer directly adjacent to GFP-positive epidermis (Fig. 6C, additional repeats in Fig. S13). By contrast, Runx2+ pObs in BMS-833923-treated fins were up to several cell layers thick and spanned the entire junction between split shha:GFP domains (Fig. 6D). At a more proximal position prior to the splitting of shha:GFP-expressing epidermis, Runx2+ pObs in BMS-833923 treated fins were four or more cell layers distant from the epidermis (Fig. 6E). In some cases, this transverse section analysis appeared to indicate BMS-833923 exposed fins had a larger population of Runx2+ pObs. However, a quantitative analyses of longitudinal sections from multiple animals revealed that overall numbers of distal Runx2+ pObs were unaffected by BMS-833923 treatment (Fig. S14A-C). Rather, the pOb pool was incompletely extended along the proximal-distal axis in the absence of Hh/Smo signaling. Furthermore, while Hh/Smo inhibition led to disorganized Runx2+ pObs, it did not alter the fraction of EdU-incorporating proliferating Runx2+ Obs (Fig. 6G), including following acute BMS-833923 treatment (Fig. S14D-F). In addition, the proliferative rate of Runx2+ pObs was not correlated with their proximity to shha:GFP-expressing epidermis (P>0.868, two-tailed Fisher's exact test, n=336 for layer 1 Runx2+ cells and n=73 for Runx2+ cells in layers 2 and over). We conclude that Shh/Smo signaling aligns underlying pObs with shha-expressing basal epidermis and then guides pObs into split pools to initiate ray branching during fin regeneration.
Shha-expressing basal epidermal cells directly contact and recruit underlying osteoblast progenitors
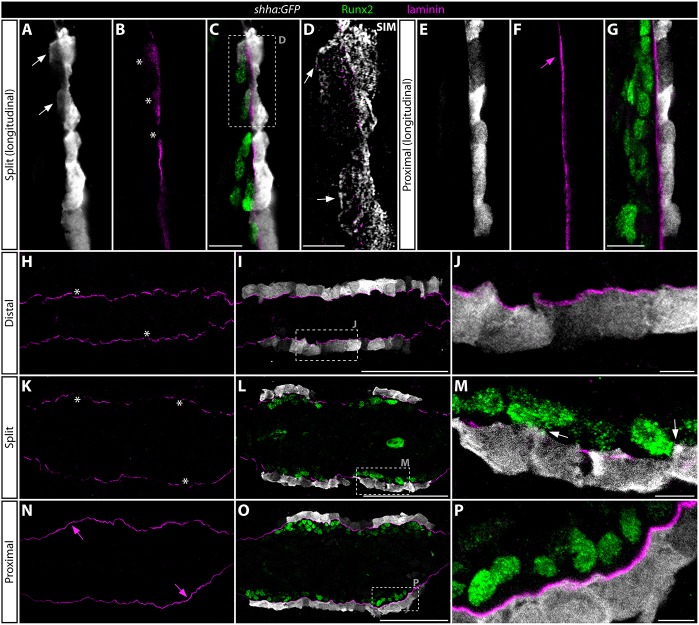
Emerging studies show that, in some contexts, Shh/Smo signaling is highly localized and even mediated by direct cell-to-cell contacts through cell surface-retained Shh protein (Sanders et al., 2013). Therefore, we used antibody staining and confocal microscopy, including structured illumination microscopy (SIM, Gustafsson, 2000) of sectioned regenerating shha:GFP fins to explore the relative positioning of basal epidermis and pObs. Strikingly, at the point of epidermal splitting, GFP+ basal epidermal cells directly appose neighboring Runx2+ pObs, including through extended cellular protrusions that contact and occasionally envelop pObs (Fig. 7A-D,K-M). Antibody staining of regenerating fin sections prepared from permanently labeled epidermal mosaic fish (Stewart and Stankunas, 2012) confirmed that basal epidermal-originating processes directly contact Runx2+ pObs (Fig. S15).
Fig. 7.
shha:GFP-expressing basal epidermal cells extend cellular protrusions through incompletely assembled basement membrane to contact Runx2+ progenitor osteoblasts. (A-P) GFP (white), laminin (magenta) and Runx2 (green) antibody stained fin sections from 96 hpa shha:GFP fish. All images are 1 airy unit (∼1 µm) single optical confocal sections, except D, which is a structured illumination microscopy (SIM) image representing an ∼100 nm section. (A-D) Longitudinal section showing the distal regenerate where the shha-expressing basal epidermis is split into two clusters and Hh/Smo signaling is activate in both pObs and basal epidermis. (C) An overlay showing Runx2 staining together with A and B. (D) A high-magnification SIM image of the boxed area in C. (E-G) A proximal field from the same longitudinal section shown in A-D. (H-P) Transverse sections representing three positions along the proximal-distal axis from a single regenerating ray. (H-J) An extreme distal section beyond the distal extent of Runx2+ pOb pools. (K-M) Section from a position where shha:GFP-expressing basal epidermis is split into two clusters on each side of the regenerating fin. (N-P) A further proximal section where shha expression has initiated in epidermal cells but prior to their division into split clusters. The dashed boxes in I, L and O mark regions shown at higher magnification in J, M and P, respectively. White arrows indicate cellular protrusions from shha:GFP+ cells that contact or enshroud Runx2+ osteoblasts. White asterisks mark gaps in the basal lamina. Magenta arrows show a continuous laminin-containing basement membrane that physically separates the basal epidermis from Runx2+ pObs. Scale bars: 5 µm in D,J,M,P; 50 µm in C,G,I,L,O.
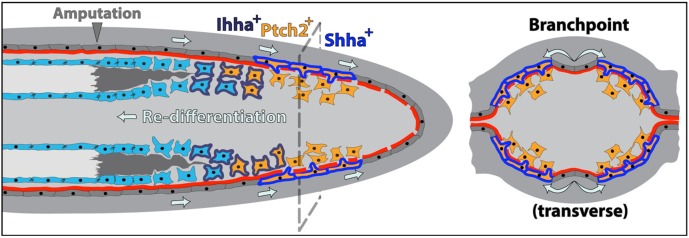
The basal epidermis and underlying mesenchyme of regenerating fins seemingly are separated by a continuous laminin β1a-containing (Lamb1a) basement membrane (Chen et al., 2015) that should impede epithelial-stromal interactions (Kelley et al., 2014). However, our confocal analysis of antibody-stained longitudinal sections revealed diffuse laminin staining in distal regions featuring basal epidermal-pOb contacts (Fig. 7B). At more proximal positions, laminin expression defined an unbroken barrier between basal epidermis and neighboring pObs (Fig. 7E-G). Immunostaining of serial transverse sections from 96 hpa shha:GFP fins confirmed extensive laminin gaps exclusively where Runx2+ pObs and shha:GFP-expressing epidermal cells interacted (Fig. 7K-P). Given the continuous distal displacement of epidermal cells and especially high lamb1a transcript levels in the distal regenerate (Chen et al., 2015), we suggest that this region is the active site of ongoing basal lamina assembly associated with fin regenerative outgrowth. Intriguingly, far distal regions had a more robust basement membrane than at the point of shha:GFP domain splitting (Fig. 7H-J). We propose that basal lamina gaps at the proximal/distal position defined by active Shha/Smo signaling enable direct interactions between Shha-positive epidermal cells and Ptch2-expressing pObs. These Hh/Smo-reinforced interactions coupled with epidermal movements then escort pObs into separated pools to initiate ray branching (Fig. 8).
Fig. 8.
Model showing how cell movements, Hh/Smo pathway dynamics and direct cell-to-cell interactions between neighboring basal epidermal cells and progenitor osteoblasts induce ray bifurcation during fin regeneration. Basal epidermal cells generated proximal to the caudal fin amputation site continuously advance distally along a mature basal lamina. During this effective migration, groups of basal epidermal cells overlying the regenerating blastema transiently upregulate shha expression (cells outlined in blue) and then split into two clusters on each side of the ray. Shha drives active Hh/Smo signaling marked by ptch2 expression (orange cells) in a narrow distal zone of shha-expressing epidermal cells and adjacent progenitor osteoblasts (pObs). As fin outgrowth proceeds, some pObs escape self-renewal signals, initiate re-differentiation and progressively extend reforming bone. Fully differentiated Obs secrete new bone matrix, enabled by the earlier non-canonical activity of Ob-expressed Ihha (cells outlined in dark blue). The interface between Hh/Smo-responsive Obs and basal epidermis is the active site of basal lamina (red) assembly associated with fin regenerative outgrowth. Coinciding with the epidermal ‘branchpoint’, this incompletely assembled basement membrane enables shha-expressing basal epidermal cells to extend cellular protrusions that contact neighboring pObs. This direct binding activates Smo-dependent signaling (including further ptch2 expression) and progressively escorts pObs into physically separated pools. The newly divided pools of Runx2+ pObs then continue regenerating independently to produce a now bifurcated ray.
DISCUSSION
Active Hh/Smo signaling during fin regeneration is tightly restricted to distally migrating basal epidermis and adjacent osteoblast progenitor cells
Our ptch2:Kaede photoconversion experiments monitoring Hh/Smo-responsive cells at the time of ray branching resolve active Hh/Smo signaling to a narrow stretch of distal basal epidermal cells and neighboring Runx2+ pObs. This highly restricted nature of Hh/Smo-active cells in the regenerating fin suggests a short-range communication mode and tight pathway control that is inconsistent with the proposition that Hh/Smo signaling is a major regulator of proliferative outgrowth (Lee et al., 2009; Quint et al., 2002; Wehner et al., 2014). These experiments also highlight the distinct advantage afforded by photoconvertible reporter proteins when monitoring gene expression and cell signaling dynamics. Without photoconversion, ptch2-driven Kaede, a relatively stable protein, misleadingly appears to be broadly expressed in both the basal epidermis and in Obs. Furthermore, by effectively enabling lineage tracing, Kaede photoconversion shows that Hh/Smo activity is continuously maintained at the same relative position in both Ob and epidermal lineages, attributable to distal-migrating epidermis that provides a constant source of new shha-expressing cells.
The distal movement of basal epidermis, concurrent with regenerative outgrowth is consistent with the observation that the fin epidermis largely proliferates proximally to the site of amputation (Poleo et al., 2001). We propose this proximal expansion pushes continuously generated basal epidermal cells to displace distally. During this effective ‘migration’, basal epidermal cells transiently upregulate shha to generate a zone of active Hh/Smo signaling in both basal epidermis and adjacent Obs. Conversely, proliferative pObs in the active zone remain ‘in place’ as the regenerate extends distally. pObs that escape self-renewing Wnt signals similarly lose Hh/Smo activity while upregulating BMP and other differentiation pathways that promote them to progressively extend reforming bone (Stewart et al., 2014).
Ihha promotes mineralization of regenerated bone through non-canonical signaling
Ihh promotes Ob proliferation and/or differentiation during bone development (Abzhanov et al., 2007; Huycke et al., 2012; Lenton et al., 2011; Long, 2012). Expanding on previous fin regeneration studies (Avaron et al., 2006), we found that ihha is expressed distinctly in pObs as they re-differentiate and acquire sp7 expression but not in Runx2+/sp7− self-renewing pObs or fully differentiated Runx2−/sp7+ Obs. This unique pattern implies a role for Ihha in early steps of bone re-differentiation. However, our analysis of fin regeneration in viable homozygous null ihha zebrafish demonstrates that Ihha does not control pOb proliferation, sp7 expression, BMP signaling or organization of re-differentiated Obs. Instead, the deficient accumulation of calcified bone in ihha-null fish suggests Ihha functions in re-differentiating pObs to prime their later mineralization, likely acting in parallel with the BMP/sp7 differentiation network.
Ihha−/−;ptch2:Kaede regenerating fins did not show decreased Kaede expression, indicating that Hh/Smo activity in the basal epidermis and Obs is driven solely by Shha, the only other appreciably expressed Hh ligand. Furthermore, given ihha-deficient zebrafish did show a bone maturation defect during fin regeneration that was not recapitulated by BMS-833923 exposure, Ihha may act by Smo-independent non-canonical Hh signaling (Jenkins, 2009). Finally, the modest expansion of ptch2:Kaede in ihha−/− regenerating fins suggests that upregulation of Ihha in differentiating Obs negatively regulates Hh/Smo output in surrounding cells to constrain pathway activity to the observed narrow distal zone. Expanded shha:GFP expression in BMS-833923-treated animals further indicates that negative-feedback networks restrict epidermal shha expression and therefore Hh/Smo output.
The Smo inhibitor BMS-833923 avoids the off-target anti-proliferative effects of cyclopamine in zebrafish
Cyclopamine and smoothened agonist exposure experiments suggest mitogenic Hh/Smo signaling supports regenerative fin outgrowth (Blum and Begemann, 2015; Lee et al., 2009; Quint et al., 2002; Wehner et al., 2014). However, our observation that only two small distal cell populations undergo active Hh/Smo signaling during fin regeneration is incongruous with this model. By screening recently developed Smo inhibitors, we discovered that BMS-833923 (Akare et al., 2014) is as effective as cyclopamine in blocking zebrafish Hh/Smo activity. However, BMS-833923 more faithfully recapitulates smo mutant phenotypes during development, including blocking development of muscle pioneer cells (Aanstad et al., 2009; Barresi et al., 2000). Crucially, unlike cyclopamine, BMS-833923 did not inhibit cell proliferation during fin regeneration and, instead, dramatically and specifically abrogates ray branching. We conclude that the widespread anti-proliferative effects of cyclopamine on regenerating fins is not due to Hh/Smo inhibition but rather reflects unknown off-target effects of the compound.
A non-specific anti-proliferative effect of cyclopamine is consistent with the observation that ablation of shha-expressing basal epidermal cells does not affect overall regenerative outgrowth of the fin (Zhang et al., 2012). Consistent with cyclopamine having off-target effects in zebrafish, cyclopamine-induced aberrant migration of primordial germ cells occurs in a Smo-independent manner (Mich et al., 2009). Furthermore, high doses of cyclopamine (≥10 μM) inhibit proliferation in cells that lack detectable Smo expression (Zhang et al., 2009). We advocate that the many studies using cyclopamine in zebrafish, including those that have led to fin regeneration models incorporating mitogenic roles of Hh/Smo signaling, be revisited and/or interpreted with caution.
Shha-expressing basal epidermal cells directly contact and recruit Runx2+ osteoblast progenitors to promote ray branching
Inhibition of Hh/Smo signaling using BMS-833923 from 48-96 hpa strikingly blocks ray branching without disrupting regenerative bone growth or maturation. Given the distinct bone maturation defects in ihha−/− mutants and the unappreciable expression of other Hh ligands, we conclude that Shha-driven Hh/Smo signaling induces ray bifurcation. Consistent with this notion, ablation of shha-expressing basal epidermal cells significantly delays ray branching (Zhang et al., 2012). However, using PCNA staining to identify cycling cells, Zhang et al. concluded that the shha:GFP-expressing basal epidermis induces localized Ob proliferation to direct ray branching. In contrast, by quantitative EdU incorporation studies, we found that Runx2+ pOb proliferation at the ray branching site is unaffected by Hh/Smo inhibition. Furthermore, pOb proliferative rates are independent of proximity to Shha-expressing epidermal cells. However, in the absence of Shha/Smo signaling, pObs, but not shha:GFP+ basal epidermal cells, fail to form two spatially distinct pools that precede branching events. Therefore, Shha/Smo signaling promotes ray branching by directing the cellular migration rather than localized proliferation of pObs.
Our observation that shha:GFP-expressing basal epidermal cells directly contact underlying pObs, including through cellular protrusions, suggests Shh/Smo-promoted pOb migration is driven by simple and direct intercellular interactions. A model that very short-range signaling by Shha drives ray branching conflicts with the widely held paradigm that Shh works as a long-range morphogen [e.g. to pattern the spinal cord (Briscoe and Thérond, 2013)]. However, several other recent studies demonstrate exceptions to this rule. In the embryonic chick limb, Shh remains tightly associated with its producing cells and likely acts through its retention on long cytoplasmic extensions (Sanders et al., 2013). In fly imaginal discs, localized Hh acts as a short-range signal (Ayers et al., 2010). At the simplest level, membrane-retained epidermal Shha may interact with Ptch2 on pObs to produce the observed cell-cell adhesion that promotes bone branching. As such, ‘positive’-feedback activation of ptch2 to reinforce cell interactions could be the major or even only relevant target gene of Shha/Smo signal transduction. Alternatively, Hh/Smo activation in both epidermal and pObs could direct the transcriptional upregulation of more traditional cell-adhesion molecules. Regardless, our demonstration of direct cell-cell Shh-promoted interactions during osteoblast patterning suggests similar mechanisms underlie Hh/Smo signaling roles in other regeneration, developmental or disease contexts.
Incompletely assembled basement membrane enables localized Ob recruitment by epidermal cellular protrusions
The physical separation of epidermis and mesenchyme by basement membrane in the zebrafish fin is thought to provide spatially restricted and efficient epithelial-mesenchymal signaling during development and regeneration (Lee et al., 2009; Tornini and Poss, 2014). Recent work establishes that Lamb1a is a major component of the basal lamina in the regenerating caudal fin and its function is required to ensure polarity of the basal epithelium during regeneration (Chen et al., 2015). We demonstrate that direct basal epidermis-pOb interactions are enabled by the lack of a robust Lamb1a-containing basement membrane at the forming branch site. In further support, electron microscopy studies show an irregular epidermal-blastemal interface towards the distal tip of regenerating fins where basal epidermal cells extend cellular processes (or ‘digitations’) that contact underlying mesenchyme (Becerra et al., 1996; Géraudie and Singer, 1992). We suggest these distal basal lamina gaps reflect the active site of basement membrane extension that is associated with regenerative outgrowth. Newly synthesized Lamb1a progressively self-organizes to help establish a robust basement membrane that precludes prolonged contacts between the basal epidermis and re-differentiating pObs. Notably, the relatively more robust distal-most basal lamina could reflect persistent material established by the wound epidermis at the onset of regeneration.
Modular and orthogonal signaling networks cooperate to regenerate functional bone
Our study of Hh/Smo signaling during zebrafish fin regeneration illustrates how distinct signaling networks can produce independent modules that cooperate to regenerate a bone of the proper size and shape. A Wnt/BMP network establishes a system of balanced pOb growth and differentiation that allows the progressive reformation of mature bone (Stewart et al., 2014). Simultaneously, the Shh/Ptch2/Smo network acts orthogonally to control the shape of the bone by periodically splitting regenerating pObs into physically separated pools. Of relevance for regenerative medicine, these signaling networks are likely conserved but tightly restrained in adult mammals, including humans. Notably, Shh signaling has long been appreciated to control bone patterning during chick and mouse limb development (Capdevila and Izpisúa Belmonte, 2001; Riddle et al., 1993). Therefore, the localized delivery of Shh, perhaps immobilized on scaffolds, could guide therapeutic osteoblast stem cells expanded by Wnt signaling to reshape severely damaged or diseased bone.
MATERIALS AND METHODS
Zebrafish
Danio rerio wild-type AB, ihhahu2131 (Hammond and Schulte-Merker, 2009), Tg(sp7:EGFP)b1212 (DeLaurier et al., 2010), TgBAC(ptch2:Kaede)a4596 (Huang et al., 2012) and Tg(-2.4shha:gfp:ABC)sb15 [previously known as Tg(-2.2shha:gfp:ABC) (Ertzer et al., 2007; Shkumatava et al., 2004) lines were maintained at 28-29°C (Westerfield, 2007). All experiments were approved by the University of Oregon Institutional Animal Care and Use Committee (IACUC). The ihhahu2131 allele was genotyped by PCR (primers: 5′-CTGTGCCACCGTACCACTC-3′ and 5′-GCTACATTTGGACTAAACTGCAT-3′) with subsequent NspI-mediated digestion of the PCR products. Adult zebrafish were anesthetized by immersion in water containing 0.6 mM Tricaine-s (Western Chemical). Caudal fins were amputated two segments proximal to the first branch point of the fin rays using a razor blade.
Kaede photoconversion and imaging
ptch2:Kaede fish were anesthetized and immediately viewed on a glass slide using a Nikon Eclipse Ti wide-field inverted microscope. A desired field containing Kaede-expressing tissue was photoconverted by a 2-min illumination using a DAPI filter set. For the experiments in Fig. 1A-E, Kaede was photoconverted using a 405 nm laser for 2 min. Fish were left in the dark for indicated periods and then re-imaged for both green and red Kaede fluorescence. Imaging used either a Nikon Eclipse Ti wide-field inverted microscope or a Leica M165 FC stereomicroscope, except for the experiments in Fig. S2A-H, where optical section z-stacks were collected using a Zeiss LSM 880 laser scanning confocal microscope. For further details, see the supplementary Materials and methods.
Small-molecule treatments
BMS-833923 (Cayman Chemical), cyclopamine (LC Labs), Vismodegib (Chemie Tek), Erismodegib (Chemie Tek), SANT-1 (Cayman Chemical), Taladegib (Cayman Chemical), Glasdegib (Selleck Chemicals) and PF-05274857 (Cayman Chemical) were dissolved in DMSO. Immediately prior to intraperitoneal injections, stock solutions were diluted 1:10 in injection buffer (50% PEG-400, 5% Propylene Glycol, 0.5% Tween-80). Unless otherwise noted, doses were: BMS-833923, 40 mg/kg; cyclopamine, 20 mg/kg; Vismodegib, 75 mg/kg; Erismodegib, 50 mg/kg; SANT-1, 50 mg/kg; Taladegib, 8.25 mg/kg; Glasdegib, 50 mg/kg; and PF-05274857, 100 mg/kg. For each treatment, cohorts of at least five animals were used.
Section immunostaining
Immunostaining was performed on frozen or paraffin wax-embedded sections using the indicated antibodies and then visualized by confocal microscopy. For further details, see the supplementary Materials and methods.
Whole-mount fin fluorescent imaging
EGFP and Kaede expression in intact fins was detected by epifluorescence on a stereo or inverted wide-field microscope. For further details, see the supplementary Materials and methods.
Quantitative RT-PCR
Quantitative changes in gene expression were determined by real-time PCR using cDNA prepared from total RNA extracted from regenerating fins. For further details, see the supplementary Materials and methods.
Fluorescent in situ hybridization
RNA in situ hybridizations used digoxigenin-labeled antisense RNA probes and frozen fin sections. For further details, see the supplementary Materials and methods.
Histological staining
Histological staining was performed on frozen or paraffin wax-embedded fin sections followed by imaging using bright-field microscopy. For further details, see the supplementary Materials and methods.
Embryo small molecule treatments
ptch2:Kaede embryos (1.5 hpf) were exposed to compounds or to vehicle added directly to the fish water. For further details, see the supplementary Materials and methods.
Embryo immunostaining
Fixed and processed embryos were incubated overnight with the indicated antibodies, developed and imaged by confocal microscopy. For further details, see the supplementary Materials and methods.
In vivo 5-ethynyl-2′-deoxyuridine (EdU) labeling
Fish were injected intraperitoneally with EdU, which was subsequently detected on fin sections using the Click-iT proliferation assay kit (Thermo Fisher). For further details, see the supplementary Materials and methods.
Statistical analyses and replicates
Student's t-tests and Fisher's exact tests were used to determine statistically significant differences between sample sets. For further details, see the supplementary Materials and methods.
Structured illumination microscopy (SIM)
Super-resolution structured illumination (SR-SIM) on antibody stained fin sections was performed using a Zeiss ELYRA S.1. microscope. For further details, see the supplementary Materials and methods.
Cre/lox-labeled epidermal mosaic fish
For further details, see the supplementary Materials and methods.
Acknowledgements
We thank the University of Oregon Zebrafish Facility for animal care; C. Kimmel for Tg(sp7:EGFP)b1212 and ihhahu2131 fish; S. Megason for providing the TgBAC(ptch2:Kaede)a4596 fish developed in A. Schier's lab and the Tg(-2.4shha:gfp:ABC)sb15 fish developed in U. Strahle's lab; J. Postlethwait for shha and ihha cDNA plasmids; K. Poss for the Lamb1a antibody staining protocol; G. Yette for generating epidermal mosaic fish; and J. Nichols, C. Kimmel, C. Doe and the Stankunas lab for input.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
B.E.A., S.S. and K.S. designed experiments; B.E.A., A.H. and S.S. performed experiments; B.E.A., S.S. and K.S. prepared and wrote the manuscript.
Funding
B.E.A. was supported by a National Institutes of Health training grant from the National Institute of General Medical Sciences (5T32GM007413). The National Institutes of Health provided research funding (1R01HL115294 from the National Heart, Lung, and Blood Institute to K.S. and 5R03AR067522 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases to S.S.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.143792.supplemental
References
- Aanstad P., Santos N., Corbit K. C., Scherz P. J., Trinh L. A., Salvenmoser W., Huisken J., Reiter J. F. and Stainier D. Y. R. (2009). The extracellular domain of Smoothened regulates ciliary localization and is required for high-level Hh signaling. Curr. Biol. 19, 1034-1039. 10.1016/j.cub.2009.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abzhanov A., Rodda S. J., McMahon A. P. and Tabin C. J. (2007). Regulation of skeletogenic differentiation in cranial dermal bone. Development 134, 3133-3144. 10.1242/dev.002709 [DOI] [PubMed] [Google Scholar]
- Akare U. R., Bandaru S., Shaheen U., Singh P. K., Tiwari G., Singare P., Nayarisseri A. and Banerjee T. (2014). Molecular docking approaches in identification of High affinity inhibitors of Human SMO receptor. Bioinformation 10, 737-742. 10.6026/97320630010737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando R., Hama H., Yamamoto-Hino M., Mizuno H. and Miyawaki A. (2002). An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 12651-12656. 10.1073/pnas.202320599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avaron F., Hoffman L., Guay D. and Akimenko M. A. (2006). Characterization of two new zebrafish members of the hedgehog family: atypical expression of a zebrafish Indian hedgehog gene in skeletal elements of both endochondral and dermal origins. Dev. Dyn. 235, 478-489. 10.1002/dvdy.20619 [DOI] [PubMed] [Google Scholar]
- Ayers K. L., Gallet A., Staccini-Lavenant L. and Thérond P. P. (2010). The long-range activity of Hedgehog is regulated in the apical extracellular space by the glypican Dally and the hydrolase Notum. Dev. Cell 18, 605-620. 10.1016/j.devcel.2010.02.015 [DOI] [PubMed] [Google Scholar]
- Azevedo A. S., Sousa S., Jacinto A. and Saúde L. (2012). An amputation resets positional information to a proximal identity in the regenerating zebrafish caudal fin. BMC Dev. Biol. 12, 24 10.1186/1471-213X-12-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barresi M. J., Stickney H. L. and Devoto S. H. (2000). The zebrafish slow-muscle-omitted gene product is required for Hedgehog signal transduction and the development of slow muscle identity. Development 127, 2189-2199. [DOI] [PubMed] [Google Scholar]
- Becerra J., Junqueira L. C. U., Bechara I. J. and Montes G. S. (1996). Regeneration of fin rays in teleosts: a histochemical, radioautographic, and ultrastructural study. Arch. Histol. Cytol. 59, 15-35. 10.1679/aohc.59.15 [DOI] [PubMed] [Google Scholar]
- Blum N. and Begemann G. (2015). Retinoic acid signaling spatially restricts osteoblasts and controls ray-interray organization during zebrafish fin regeneration. Development 142, 2888-2893. 10.1242/dev.120212 [DOI] [PubMed] [Google Scholar]
- Briscoe J. and Thérond P. P. (2013). The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14, 418-431. 10.1038/nrm3598 [DOI] [PubMed] [Google Scholar]
- Capdevila J. and Izpisúa Belmonte J. C. (2001). Patterning mechanisms controlling vertebrate limb development. Annu. Rev. Cell Dev. Biol. 17, 87-132. 10.1146/annurev.cellbio.17.1.87 [DOI] [PubMed] [Google Scholar]
- Chen Y. and Struhl G. (1996). Dual roles for patched in sequestering and transducing Hedgehog. Cell 87, 553-563. 10.1016/S0092-8674(00)81374-4 [DOI] [PubMed] [Google Scholar]
- Chen W., Burgess S. and Hopkins N. (2001). Analysis of the zebrafish smoothened mutant reveals conserved and divergent functions of hedgehog activity. Development 128, 2385-2396. [DOI] [PubMed] [Google Scholar]
- Chen C.-H., Merriman A. F., Savage J., Willer J., Wahlig T., Katsanis N., Yin V. P. and Poss K. D. (2015). Transient laminin beta 1a Induction Defines the Wound Epidermis during Zebrafish Fin Regeneration. PLoS Genet. 11, e1005437 10.1371/journal.pgen.1005437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concordet J. P., Lewis K. E., Moore J. W., Goodrich L. V., Johnson R. L., Scott M. P. and Ingham P. W. (1996). Spatial regulation of a zebrafish patched homologue reflects the roles of sonic hedgehog and protein kinase A in neural tube and somite patterning. Development 122, 2835-2846. [DOI] [PubMed] [Google Scholar]
- DeLaurier A., Eames B. F., Blanco-Sánchez B., Peng G., He X., Swartz M. E., Ullmann B., Westerfield M. and Kimmel C. B. (2010). Zebrafish sp7:EGFP: a transgenic for studying otic vesicle formation, skeletogenesis, and bone regeneration. Genesis 48, 505-511. 10.1002/dvg.20639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertzer R., Müller F., Hadzhiev Y., Rathnam S., Fischer N., Rastegar S. and Strähle U. (2007). Cooperation of sonic hedgehog enhancers in midline expression. Dev. Biol. 301, 578-589. 10.1016/j.ydbio.2006.11.004 [DOI] [PubMed] [Google Scholar]
- Fuse N., Maiti T., Wang B., Porter J. A., Hall T. M. T., Leahy D. J. and Beachy P. A. (1999). Sonic hedgehog protein signals not as a hydrolytic enzyme but as an apparent ligand for patched. Proc. Natl. Acad. Sci. USA 96, 10992-10999. 10.1073/pnas.96.20.10992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Géraudie J. and Singer M. (1992). The fish fin regenerate. Monogr. Dev. Biol. 23, 62-72. [PubMed] [Google Scholar]
- Gustafsson M. G. L. (2000). Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 198, 82-87. 10.1046/j.1365-2818.2000.00710.x [DOI] [PubMed] [Google Scholar]
- Hammond C. L. and Schulte-Merker S. (2009). Two populations of endochondral osteoblasts with differential sensitivity to Hedgehog signalling. Development 136, 3991-4000. 10.1242/dev.042150 [DOI] [PubMed] [Google Scholar]
- Huang P., Xiong F., Megason S. G. and Schier A. F. (2012). Attenuation of Notch and Hedgehog signaling is required for fate specification in the spinal cord. PLoS Genet. 8, e1002762-e13 10.1371/journal.pgen.1002762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huycke T. R., Eames B. F. and Kimmel C. B. (2012). Hedgehog-dependent proliferation drives modular growth during morphogenesis of a dermal bone. Development 139, 2371-2380. 10.1242/dev.079806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P. W., Taylor A. M. and Nakano Y. (1991). Role of the Drosophila patched gene in positional signalling. Nature 353, 184-187. 10.1038/353184a0 [DOI] [PubMed] [Google Scholar]
- Jenkins D. (2009). Hedgehog signalling: emerging evidence for non-canonical pathways. Cell Signal. 21, 1023-1034. 10.1016/j.cellsig.2009.01.033 [DOI] [PubMed] [Google Scholar]
- Kelley L. C., Lohmer L. L., Hagedorn E. J. and Sherwood D. R. (2014). Traversing the basement membrane in vivo: a diversity of strategies. J. Cell Biol. 204, 291-302. 10.1083/jcb.201311112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf F., Hammond C., Chekuru A., Kurth T., Hans S., Weber C. W., Mahatma G., Fisher S., Brand M., Schulte-Merker S. et al. (2011). Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev. Cell 20, 713-724. 10.1016/j.devcel.2011.04.014 [DOI] [PubMed] [Google Scholar]
- Korzh S., Winata C. L., Zheng W., Yang S., Yin A., Ingham P., Korzh V. and Gong Z. (2011). The interaction of epithelial Ihha and mesenchymal Fgf10 in zebrafish esophageal and swimbladder development. Dev. Biol. 359, 262-276. 10.1016/j.ydbio.2011.08.024 [DOI] [PubMed] [Google Scholar]
- Koudijs M. J., den Broeder M. J., Groot E. and van Eeden F. J. M. (2008). Genetic analysis of the two zebrafish patched homologues identifies novel roles for the hedgehog signaling pathway. BMC Dev. Biol. 8, 15 10.1186/1471-213X-8-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforest L., Brown C. W., Poleo G., Géraudie J., Tada M., Ekker M. and Akimenko M. A. (1998). Involvement of the sonic hedgehog, patched 1 and bmp2 genes in patterning of the zebrafish dermal fin rays. Development 125, 4175-4184. [DOI] [PubMed] [Google Scholar]
- Lee Y., Hami D., De Val S., Kagermeier-Schenk B., Wills A. A., Black B. L., Weidinger G. and Poss K. D. (2009). Maintenance of blastemal proliferation by functionally diverse epidermis in regenerating zebrafish fins. Dev. Biol. 331, 270-280. 10.1016/j.ydbio.2009.05.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenton K., James A. W., Manu A., Brugmann S. A., Birker D., Nelson E. R., Leucht P., Helms J. A. and Longaker M. T. (2011). Indian hedgehog positively regulates calvarial ossification and modulates bone morphogenetic protein signaling. Genesis 49, 784-796. 10.1002/dvg.20768 [DOI] [PubMed] [Google Scholar]
- Lewis K. E. and Eisen J. S. (2001). Hedgehog signaling is required for primary motoneuron induction in zebrafish. Development 128, 3485-3495. [DOI] [PubMed] [Google Scholar]
- Long F. (2012). Building strong bones: molecular regulation of the osteoblast lineage. Nat. Rev. Mol. Cell Biol. 13, 27-38. 10.1038/nrm3254 [DOI] [PubMed] [Google Scholar]
- Loucks E. J., Schwend T. and Ahlgren S. C. (2007). Molecular changes associated with teratogen-induced cyclopia. Birth Defect Res. A Clin. Mol. Teratol. 79, 642-651. 10.1002/bdra.20387 [DOI] [PubMed] [Google Scholar]
- Marí-Beffa M., Palmqvist P., Marín-Girón F., Montes G. S. and Becerra J. (1999). Morphometric study of the regeneration of individual rays in teleost tail fins. J. Anat. 195, 393-405. 10.1017/S0021878299005464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marigo V., Davey R. A., Zuo Y., Cunningham J. M. and Tabin C. J. (1996). Biochemical evidence that patched is the Hedgehog receptor. Nature 384, 176-179. 10.1038/384176a0 [DOI] [PubMed] [Google Scholar]
- Mich J. K., Blaser H., Thomas N. A., Firestone A. J., Yelon D., Raz E. and Chen J. K. (2009). Germ cell migration in zebrafish is cyclopamine-sensitive but Smoothened-independent. Dev. Biol. 328, 342-354. 10.1016/j.ydbio.2009.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murciano C., Fernández T. D., Durán I., Maseda D., Ruiz-Sánchez J., Becerra J., Akimenko M. A. and Marí-Beffa M. (2002). Ray-interray interactions during fin regeneration of Danio rerio. Dev. Biol. 252, 214-224. 10.1006/dbio.2002.0848 [DOI] [PubMed] [Google Scholar]
- Murciano C., Pérez-Claros J., Smith A., Avaron F., Fernández T. D., Durán I., Ruiz-Sánchez J., García F., Becerra J., Akimenko M.-A. et al. (2007). Position dependence of hemiray morphogenesis during tail fin regeneration in Danio rerio. Dev. Biol. 312, 272-283. 10.1016/j.ydbio.2007.09.026 [DOI] [PubMed] [Google Scholar]
- Parkin C. A., Allen C. E. and Ingham P. W. (2009). Hedgehog signalling is required for cloacal development in the zebrafish embryo. Int. J. Dev. Biol. 53, 45-57. 10.1387/ijdb.082669cp [DOI] [PubMed] [Google Scholar]
- Poleo G., Brown C. W., Laforest L. and Akimenko M.-A. (2001). Cell proliferation and movement during early fin regeneration in zebrafish. Dev. Dyn. 221, 380-390. 10.1002/dvdy.1152 [DOI] [PubMed] [Google Scholar]
- Quint E., Smith A., Avaron F., Laforest L., Miles J., Gaffield W. and Akimenko M.-A. (2002). Bone patterning is altered in the regenerating zebrafish caudal fin after ectopic expression of sonic hedgehog and bmp2b or exposure to cyclopamine. Proc. Natl. Acad. Sci. USA 99, 8713-8718. 10.1073/pnas.122571799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle R. D., Johnson R. L., Laufer E. and Tabin C. (1993). Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 75, 1401-1416. 10.1016/0092-8674(93)90626-2 [DOI] [PubMed] [Google Scholar]
- Sanders T. A., Llagostera E. and Barna M. (2013). Specialized filopodia direct long-range transport of SHH during vertebrate tissue patterning. Nature 497, 628-632. 10.1038/nature12157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkumatava A., Fischer S., Müller F., Strähle U. and Neumann C. J. (2004). Sonic hedgehog, secreted by amacrine cells, acts as a short-range signal to direct differentiation and lamination in the zebrafish retina. Development 131, 3849-3858. 10.1242/dev.01247 [DOI] [PubMed] [Google Scholar]
- Singh S. P., Holdway J. E. and Poss K. D. (2012). Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev. Cell 22, 879-886. 10.1016/j.devcel.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa S., Afonso N., Bensimon-Brito A., Fonseca M., Simões M., Leon J., Roehl H., Cancela M. L. and Jacinto A. (2011). Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development 138, 3897-3905. 10.1242/dev.064717 [DOI] [PubMed] [Google Scholar]
- Stewart S. and Stankunas K. (2012). Limited dedifferentiation provides replacement tissue during zebrafish fin regeneration. Dev. Biol. 365, 339-349. 10.1016/j.ydbio.2012.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S., Gomez A. W., Armstrong B. E., Henner A. and Stankunas K. (2014). Sequential and opposing activities of Wnt and BMP coordinate zebrafish bone regeneration. Cell Rep. 6, 482-498. 10.1016/j.celrep.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone D. M., Hynes M., Armanini M., Swanson T. A., Gu Q., Johnson R. L., Scott M. P., Pennica D., Goddard A., Phillips H. et al. (1996). The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384, 129-134. 10.1038/384129a0 [DOI] [PubMed] [Google Scholar]
- Tornini V. A. and Poss K. D. (2014). Keeping at arm's length during regeneration. Dev. Cell 29, 139-145. 10.1016/j.devcel.2014.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S. and Johnson S. L. (2011). Fate restriction in the growing and regenerating zebrafish fin. Dev. Cell 20, 725-732. 10.1016/j.devcel.2011.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z. M., Amores A., Lewis K. E., Yan Y. L., Postlethwait J. H., Eisen J. S. and Westerfield M. (2001). Zebrafish smoothened functions in ventral neural tube specification and axon tract formation. Development 128, 3497-3509. [DOI] [PubMed] [Google Scholar]
- Wehner D., Cizelsky W., Vasudevaro M. D., Özhan G., Haase C., Kagermeier-Schenk B., Röder A., Dorsky R. I., Moro E., Argenton F. et al. (2014). Wnt/β-catenin signaling defines organizing centers that orchestrate growth and differentiation of the regenerating zebrafish caudal fin. Cell Rep. 6, 467-481. 10.1016/j.celrep.2013.12.036 [DOI] [PubMed] [Google Scholar]
- Westerfield M. (2007). The Zebrafish Book. Eugene, OR: University of Oregon Press. [Google Scholar]
- Wolff C., Roy S. and Ingham P. W. (2003). Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr. Biol. 13, 1169-1181. 10.1016/S0960-9822(03)00461-5 [DOI] [PubMed] [Google Scholar]
- Zhang X., Harrington N., Moraes R. C., Wu M.-F., Hilsenbeck S. G. and Lewis M. T. (2009). Cyclopamine inhibition of human breast cancer cell growth independent of Smoothened (Smo). Breast Cancer Res. Treat. 115, 505-521. 10.1007/s10549-008-0093-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Jeradi S., Strähle U. and Akimenko M.-A. (2012). Laser ablation of the sonic hedgehog-a-expressing cells during fin regeneration affects ray branching morphogenesis. Dev. Biol. 365, 424-433. 10.1016/j.ydbio.2012.03.008 [DOI] [PubMed] [Google Scholar]