Abstract
Animals change developmental fates in response to external cues. In the nematode Caenorhabditis elegans, unfavorable environmental conditions induce a state of diapause known as dauer by inhibiting the conserved DAF-2 insulin-like signaling (ILS) pathway through incompletely understood mechanisms. We have previously established a role for the C. elegans dosage compensation protein DPY-21 in the control of dauer arrest and DAF-2 ILS. Here, we show that the histone H4 lysine 20 methyltransferase SET-4, which also influences dosage compensation, promotes dauer arrest in part by repressing the X-linked ins-9 gene, which encodes a new agonist insulin-like peptide (ILP) expressed specifically in the paired ASI sensory neurons that are required for dauer bypass. ins-9 repression in dauer-constitutive mutants requires DPY-21, SET-4 and the FoxO transcription factor DAF-16, which is the main target of DAF-2 ILS. By contrast, autosomal genes encoding major agonist ILPs that promote reproductive development are not repressed by DPY-21, SET-4 or DAF-16/FoxO. Our results implicate SET-4 as a sensory rheostat that reinforces developmental fates in response to environmental cues by modulating autocrine and paracrine DAF-2 ILS.
KEY WORDS: C. elegans, Dauer, Dosage compensation, H4K20, Insulin-like peptides, FoxO
Summary: The C. elegans histone methyltransferase SET-4 acts to link environmental signals to diapause through regulation of the X-linked insulin-like peptide gene ins-9 in sensory neurons.
INTRODUCTION
To maintain evolutionary fitness, organisms must react appropriately to environmental cues. The free-living nematode Caenorhabditis elegans has evolved a developmental strategy to optimize survival in changing environments. Under replete conditions, larvae progress through four stages (L1-L4) to become reproductive adults. In adverse conditions such as overcrowding, heat or food scarcity, larvae arrest in an alternative stage known as dauer. Adapted for survival in harsh environments, dauers are morphologically, metabolically and behaviorally distinct from reproductive L3 larvae. Improvement of ambient conditions induces dauer exit and resumption of reproductive development (Fielenbach and Antebi, 2008; Riddle, 1988). The dominant environmental cue that influences dauer arrest is a constitutively elaborated pheromone that indicates population density (Butcher et al., 2008; Golden and Riddle, 1982).
C. elegans dauer arrest has served as a useful paradigm for understanding the molecular basis of developmental plasticity. Genetic analysis has defined four conserved signaling pathways that promote reproductive development in favorable environments. The DAF-11 transmembrane guanylyl cyclase acts in chemosensory neurons to regulate dauer arrest through the cyclic nucleotide-gated channel subunits TAX-2 and TAX-4. Downstream of DAF-11, DAF-2 insulin receptor (InsR)- and DAF-7 transforming growth factor-β (TGFβ)-like pathways act in parallel to promote reproductive development by inhibiting the activities of the FoxO transcription factor DAF-16 and the SMAD transcription factor DAF-3, respectively. Distal to DAF-16/FoxO and DAF-3/SMAD, bile-acid-like steroid hormones known as dafachronic acids (DAs) promote reproductive development by regulating the activity of the conserved nuclear receptor DAF-12 (Fielenbach and Antebi, 2008).
Although genetic analysis has identified how components of the DAF-11, DAF-2/InsR, DAF-7/TGFβ and DAF-12 pathways interact to promote reproductive development in favorable conditions (Gottlieb and Ruvkun, 1994; Riddle et al., 1981; Thomas et al., 1993; Vowels and Thomas, 1992), the molecular nature of the upstream events that couple external cues to the activities of these pathways remains poorly understood. Laser ablation experiments demonstrated that the amphid sensory neurons are required for induction of dauer arrest by pheromone (Schackwitz et al., 1996; Vowels and Thomas, 1994). Indeed, the dauer-inhibitory ASI sensory neurons (Bargmann and Horvitz, 1991) are specific sites of expression of three insulin-like peptides (ILPs) that promote reproductive development through DAF-2/InsR (INS-4, INS-6 and DAF-28) (Chen and Baugh, 2014; Cornils et al., 2011; Hung et al., 2014; Li et al., 2003), as well as the DAF-7 TGFβ-like ligand that promotes reproductive development (Ren et al., 1996; Schackwitz et al., 1996). Furthermore, crude dauer pheromone reduces the expression of DAF-28 and DAF-7 in ASI (Li et al., 2003; Schackwitz et al., 1996), suggesting that pheromone induces dauer arrest at least in part by reducing the expression of agonist ligands in sensory neurons that regulate DAF-2/InsR and TGFβ-like signaling. How pheromone represses these ligands remains a mystery.
We have previously reported an unforeseen role for the C. elegans dosage compensation protein DPY-21 in promoting dauer arrest through inhibition of the DAF-2/InsR pathway (Dumas et al., 2013). DPY-21 is a component of the condensin-like dosage compensation complex (DCC) that equalizes X-linked gene expression between males and hermaphrodites by binding to both hermaphrodite X chromosomes during embryogenesis and repressing gene expression approximately twofold (Meyer, 2010; Yonker and Meyer, 2003). Here, we show that the conserved histone H4 lysine 20 (H4K20) methyltransferase SET-4, which also influences dosage compensation (Kramer et al., 2015; Vielle et al., 2012; Wells et al., 2012), promotes dauer arrest in a sex-specific manner by synergizing with DAF-16/FoxO to repress ins-9, an X-linked gene that encodes an ILP expressed specifically in ASI neurons (Chen and Baugh, 2014; Pierce et al., 2001). These findings reveal a sexually dimorphic role for regulators of histone H4K20 methylation in broadening the dynamic range of sensory responses to environmental cues that control developmental plasticity.
RESULTS
SET-4 acts through DAF-2 ILS to promote dauer arrest in a sex-specific manner
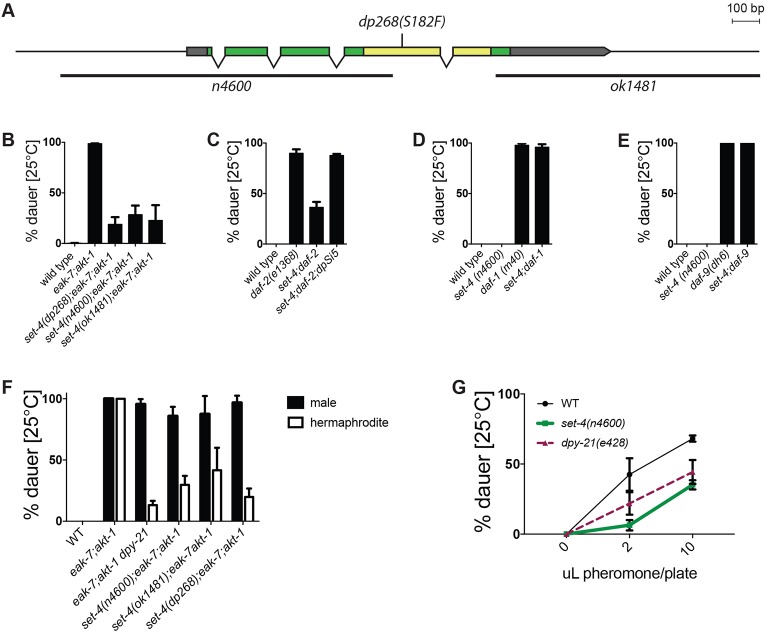
DAF-2/InsR promotes reproductive development by activating a conserved phosphoinositide 3-kinase (PI3K)/Akt pathway to inhibit DAF-16/FoxO (Murphy and Hu, 2013). The conserved protein EAK-7 acts in parallel to AKT-1 to inhibit nuclear DAF-16/FoxO activity (Alam et al., 2010). In contrast to eak-7 and akt-1 single mutants, which develop reproductively, eak-7;akt-1 double mutant animals arrest as dauers in a DAF-16/FoxO-dependent manner (Alam et al., 2010). To identify new DAF-16/FoxO regulators, we performed a forward genetic screen for suppressors of the eak-7;akt-1 dauer-constitutive phenotype (seak). The first seak mutants characterized harbored loss-of-function mutations in the dosage compensation gene dpy-21 (Dumas et al., 2013). dpy-21 encodes a conserved component of the condensin-like dosage compensation complex (DCC) that binds to X chromosomes and represses X-linked gene expression (Meyer, 2010; Yonker and Meyer, 2003). One seak mutant strain contained a point mutation in set-4, which encodes a histone H4K20 methyltransferase homolog that influences dosage compensation (Vielle et al., 2012; Wells et al., 2012). set-4(dp268) is predicted to change the conserved SET domain catalytic residue serine 182 (Southall et al., 2014) to phenylalanine (Fig. 1A, Fig. S1A). In light of our findings on dpy-21 (Dumas et al., 2013), we tested the possibility that set-4(dp268) was the causative seak mutation in this strain. After outcrossing removed all but two closely linked single nucleotide variants, set-4(dp268) suppressed dauer arrest to a similar extent to two independently derived set-4 deletions, n4600 (Andersen and Horvitz, 2007) and ok1481 (Fig. 1B). Furthermore, an integrated single-copy HA::set-4 transgene rescued dauer arrest in set-4(n4600) animals (Fig. 1C and Fig. S1B). Therefore, SET-4 promotes dauer arrest.
Fig. 1.
SET-4 promotes dauer arrest. (A) Schematic of the set-4 locus and three mutant alleles. Exons are indicated as boxes, separated by introns. Gray, green and yellow denote untranslated regions, coding sequence and SET domain coding sequence, respectively. Deletions are indicated by black bars. (B) set-4 mutations suppress the dauer-constitutive phenotype of eak-7;akt-1 mutants [n (left to right)=236, 627, 389, 521, 638]. (C) set-4(n4600) suppresses the dauer-constitutive phenotype of daf-2(e1368) mutants and is rescued by the single-copy set-4 transgene dpSi5 (n=913, 1668, 1568, 1785). (D,E) set-4 is dispensable for dauer arrest in (D) daf-1(m40) (n=2215, 2189, 1816, 1899) and (E) daf-9(dh6) mutants (n=1753, 1561, 2188, 2486). (F) set-4 and dpy-21 mutations suppress dauer arrest in XX hermaphrodites but not in XO males (n=567, 1330, 579, 1003, 1012, 700). (G) set-4 and dpy-21 mutations attenuate the response of wild-type animals to dauer pheromone [n (0 μl, 2 μl, 10 μl): wild type=542, 461, 544; set-4=413, 246, 387; dpy-21=334, 220, 275]. set-4 versus wild type: P<0.01 by two-way ANOVA.
DPY-21 enhances dauer arrest by activating DAF-16/FoxO, indicating that it acts in the DAF-2/InsR pathway to regulate dauer formation (Dumas et al., 2013). To determine whether SET-4 functions in the DAF-2/InsR pathway, we tested the effect of set-4 mutation on dauer-constitutive phenotypes caused by mutations in the daf-2/InsR, daf-7/TGFβ and daf-12 pathways. set-4 mutation suppressed the dauer-constitutive phenotypes of daf-2(e1368) mutants (Fig. 1C) as well as akt-1(ok525) and eak-7(tm3188) single mutants, which develop reproductively at 25°C but arrest as dauers at 27°C (Fig. S1C,D) (Ailion and Thomas, 2003; Alam et al., 2010; Hu et al., 2006). By contrast, set-4 mutation had no effect on the dauer-constitutive phenotypes caused by mutations in daf-1, which encodes a type 1 TGFβ receptor homolog (Georgi et al., 1990), or daf-8, which encodes a SMAD homolog (Park et al., 2010) (Fig. 1D and Fig. S1E). Similarly, set-4 mutation did not suppress dauer arrest in animals harboring mutations in daf-9 or daf-36, which encode DA biosynthesis pathway components (Fig. 1E and Fig. S1F) (Gerisch et al., 2001; Jia et al., 2002; Rottiers et al., 2006). Therefore, SET-4 acts specifically in the DAF-2/InsR pathway to promote dauer arrest.
As previous reports link H4K20 methylation status to dosage compensation (Vielle et al., 2012; Wells et al., 2012) and DPY-21 promotes dauer arrest through dosage compensation (Dumas et al., 2013), we hypothesized that SET-4 may also regulate dauer arrest through dosage compensation. To test this, we determined the effect of set-4 mutation on the dauer-constitutive phenotype of eak-7;akt-1 double mutant hermaphrodites and males. If SET-4 promotes dauer arrest through the same mechanism as dosage compensation, then set-4 mutation should suppress dauer in hermaphrodites but not in males, as the DCC is inactive in males (Meyer, 2010). dpy-21 and set-4 mutations suppressed the dauer-constitutive phenotype of eak-7;akt-1 hermaphrodites but did not affect dauer arrest in males (Fig. 1F). Therefore, SET-4 may act through dosage compensation to control dauer arrest. We verified the role of SET-4 in dosage compensation by showing that set-4 mutation suppressed lethality in xol-1 sex-1 mutant males, which die due to inappropriate activation of dosage compensation (Dawes et al., 1999) (Fig. S1G).
In order to determine whether SET-4 plays a role in regulating dauer entry in wild-type animals in response to physiologic stimuli, we tested the ability of dauer pheromone to induce dauer arrest in wild-type and set-4 mutant animals. Mutation of either set-4 or dpy-21 decreased the sensitivity of wild-type animals to pheromone (Fig. 1G). Therefore, SET-4 and DPY-21 promote dauer arrest in wild-type animals in response to increases in population density.
SET-4 is a H4K20 methyltransferase
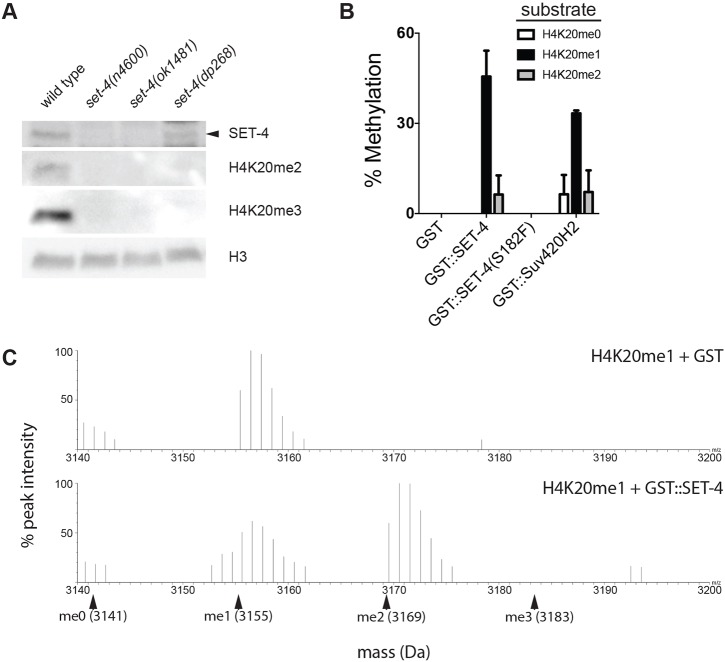
The mammalian SET-4 ortholog SUV420H2 is a H4K20 methyltransferase (Schotta et al., 2004), and C. elegans SET-4 promotes H4K20 trimethylation (Vielle et al., 2012; Webster et al., 2013; Wells et al., 2012). We confirmed the requirement of SET-4 for H4K20 di- and trimethylation in vivo (Fig. 2A). Immunoblots showed no detectable SET-4 protein in set-4(n4600) and set-4(ok1481) backgrounds, consistent with these being strong loss-of-function alleles. SET-4 protein levels in set-4(dp268) are comparable with wild type (Fig. 2A). H4K20me2 and H4K20me3 levels are undetectable in all three set-4 mutant backgrounds (Fig. 2A), suggesting that the S182F substitution in the SET domain abrogates catalytic activity. To test this possibility directly, we purified recombinant wild-type and mutant GST-SET-4 fusion proteins and tested their ability to methylate modified H4 peptides (H4K20me0, H4K20me1 and H4K20me2) in vitro. Mass spectrometry analysis revealed that both wild-type GST-SET-4 and GST-SUV420H2 were capable of converting H4K20me1 to H4K20me2 (Fig. 2B,C). Consistent with in vitro experiments using human SET-4 orthologs SUV420H1 and SUV420H2 (Southall et al., 2014; Wu et al., 2013), methylation was not detected with unmethylated or dimethylated substrates, nor were trimethylated products detected in any assay (Fig. 2B,C). It is possible that an enzyme distinct from SET-4 catalyzes H4K20 trimethylation in vivo. Alternatively, conversion of H4K20me2 to H4K20me3 by SET-4 in vivo may require a co-factor that is absent from our in vitro reactions.
Fig. 2.
SET-4 is a histone H4K20 methyltransferase. (A) SET-4 promotes H4K20 methylation in vivo. Anti-SET-4, H4K20me2, H4K20me3 and H3 immunoblots of lysates from wild-type and set-4 mutant animals are shown. SET-4 protein is indicated by the arrowhead. Images are representative of four biological replicates. (B) Recombinant GST-SET-4 fusion protein methylates H4K20me1 in vitro. Percentage methylation of H4K20 peptide substrates by GST proteins fused to wild-type SET-4, mutant SET-4(S182F) or human SET-4 ortholog SUV420H2 is shown. Data represent mean values from three biological replicates. (C) MALDI spectra illustrating conversion of H4K20me1 to H4K20me2 by GST-SET-4. Monoisotopic masses (protonated) of peptide substrates are indicated with arrowheads. Spectra are representative of three independent experiments.
GST-SET-4(S182F) did not methylate H4K20me1, indicating that the S182F mutation abolishes catalytic activity (Fig. S2). Given that set-4(dp268) suppresses dauer to a similar extent to the two deletion alleles (Fig. 1B), these data are consistent with SET-4 influencing dauer arrest through its conserved role in methylating H4K20.
SET-4 acts in neurons to promote dauer arrest
To determine where SET-4 is expressed, we generated strains expressing reporter genes under the control of set-4 regulatory elements. Because two independent C-terminal SET-4::GFP translational fusions failed to rescue dauer arrest in set-4 mutants, we generated strains expressing a set-4p::GFP promoter fusion to determine the spatiotemporal activity of the set-4 promoter. set-4p::GFP transgenic animals expressed GFP broadly in embryos (Fig. S3). Post-embryonically, we detected fluorescence predominantly in the head and tail regions of the animal, in a pattern consistent with neuronal expression. To confirm this, we established a transgenic strain that co-expressed set-4p::GFP and mCherry driven by the pan-neuronal rab-3 promoter (Nonet et al., 1997). At all developmental stages interrogated, we observed colocalization of green and red fluorescence (Fig. 3A), consistent with somatic set-4p::GFP expression being predominantly neuronal. We did not observe significant GFP expression in intestine, body wall muscle or hypodermis.
Fig. 3.
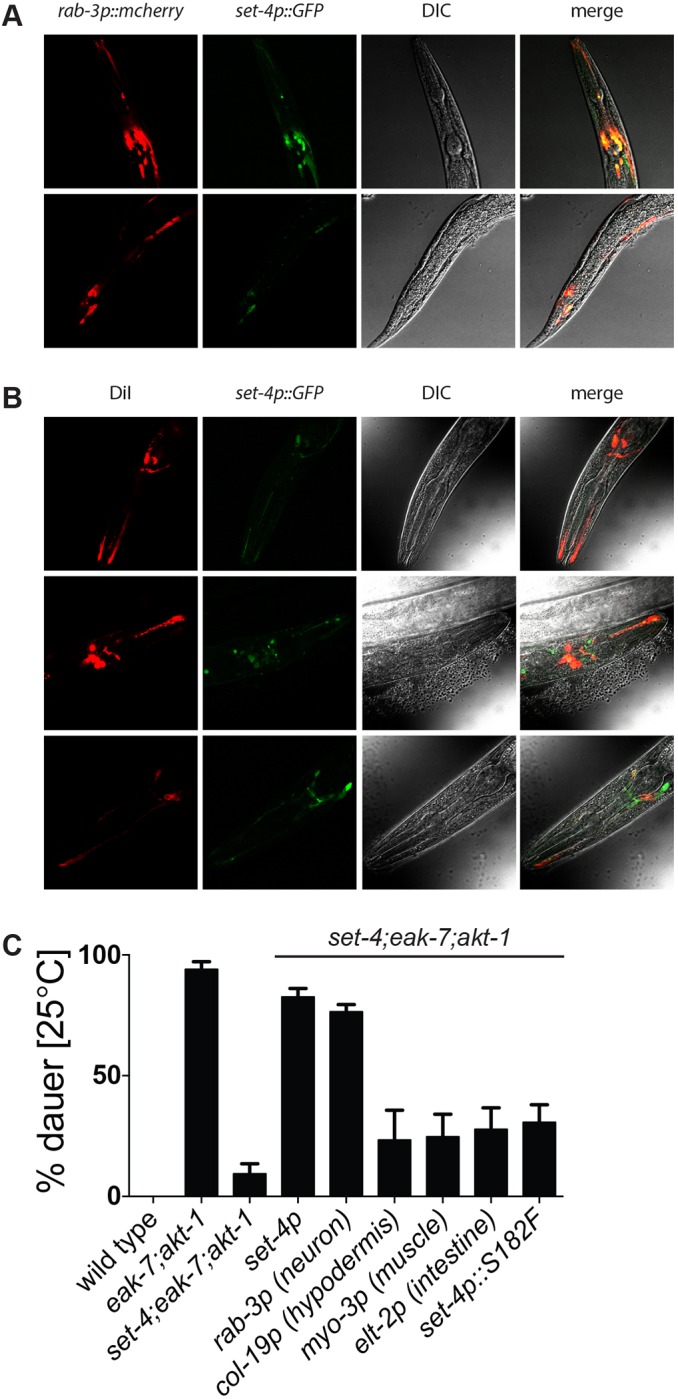
set-4 acts in the nervous system to promote dauer arrest. (A) Representative photomicrographs showing colocalization of GFP and mCherry in animals co-expressing set-4p::gfp and the pan-neuronal reporter rab-3p::mcherry (n=15). (B) Representative photomicrographs showing colocalization of red and green fluorescence in amphid neurons of set-4p::gfp transgenic animals exposed to DiI (n=21). (C) Rescue of dauer arrest in set-4;eak-7;akt-1 mutants by set-4 transgenes driven by native set-4 or tissue-specific promoters (n=2328, 2143, 2058, 662, 811, 705, 936, 516, 541).
As the amphid sensory neurons play a crucial role in regulating dauer arrest (Bargmann and Horvitz, 1991; Schackwitz et al., 1996; Vowels and Thomas, 1994), we interrogated them for expression of set-4p::GFP. Amphid neurons possess ciliary dendrites that are in direct contact with the environment and can be labeled with the lipophilic dye DiI (Starich et al., 1995). The extent of colocalization of green and red fluorescence in set-4p::GFP transgenic animals exposed to DiI (Fig. 3B) reveals that the set-4 promoter is active in amphid sensory neurons as well as in other cells.
To test whether neuronal expression of set-4 is functionally important for dauer arrest, we generated tissue-specific set-4 transgenes and tested them for the ability to rescue dauer arrest in set-4 mutants. The neuronal rab-3p::set-4 transgene rescued dauer formation to a similar extent to a set-4 transgene driven by its native promoter (Fig. 3C). By contrast, intestine-, hypodermis- and muscle-specific set-4 transgenes did not rescue dauer arrest to a greater extent than a transgene expressing the set-4(dp268) mutant. Taken together, these data indicate that SET-4 functions in the nervous system to promote dauer arrest.
Transcriptome-wide influences of DPY-21 and SET-4 on dauer regulation
We previously showed that the DCC component DPY-21 promotes DAF-16/FoxO activity (Dumas et al., 2013). To gain broader insight into how DPY-21 and SET-4 control dauer arrest, we performed whole-transcriptome profiling to compare genome-wide regulatory influences (henceforth referred to as the ‘regulome’) of DPY-21 and SET-4 to those of the key transcription factors controlling dauer arrest in eak-7;akt-1 animals, DAF-16/FoxO and the nuclear receptor DAF-12 (Alam et al., 2010). We identified genes differentially expressed between wild-type and eak-7;akt-1 double mutant animals [fold change ≥1.5 and false discovery rate (FDR) <0.05]. We then compared the transcriptomes of eak-7;akt-1 double mutants with those of eak-7;akt-1 animals harboring mutations in dpy-21, set-4, daf-16/FoxO or daf-12, and identified genes that are differentially expressed in the opposite direction as in wild-type relative to eak-7;akt-1 (Table S1). Regulomes were validated by comparison with published data where possible (see below).
We defined the SET-4 dauer regulome by identifying 333 genes common to set-4(n4600) and set-4(dp268) regulomes (Table S1). Comparison of this gene set with SET-4-regulated genes identified in wild-type embryos and L3 larvae (Kramer et al., 2015) revealed minimal overlap [one gene (MTCE.34) among 94 regulated by SET-4 in embryos, and one gene (B0511.11) among 18 SET-4-regulated genes in L3 larvae]. This lack of concordance could be due to differences in genetic background (eak-7;akt-1 versus wild-type), developmental stage (early L2 larvae versus embryos/L3 larvae) and/or ambient temperature (25°C versus 20°C).
A similar analysis with eak-7;akt-1 dpy-21 mutants revealed 2431 genes that comprise the DPY-21 dauer regulome (Table S1). To validate the DPY-21 regulome, we found significant overlap between the set of 700 X-linked genes differentially expressed in eak-7;akt-1 dpy-21 versus eak-7;akt-1 animals with the 374 X-linked genes subject to dosage compensation in embryos (Jans et al., 2009) (119 genes; Fig. S4A and Table S2; P=2.1e−26). Three hundred and eight of the 333 genes that make up the SET-4 dauer regulome (92.5%) are also part of the DPY-21 dauer regulome (Fig. 4A and Table S1), suggesting that a functional relationship between DPY-21 and SET-4 may exist in post-embryonic dauer regulation.
Fig. 4.
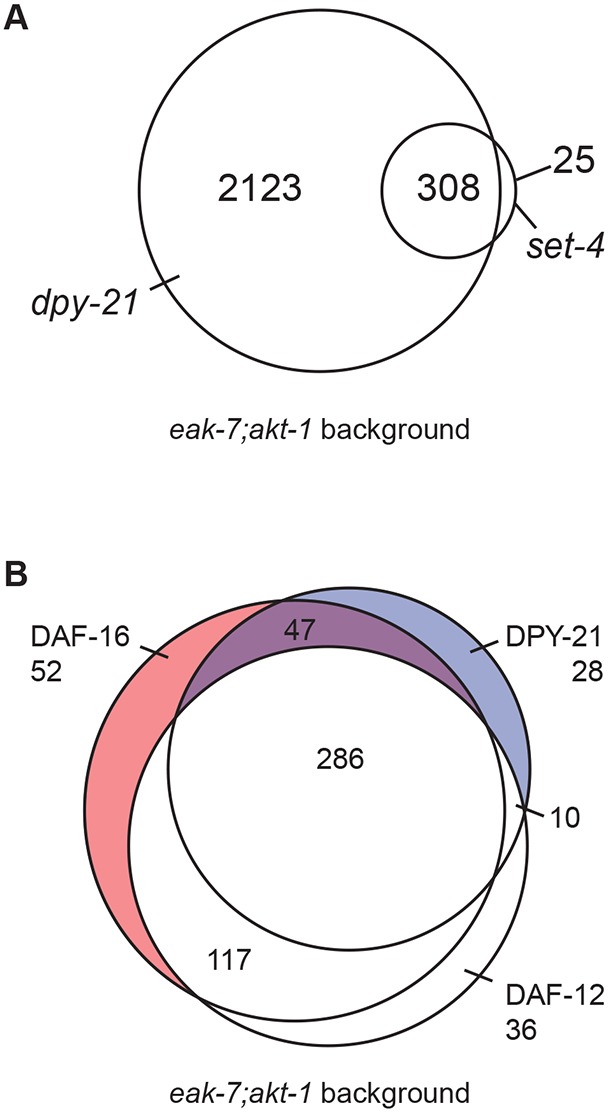
Whole-transcriptome profiling defines genes coordinately regulated by DPY-21/SET-4 and DAF-16/FoxO. (A) Venn diagram of genes regulated by SET-4 and DPY-21. Three-hundred and eight genes are coordinately regulated by SET-4 and DPY-21. (B) Venn diagram of X-linked genes regulated by DPY-21, DAF-16/FoxO and DAF-12. Forty-seven genes coordinately regulated by DPY-21 and DAF-16/FoxO but not by DAF-12 are depicted in purple and listed in Table S5. Data represent the aggregate of five biological replicate samples, each from thousands of progeny with no fewer than 200 animals per sample.
The DAF-16/FoxO regulome consists of 2957 genes (Table S1). This gene set overlapped significantly with both the 469-gene young adult DAF-16/FoxO regulome (Chen et al., 2015) (203 common genes; Fig. S4B and Table S3, P=3.2e−115) as well as the 1116-gene dauer regulome defined using daf-7 TGFβ-like pathway mutants (Liu et al., 2004) (522 common genes; Fig. S4C and Table S4, P<1e−100). Furthermore, 65 of the 132 genes regulated by the DAF-7 TGFβ-like pathway that contained at least one upstream DAF-16/FoxO-binding motif (Liu et al., 2004) were part of the DAF-16/FoxO regulome [Fig. S4D and Table S4 (bold text), P=6.5e−42]. Most genes in the DAF-16/FoxO regulome (2282 of 2957 genes; 77.2%) are also regulated by DAF-12 (Fig. 4B and Table S1). Over two-thirds of genes that comprise both DAF-16/FoxO (2041 of 2957 genes; 69.0%; Table S1) and DAF-12 (1804 of 2556 genes; 70.6%; Table S1) regulomes are concordantly regulated by DPY-21.
The X-linked ins-9 gene is repressed by DPY-21, SET-4 and DAF-16/FoxO
Based on genetic epistasis experiments (Dumas et al., 2013) (Fig. 1), we hypothesized that DPY-21 and SET-4 influence DAF-16/FoxO activity through repression of X-linked genes. Moreover, in light of the neuronal site of action of SET-4 (Fig. 3C) and its expression in amphid sensory neurons (Fig. 3B), we speculated that key dauer regulatory genes subject to dosage compensation might function in a signaling capacity in the nervous system, upstream of DAF-12. Therefore, we examined the set of X-linked genes common to DPY-21 and DAF-16/FoxO regulomes that were not regulated by DAF-12, which acts downstream in the dauer regulatory cascade (Fielenbach and Antebi, 2008; Schaedel et al., 2012). This filtering strategy defined a set of 47 X-linked genes coordinately regulated by DPY-21 and DAF-16/FoxO but not influenced by daf-12 mutation (Fig. 4B; Table S5).
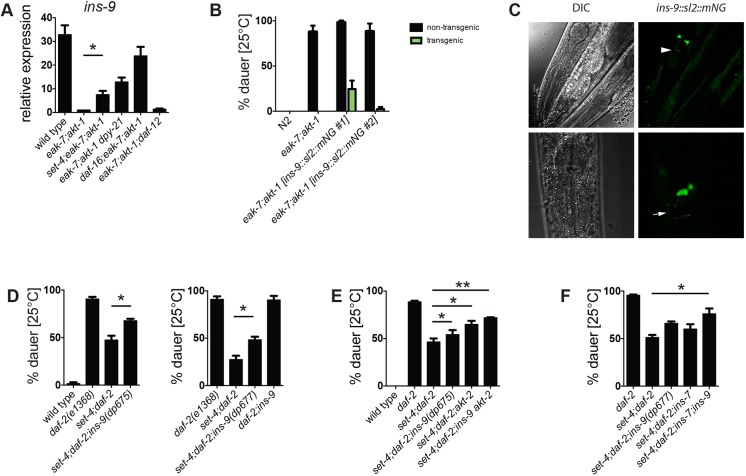
Among these X-linked genes was ins-9, which encodes one of 40 C. elegans ILPs (Pierce et al., 2001). Transcriptome profiling revealed that ins-9 is silenced in eak-7;akt-1 animals in a manner that requires both daf-16/FoxO and dpy-21 (Table S2). We verified this by qPCR; ins-9 expression was reduced more than 30-fold in eak-7;akt-1 double mutants compared with wild-type animals (Fig. 5A). Full repression required DAF-16/FoxO as well as DPY-21 and SET-4, but was independent of daf-12. Notably, a daf-16/FoxO-null mutation did not restore ins-9 transcript levels to wild-type levels. Furthermore, mutation of either dpy-21 or set-4 increased ins-9 expression by substantially greater than twofold (Fig. 5A; 7.5-fold increase in set-4;eak-7;akt-1 versus eak-7;akt-1; 13.5-fold increase in eak-7;akt-1 dpy-21 versus eak-7;akt-1). Taken together, these observations suggest that DAF-16/FoxO and DPY-21/SET-4 act synergistically to repress ins-9.
Fig. 5.
SET-4 promotes dauer arrest by repressing the X-linked ins-9 gene. (A) ins-9 is repressed by DPY-21, SET-4 and DAF-16/FoxO but not DAF-12. Results are the mean±s.e.m. of five experiments. (B) ins-9 overexpression suppresses dauer arrest in eak-7;akt-1 animals. Non-transgenic (NT) and transgenic (T) progeny of transgenic hermaphrodites are shown. n=1200, 1522, 718/156 (NT/T), 702/210 (NT/T). (C) An ins-9::SL2::mNG transgene is expressed specifically in ASI sensory neurons. The arrowhead indicates the top of the nerve ring; the arrow indicates the ASI axon. Representative images are shown (n=20). (D) Two ins-9 nonsense mutations partially rescue dauer arrest in set-4;daf-2 mutants. n=957, 1205, 1398 (left); n=892, 1028, 897 (right). (E) ins-9 and akt-2 mutations rescue dauer arrest in an additive fashion in set-4;daf-2 mutants (n=949, 1281, 1334, 1524, 1333, 1478). (F) ins-9 and ins-7 mutations rescue dauer arrest in an additive fashion in set-4;daf-2 mutants (n=1396, 1251, 754, 827, 1327). *P<0.05, **P<0.01.
ins-9 overexpression phenocopies dauer suppression caused by dpy-21 or set-4 mutation
INS-9 was an attractive candidate regulator of dauer arrest and DAF-2 ILS, as multiple ILPs have been shown to control dauer arrest through DAF-2/InsR (Cornils et al., 2011; Fernandes de Abreu et al., 2014; Hung et al., 2014; Li et al., 2003; Murphy et al., 2003; Pierce et al., 2001). If derepression of ins-9 contributes to suppression of eak-7;akt-1 dauer arrest in dosage compensation mutants, then ins-9 overexpression should phenocopy dauer suppression caused by set-4 or dpy-21 mutation. We tested this by establishing transgenic strains harboring a polycistronic construct that permitted expression of both ins-9 and mNeonGreen (Shaner et al., 2013) fused to an SL2 leader sequence (Blumenthal, 2005) under the control of native ins-9 5′ and 3′ regulatory elements (ins-9::SL2::mNG). In two independent transgenic lines, ins-9 overexpression (indicated by mNeonGreen detection) suppressed the dauer-constitutive phenotype of eak-7;akt-1 double mutants (Fig. 5B). Therefore, ins-9 overexpression recapitulated the phenotype caused by set-4 and dpy-21 mutations (Dumas et al., 2013) (Fig. 1B). These data are consistent with INS-9 acting as an agonist DAF-2/InsR ligand.
ins-9 is expressed specifically in a single pair of amphid neurons
Previous studies using reporters driven by the ins-9 promoter suggested that ins-9 is expressed in the ASI and ASJ amphid neurons, as well as in additional tissues (Chen and Baugh, 2014; Pierce et al., 2001; Ritter et al., 2013). By contrast, in transgenic L2 larvae expressing ins-9::SL2::mNG, we consistently observed green fluorescence solely in one pair of sensory neurons. In animals in which neuronal morphology was discernable, we identified the fluorescent cells as the ASI amphid neurons (Fig. 5C). We did not observe fluorescence in more than one pair of amphid neurons in any animal, nor did we detect fluorescence in other neurons or tissues. As ins-9::SL2::mNG contains genomic elements from the ins-9 locus that are missing from other reporters in the literature (Chen and Baugh, 2014; Pierce et al., 2001; Ritter et al., 2013), these observed patterns of expression are likely to be physiologically relevant.
ins-9 and akt-2 are required for suppression of dauer arrest by set-4 mutation
To determine the extent to which ins-9 derepression contributes to dauer suppression in set-4 mutants, we tested the ability of set-4 to suppress the dauer-constitutive phenotype of daf-2/InsR mutants in wild-type and ins-9 loss-of-function backgrounds. We generated strong loss-of-function ins-9 alleles using CRISPR/Cas9 genome editing (Paix et al., 2015). Two probable null alleles, dp675 and dp677, have nonsense mutations in the F-peptide region of ins-9 that lies N-terminal to the functional B and A peptides (Pierce et al., 2001) (Fig. S5). Although neither allele induced dauer arrest in a wild-type background, both ins-9(dp675) and ins-9(dp677) partially rescued dauer arrest in set-4;daf-2 double mutants (Fig. 5D), indicating that dauer suppression caused by set-4 mutation is due in part to de-repression of ins-9.
We previously showed that the X-linked gene akt-2 is required for dauer suppression caused by dpy-21 mutation (Dumas et al., 2013). We verified that akt-2 transcripts increase two-fold in eak-7;akt-1 double mutants when set-4 or dpy-21 is mutated (Fig. S6A). Similar to ins-9 mutation, akt-2 mutation also partially rescued dauer arrest in animals lacking set-4 (Fig. 5E), and the phenotypic effects of ins-9 and akt-2 mutations on dauer arrest may be additive (Fig. 5E). However, set-4;daf-2;ins-9 akt-2 compound mutant animals still did not undergo dauer arrest to the same extent as daf-2 single mutant animals (Fig. 5E), indicating that regulation of additional genes may contribute to dauer arrest.
The autosomal ins-7 gene contributes to suppression of dauer arrest by set-4 mutation
As the only X-linked ins gene, ins-9 is the sole ins gene subject to direct regulation by dosage compensation or other X-chromosome-specific mechanisms of gene regulation. However, it is conceivable that other ins genes could contribute to dauer regulation through indirect effects on their expression. Genes encoding three agonist ILPs, INS-4, INS-6 and DAF-28, are expressed in the ASI sensory neurons and have established roles in inhibiting dauer arrest and promoting reproductive development (Cornils et al., 2011; Hung et al., 2014; Li et al., 2003). To determine whether regulation of ins-4, ins-6 and/or daf-28 contributes to dauer suppression in this context, we measured ins-4, ins-6 and daf-28 transcript levels in wild-type, eak-7;akt-1 double mutant and eak-7;akt-1 triple mutants with reduced DPY-21 or SET-4 activity. None of these genes was repressed in eak-7;akt-1 double mutants compared with wild-type animals, nor did loss of set-4 or dpy-21 cause significant increases in their expression (Fig. S6B-D). Therefore, neither DPY-21 nor SET-4 influences dauer arrest through regulation of ins-4, ins-6 and daf-28 expression.
To determine whether other ILPs could contribute to dauer regulation by DPY-21 or SET-4, we searched the set of genes common to DAF-16/FoxO and DPY-21 regulomes for ins genes. Seven ins genes are concordantly regulated by DAF-16/FoxO and DPY-21 based on FPKM counts from whole-transcriptome data (Table S6). Two genes encoding putative antagonist ILPs, ins-20 and ins-11 (Fernandes de Abreu et al., 2014), are repressed by DAF-16/FoxO and DPY-21; however, an increase in their expression in dpy-21 mutants would be expected to enhance, rather than suppress, dauer arrest. Similarly, two ins genes encoding agonist ILPs, ins-33 and ins-35 (Fernandes de Abreu et al., 2014; Michaelson et al., 2010), are induced by DAF-16/FoxO and DPY-21; a decrease in their expression in dpy-21 mutants would also be expected to enhance dauer arrest if changes in their expression were functionally important in dauer regulation. DAF-16/FoxO and DPY-21 induce the expression of two ins genes of unknown function, ins-16 and ins-29 (Fernandes de Abreu et al., 2014). Finally, ins-7, which encodes an agonist ILP (Murphy et al., 2007, 2003), is repressed by DAF-16/FoxO and DPY-21 (Murphy et al., 2007, 2003) (Table S6). As ins-7 and ins-9 both encode agonist ILPs, are concordantly regulated by DAF-16/FoxO and DPY-21, and have been shown to influence dauer arrest (Murphy et al., 2007, 2003) (Fig. 5C-E), we tested ins-7 for a role in promoting reproductive development in dpy-21 and set-4 mutants. We verified ins-7 repression by DAF-16/FoxO, SET-4 and DPY-21 using qPCR (Fig. S6E). The ins-7(tm1907) deletion allele partially rescued dauer in set-4;daf-2 animals and may have an additive effect with ins-9 mutation on dauer suppression (Fig. 5F). Thus, SET-4 may influence dauer arrest through both the direct repression of X-linked genes such as ins-9 and akt-2 and perhaps the indirect regulation of autosomal dauer regulatory genes such as ins-7.
DISCUSSION
Although much is known about the conserved signaling pathways that control C. elegans dauer arrest, how these pathways are regulated by upstream inputs is poorly understood. In the present study, we have established a framework for understanding how DPY-21 and SET-4 promote dauer arrest in the context of reduced DAF-2 ILS. Specifically, we have discovered that the conserved H4K20 methyltransferase SET-4 acts in the nervous system to promote dauer arrest. It does so, in part, by synergizing with DAF-16/FoxO to repress the X-linked insulin-like peptide gene ins-9. We hypothesize that SET-4 and DPY-21 act similarly to repress ins-9 and akt-2 directly, thus attenuating DAF-2 ILS and promoting dauer arrest through activation of DAF-16/FoxO (Fig. 6).
Fig. 6.
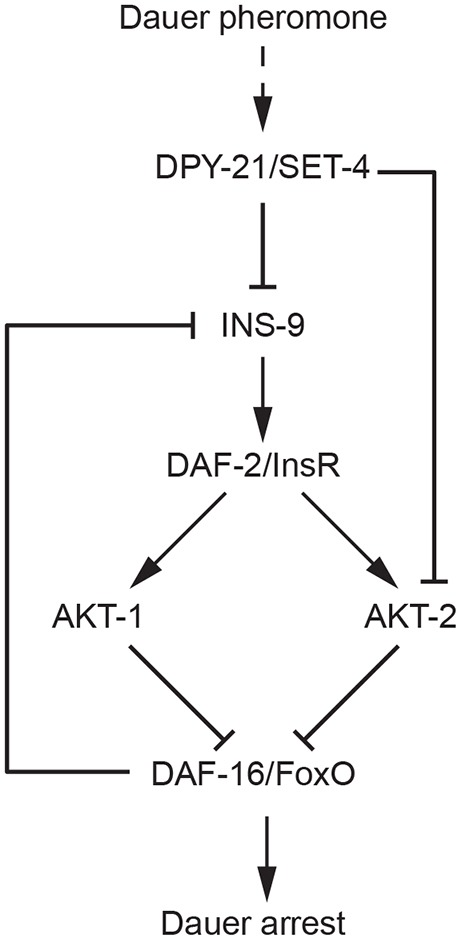
Hypothetical model of dauer regulation by pheromone through DPY-21/SET-4 and DAF-2 ILS. DPY-21 and SET-4 act in concert to promote transduction of pheromone cues by repressing X-linked genes encoding the DAF-2/InsR agonist INS-9 and the serine/threonine kinase AKT-2, resulting in increased DAF-16 activation and subsequent dauer arrest.
Although INS-9 has been predicted to function as an agonist ILP based both on structural models that indicate similarity to the agonist ILPs INS-4, INS-6 and DAF-28 (Pierce et al., 2001) and on expression changes upon starvation and feeding of larvae (Chen and Baugh, 2014), analysis of existing ins-9 mutants has not revealed phenotypes consistent with this (Fernandes de Abreu et al., 2014). This may be due to ins-9(tm3618) not being a strong loss-of-function allele. By contrast, our analysis of transgenic animals overexpressing ins-9 (Fig. 5B) and mutant animals harboring nonsense ins-9 alleles (Fig. 5D-F) provides the first experimental evidence demonstrating that INS-9 is an agonist ILP.
Several ILPs have been implicated in dauer regulation (Cornils et al., 2011; Fernandes de Abreu et al., 2014; Hung et al., 2014; Li et al., 2003; Murphy et al., 2003; Pierce et al., 2001). However, the mechanistic basis for how environmental cues regulate ILPs remains obscure. The initial events that control dauer arrest through DAF-2 ILS likely take place in the amphid sensory neurons, which are required for dauer formation in response to pheromone (Schackwitz et al., 1996; Vowels and Thomas, 1994). Indeed, genes encoding INS-4, INS-6 and DAF-28, which collectively play a major role in promoting reproductive development through DAF-2 ILS (Cornils et al., 2011; Hung et al., 2014; Li et al., 2003), are expressed in ASI (Chen and Baugh, 2014; Cornils et al., 2011; Hung et al., 2014; Li et al., 2003), and the transcription of ins-6 and daf-28 is inhibited by dauer pheromone through unknown mechanisms (Cornils et al., 2011; Li et al., 2003). Our finding that DPY-21 and SET-4 influence dauer arrest in part by repressing the X-linked ins-9 gene provides a potential mechanistic link between a dauer-regulatory environmental cue and an ILP expressed in sensory neurons that control the dauer decision.
An intriguing but incompletely understood aspect of dauer morphogenesis is the mechanistic basis for the commitment of larvae to either the reproductive or dauer developmental fate. Assays in which larvae are shifted between favorable and unfavorable conditions at different times after hatching indicate the existence of commitment points beyond which animals develop reproductively or arrest as dauers regardless of ambient conditions (Golden and Riddle, 1984; Schaedel et al., 2012). Commitment to reproductive development correlates temporally with the activation of a feed-forward loop amplifying organismal DA biosynthesis through DA-dependent induction of DAF-9 expression in the hypodermis (Schaedel et al., 2012). However, the XXX cells (Ohkura et al., 2003), which are thought to be the sole source of DA biosynthesis prior to the commitment point (Schaedel et al., 2012), are not in direct contact with the environment. Therefore, they must receive upstream inputs from sensory neurons that convey information about ambient conditions.
Our finding that DPY-21 and SET-4 synergize with DAF-16/FoxO to repress ins-9 is consistent with a hypothetical model in which INS-9 may function as a key node in an autocrine feed-forward loop in the ASI sensory neurons that reinforces levels of its own expression in response to changing environments, upstream of DA biosynthesis in the XXX cells and hypodermis. In replete conditions, ins-9 expression in ASI is expected to lead to activation of DAF-2 ILS and inhibition of DAF-16/FoxO. As DAF-16/FoxO inhibits ins-9 expression, decreased DAF-16/FoxO activity would lead to increased ins-9 expression, which would presumably lead to further activation of DAF-2 ILS and inhibition of DAF-16/FoxO, both in an autocrine fashion in ASI as well as in other cells that express DAF-2/InsR. In the context of increased population density, pheromone would promote ins-9 repression through DPY-21 and SET-4, and reduce autocrine and paracrine engagement of DAF-2/InsR, resulting in DAF-16/FoxO activation, further repression of ins-9 and dauer arrest (Fig. 6). The effect of DPY-21 and SET-4 would not be limited to sensory neurons, as they would also act in other cells responding to INS-9 to control their sensitivity to ILPs by repressing akt-2 (Dumas et al., 2013) (Fig. 6 and Fig. S6A). In addition, ins-9 regulation may also be amplified through other ILPs such as INS-7, which functions in a feed-forward loop in adults to coordinate DAF-16/FoxO activity throughout the animal (Murphy et al., 2007).
MATERIALS AND METHODS
C. elegans strains and maintenance
Mutant alleles are listed in the supplementary Materials and Methods. Compound mutants were generated using standard protocols. All animals were maintained on nematode growth media (NGM) plates seeded with E. coli OP50 using standard techniques. Strains are available upon request.
Dauer arrest assays
Dauer arrest assays were performed as previously described (Hu et al., 2006). daf-9(dh6) mutant animals were propagated on NGM plates supplemented with 10 nM Δ7-DA, then transferred to NGM plates for egglays as previously described (Dumas et al., 2013). For male dauer assays, males were crossed to isogenic L4 hermaphrodites, and the gender of dauer progeny was determined after dauer exit.
Dauer pheromone assays
Dauer pheromone was prepared as previously described (Golden and Riddle, 1982; Schroeder and Flatt, 2014). Details are provided in the supplementary Materials and Methods.
Generation of transgenic strains
Details pertaining to the generation of reporter constructs and transgenic strains are provided in the supplementary Materials and Methods.
CRISPR/Cas9-based mutagenesis
ins-9(dp675) and ins-9(dp677) were generated using recombinant crRNA and tracrRNA (Dharmacon) and Cas9 (PNA Bio) as described previously (Paix et al., 2015). See Table S7 for sequences of guide RNAs and repair oligonucleotides.
RNA isolation
Greater than 200 gravid hermaphrodites were allowed to lay eggs for 6 h at 20°C and then removed. Eggs were transferred to 25°C for 24 h. Larvae were harvested, washed once in M9 buffer and once in water, and resuspended in TRIzol (Invitrogen). After five sequential freeze-thaws, RNA was extracted using chloroform. Extracted RNA was purified using a Direct-zol RNA Miniprep Kit (Zymo Research).
qPCR
cDNA was synthesized with oligo-dT priming using the SuperScript III First Strand Synthesis Kit (Invitrogen). The equivalent of 10 ng of starting RNA was used as template in a 15 μl reaction using the Quantitect SYBRgreen qPCR Kit (Qiagen). Reactions were performed in a RotorGene 6000 (Corbett Research) and results analyzed using RotorGene 6000 Software (version 1.7). Samples were normalized to pmp-3 expression prior to comparison between groups (Hoogewijs et al., 2008). See Table S8 for primer sequences. Relative expression was calculated as described (Nolan et al., 2006).
Confocal microscopy
Animals were mounted on slides layered with a thin 3% agarose pad containing 25 mM sodium azide. Images were captured on a Leica Inverted SP5X Confocal Microscope (Leica) using LAS AF software.
RNA-seq analysis
Whole-transcriptome profiling was performed by the University of Michigan DNA Sequencing Core as previously described (Chen et al., 2015) using 100 ng input RNA per sample. Samples were barcoded and multiplexed, and 100-nucleotide paired-end sequencing was performed using an Illumina HiSeq 2000 sequencer and Version 4 reagents. Five experimental replicates were analyzed. Correlation coefficients between replicates and genotypes are shown in Table S9.
Annotated gene expression data output from CuffDiff v2.2.1 (Trapnell et al., 2013) was read into R version 3.2.1 (2015-06-18; The R Foundation for Statistical Computing; http://www.r-project.org/) for six comparisons: eak-7;akt-1 compared with: (1) wild type, (2) daf-16(mu86);eak-7;akt-1, (3) daf-12;eak-7;akt-1, (4) set-4(n4600);eak-7;akt-1, (5) set-4(dp268);eak-7;akt-1 and (6) dpy-21;eak-7;akt-1. We filtered genes using the following criteria: (1) status=‘OK’ for wild type versus eak-7;akt-1, (2) fold change (FC) ≥1.5 or FC ≤1/1.5 for wild type versus eak-7;akt-1 and (3) FDR <0.05 for at least two separate comparisons.
DAF-16 targets were defined as those filtered genes that also met (1) status=‘OK’ for eak-7;akt-1 versus daf-16;eak-7;akt-1 and (2) FC ≥1.5 for eak-7;akt-1 versus daf-16;eak-7;akt-1 in the opposite direction to wild type versus eak-7;akt-1. DPY-21, SET-4 and DAF-12 targets were determined in an analogous fashion. For SET-4, we generated a list of SET-4 targets that showed FC ≥1.5 or FC ≤1/1.5 for both set-4 alleles, and a list that showed these changes for either one or both set-4 alleles. Lists of overlapping targets were then generated from these target lists.
The significance of overlap with dosage-compensated X-linked genes (Jans et al., 2009), strongly regulated dauer genes (Liu et al., 2004) and DAF-16 targets in the daf-2(e1370) background (Chen et al., 2015) was calculated using a hypergeometric distribution, assuming 5863 X-linked transcripts and 46233 genome-wide transcripts in C. elegans detected in our RNA-seq analysis (by either the UCSCce10 reference transcriptome or de novo transcript assembly). If necessary, common WormBase Gene identifiers were downloaded from WormBase version WS250 (intermine.wormbase.org).
Immunoblotting and antibodies
To generate protein lysates, animals were washed in M9, then in sterile water. Pelleted animals were resuspended in equal volumes of worm lysis buffer (Webster et al., 2013), incubated at 85°C for 5 min, then sonicated on ice for two cycles of 30 s each at 70% power using a Sonic Dismembrator Model 100 (Fisher Scientific). Homogenates were quantified using a DC Protein Quantification Kit (BioRad). Protein (50 μg per lane) was loaded using Criterion systems (BioRad) and transferred to Immobilon Psq (Millipore). Details pertaining to antibodies are provided in the supplementary Materials and Methods. Membranes were blocked with 5% milk in TBS+0.5% Tween 20. Antibodies were diluted in Western Blocking Solution (Sigma) prior to incubation with membranes. Blots were washed with TBS+0.5% Tween 20. Signal was detected by ECL (Pierce).
Histone methyltransferase assay
set-4 cDNAs were amplified from RNA isolated from wild-type or set-4(dp268) mutant animals. Human Suv420H2 cDNA was obtained from Origene. Clones were ligated into pGEX4T1 vector (GE Healthcare). Protein expression was induced overnight at 16°C with 0.1 mM IPTG in BL21-CodonPlus(DE3)-RIPL cells (Agilent Technologies) grown in Terrific Broth (Invitrogen)+4% glycerol (Sigma). Cells were disrupted using a Sonic Dismembrator Model 100 sonicator (Fisher Scientific), with four cycles of ten 1 s on/off pulses of 10-30% intensity. Lysates were cleared by centrifugation at 20,000 g for 5 min and incubated with Glutathione-Sepharose beads (GE Healthcare) rotating overnight at 4°C. Expression of recombinant protein was confirmed by Coomassie staining and anti-GST immunoblot. Beads bound to recombinant protein were incubated in 10 mM Tris (pH 8.0), 2 μM β-mercaptoethanol, and 7 mM S-adenosylmethionine (Sigma) in the presence of 2 mM substrate peptide corresponding to amino acids 8-30 of C. elegans histone H4 (AnaSpec) rotating for 4 h at 30°C. Reactions were analyzed using a Waters MicroMass MALDI-TOF mass spectrometer and analyzed with MassLynx software. Ratios of peak heights corresponding to reactant and product peptide were calculated to define percent conversion.
Male rescue assay
Mated gravid hermaphrodites were placed on NGM plates to lay eggs for 24 h at 20°C. Egglayers were removed, and the numbers of hatchlings and eggs were counted. After 72 h, the animals were moved to 4°C to slow their movement. Male rescue was calculated as the ratio of live males to total eggs laid. Each experiment was performed in triplicate.
Statistics
Two-tailed Student's t-test was used to measure significance in experiments unless otherwise noted. Data are presented as the average±standard error of the mean (s.e.m.) of at least three biological replicates, each replicate performed in triplicate. n values for dauer assays are listed from left to right.
Acknowledgements
Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), and by Shohei Mitani at the National Bioresource Project for the Nematode C. elegans. We thank Stephane Flibotte for analysis of raw genome sequencing data; Yali Dou and Shirley Lee for reagents and advice with methyltransferase assays; Scott Rothbart for validation of histone antibody specificity; Brian Shay for training and assistance with mass spectrometry; Frank Schroeder for Δ7-DA; the University of Michigan Medical School Bioinformatics Core for assistance with RNA-seq analysis; Andrew Fire for pPD95.75; Jeremy Nance for pSA120; Daniel Dickinson for pDD268; John Kim for pJK343; Erik Andersen, Xantha Karp, Liberta Nika, David Sherman and Ashootosh Tripathi for assistance with pheromone preparation; and Gyorgyi Csankovszki for critical reading of the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
C.E.D. conceived the project, designed and performed experiments, interpreted results and wrote the manuscript. A.T.C. performed bioinformatic analysis of RNA-seq data. J.V.G. designed and performed experiments. K.J.D. designed and performed experiments. P.J.H. conceived the project, designed experiments, interpreted results and wrote the manuscript
Funding
This work was funded by grants from the National Institutes of Health (R01AG041177 to P.J.H.), the American Cancer Society (119640-RSG-10-132-01-DDC to P.J.H.), the American Heart Association (11IRG5170009 to P.J.H.) and the National Science Foundation (DGE 1256260 to C.E.D.); and by a pilot grant from the Michigan Diabetes Research Center (to P.J.H.). Deposited in PMC for immediate release.
Data availability
Whole-transcriptome profiling data are available at GEO with the Accession Number GSE89295.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.145722.supplemental
References
- Ailion M. and Thomas J. H. (2003). Isolation and characterization of high-temperature-induced Dauer formation mutants in Caenorhabditis elegans. Genetics 165, 127-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam H., Williams T. W., Dumas K. J., Guo C., Yoshina S., Mitani S. and Hu P. J. (2010). EAK-7 controls development and life span by regulating nuclear DAF-16/FoxO activity. Cell Metab. 12, 30-41. 10.1016/j.cmet.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen E. C. and Horvitz H. R. (2007). Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development 134, 2991-2999. 10.1242/dev.009373 [DOI] [PubMed] [Google Scholar]
- Bargmann C. I. and Horvitz H. R. (1991). Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science 251, 1243-1246. 10.1126/science.2006412 [DOI] [PubMed] [Google Scholar]
- Blumenthal T. (2005). Trans-splicing and operons. WormBook ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.5.1, http://www.wormbook.org 10.1895/wormbook.1.5.1 [DOI] [PubMed] [Google Scholar]
- Butcher R. A., Ragains J. R., Kim E. and Clardy J. (2008). A potent dauer pheromone component in Caenorhabditis elegans that acts synergistically with other components. Proc. Natl. Acad. Sci. USA 105, 14288-14292. 10.1073/pnas.0806676105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. and Baugh L. R. (2014). Ins-4 and daf-28 function redundantly to regulate C. elegans L1 arrest. Dev. Biol. 394, 314-326. 10.1016/j.ydbio.2014.08.002 [DOI] [PubMed] [Google Scholar]
- Chen A. T.-Y., Guo C., Itani O. A., Budaitis B. G., Williams T. W., Hopkins C. E., McEachin R. C., Pande M., Grant A. R., Yoshina S. et al. (2015). Longevity genes revealed by integrative analysis of isoform-specific daf-16/FoxO mutants of Caenorhabditis elegans. Genetics 201, 613-629. 10.1534/genetics.115.177998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornils A., Gloeck M., Chen Z., Zhang Y. and Alcedo J. (2011). Specific insulin-like peptides encode sensory information to regulate distinct developmental processes. Development 138, 1183-1193. 10.1242/dev.060905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes H. E., Berlin D. S., Lapidus D. M., Nusbaum C., Davis T. L. and Meyer B. J. (1999). Dosage compensation proteins targeted to X chromosomes by a determinant of hermaphrodite fate. Science 284, 1800-1804. 10.1126/science.284.5421.1800 [DOI] [PubMed] [Google Scholar]
- Dumas K. J., Delaney C. E., Flibotte S., Moerman D. G., Csankovszki G. and Hu P. J. (2013). Unexpected role for dosage compensation in the control of dauer arrest, insulin-like signaling, and FoxO transcription factor activity in Caenorhabditis elegans. Genetics 194, 619-629. 10.1534/genetics.113.149948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes de Abreu D. A., Caballero A., Fardel P., Stroustrup N., Chen Z., Lee K., Keyes W. D., Nash Z. M., Lopez-Moyado I. F., Vaggi F. et al. (2014). An insulin-to-insulin regulatory network orchestrates phenotypic specificity in development and physiology. PLoS Genet. 10, e1004225 10.1371/journal.pgen.1004225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N. and Antebi A. (2008). C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 22, 2149-2165. 10.1101/gad.1701508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgi L. L., Albert P. S. and Riddle D. L. (1990). daf-1, a C. elegans gene controlling dauer larva development, encodes a novel receptor protein kinase. Cell 61, 635-645. 10.1016/0092-8674(90)90475-T [DOI] [PubMed] [Google Scholar]
- Gerisch B., Weitzel C., Kober-Eisermann C., Rottiers V. and Antebi A. (2001). A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev. Cell 1, 841-851. 10.1016/S1534-5807(01)00085-5 [DOI] [PubMed] [Google Scholar]
- Golden J. W. and Riddle D. L. (1982). A pheromone influences larval development in the nematode Caenorhabditis elegans. Science 218, 578-580. 10.1126/science.6896933 [DOI] [PubMed] [Google Scholar]
- Golden J. W. and Riddle D. L. (1984). The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev. Biol. 102, 368-378. 10.1016/0012-1606(84)90201-X [DOI] [PubMed] [Google Scholar]
- Gottlieb S. and Ruvkun G. (1994). daf-2, daf-16 and daf-23: genetically interacting genes controlling Dauer formation in Caenorhabditis elegans. Genetics 137, 107-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogewijs D., Houthoofd K., Matthijssens F., Vandesompele J. and Vanfleteren J. R. (2008). Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 9, 9 10.1186/1471-2199-9-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P. J., Xu J. and Ruvkun G. (2006). Two membrane-associated tyrosine phosphatase homologs potentiate C. elegans AKT-1/PKB signaling. PLoS Genet. 2, e99 10.1371/journal.pgen.0020099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung W. L., Wang Y., Chitturi J. and Zhen M. (2014). A Caenorhabditis elegans developmental decision requires insulin signaling-mediated neuron-intestine communication. Development 141, 1767-1779. 10.1242/dev.103846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans J., Gladden J. M., Ralston E. J., Pickle C. S., Michel A. H., Pferdehirt R. R., Eisen M. B. and Meyer B. J. (2009). A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev. 23, 602-618. 10.1101/gad.1751109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K., Albert P. S. and Riddle D. L. (2002). DAF-9, a cytochrome P450 regulating C. elegans larval development and adult longevity. Development 129, 221-231. [DOI] [PubMed] [Google Scholar]
- Kramer M., Kranz A.-L., Su A., Winterkorn L. H., Albritton S. E. and Ercan S. (2015). Developmental dynamics of X-Chromosome dosage compensation by the DCC and H4K20me1 in C. elegans. PLoS Genet. 11, e1005698 10.1371/journal.pgen.1005698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Kennedy S. G. and Ruvkun G. (2003). daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. 17, 844-858. 10.1101/gad.1066503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Zimmerman K. K. and Patterson G. I. (2004). Regulation of signaling genes by TGFbeta during entry into dauer diapause in C. elegans. BMC Dev. Biol. 4, 11 10.1186/1471-213X-4-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B. J. (2010). Targeting X chromosomes for repression. Curr. Opin. Genet. Dev. 20, 179-189. 10.1016/j.gde.2010.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D., Korta D. Z., Capua Y. and Hubbard E. J. (2010). Insulin signaling promotes germline proliferation in C. elegans. Development 137, 671-680. 10.1242/dev.042523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C. T. and Hu P. J. (2013). Insulin/insulin-like growth factor signaling in C. elegans. WormBook ed. The C. elegans Research Community, WormBook, 10.1895/wormbook.1.164.1, http://www.wormbook.org 10.1895/wormbook.1.164.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S., Ahringer J., Li H. and Kenyon C. (2003). Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277-283. 10.1038/nature01789 [DOI] [PubMed] [Google Scholar]
- Murphy C. T., Lee S.-J. and Kenyon C. (2007). Tissue entrainment by feedback regulation of insulin gene expression in the endoderm of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 104, 19046-19050. 10.1073/pnas.0709613104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T., Hands R. E. and Bustin S. A. (2006). Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 1, 1559-1582. 10.1038/nprot.2006.236 [DOI] [PubMed] [Google Scholar]
- Nonet M. L., Staunton J. E., Kilgard M. P., Fergestad T., Hartwieg E., Horvitz H. R., Jorgensen E. M. and Meyer B. J. (1997). Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J. Neurosci. 17, 8061-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura K., Suzuki N., Ishihara T. and Katsura I. (2003). SDF-9, a protein tyrosine phosphatase-like molecule, regulates the L3/dauer developmental decision through hormonal signaling in C. elegans. Development 130, 3237-3248. 10.1242/dev.00540 [DOI] [PubMed] [Google Scholar]
- Paix A., Folkmann A., Rasoloson D. and Seydoux G. (2015). High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics 201, 47-54. 10.1534/genetics.115.179382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D., Estevez A. and Riddle D. L. (2010). Antagonistic Smad transcription factors control the dauer/non-dauer switch in C. elegans. Development 137, 477-485. 10.1242/dev.043752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce S. B., Costa M., Wisotzkey R., Devadhar S., Homburger S. A., Buchman A. R., Ferguson K. C., Heller J., Platt D. M., Pasquinelli A. A. et al. (2001). Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 15, 672-686. 10.1101/gad.867301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren P., Lim C. S., Johnsen R., Albert P. S., Pilgrim D. and Riddle D. L. (1996). Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science 274, 1389-1391. 10.1126/science.274.5291.1389 [DOI] [PubMed] [Google Scholar]
- Riddle D. L. (1988). The dauer larva. In The Nematode Caenorhabditis elegans (ed. Wood W. B.), pp. 393-412. Plainview, New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Riddle D. L., Swanson M. M. and Albert P. S. (1981). Interacting genes in nematode dauer larva formation. Nature 290, 668-671. 10.1038/290668a0 [DOI] [PubMed] [Google Scholar]
- Ritter A. D., Shen Y., Fuxman Bass J., Jeyaraj S., Deplancke B., Mukhopadhyay A., Xu J., Driscoll M., Tissenbaum H. A. and Walhout A. J. (2013). Complex expression dynamics and robustness in C. elegans insulin networks. Genome Res. 23, 954-965. 10.1101/gr.150466.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottiers V., Motola D. L., Gerisch B., Cummins C. L., Nishiwaki K., Mangelsdorf D. J. and Antebi A. (2006). Hormonal control of C. elegans dauer formation and life span by a Rieske-like oxygenase. Dev. Cell 10, 473-482. 10.1016/j.devcel.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Schackwitz W. S., Inoue T. and Thomas J. H. (1996). Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron 17, 719-728. 10.1016/S0896-6273(00)80203-2 [DOI] [PubMed] [Google Scholar]
- Schaedel O. N., Gerisch B., Antebi A. and Sternberg P. W. (2012). Hormonal signal amplification mediates environmental conditions during development and controls an irreversible commitment to adulthood. PLoS Biol. 10, e1001306 10.1371/journal.pbio.1001306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G., Lachner M., Sarma K., Ebert A., Sengupta R., Reuter G., Reinberg D. and Jenuwein T. (2004). A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18, 1251-1262. 10.1101/gad.300704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder N. E. and Flatt K. M. (2014). In vivo imaging of Dauer-specific neuronal remodeling in C. elegans. J. Vis. Exp. 91, e51834 10.3791/51834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Lambert G. G., Chammas A., Ni Y., Cranfill P. J., Baird M. A., Sell B. R., Allen J. R., Day R. N., Israelsson M. et al. (2013). A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407-409. 10.1038/nmeth.2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall S. M., Cronin N. B. and Wilson J. R. (2014). A novel route to product specificity in the Suv4-20 family of histone H4K20 methyltransferases. Nucleic Acids Res. 42, 661-671. 10.1093/nar/gkt776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starich T. A., Herman R. K., Kari C. K., Yeh W. H., Schackwitz W. S., Schuyler M. W., Collet J., Thomas J. H. and Riddle D. L. (1995). Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics 139, 171-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. H., Birnby D. A. and Vowels J. J. (1993). Evidence for parallel processing of sensory information controlling dauer formation in Caenorhabditis elegans. Genetics 134, 1105-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L. and Pachter L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46-53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle A., Lang J., Dong Y., Ercan S., Kotwaliwale C., Rechtsteiner A., Appert A., Chen Q. B., Dose A., Egelhofer T. et al. (2012). H4K20me1 contributes to downregulation of X-linked genes for C. elegans dosage compensation. PLoS Genet. 8, e1002933 10.1371/journal.pgen.1002933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels J. J. and Thomas J. H. (1992). Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics 130, 105-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels J. J. and Thomas J. H. (1994). Multiple chemosensory defects in daf-11 and daf-21 mutants of Caenorhabditis elegans. Genetics 138, 303-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster C. M., Wu L., Douglas D. and Soukas A. A. (2013). A non-canonical role for the C. elegans dosage compensation complex in growth and metabolic regulation downstream of TOR complex 2. Development 140, 3601-3612. 10.1242/dev.094292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells M. B., Snyder M. J., Custer L. M. and Csankovszki G. (2012). Caenorhabditis elegans dosage compensation regulates histone H4 chromatin state on X chromosomes. Mol. Cell. Biol. 32, 1710-1719. 10.1128/MCB.06546-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Siarheyeva A., Zeng H., Lam R., Dong A., Wu X.-H., Li Y., Schapira M., Vedadi M. and Min J. (2013). Crystal structures of the human histone H4K20 methyltransferases SUV420H1 and SUV420H2. FEBS Lett. 587, 3859-3868. 10.1016/j.febslet.2013.10.020 [DOI] [PubMed] [Google Scholar]
- Yonker S. A. and Meyer B. J. (2003). Recruitment of C. elegans dosage compensation proteins for gene-specific versus chromosome-wide repression. Development 130, 6519-6532. 10.1242/dev.00886 [DOI] [PubMed] [Google Scholar]