Abstract
The T-box transcription factor (TF) Eomes is a key regulator of cell fate decisions during early mouse development. The cis-acting regulatory elements that direct expression in the anterior visceral endoderm (AVE), primitive streak (PS) and definitive endoderm (DE) have yet to be defined. Here, we identified three gene-proximal enhancer-like sequences (PSE_a, PSE_b and VPE) that faithfully activate tissue-specific expression in transgenic embryos. However, targeted deletion experiments demonstrate that PSE_a and PSE_b are dispensable, and only VPE is required for optimal Eomes expression in vivo. Embryos lacking this enhancer display variably penetrant defects in anterior-posterior axis orientation and DE formation. Chromosome conformation capture experiments reveal VPE-promoter interactions in embryonic stem cells (ESCs), prior to gene activation. The locus resides in a large (500 kb) pre-formed compartment in ESCs and activation during DE differentiation occurs in the absence of 3D structural changes. ATAC-seq analysis reveals that VPE, PSE_a and four additional putative enhancers display increased chromatin accessibility in DE that is associated with Smad2/3 binding coincident with transcriptional activation. By contrast, activation of the Eomes target genes Foxa2 and Lhx1 is associated with higher order chromatin reorganisation. Thus, diverse regulatory mechanisms govern activation of lineage specifying TFs during early development.
KEY WORDS: Eomesodermin, Enhancer, Capture-C, Nodal signalling, Definitive endoderm
Summary: Expression of the mouse T-box factor Eomes is controlled by a key gene-proximal enhancer-like element, with changes in chromatin accessibility influencing its activity in definitive endoderm.
INTRODUCTION
Reciprocal signalling cues between the pluripotent epiblast and adjacent tissues, namely the extra-embryonic ectoderm (ExE) and visceral endoderm (VE), precisely coordinate cell fate decisions during gastrulation. Nodal/Smad signals from the epiblast are required for specification of the AVE, a discrete signalling centre that establishes anterior-posterior (A-P) polarity (Brennan et al., 2001; Robertson, 2014; Stower and Srinivas, 2014). The A-P axis initially becomes visible at gastrulation, when proximal posterior cells undergo an epithelial-to-mesenchymal transition (EMT) at the PS to form nascent mesoderm. Slightly later, following distal extension of the streak, endoderm progenitors delaminate and emerge onto the surface of the embryo (Kwon et al., 2008).
The T-box transcription factor (TF) eomesodermin (Eomes), acting downstream of Nodal/Smad signals, is required to promote AVE formation and orientation of the A-P axis (Arnold et al., 2008a; Ciruna and Rossant, 1999; Nowotschin et al., 2013), as well as EMT of nascent mesoderm cells (Arnold et al., 2008a; Costello et al., 2011; Russ et al., 2000; van den Ameele et al., 2012). At post-implantation stages, Eomes is expressed in the ExE and embryonic-VE, robustly induced at the onset of gastrulation in the PS and maintained in the anterior PS as it extends, before being abruptly lost (coincident with node formation) (Kwon and Hadjantonakis, 2007). Fate-mapping experiments demonstrate that transient Eomes expression marks progenitors of the cardiovascular lineage, definitive endoderm (DE), node and midline (Costello et al., 2011).
Transgenic and targeted deletion approaches have provided insight into cell type-specific developmental enhancers that govern expression of key genes responsible for partitioning the pluripotent epiblast into discrete cell lineages. Proximal cis-regulatory regions within 20 kb of the transcriptional start sites (TSS) directing spatiotemporally restricted expression of Nodal, Mesp1/2 and Lhx1 have been identified. Both the ASE, an intronic autoregulatory enhancer (Adachi et al., 1999; Norris and Robertson, 1999), and the Wnt signalling responsive 5′ PEE (Ben-Haim et al., 2006) cooperatively regulate Nodal expression. Mutant embryos lacking these genomic sequences display dose-dependent defects in specification of mesoderm and DE/midline progenitors (Norris et al., 2002; Vincent et al., 2003). Similarly, the Mesp1/2 genes, which are essential for formation of nascent mesoderm, are jointly regulated by the EME, an Eomes-dependent enhancer (Costello et al., 2011; Haraguchi et al., 2001). Our recent work demonstrates that Lhx1, which is required for AVE and anterior mesendoderm specification (Barnes et al., 1994; Shawlot and Behringer, 1995), is directly controlled by Eomes binding to a proximal promoter element (Nowotschin et al., 2013).
Eomes, which is rapidly induced in the proximal-posterior epiblast coincident with the acquisition of A-P polarity (Ciruna and Rossant, 1999), is widely viewed as a master regulator of mesendodermal lineages (Costello et al., 2011; Izumi et al., 2007; Teo et al., 2011; van den Ameele et al., 2012). Thus, Eomes represents the earliest lineage-specifying gene in the embryo proper. However, relatively little is known about the cis-acting regulatory elements controlling its dynamic pattern of expression. Recent studies of mouse and human ESCs have identified a conserved switch enhancer −7 kb upstream of the TSS (Beyer et al., 2013; Kartikasari et al., 2013; Rada-Iglesias et al., 2011) that is repressed under self-renewing conditions (Teo et al., 2011), and becomes activated during mesoderm and endoderm differentiation. However, possible functional contributions made by this genomic region have yet to be assessed in vivo.
Here, we investigate the structural features of the locus that govern Eomes expression during early mouse development. Gain-of-function transgenic reporter assays identified three gene-proximal Eomes enhancer-like sequences (PSE_a, PSE_b and VPE). However, when we engineered germline deletions to evaluate their functional contributions in vivo, surprisingly, only the VPE was found to influence expression in the early embryo. We also exploited Next Generation (NG) Capture-C technology (Davies et al., 2016) to describe the 3D structural features of the locus. The Eomes promoter occupies a discrete 500 kb regulatory compartment in ESCs, and this chromatin conformation is not appreciably altered during DE differentiation. However, our ATAC-seq analysis revealed that the VPE, PSE_a and four additional distal regulatory elements located within this pre-formed compartment display increased chromatin accessibility and acquire Smad2/3 occupancy during DE differentiation. This mode of 3D genome organisation probably serves to facilitate rapid Nodal/Smad-dependent activation of the locus. By contrast, developmentally regulated Foxa2 and Lhx1 promoter-promoter and promoter-enhancer interactions seem to require substantial structural changes during the shift from a transcriptionally inactive to active conformation.
RESULTS
Identification of proximal Eomes enhancers that are active during gastrulation
Putative enhancer elements containing DNase I hypersensitive sites and marked by H3K4me1 are considered to be active if also enriched for H3K27ac or, alternatively, viewed as poised if enriched for H3K27me3 (Rada-Iglesias et al., 2011; Zentner et al., 2011). To identify candidate enhancers at the Eomes locus, we examined ChIP-seq datasets from undifferentiated ESC, epiblast-like cells (EpiLC) and mesodermal precursors (MES) (Alexander et al., 2015; Buecker et al., 2014; ENCODE Project Consortium, 2012) corresponding to the E4.5 epiblast (ESC), the E5.5 epiblast (EpiLC) or E6.5 primitive streak (MES) cell populations.
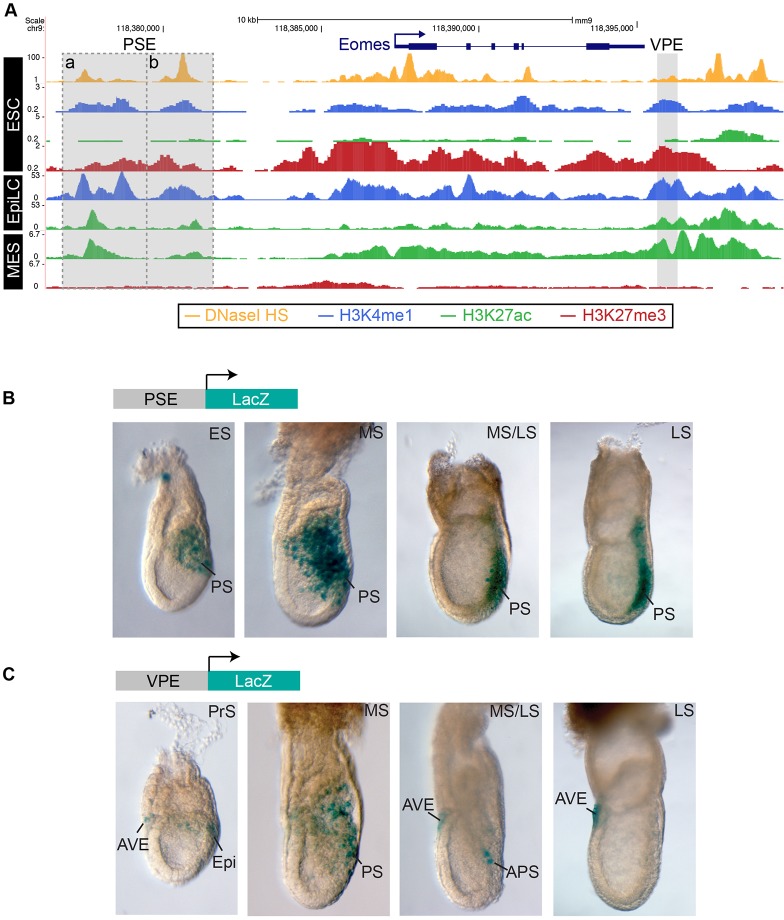
We identified three DNase I hypersensitive sites close to the Eomes promoter marked by H3K4me1 that show increased H3K27ac upon differentiation, including two sites (PSE_a and PSE_b) located close together, spanning a 5 kb region between −11 kb to −6 kb upstream of the transcriptional start site (TSS), and a third candidate region (VPE) lying +8 kb downstream of the TSS (Fig. 1A, Fig. S1A). Notably, the upstream cluster contains the previously described switch enhancer (PSE_b) activated during ESC differentiation to DE and mesendoderm (Beyer et al., 2013; Kartikasari et al., 2013). Additionally, two downstream DNaseI hypersensitive sites bound by CCCTC-binding factor (CTCF) were identified in ESCs (Fig. S1A). The three proximal regions are highly conserved among mammals (Fig. S1A), associated with H3K4me1/H3K27me3 in ESCs and, thus, probably represent poised enhancers that are primed for activation. Consistent with a shift to the active state during the transition from pluripotency to lineage commitment, these regions contain increased H3K27ac and decreased H3K27me3 in EpiLC and MES. The homologous regions are also associated with active enhancer marks in human DE cultures (Fig. S1B).
Fig. 1.
Mapping proximal Eomes enhancers active at gastrulation. (A) ChIP-seq of H3K4me1, H3K27me3 and H3K27ac, and DNaseI hypersensitivity (HS) in ESCs, epiblast-like cells (EpiLC) and mesoderm (MES) (Alexander et al., 2015; Buecker et al., 2014; ENCODE Project Consortium, 2012) identify potential proximal Eomes enhancers that are activated during differentiation. The PSE cluster and VPE regions are highlighted in grey. (B,C) X-gal-stained transgenic embryos expressing enhancer-driven LacZ reporters. (B) PSE reporter activity is confined to the primitive streak (PS) at early- (ES), mid- (MS) and late-streak (LS) stages of gastrulation (2/4 transgenic mouse lines). (C) VPE reporter activity detectable in the proximal posterior epiblast (Epi) at the pre-streak (PrS) stage and in the PS at the MS stage, becomes restricted to the anterior PS (APS) and is lost at LS stage. Between the PrS stage and the LS stage, VPE activity is also detectable in the anterior visceral endoderm (AVE) (2/6 transgenic mouse lines).
To test the activities of these candidate enhancers, we generated transgenic strains carrying LacZ reporter constructs and subsequently examined embryonic expression at early post-implantation stages (Kothary et al., 1989). The 5 kb upstream region was designated the PSE (primitive streak enhancer) because PSE-LacZ activity is restricted to the PS at early (ES), mid- (MS) and late-streak (LS) stages (Fig. 1B). There was no detectable LacZ expression in the ExE or VE. However, the 0.7 kb downstream enhancer, designated the VPE (visceral endoderm and primitive streak enhancer), showed activity in the proximal-posterior epiblast, and also in the AVE at pre-streak (PrS) stages (Fig. 1C). Slightly later, LacZ expression was detectable in the PS, nascent mesendoderm and the AVE, subsequently became restricted to the anterior PS, and was lost by LS stages. Collectively, these three enhancers faithfully recapitulate the endogenous Eomes expression patterns within both the VE and embryo proper.
The PSE is dispensable for normal embryonic development
The 5 kb PSE contains both an upstream element, PSE_a, as well as the previously described PSE_b switch enhancer reported to interact with the Eomes promoter during DE differentiation (Fig. S1A) (Beyer et al., 2013; Kartikasari et al., 2013). To investigate their functional activities in the context of the developing embryo, we generated discrete germline targeted deletions (Fig. 2A, Fig. S2). Surprisingly, homozygous mice lacking the 2 kb PSE_b genomic fragment ∼8 kb to ∼6 kb upstream of the TSS (ΔPSE_b) were recovered at Mendelian ratios and are indistinguishable from wild-type littermates (Table 1A). These results demonstrate that the PSE_b is dispensable in vivo. It is well known that heterozygous mice carrying null alleles (EomesGFP/+, EomesLacZ/+ or EomesΔexon2-5/+) are fully viable (Arnold et al., 2008a, 2009; Russ et al., 2000). To investigate whether the PSE_b deletion may compromise transcriptional output, we crossed EomesΔPSE_b/ΔPSE_b mice to those carrying the EomesGFP/+ allele (hereafter referred to as Eomes null; Eomes+/−). The resulting EomesΔPSE_b/− compound mutants develop normally (Table 1B).
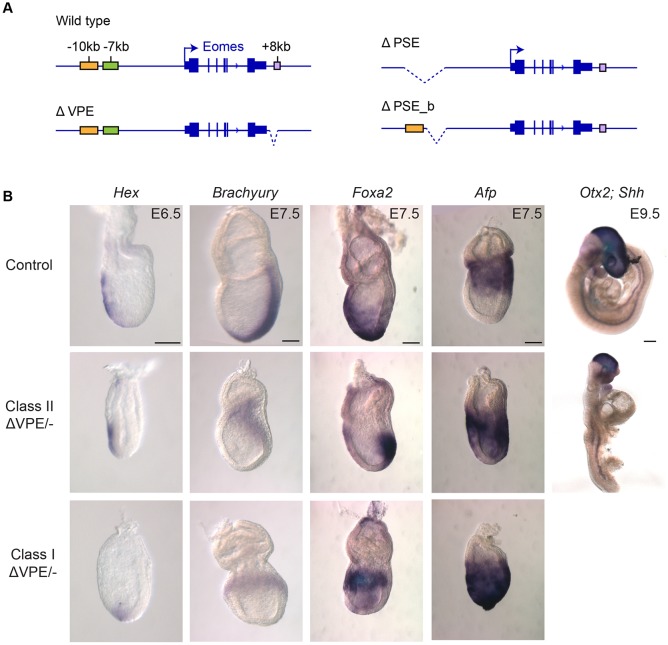
Fig. 2.
Targeted deletions of proximal enhancers show that only the VPE is required for proper gastrulation. (A) Targeted deletions of the 5 kb ΔPSE, 2 kb ΔPSE_b and 0.7 kb ΔVPE generated by homologous recombination (Figs S2-S4). (B) Whole-mount in situ hybridisation of EomesΔVPE/− embryos. Class I mutants exhibit failure in A-P axis specification; class II display APS defects. At E6.5 in class I mutants, expression of the AVE marker Hex is confined to the distal VE (n=4/10 EomesΔVPE/− embryos analysed). At E7.5, the mesoderm marker Brachyury (n=2/5) and the DE marker Foxa2 (n=3/7) are mislocalised proximally. In class II mutants, Hex marks the AVE, Brachyury expression fails to extend distally (n=3/5), whereas the Foxa2 domain is confined to the APS and the DE domain is lost (n=3/7). Consistent with failure to specify DE in both mutant classes, expression of Afp+ VE cells fails to disperse proximally (for class I and class II, n=2 and n=2 out of 7 EomesΔVPE/− embryos analysed, respectively). At E9.5, class II mutants display venture closure and neural tube defects, fused or malformed somites, loss of Otx2+ forebrain tissue and an anterior truncation of the Shh midline (n=3/3 viable morphologically abnormal EomesΔVPE/− embryos recovered). Scale bars: 100 μm.
Table 1.
Genotypes of mutant weanlings

Next, we engineered a deletion that eliminates the entire 5 kb PSE cluster (referred to as ΔPSE, Fig. S3). However, as for the PSE_b, removal of the entire PSE region in EomesΔPSE/ΔPSE mice has no noticeable effect on viability (Table 1A). Finally, crossing these deletion mutants with mice carrying the Eomes null allele also failed to perturb embryonic development (Table 1B). Thus, it appears that the PSE can activate expression in gain-of-function transgenic embryos. Nonetheless, this genomic region is clearly dispensable for Eomes expression in vivo.
Targeted deletion of the VPE leads to defective gastrulation
To investigate functional contributions made by the VPE, we generated a targeted deletion lacking this 0.7 kb region (Fig. S4). Homozygous ΔVPE mutants are viable and fertile (Table 1A). However, when we crossed EomesΔVPE/ΔVPE mice with Eomes+/− heterozygous animals carrying the null allele, we observed a significant under-representation of viable EomesΔVPE/− compound heterozygotes (Table 1B), with ∼40% (n=18) of the expected numbers recovered at weaning (equivalent to EomesΔVPE/+, n=44). These results strongly suggest that EomesΔVPE acts as a hypomorphic allele.
Next, to determine the onset of lethality, we examined embryos from E6.5 onwards. Approximately one-third of EomesΔVPE/− embryos are morphologically normal. However, two distinct classes of abnormal embryos were recovered at roughly equivalent numbers. The most severely affected (class I) mutants arrest at early gastrulation stages, while a second group (class II) progress to mid-gestation (Fig. 2B).
In class I embryos, the AVE marker Hex is induced at E6.5 but remains localised to the distal tip. Thus, the AVE is specified but fails to migrate towards the prospective anterior side of the embryo. These embryos fail to correctly orient the A-P axis and lack a discrete PS. At E7.5, mesoderm (Brachyury) and DE (Foxa2) markers are restricted proximally. Class I mutant embryos, distinguished by the accumulation of disorganised mesenchymal cells in the epiblast cavity and a constriction at the embryonic and extra-embryonic boundary, phenocopy those selectively lacking Eomes activity in the VE (Nowotschin et al., 2013). Taken together with results above that demonstrate VPE-LacZ expression in the VE, the simplest explanation is that these abnormalities are caused by loss of Eomes function in the VE.
The class II embryos, which represent approximately one-third of the EomesΔVPE/− embryos, successfully establish normal A-P polarity. However, as gastrulation proceeds they display focal defects in the anterior PS (APS) and its derivatives the DE, midline, node and notochord. Brachyury (T) expression in the PS fails to extend to the distal tip of the streak at E7.5. Foxa2-positive DE progenitors are specified but fail to migrate anteriorly. As judged by Afp expression, the VE is retained over the epiblast and fails to become distally restricted. These tissue disturbances probably reflect the functional loss of Eomes within the APS (Arnold et al., 2008a; Teo et al., 2011). APS derivatives are known to provide essential trophic signals required for patterning the anterior neurectoderm (Arkell and Tam, 2012). Consistent with this, at E9.5, class II mutant embryos display ventral closure and neural tube defects, fused or malformed somites, and loss of forebrain tissue.
The VPE is required for optimal Eomes expression levels
To test directly whether targeted loss of the VPE compromises Eomes transcriptional output, we eliminated the VPE in the context of our EomesGFP reporter allele containing an EGFP-pA cassette inserted in-frame at the translational start site in exon 1 (Fig. 3A, Fig. S5) (Arnold et al., 2009) and performed flow cytometry analysis to quantify expression levels. The EomesGFP reporter is robustly activated during ESC differentiation to embryoid bodies (EBs) (Costello et al., 2011) (Fig. 3B). As shown in Fig. 3C, GFP expression is dramatically reduced in EomesGFPΔVPE/+ EBs when compared with EomesGFP/+ EBs. The VPE deletion results in markedly reduced expression to 42% of the control EomesGFP/+ EBs (Student's t-test P=0.05) (Fig. 3D).
Fig. 3.
VPE deletion profoundly reduces the level of EomesGFP reporter expression. (A) Configuration of the EomesGFP and EomesGFPΔVPE alleles (Fig. S5). (B) Schematic of the embryoid body (EB) differentiation protocol. (C,D) Flow cytometry analysis of wild-type, EomesGFP/+ and EomesGFPΔVPE/+ day 4 EBs. (C) Representative histograms showing wild-type, two independently targeted EomesGFP/+ and two EomesGFPΔVPE/+ clones. (D) Average GFP intensity in EomesGFP/+ (n=4) and EomesGFPΔVPE/+ (n=4) cultures. Deletion of the VPE significantly reduces expression to 42% of the intact EomesGFP reporter (P=0.05, Student's t-test). Error bars represent the s.e.m. (E,F) Confocal images of EomesGFP and EomesGFPΔVPE reporter expression in E6.5 embryos stained with anti-GFP antibody, DAPI (DNA) and phalloidin (F-actin). Domains of reporter expression are not perturbed by VPE deletion.
These heterogenous EB cultures contain mixtures of cardiac mesoderm, DE and VE Eomes+ cell populations. To investigate the impact of the VPE deletion in vivo, we generated EomesGFPΔVPE/+ mice and examined expression during gastrulation. GFP expression in EomesGFPΔVPE/+ embryos recapitulates domains of the EomesGFP/+ control embryos at E6.5, in the ExE, PS, nascent mesoderm and VE (Fig. 3E,F). The VPE deletion reduced expression levels but tissue-specific expression patterns were unperturbed. Similar conclusions were reached by whole-mount in situ hybridisation experiments examining Eomes mRNA expression in EomesΔVPE/ΔVPE embryos (Fig. S4E). Thus, reduced Eomes transcription (∼50%) as in Eomes+/− or EomesΔVPE/ΔVPE embryos is sufficient to promote A-P axis specification and gastrulation. However, as shown above, further reduced expression (∼25%) in EomesΔVPE/− embryos results in gastrulation defects.
Foxh1-independent Nodal/Smad2/3 signals regulate VPE activity
Eomes activation in the VE and PS depends on Nodal/Smad signals (Brennan et al., 2001; Nowotschin et al., 2013). To investigate Nodal/Smad requirements in cultured EBs, we used the small molecule SB-431542 (SB), a potent inhibitor of type 1 activin receptor like kinases 4, 5 and 7. As expected, in control cultures, maximal Eomes expression was detectable between day (d)3.5 and d4 (Fig. 4A). Eomes expression was dramatically reduced in cultures treated with the SB inhibitor from d3, and by d4 is severely compromised to only 2% of that seen in controls (Fig. 4A). These results confirm that Nodal signalling is required to induce Eomes expression during the transition from pluripotency to lineage commitment. Additionally, when we compared Smad2/3 ChIP-seq datasets in ESC and DE cultures (Yoon et al., 2015), we found evidence for Smad2/3 occupancy at the VPE specifically in DE cultures (Fig. 4B). These observations strengthen the idea that Nodal/Smad signals controlling Eomes expression activate transcription via the VPE.
Fig. 4.
VPE expression is regulated by Smad2 and independently of Foxh1. (A) RT-qPCR analysis of Eomes mRNA expression during EB differentiation. SB-431542 (SB) inhibition of Nodal/Smad2 signalling from day 3 onwards significantly reduces Eomes expression at d3.5 and d4 of differentiation (*P<0.05, ***P<0.001, Student's t-test, n=3). Error bars represent s.e.m. (B) ChIP-seq of Smad2/3 in definitive endoderm (DE) reveals binding to the VPE (Yoon et al., 2015), overlapping a predicted and conserved binding site for Foxh1, identified with JASPAR at >80% confidence (Mathelier et al., 2016). (C) Whole-mount in situ hybridisation of Eomes mRNA in control and Foxh1-null embryos. Eomes is expressed in both AVE- and APS-defective Foxh1 mutant subtypes at E6.5 and E7.5. (D) VPE-LacZ reporter activity both in the VE and epiblast is retained in Foxh1 mutant embryos at E6.5. Scale bars: 100 μm.
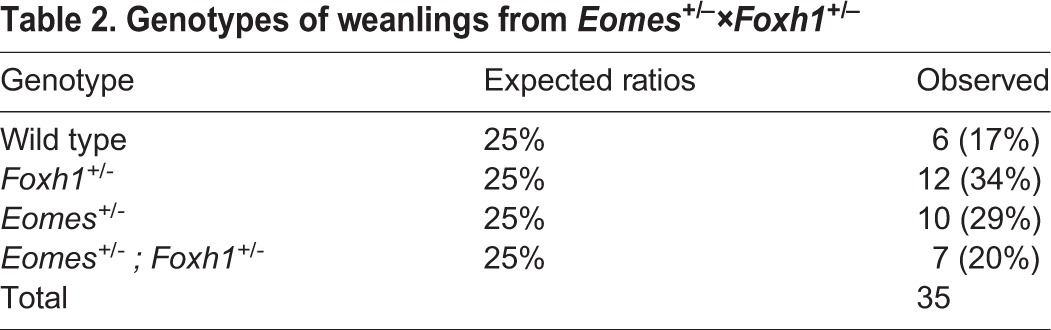
It is well known that the forkhead transcription factor Foxh1 functions as a Smad2/3 co-factor governing Nodal/Smad target gene expression (Attisano et al., 2001; Izzi et al., 2007). Foxh1 has been proposed to act as a pioneer factor and to recruit Smad2/3 complexes to switch enhancers, activated as ESCs transition to DE fates (Beyer et al., 2013; Cirillo et al., 2002; Cirillo and Zaret, 1999; Kim et al., 2011). Interestingly, the VPE Smad2/3 peak also contains a conserved Foxh1-binding motif. Moreover, the VPE region is co-bound by FOXH1, SMAD2/3 and SMAD4 in human DE cultures (Fig. S6) (Beyer et al., 2013; Brown et al., 2011; Kim et al., 2011; Teo et al., 2011). Consistent with the idea that Foxh1 cooperatively activates Eomes expression via the VPE, homozygous null Foxh1−/− embryos phenocopy the EomesΔVPE/− embryos, displaying either defective AVE formation prior to gastrulation or disturbances in APS specification at later stages (Hoodless et al., 2001; Yamamoto et al., 2001).
To evaluate directly Foxh1 functional contributions, we analysed Eomes expression at E6.5 and E7.5 in the context of Foxh1−/− mutant embryos (Fig. 4C). In mutants with AVE/DVE defects at E6.5, Eomes is expressed in the thickened VE at the distal tip of the embryo, and at E7.5 in the chorion and proximal epiblast. Foxh1 mutants with APS defects express Eomes in the ExE and PS. Eomes is clearly expressed in both classes of Foxh1 mutant embryos. Slightly reduced levels in the PS can be explained by the loss of Foxh1-dependent activation of the auto-regulatory ASE Nodal enhancer (Norris et al., 2002). In striking contrast to Eomes/Nodal double heterozygotes (Arnold et al., 2008a), we found no evidence here for Eomes and Foxh1 genetic interactions. Indeed, Eomes and Foxh1 compound mutant mice are fully viable (Table 2). Finally, to confirm that VPE activity is Foxh1 independent, we examined expression of the VPE-LacZ transgene in Foxh1 mutant embryos. LacZ staining is detectable throughout the epiblast at E6.5 (Fig. 4E), and also in the thickened VE at the distal tip. Foxh1 function is nonessential for VPE-LacZ reporter activity. Thus, we conclude that Nodal/Smad signals activate Eomes expression in a Foxh1-independent manner, raising the possibility that other forkhead family members may recruit Smad2/3 complexes during Eomes induction in vivo.
Table 2.
Genotypes of weanlings from Eomes+/–×Foxh1+/–
Characterisation of the Eomes 3D regulatory chromatin compartment during endoderm differentiation
The finding that the VPE targeted deletion partially reduces but fails to completely eliminate Eomes expression, strongly suggests that additional regulatory elements contribute to transcriptional output of the locus. Enhancer interactions with target promoters have been analysed by chromatin conformation capture techniques (de Wit and de Laat, 2012). We took advantage of the recently developed Next Generation (NG) Capture-C methodology (Davies et al., 2016) to screen for Eomes regulatory enhancer elements. During DE differentiation, Eomes expression increased by ∼600 fold (Fig. S7B) resulting in activation of the Eomes target genes, Lhx1 and Foxa2 (Fig. S7C) (Nowotschin et al., 2013; Teo et al., 2011).
NG Capture-C using viewpoints from the PSE_a and PSE_b exhibited promoter interactions in ESC (Fig. S8) when analysed with FourCseq (Klein et al., 2015). These interactions were marginally reduced in DE. However, the overall change was not statistically significant. By contrast, NG Capture-C revealed significant interactions between the VPE and the Eomes promoter in both ESC and DE cells (Fig. S8). Thus, the locus appears to be primed for activation prior to expression.
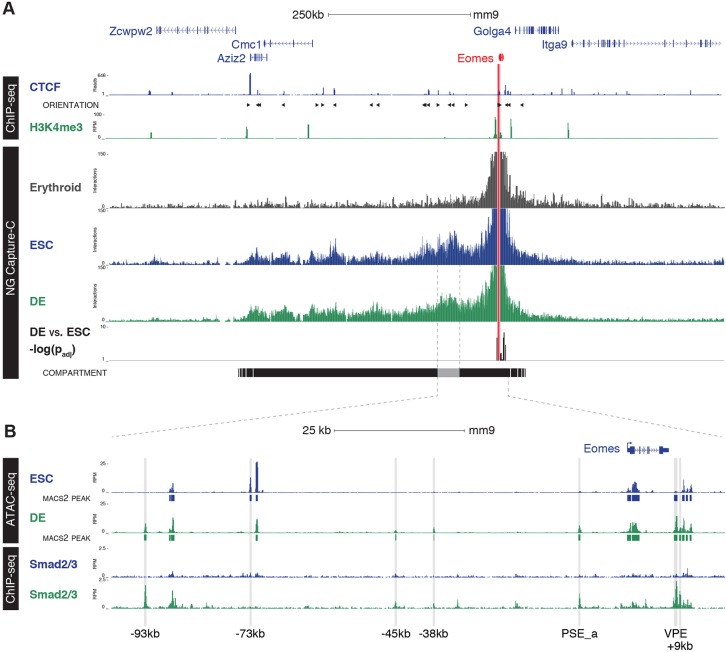
Next, performing Capture-C using a viewpoint from the Eomes promoter revealed that the Eomes locus, together with an upstream 300 kb gene desert and its neighbouring genes Azi2 and Cmc1, occupies a discrete ∼500 kb chromatin compartment (Fig. 5A). This region contains numerous CTCF-binding sites (Handoko et al., 2011). Consistent with CTCF-mediated chromatin loops forming the compartment boundaries, motif analysis suggests that the outermost binding sites face inwards (Fig. 5A). This compartment structure is readily detectable in both ESC and DE cells but is completely absent in control terminally differentiated erythrocytes lacking Eomes expression (Fig. 5A, Fig. S9). Comparison of the NG Capture-C data from ESC and DE, in which the Eomes locus is transcriptionally silent or active, respectively, demonstrates that the compartment is highly stable. Moreover, there were no detectable changes in long-range promoter interactions within the compartment (Fig. S10).
Fig. 5.
Eomes is regulated by Smad2/3 binding in a preformed compartment. (A) NG Capture-C interaction profiles of the Eomes promoter (chr9:117,683,476-118,771,067) from erythrocytes (grey), ESC (blue) and DE (green). Tracks show mean interactions of normalised biological replicates (n=3) and DESeq2 significant differences between DE and ESC [−log(Padj); P≤0.05]. The Eomes compartment, as determined by boundaries of strong promoter interactions with CTCF orientation (arrowheads), is based upon binding in ESCs (Handoko et al., 2011). Histone modifications for H3K4me3 (DE, n=3) show promoter regions. (B) Enlargement of the region of the Eomes compartment showing highest association with the promoter, from chr9: 118,252,500-118,405,500. Open chromatin was generated using ATAC-seq in ESC and DE (n=3), with the addition of MACS2 called peaks annotated beneath each ATAC-seq track and Smad2/3 ChIP-seq in ESCs (blue) and DE (green) (Yoon et al., 2015). Regions of chromatin accessibility unique to ESCs (−73 kb) and those associated with Smad2/3 occupancy in DE (−93 kb, −45 kb, −38 kb, PSE_a, VPE and +9 kb) are indicated.
To map changes in regions of open chromatin associated with Eomes activation and identify potential novel DE enhancers within the compartment, we performed ATAC-seq. We identified 85,581 total peaks in ESC and DE, and of these 19% were gained and 32.5% lost during differentiation (Fig. S9). Within the Eomes compartment we identified six regions that show increased accessibility in DE, including the VPE and the PSE_a, as well as four additional sites at −93 kb, −45 kb, −38 kb and +9 kb relative to the Eomes TSS (Fig. 5B).
Next, we examined Smad2/3 binding across the compartment (Yoon et al., 2015). Smad2/3 occupancy was detectable in DE but not in ESCs at all six of the differentially accessible sites (Fig. 5B). These findings demonstrate the Eomes locus is organised into a large 3D regulatory chromatin compartment in pluripotent ESCs that is maintained upon DE differentiation. Global structural changes are not required for Eomes induction during DE differentiation. Rather, transcriptional activation seems to reflect increased chromatin accessibility and Smad2/3 recruitment at DE enhancers. The −95 kb and −45 kb regions, and to a lesser extent the −38 kb region, are associated with poised and active enhancer marks as cells transition from ES to Epi to MES states, respectively (Fig. S11). Additionally, recently published TF ChIP-seq data demonstrate that the −45 kb ATAC-seq peak, together with the PSE_a and VPE, are co-bound by Tcf3 in DE (Wang et al., 2017), suggesting that both Nodal and Wnt signalling converge on these enhancer regions during gastrulation (Ben-Haim et al., 2006). Consistent with its activities as a key Eomes regulatory element during DE specification, the VPE is also bound by Otx2 and Lhx1 in EpiLC and mesendoderm cultures, respectively (Buecker et al., 2014; Costello et al., 2015).
Foxa2 and Lhx1 promoters form long-range interactions in polycomb bodies
The forkhead TF Foxa2 and the LIM domain homeobox TF Lhx1 function together with Eomes as master regulators of APS cell fates (Ang and Rossant, 1994; Costello et al., 2015; Perea-Gomez et al., 1999; Shawlot and Behringer, 1995). One possible model is that this pre-configured genomic structure might be a common feature shared by endoderm-specific transcriptional factors (Fig. S7C). As for Eomes, Capture-C of the Foxa2 and Lhx1 promoters demonstrates localisation within pre-formed compartments (both ∼350 kb) in ESCs, but not in erythrocytes where the genes are inactive (Fig. 6A,B). However, these Foxa2 and Lhx1 compartments were found to undergo significant rearrangements during DE differentiation (Fig. 6A,B). Unlike Eomes, Lhx1 and Foxa2 promoters both make long-range contacts with neighbouring developmental genes lying outside the compartment boundaries in ESCs (Fig. 6A,B). These long-range interactions range from 370 kb to 1.8 Mb in size and are almost entirely specific to gene promoters (Table S3); they are lost as cells acquire a DE fate (Fig. 6A,B).
Fig. 6.
Foxa2 and Lhx1 form long-range interactions with polycomb-repressed promoters. (A,B) NG Capture-C interaction profiles of the Foxa2 (A) and Lhx1 (B) promoters from erythrocytes (grey), ESC (blue) and DE (green) with chr2: 146,001,500-148,328,000 (A) and chr11: 82,700,000-85,808,000 (B) shown. Tracks show mean interactions of normalised biological replicates (n=3), subtraction of ESCs from DE (Subtr.) and DESeq2 significant differences between DE and ESC [−log(Padj); P≤0.05]. Peaks of the strongest interactions in ESCs (shaded boxes) were manually identified and highlighted. Compartments were determined by boundaries of strong (continuous) promoter interactions. Location of the Polycomb Repressor Complexes components (Ezh2, Suz12, Ring1b) and associated histone modification (H3K27me3) in ESCs are shown (Ku et al., 2008; Mikkelsen et al., 2007).
Both Foxa2 and Lhx1 are repressed by polycomb in ESC (Leeb et al., 2010). Examination of published ESC ChIP-seq data-sets for Polycomb components Ezh2, Suz12 (PRC2) and Ring1b (PRC1) (Chen et al., 2008; Ku et al., 2008), as well as the polycomb repressive mark H3K27me3 (Yue et al., 2014), showed they are present at all of the promoters of the adjacent genes with which Lhx1 and Foxa2 interact (Fig. 6), suggesting that these genes are present in Polycomb bodies (Pirrotta and Li, 2012). Interestingly, these Polycomb repressive components are also present at the Eomes promoter in ESC, but we found no evidence for long-range interactions with gene promoters lying outside the compartment (Fig. S10). Collectively, the results above demonstrate that three essential TFs required for cell fate specification, Eomes, Foxa2 and Lhx1, were found to exhibit distinct modes of 3D chromatin organisation during differentiation.
DISCUSSION
The spatiotemporal expression of key lineage-specifying transcription factors (TF) is tightly controlled during early mouse development to ensure correct cell fate decisions. Interactions of cell type-specific cis-acting enhancer elements with gene promoters, within topologically discrete chromatin compartments, directs developmentally regulated patterns of expression (de Laat and Duboule, 2013). Our recent studies demonstrate that the T-box TF Eomes, dynamically expressed in the VE, ExE and PS during gastrulation, acts downstream of the Nodal signalling pathway as an essential master-regulator of the DE and cardiac mesoderm cell lineages. Here, we exploit transgenic reporter assays, targeted deletion and NG Capture-C strategies to investigate the regulatory landscape at the Eomes locus.
We demonstrate using gain-of-function experiments that conserved proximal cis-regulatory elements, namely the so-called PSE (comprising PSE_a and PSE_b) and the VPE, have the ability to drive reporter activity in the PS, or VE and PS, respectively. The conserved Eomes PSE_b region, which represents an archetypal poised developmental enhancer in both human and mouse ESC, was recently shown to be activated upon mesendoderm induction in response to Nodal (Smad2/3, Foxh1) and Wnt (β-cat) signalling pathways (Beyer et al., 2013; Brown et al., 2011; Buecker and Wysocka, 2012; Estarás et al., 2015; Funa et al., 2015; Kartikasari et al., 2013; Kim et al., 2011; Rada-Iglesias et al., 2011). However, surprisingly our targeted deletion experiments demonstrate that this switch enhancer, and the adjacent PSE_a, are dispensable for correct developmentally regulated Eomes expression in the early embryo. Moreover, mutant mice that entirely lack this genomic region develop normally and are viable and fertile.
Eomes is required for the maintenance and migration of the AVE (Nowotschin et al., 2013). Additionally, robust expression in the PS is essential for formation of APS progenitors (Arnold et al., 2008a). The present results demonstrate that the VPE activates expression in both the AVE and PS, and makes important functional contributions that govern Eomes activities during gastrulation. We found that removal of this element halves transcriptional output from the locus as assessed in vitro. Moreover, EomesΔVPE/− embryos exhibit pleiotropic tissue defects, due to compromised specification of AVE or APS, that closely resemble those caused by defective Nodal signalling or loss of the Smad2/3/4 co-factor Foxh1 (Arnold et al., 2008a; Hoodless et al., 2001; Norris et al., 2002; Yamamoto et al., 2001).
Our NG Capture-C experiments revealed that the VPE directly interacts with the Eomes promoter in both ESC and DE. Moreover the Eomes locus lies within a large pre-formed 3D regulatory chromatin compartment in pluripotent ESCs that is maintained upon differentiation to DE. Thus, activation of the locus occurs in the absence of remodelling long-range interactions. By contrast, previous studies of mouse and human ESC implicate de novo enhancer-promoter interactions during DE and mesendoderm differentiation (Estarás et al., 2015; Kartikasari et al., 2013). These inconsistencies probably reflect technical differences because a target-led (one-versus-some) 3C PCR technique was used previously, when compared with the unbiased (one-versus-all) NG Capture-C sequencing approach exploited here.
NG Capture-C analysis of the direct Eomes targets Foxa2 and Lhx1, which are known to regulate APS fates, demonstrates they similarly occupy discrete regulatory compartments in transcriptionally silent ESC. However, in contrast to Eomes, Foxa2 and Lhx1 promoters display contacts with polycomb-associated gene promoters that lie far outside their compartments. These associations are specifically lost during DE differentiation (Fig. 7). Promoter-promoter interactions within ESCs are often occupied by polycomb repressive complexes (PRC) that organise the 3D chromatin structure into polycomb bodies to silence gene expression (Denholtz et al., 2013; Schoenfelder et al., 2015; Sexton et al., 2012; Williamson et al., 2014). These epigenetic barriers are thought to block lineage-specifying gene activation and thus prevent precocious differentiation. We demonstrate here that, in contrast to Foxa2 and Lhx1, the Eomes locus exhibits a distinct mode of regulation. Rather, in the absence of polycomb-mediated repressive contacts, the Eomes promoter can rapidly respond to dynamic signalling cues during gastrulation (Fig. 7).
Fig. 7.
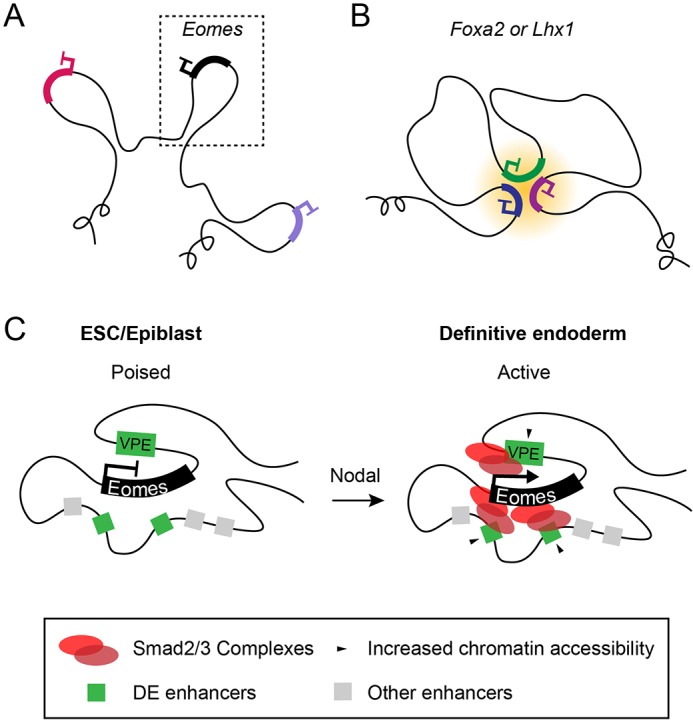
Eomes, Foxa2 and Lhx1 exhibit distinct modes of 3D chromatin organisation during differentiation. (A) In ESCs, Eomes, Foxa2 and Lhx1 are organised into pre-formed chromatin compartments. (B) Unlike Eomes, both Foxa2 and Lhx1 promoters form extra-compartmental contacts with other polycomb-repressed gene promoters. (C) Model for Eomes activation. The poised chromatin architecture at the Eomes locus is permissive for rapid transcriptional induction in response to localised Nodal signalling during gastrulation, primarily via enhancer binding of Smad2/3 complexes.
Considerable evidence suggests that stable enhancer-promoter interactions within pre-formed chromatin compartments initiate transcription through the release of paused polymerase (de Laat and Duboule, 2013; Ghavi-Helm et al., 2014; Jin et al., 2013; Williamson et al., 2016). We found that promoter-enhancer interactions are relatively stable. However our ATAC-seq experiments reveal significant changes in open chromatin regions during DE differentiation. We identified several candidate enhancers within the Eomes compartment that display increased chromatin accessibility and are greatly enriched for Smad2/3 occupancy upon DE differentiation (Yoon et al., 2015). Moreover, we confirm that Smad2/3 is required for Eomes activation, as inhibition of receptor-mediated Nodal/Smad2/3 signalling blocks transcription. Smad2/3 associations with the histone demethylase Jmjd3 are known to be required for the activation of Nodal target genes (Dahle et al., 2010; Kartikasari et al., 2013). Jmjd3 activates poised developmental genes by removing promoter-proximal H3K27me3 and releasing paused polymerase (Chen et al., 2012). We propose that the poised chromatin architecture at the Eomes locus is permissive for rapid transcriptional induction in response to localised Nodal signalling during gastrulation, primarily via enhancer binding of Smad2/3/Jmjd3 complexes to release promoter-paused polymerase.
The 3C technologies developed over the past two decades have provided important new insights into the regulatory chromatin landscapes that orchestrate tissue-specific transcription. Here, we characterise for the first time cis-regulatory elements that activate Eomes expression during gastrulation, and describe the higher order chromatin architecture of the locus. We speculate that the pre-formed chromatin compartment and the absence of additional epigenetic safeguards prior to expression facilitates the rapid induction of Eomes expression in response to dynamic signalling cues at the onset of gastrulation. However, the stage of embryonic development during which these compartments are established, and later dismantled, remains elusive. Future studies will investigate whether these enhancers and permissive chromatin configuration are tissue invariant and can also control cell type-specific Eomes expression governing cell fate decisions at other sites such as the developing cortex, and adult NK and CD8+ T-cell lineages (Arnold et al., 2008b; Gordon et al., 2012; Pearce et al., 2003).
MATERIALS AND METHODS
Animals and PCR genotyping
EomesGFP/+ (Arnold et al., 2009) and Foxh1+/− (Hoodless et al., 2001) strains were genotyped as described. EomesΔPSE/+, EomesΔPSE_b/+, EomesΔVPE/+ and EomesGFPΔVPE/+ strains were generated from targeted ESC clones using standard methods (Arnold et al., 2009) (Figs S2-S5, see supplementary Materials and Methods) and maintained on a mixed 129Sv/Ev/C57BL/6 background. To generate PSE.LacZ and VPE.LacZ transgenic constructs, the 4.6 kb HincII-KpnI PSE fragment and a 696 bp PCR-amplified VPE sequence (Table S1), were cloned upstream of a hsp68 promoter, LacZ cassette and SV40 polyA signal (Sasaki and Hogan, 1996). Zygotes were injected with NotI linearised plasmid and transferred into pseudo-pregnant foster females. Embryos were either collected at E6.5-E7.5 or used to establish stable transgenic mouse lines. PCR genotyping primers are listed in Table S1. All animal experiments were performed in accordance with Home Office (UK) regulations and approved by the University of Oxford Local Ethical Committee.
ESC differentiation
ESC lines were maintained in DMEM (Invitrogen) supplemented with 15% fetal calf serum (Gibco), 1% penicillin/streptomycin (Invitrogen), 0.1 mM 2-mercaptoethanol (Sigma), 1% glutamine (Invitrogen), 1% MEM non-essential amino acids (Gibco), 1 mM sodium pyruvate (Sigma), 1000 U/ml LIF (ESGRO) on gelatin-coated plates.
For analysis of GFP reporter expression, wild-type (CCE), EomesGFPΔVPE/+ and EomesGFP/+ ESCs were seeded as 10 μl hanging drops (1×104 cells/ml) in the absence of LIF to induce EB formation. After 2 days, EBs were transferred to suspension culture. For SB inhibition experiments, ES cells were seeded in suspension at low density (1×104 cells/ml) in the absence of LIF to form EBs. On day 3, EBs were cultured in the presence or absence of 10 μM SB431542 inhibitor (Tocris). For DE differentiation, ES cells were induced to form EBs in suspension, as described above, but were transferred on day 2 into N2B27 medium (Cellartis) supplemented with 20 ng/ml activin A (R&D systems) and 20 ng/ml EGF (Peprotech) to induce DE differentiation (Morrison et al., 2008). For Capture-C, ChIP-seq and ATAC-seq experiments, EBs were dissociated by incubation with 0.25% trypsin (Gibco) for 3 min at 37°C with constant agitation followed by gentle pipetting to obtain a single cell suspension.
RNA analysis
RNA was isolated from using Qiashredder homogeniser (Qiagen), RNeasy mini kit (Qiagen) and RNase-Free DNase Set (Qiagen). RNA was reverse transcribed to cDNA using Superscript III First Strand Synthesis System (Life Technologies) and qRT-PCR was carried out in triplicate using SYBR-green kit (Qiagen) on a Rotagene cycler (Qiagen) with primers listed in Table S1. Relative gene expression was normalised to Gapdh and calculated as 2ΔΔCt.
In situ hybridisation, X-gal staining and immunofluorescence
Whole-mount in situ hybridisation was performed according to published protocols (Behringer et al., 2013). LacZ activity was visualised using whole-mount X-gal staining as described previously (Behringer et al., 2013). Whole-mount in situ hybridisation and X-gal-stained embryos were photographed after clearing in 80% glycerol.
For immunofluorescence, embryos were fixed overnight in 1% PFA. EBs were fixed in 4% PFA for 30 min at room temperature. Samples were washed in 0.1% Triton-X in PBS, permeabilised in 0.5% Triton-X in PBS for 15 min, washed in 0.1% Triton-X in PBS, then blocked in 0.1% Triton-X, 0.2% BSA and 5% donkey serum in PBS for 2 h at room temperature. Samples were incubated with primary antibodies (Table S2) overnight at 4°C, washed, incubated with secondary antibodies or phalloidin AlexaFluor 633 stain (A22284; Invitrogen) in block solution for 2 h at room temperature, counterstained with DAPI and mounted in Vectashield (Vector Laboratories) on chamber slides (LabTek). Images were acquired using an Olympus FV1000 inverted confocal microscope.
Flow cytometry
Day 4 EBs were incubated in 0.25% trypsin for 5 min at 37°C and dissociated into single cells using a 20-guage needle. FACS analysis was performed using a BD FACSCalibur 4 (BD Biosciences) and data analysed using FlowJo.
ATAC-seq
Tagmentation and indexing of single cell suspensions of ESC, DE and erythrocytes from phenylhydrazine-treated mice (Davies et al., 2016) was performed as previously described (Buenrostro et al., 2013; Hay et al., 2016). Samples were sequenced using a 75-cycle paired-end kit on the Illumina NextSeq platform.
ChIP-seq
Single cell suspensions (5×106) were cross-linked in 1% formaldehyde for 15 min at room temperature and processed using standard methods. Briefly, cells were lysed on ice for 20 min (5 mM PIPES, 85 mM KCl and 0.5% Igepal-CA 630), and pelleted nuclei were lysed (50 mM Tris-HCl, 10 mM EDTA and 1% SDS). Sonicated chromatin was incubated overnight with anti-H3K4me3 (2 μl; 07-473; Millipore) and Protein A/G Dynabeads (Invitrogen). Beads were washed with RIPA buffer variants (10 mM Tris-HCl, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% SDS and 0.1% sodium deoxycholate) – RIPA, high salt RIPA (500 mM NaCl) and RIPA with 250 mM LiCl – and with TE buffer before RNase A (Roche) and proteinase K (Thermo Fisher) treatment. Phenol-chloroform-extracted DNA was indexed using NebNext Ultra II (New England BioLabs), multiplexed and sequenced using a 75-cycle paired-end kit on the Illumina NextSeq platform.
ATAC-seq and ChIP-seq analysis
ATAC-seq and ChIP-seq data were analysed as described previously (Hay et al., 2016) using a custom pipeline (http://userweb.molbiol.ox.ac.uk/public/telenius/PipeSite.html). Sequenced reads were aligned using Bowtie to the mm9 build of the mouse genome. Genomic browser tracks were generated from pooled data from multiple replicates and normalised per million mapped reads using a custom Perl script. Peak detection was performed with the MACS2 (Feng et al., 2012). For differential analysis, a union set of peaks for each cell type generated from at least two peak calls per site. Peaks were filtered for high ploidy regions using MIG Viewer (McGowan et al., 2013). CTCF-motifs were identified using the FIMO function of MEME Suite (Bailey et al., 2009; Grant et al., 2011).
NG Capture-C and analysis
NG Capture-C was performed as described previously (Davies et al., 2016) on single cell suspensions of ESC, DE or erythrocytes. Samples were indexed for multiplexing and co-capture of enhancers or promoters using biotinylated 120-mers (Sigma, IDT) designed with the CapSequm webtool (http://apps.molbiol.ox.ac.uk/CaptureC/cgi-bin/CapSequm.cgi) (Hughes et al., 2014) and pooled to a final concentration of 2.9 nM (Table S4). Captured material was pooled and sequenced using the Illumina NextSeq platform with 150 bp paired-end reads (300 cycle kit, Illumina). Reads were mapped using Capture-C scripts (https://github.com/telenius/captureC/releases), analysed as previously described (Hay et al., 2016), and additionally with FourCSeq (Klein et al., 2015) and DESeq2 (Love et al., 2014).
Acknowledgements
We thank Julie Baker for valuable discussions during the initial stage of the project; Ben Davies and Chris Preece (Wellcome Trust Centre for Human Genetics), and Jonathan Godwin (Department of Biochemistry, University of Oxford) for generating transgenic mice; the Dunn School Bioimaging Facility; Nigel Rust for flow cytometry; Xin Sun for help with gene targeting; and Thomas Clague for genotyping assistance.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
C.S.S., I.C., E.K.B. and E.J.R. designed the project; C.S.S., D.J.D., M.E.G. and E.J.R. performed the experiments; C.S.S., D.J.D., J.T., D.R.H., J.R.H, E.K.B. and E.J.R. analysed and interpreted the data; C.S.S., D.J.D., E.K.B and E.J.R. wrote the paper.
Funding
This work was supported by a Wellcome Trust Grant (WT 102811 to E.J.R.), by an Edward Penley Abraham studentship (to C.S.S.), by a Wellcome Trust Strategic Award (WT 106130 to J.R.H. and D.R.H.) and by a Medical Research Council Award (4050189188 to J.R.H. and D.R.H.). E.J.R. is a Wellcome Trust Principal Research Fellow. Deposited in PMC for immediate release.
Data availability
ChIP-seq, ATAC-seq and NG Capture-C data have been deposited in NCBI GEO (GSE94250). Accession numbers of published ChIP-seq data sets used in this study are listed in Table S5.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.147322.supplemental
References
- Adachi H., Saijoh Y., Mochida K., Ohishi S., Hashiguchi H., Hirao A. and Hamada H. (1999). Determination of left/right asymmetric expression of nodal by a left side-specific enhancer with sequence similarity to a lefty-2 enhancer. Genes Dev. 13, 1589-1600. 10.1101/gad.13.12.1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J. M., Hota S. K., He D., Thomas S., Ho L., Pennacchio L. A. and Bruneau B. G. (2015). Brg1 modulates enhancer activation in mesoderm lineage commitment. Development 142, 1418-1430. 10.1242/dev.109496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang S.-L. and Rossant J. (1994). HNF-3 beta is essential for node and notochord formation in mouse development. Cell 78, 561-574. 10.1016/0092-8674(94)90522-3 [DOI] [PubMed] [Google Scholar]
- Arkell R. M. and Tam P. P. L. (2012). Initiating head development in mouse embryos: integrating signalling and transcriptional activity. Open Biol. 2, 120030 10.1098/rsob.120030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold S. J., Hofmann U. K., Bikoff E. K. and Robertson E. J. (2008a). Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development 135, 501-511. 10.1242/dev.014357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold S. J., Huang G.-J., Cheung A. F. P., Era T., Nishikawa S.-I., Bikoff E. K., Molnar Z., Robertson E. J. and Groszer M. (2008b). The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 22, 2479-2484. 10.1101/gad.475408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold S. J., Sugnaseelan J., Groszer M., Srinivas S. and Robertson E. J. (2009). Generation and analysis of a mouse line harboring GFP in the Eomes/Tbr2 locus. Genesis 47, 775-781. 10.1002/dvg.20562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attisano L., Silvestri C., Izzi L. and Labbé E. (2001). The transcriptional role of Smads and FAST (FoxH1) in TGFbeta and activin signalling. Mol. Cell. Endocrinol. 180, 3-11. 10.1016/S0303-7207(01)00524-X [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W. and Noble W. S. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202-W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes J. D., Crosby J. L., Jones C. M., Wright C. V. E. and Hogan B. L. M. (1994). Embryonic expression of Lim-1, the mouse homolog of Xenopus Xlim-1, suggests a role in lateral mesoderm differentiation and neurogenesis. Dev. Biol. 161, 168-178. 10.1006/dbio.1994.1018 [DOI] [PubMed] [Google Scholar]
- Behringer R., Gertsenstein M., Nagy K. and Nagy A. (2013). Manipulating the Mouse Embryo: A Laboratory Manual, 4th edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Ben-Haim N., Lu C., Guzman-Ayala M., Pescatore L., Mesnard D., Bischofberger M., Naef F., Robertson E. J. and Constam D. B. (2006). The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev. Cell 11, 313-323. 10.1016/j.devcel.2006.07.005 [DOI] [PubMed] [Google Scholar]
- Beyer T. A., Weiss A., Khomchuk Y., Huang K., Ogunjimi A. A., Varelas X. and Wrana J. L. (2013). Switch enhancers interpret TGF-beta and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep. 5, 1611-1624. 10.1016/j.celrep.2013.11.021 [DOI] [PubMed] [Google Scholar]
- Brennan J., Lu C. C., Norris D. P., Rodriguez T. A., Beddington R. S. P. and Robertson E. J. (2001). Nodal signalling in the epiblast patterns the early mouse embryo. Nature 411, 965-969. 10.1038/35082103 [DOI] [PubMed] [Google Scholar]
- Brown S., Teo A., Pauklin S., Hannan N., Cho C. H.-H., Lim B., Vardy L., Dunn N. R., Trotter M., Pedersen R. et al. (2011). Activin/Nodal signaling controls divergent transcriptional networks in human embryonic stem cells and in endoderm progenitors. Stem Cells 29, 1176-1185. 10.1002/stem.666 [DOI] [PubMed] [Google Scholar]
- Buecker C. and Wysocka J. (2012). Enhancers as information integration hubs in development: lessons from genomics. Trends Genet. 28, 276-284. 10.1016/j.tig.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buecker C., Srinivasan R., Wu Z., Calo E., Acampora D., Faial T., Simeone A., Tan M., Swigut T. and Wysocka J. (2014). Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell 14, 838-853. 10.1016/j.stem.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y. and Greenleaf W. J. (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213-1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J. et al. (2008). Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106-1117. 10.1016/j.cell.2008.04.043 [DOI] [PubMed] [Google Scholar]
- Chen S., Ma J., Wu F., Xiong L.-J., Ma H., Xu W., Lv R., Li X., Villen J., Gygi S. P. et al. (2012). The histone H3 Lys 27 demethylase JMJD3 regulates gene expression by impacting transcriptional elongation. Genes Dev. 26, 1364-1375. 10.1101/gad.186056.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo L. A. and Zaret K. S. (1999). An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol. Cell 4, 961-969. 10.1016/S1097-2765(00)80225-7 [DOI] [PubMed] [Google Scholar]
- Cirillo L. A., Lin F. R., Cuesta I., Friedman D., Jarnik M. and Zaret K. S. (2002). Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9, 279-289. 10.1016/S1097-2765(02)00459-8 [DOI] [PubMed] [Google Scholar]
- Ciruna B. G. and Rossant J. (1999). Expression of the T-box gene Eomesodermin during early mouse development. Mech. Dev. 81, 199-203. 10.1016/S0925-4773(98)00243-3 [DOI] [PubMed] [Google Scholar]
- Costello I., Pimeisl I.-M., Dräger S., Bikoff E. K., Robertson E. J. and Arnold S. J. (2011). The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat. Cell Biol. 13, 1084-1091. 10.1038/ncb2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello I., Nowotschin S., Sun X., Mould A. W., Hadjantonakis A.-K., Bikoff E. K. and Robertson E. J. (2015). Lhx1 functions together with Otx2, Foxa2, and Ldb1 to govern anterior mesendoderm, node, and midline development. Genes Dev. 29, 2108-2122. 10.1101/gad.268979.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahle O., Kumar A. and Kuehn M. R. (2010). Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci. Signal. 3, ra48 10.1126/scisignal.2000841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. O. J., Telenius J. M., McGowan S. J., Roberts N. A., Taylor S., Higgs D. R. and Hughes J. R. (2016). Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat. Methods 13, 74 10.1038/nmeth.3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W. and Duboule D. (2013). Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 502, 499-506. 10.1038/nature12753 [DOI] [PubMed] [Google Scholar]
- de Wit E. and de Laat W. (2012). A decade of 3C technologies: insights into nuclear organization. Genes Dev. 26, 11-24. 10.1101/gad.179804.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denholtz M., Bonora G., Chronis C., Splinter E., de Laat W., Ernst J., Pellegrini M. and Plath K. (2013). Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell 13, 602-616. 10.1016/j.stem.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57-74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estarás C., Benner C. and Jones K. A. (2015). SMADs and YAP Compete to Control Elongation of beta-Catenin:LEF-1-Recruited RNAPII during hESC Differentiation. Mol. Cell 58, 780-793. 10.1016/j.molcel.2015.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Liu T., Qin B., Zhang Y. and Liu X. S. (2012). Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7, 1728-1740. 10.1038/nprot.2012.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funa N. S., Schachter K. A., Lerdrup M., Ekberg J., Hess K., Dietrich N., Honoré C., Hansen K. and Semb H. (2015). beta-catenin regulates primitive streak induction through collaborative interactions with SMAD2/SMAD3 and OCT4. Cell Stem Cell 16, 639-652. 10.1016/j.stem.2015.03.008 [DOI] [PubMed] [Google Scholar]
- Ghavi-Helm Y., Klein F. A., Pakozdi T., Ciglar L., Noordermeer D., Huber W. and Furlong E. E. (2014). Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512, 96-100. 10.1038/nature13417 [DOI] [PubMed] [Google Scholar]
- Gordon S. M., Chaix J., Rupp L. J., Wu J., Madera S., Sun J. C., Lindsten T. and Reiner S. L. (2012). The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 36, 55-67. 10.1016/j.immuni.2011.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C. E., Bailey T. L. and Noble W. S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017-1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko L., Xu H., Li G., Ngan C. Y., Chew E., Schnapp M., Lee C. W. H., Ye C., Ping J. L. H., Mulawadi F. et al. (2011). CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 43, 630-638. 10.1038/ng.857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi S., Kitajima S., Takagi A., Takeda H., Inoue T. and Saga Y. (2001). Transcriptional regulation of Mesp1 and Mesp2 genes: differential usage of enhancers during development. Mech. Dev. 108, 59-69. 10.1016/S0925-4773(01)00478-6 [DOI] [PubMed] [Google Scholar]
- Hay D., Hughes J. R., Babbs C., Davies J. O. J., Graham B. J., Hanssen L. L. P., Kassouf M. T., Oudelaar A. M., Sharpe J. A., Suciu M. C. et al. (2016). Genetic dissection of the alpha-globin super-enhancer in vivo. Nat. Genet. 48, 895-903. 10.1038/ng.3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoodless P. A., Pye M., Chazaud C., Labbé E., Attisano L., Rossant J. and Wrana J. L. (2001). FoxH1 (Fast) functions to specify the anterior primitive streak in the mouse. Genes Dev. 15, 1257-1271. 10.1101/gad.881501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. R., Roberts N., McGowan S., Hay D., Giannoulatou E., Lynch M., De Gobbi M., Taylor S., Gibbons R. and Higgs D. R. (2014). Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet. 46, 205-212. 10.1038/ng.2871 [DOI] [PubMed] [Google Scholar]
- Izumi N., Era T., Akimaru H., Yasunaga M. and Nishikawa S.-I. (2007). Dissecting the molecular hierarchy for mesendoderm differentiation through a combination of embryonic stem cell culture and RNA interference. Stem Cells 25, 1664-1674. 10.1634/stemcells.2006-0681 [DOI] [PubMed] [Google Scholar]
- Izzi L., Silvestri C., von Both I., Labbé E., Zakin L., Wrana J. L. and Attisano L. (2007). Foxh1 recruits Gsc to negatively regulate Mixl1 expression during early mouse development. EMBO J. 26, 3132-3143. 10.1038/sj.emboj.7601753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F., Li Y., Dixon J. R., Selvaraj S., Ye Z., Lee A. Y., Yen C. A., Schmitt A. D., Espinoza C. A. and Ren B. (2013). A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290-294. 10.1038/nature12644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartikasari A. E. R., Zhou J. X., Kanji M. S., Chan D. N., Sinha A., Grapin-Botton A., Magnuson M. A., Lowry W. E. and Bhushan A. (2013). The histone demethylase Jmjd3 sequentially associates with the transcription factors Tbx3 and Eomes to drive endoderm differentiation. EMBO J. 32, 1393-1408. 10.1038/emboj.2013.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. W., Yoon S.-J., Chuong E., Oyolu C., Wills A. E., Gupta R. and Baker J. (2011). Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev. Biol. 357, 492-504. 10.1016/j.ydbio.2011.06.009 [DOI] [PubMed] [Google Scholar]
- Klein F. A., Pakozdi T., Anders S., Ghavi-Helm Y., Furlong E. E. M. and Huber W. (2015). FourCSeq: analysis of 4C sequencing data. Bioinformatics 31, 3085-3091. 10.1093/bioinformatics/btv335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothary R., Clapoff S., Darling S., Perry M. D., Moran L. A. and Rossant J. (1989). Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development 105, 707-714. [DOI] [PubMed] [Google Scholar]
- Ku M., Koche R. P., Rheinbay E., Mendenhall E. M., Endoh M., Mikkelsen T. S., Presser A., Nusbaum C., Xie X., Chi A. S. et al. (2008). Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 4, e1000242 10.1371/journal.pgen.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon G. S. and Hadjantonakis A.-K. (2007). Eomes::GFP-a tool for live imaging cells of the trophoblast, primitive streak, and telencephalon in the mouse embryo. Genesis 45, 208-217. 10.1002/dvg.20293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon G. S., Viotti M. and Hadjantonakis A.-K. (2008). The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev. Cell 15, 509-520. 10.1016/j.devcel.2008.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M., Pasini D., Novatchkova M., Jaritz M., Helin K. and Wutz A. (2010). Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 24, 265-276. 10.1101/gad.544410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W. and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A., Fornes O., Arenillas D. J., Chen C.-Y., Denay G., Lee J., Shi W., Shyr C., Tan G., Worsley-Hunt R. et al. (2016). JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 44, D110-D115. 10.1093/nar/gkv1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan S. J., Hughes J. R., Han Z.-P. and Taylor S. (2013). MIG: Multi-Image Genome viewer. Bioinformatics 29, 2477-2478. 10.1093/bioinformatics/btt406 [DOI] [PubMed] [Google Scholar]
- Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.-K., Koche R. P. et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553-560. 10.1038/nature06008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison G. M., Oikonomopoulou I., Migueles R. P., Soneji S., Livigni A., Enver T. and Brickman J. M. (2008). Anterior definitive endoderm from ESCs reveals a role for FGF signaling. Cell Stem Cell 3, 402-415. 10.1016/j.stem.2008.07.021 [DOI] [PubMed] [Google Scholar]
- Norris D. P. and Robertson E. J. (1999). Asymmetric and node-specific nodal expression patterns are controlled by two distinct cis-acting regulatory elements. Genes Dev. 13, 1575-1588. 10.1101/gad.13.12.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris D. P., Brennan J., Bikoff E. K. and Robertson E. J. (2002). The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development 129, 3455-3468. [DOI] [PubMed] [Google Scholar]
- Nowotschin S., Costello I., Piliszek A., Kwon G. S., Mao C.-A., Klein W. H., Robertson E. J. and Hadjantonakis A.-K. (2013). The T-box transcription factor Eomesodermin is essential for AVE induction in the mouse embryo. Genes Dev. 27, 997-1002. 10.1101/gad.215152.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce E. L., Mullen A. C., Martins G. A., Krawczyk C. M., Hutchins A. S., Zediak V. P., Banica M., DiCioccio C. B., Gross D. A., Mao C.-A. et al. (2003). Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science 302, 1041-1043. 10.1126/science.1090148 [DOI] [PubMed] [Google Scholar]
- Perea-Gomez A., Shawlot W., Sasaki H., Behringer R. R. and Ang S. (1999). HNF3beta and Lim1 interact in the visceral endoderm to regulate primitive streak formation and anterior-posterior polarity in the mouse embryo. Development 126, 4499-4511. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. and Li H.-B. (2012). A view of nuclear Polycomb bodies. Curr. Opin. Genet. Dev. 22, 101-109. 10.1016/j.gde.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S. A., Flynn R. A. and Wysocka J. (2011). A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279-283. 10.1038/nature09692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson E. J. (2014). Dose-dependent Nodal/Smad signals pattern the early mouse embryo. Semin. Cell Dev. Biol. 32, 73-79. 10.1016/j.semcdb.2014.03.028 [DOI] [PubMed] [Google Scholar]
- Russ A. P., Wattler S., Colledge W. H., Aparicio S. A. J. R., Carlton M. B. L., Pearce J. J., Barton S. C., Surani M. A., Ryan K., Nehls M. C. et al. (2000). Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 404, 95-99. 10.1038/35003601 [DOI] [PubMed] [Google Scholar]
- Sasaki H. and Hogan B. L. M. (1996). Enhancer analysis of the mouse HNF-3 beta gene: regulatory elements for node/notochord and floor plate are independent and consist of multiple sub-elements. Genes Cells 1, 59-72. 10.1046/j.1365-2443.1996.04004.x [DOI] [PubMed] [Google Scholar]
- Schoenfelder S., Sugar R., Dimond A., Javierre B.-M., Armstrong H., Mifsud B., Dimitrova E., Matheson L., Tavares-Cadete F., Furlan-Magaril M. et al. (2015). Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat. Genet. 47, 1179-1186. 10.1038/ng.3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A. and Cavalli G. (2012). Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458-472. 10.1016/j.cell.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Shawlot W. and Behringer R. R. (1995). Requirement for Lim1 in head-organizer function. Nature 374, 425-430. 10.1038/374425a0 [DOI] [PubMed] [Google Scholar]
- Stower M. J. and Srinivas S. (2014). Heading forwards: anterior visceral endoderm migration in patterning the mouse embryo. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130546 10.1098/rstb.2013.0546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo A. K. K., Arnold S. J., Trotter M. W. B., Brown S., Ang L. T., Chng Z., Robertson E. J., Dunn N. R. and Vallier L. (2011). Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 25, 238-250. 10.1101/gad.607311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ameele J., Tiberi L., Bondue A., Paulissen C., Herpoel A., Iacovino M., Kyba M., Blanpain C. and Vanderhaeghen P. (2012). Eomesodermin induces Mesp1 expression and cardiac differentiation from embryonic stem cells in the absence of Activin. EMBO Rep. 13, 355-362. 10.1038/embor.2012.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent S. D., Dunn N. R., Hayashi S., Norris D. P. and Robertson E. J. (2003). Cell fate decisions within the mouse organizer are governed by graded Nodal signals. Genes Dev. 17, 1646-1662. 10.1101/gad.1100503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zou Y., Nowotschin S., Kim S. Y., Li Q. V., Soh C.-L., Su J., Zhang C., Shu W., Xi Q. et al. (2017). The p53 family coordinates Wnt and nodal inputs in mesendodermal differentiation of embryonic stem cells. Cell Stem Cell 20, 70-86. 10.1016/j.stem.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson I., Berlivet S., Eskeland R., Boyle S., Illingworth R. S., Paquette D., Dostie J. and Bickmore W. A. (2014). Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes Dev. 28, 2778-2791. 10.1101/gad.251694.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson I., Lettice L. A., Hill R. E. and Bickmore W. A. (2016). Shh and ZRS enhancer colocalisation is specific to the zone of polarising activity. Development 143, 2994-3001. 10.1242/dev.139188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Meno C., Sakai Y., Shiratori H., Mochida K., Ikawa Y., Saijoh Y. and Hamada H. (2001). The transcription factor FoxH1 (FAST) mediates Nodal signaling during anterior-posterior patterning and node formation in the mouse. Genes Dev. 15, 1242-1256. 10.1101/gad.883901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S.-J., Foley J. W. and Baker J. C. (2015). HEB associates with PRC2 and SMAD2/3 to regulate developmental fates. Nat. Commun. 6, 6546 10.1038/ncomms7546 [DOI] [PubMed] [Google Scholar]
- Yue F., Cheng Y., Breschi A., Vierstra J., Wu W., Ryba T., Sandstrom R., Ma Z., Davis C., Pope B. D. et al. (2014). A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355-364. 10.1038/nature13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner G. E., Tesar P. J. and Scacheri P. C. (2011). Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 21, 1273-1283. 10.1101/gr.122382.111 [DOI] [PMC free article] [PubMed] [Google Scholar]